Abstract
SRP is essential for targeting nascent chains to the endoplasmic reticulum, and it delays nascent chain elongation in cell-free translation systems. However, the significance of this function has remained unclear. We show that efficient protein translocation into the ER is incompatible with normal cellular translation rates due to rate-limiting concentrations of SRP receptor (SR). We complemented mammalian cells depleted of SRP14 by expressing mutant versions of the protein lacking the elongation arrest function. The absence of a delay caused inefficient targeting of preproteins leading to defects in secretion, depletion of proteins in the endogenous membranes and reduced cell growth. The detrimental effects were reversed by either reducing the cellular protein synthesis rate or by increasing SR expression. SRP therefore ensures that nascent chains remain translocation-competent during the targeting time window dictated by SR. Since SRP-signal sequence affinities vary, the delay may also regulate which proteins are preferentially targeted.
Introduction
Efficient delivery of proteins to their subcellular locations and to the outside of the cell is important to maintain cell function and organization. Thus, cells have developed competent mechanisms to deliver proteins to their target sites. The universally conserved signal recognition particle (SRP) and its membrane-associated receptor (SRP receptor (SR) or docking protein) are responsible for cotranslational targeting of secretory and membrane proteins to the endoplasmic reticulum (ER).
SRP-mediated targeting is achieved via a series of ordered steps that are closely coordinated (for review, see Keenan et al., 2001; Pool, 2005). The hydrophobic signal sequence, a common hallmark of ER-targeted proteins, is first recognized by the SRP54 subunit of SRP, and their association causes a delay in the elongation of the nascent chain that is termed the elongation arrest (Walter and Blobel, 1981). The ribosome-nascent chain-SRP complex (RNC-SRP) is then docked to the ER membrane through the interaction of SRP with SR (Gilmore et al., 1982; Meyer et al., 1982), both in their GTP-bound form (Connolly and Gilmore, 1986). After docking of the ribosome onto the protein-conducting channel (translocon), SRP and SR dissociate from the ribosome and from each other by hydrolyzing their bound GTPs (Connolly et al., 1991) and translation resumes at its normal speed. Such a coordinated mechanism requires SRP and SR to switch dynamically between multiple functional states in response to cargo binding (Shan et al., 2004).
Mammalian SRP is composed of a single RNA and six protein subunits and can be divided into two domains. The heterodimer SRP9/14 and the 5′ and 3′ ends of the SRP RNA form the Alu domain, which holds the elongation arrest function (Siegel and Walter, 1985). SRP lacking the Alu domain or SRP9/14 can still promote translocation in cell-free assays, albeit at reduced efficiency. However, it lacks the capacity to bind RNCs lacking signal sequences (Hauser et al., 1995; Powers and Walter, 1996). A C-terminal truncation of murine SRP14 abrogates the elongation arrest function, but does not interfere with its ribosome binding capacity (Thomas et al., 1997). A similar truncation in S. cerevisiae SRP14 (Srp14pΔ29) also leads to the loss of the elongation arrest function (Mason et al., 2000). These results suggest an elongation arrest-specific role for the C-terminal region in SRP14.
Cryo-electron microscopy images of SRP bound to artificially arrested ribosomes showed that the Alu domain of SRP is located in the elongation factor-binding site (Halic et al., 2004). Furthermore, SRP was found to interact with the ribosome at the step of the EF2-catalyzed translocation of the tRNA from the A to the P site in yeast and in mammalian cells (Ogg and Walter, 1995; Lakkaraju et al., 2007). The Alu domain might then delay the elongation cycle by preventing the binding of EF2. Cross-linking studies in ongoing translation revealed that the interactions between the Alu domain and the ribosome are dynamic and change upon signal sequence recognition (Terzi et al., 2004).
The elongation arrest function has been studied mostly in cell-free translation/translocation systems. In the heterologous translation/translocation assay using wheat germ lysate and canine microsomes and SRP, signal sequence recognition by SRP induces an arrest in the elongation of several ER-targeted proteins at one or multiple sites in the nascent chain (Walter and Blobel, 1981; Meyer et al., 1982; Lipp et al., 1987; Okun and Shields, 1992; Wolin and Walter, 1993). In the homologous mammalian and yeast translation/translocation systems, SRP causes a delay in the accumulation of full-length protein rather than an accumulation of arrested fragments (Wolin and Walter, 1989; Mason et al., 2000), although specific pause sites of the ribosome could be revealed at the level of the mRNA. The delay in elongation of the nascent chain by SRP leads to the stacking of ribosomes at the pause sites (Wolin and Walter, 1988; Wolin and Walter, 1989; Wolin and Walter, 1993).
Abrogating the elongation arrest function of SRP reduces the translocation efficiency in the heterologous as well as in the yeast homologous translation/translocation systems (Siegel and Walter, 1986; Thomas et al., 1997; Mason et al., 2000) leading to the hypothesis that the elongation arrest activity may increase the time window of opportunity for the SRP-RNC complex to interact with SR (Siegel and Walter, 1985). A time window for SRP-mediated targeting also served as a parameter to develop a mathematical model of the translation/translocation process, which predicted that translation inhibition is not required for efficient translocation in vivo unless SR concentrations would be strongly limiting (Rapoport et al., 1987). The mutant S. cerevisiae strain expressing an elongation arrest-defective version of the SRP14 subunit failed to reveal growth and translocation defects under normal conditions (Mason et al., 2000). The strain is temperature-sensitive for growth and, at non-permissive temperature, small amounts of the untargeted precursor protein of Pho8p could be detected whereas DPAPB was still translocated efficiently even though both proteins are SRP-dependent for ER targeting (Ng et al., 1996). The ubiquitin-assisted translocation assay (Johnsson and Varshavsky, 1994) also revealed a defect in the tight coupling of translation and translocation.
Although absent in many bacteria, the Alu domain of SRP is otherwise highly conserved in evolution, consistent with its having an important function. It therefore remained a key task to understand the significance of the function for protein translocation into the ER. Based on previous SRP protein depletion experiments (Lakkaraju et al., 2007), we developed a complementation assay in mammalian cells. It was used to investigate the phenotypes caused by mutations in human SRP14 that abrogate exclusively the elongation arrest function of SRP in cell-free assays without interfering with its signal recognition and targeting activities. The mutant versions of the h14 subunit assembled well and restored normal SRP levels, but caused significant growth and translocation defects. The defects could be rescued i) by specifically slowing down the elongation step in protein synthesis or ii) by increasing the cellular levels of the receptor subunits SRα and SRβ. Our results demonstrate that the elongation arrest activity has an essential function in mammalian cells. SRP reduces the elongation rate of nascent chains to maximize the in vivo efficiency of protein translocation into the ER through a limited number of SR targeting sites. In doing so, SRP may also function in a regulatory role by favoring the targeting of RNCs whose signal sequences bind to SRP with high affinity.
Results
A short basic region in SRP14 is essential for the elongation arrest function of SRP
The C-terminal region of human SRP14 (h14) is composed mainly of highly conserved basic amino acid residues in positions 96-107, followed by an alanine tail which is unique to primates (Figure 1A) and dispensable for elongation arrest activity (Bovia et al., 1997). In murine SRP14, truncation of the C-terminal residues 91 to 110 abrogated elongation arrest activity of SRP (Thomas et al., 1997). Crystal structures of the protein-RNA complex revealed that amino acid residues 93 to 95 make contacts with SRP9 (Weichenrieder et al., 2000) whereas residues after amino acid 95 could not be traced. This suggested that residues past amino acid 95 might be important for elongation arrest activity. We therefore decided to change the basic amino acid residues 96-107 in h14 (Figure 1A) either completely (h14A12) or partially (h14A5, h14A6-12). Since h14 functions in complex with h9, we purified the recombinant h14 proteins as heterodimeric complex with recombinant h9 as described (Terzi et al., 2004).
Figure 1. Amino acid residues 96-100 in SRP14 are critical for elongation arrest activity of SRP.

(A) Sequence alignment of the C-terminal portions of wild type and mutated human SRP14 proteins (h14).
(B) and (C) Elongation arrest (black) and translocation (gray) activities of SRPs (RCs) reconstituted with different h14 proteins. The right panels: quantification of the results (n≥3). EKRM: 0.2 eq./reaction. Activities were normalized to RCwt (100%.) The elongation arrest and the translocation activities of RCwt were 72±7% and 79±10%, respectively.
(D) Targeting of pPL86-RNCs to microsomes. Targeting efficiencies (T [%]) were monitored by sedimenting microsomes through a sucrose cushion. EKRM: 1 eq./reaction; S: Supernatant, P: Pellet. Right panel: Quantification of the assays (n=3). T [%] was normalized to RCwt (100%).
–RC: Buffer. RC(-14): SRP without h9/14. EKRM: SRP-depleted microsomes.
To analyze the effects of the mutations, it was necessary to reconstitute particles from wild type and mutated h9/14 proteins together with all other SRP proteins and synthetic SRP RNA. The activities of the particles were assayed by adding the reconstitution reactions directly to wheat germ lysate programmed for translation with synthetic mRNAs encoding preprolactin (a secreted protein) and a truncated form of cyclin D (a cytosolic protein). The relative inhibition of preprolactin synthesis as compared to cyclin D synthesis was monitored to determine elongation arrest activity.
As expected, particles reconstituted with h9/14 (RCwt) showed maximal elongation arrest activity confirming the assembly of active SRP in vitro (Figure 1B) whereas the negative controls (-RC and RC(-14)) displayed strongly reduced elongation arrest activities. The results of the negative controls confirmed that our assay system was dependent on exogenous SRP and that h9/14 is essential for the activity. Of the three mutated proteins we analyzed, h9/14A12 and h9/14A5 lacked the capacity to reconstitute elongation arrest-competent particles whereas h9/14A6-12 was able to do so, albeit slightly less efficiecently than the wild-type protein. With insulin as a secretory protein in the elongation arrest assay, we obtained the same results (not shown).
Although unlikely based on structure information, h14A12 and h14A5 might fail to assemble into SRP. To examine this possibility, we fractionated the reconstitution reactions on glycerol gradients. The fractionation profiles of h14 and the synthetic SRP RNA were comparable in the three reconstitution reactions (Figure S1). Hence, h9/14A12 and h9/14A5 are assembly competent.
Next, we examined processing of preprolactin into prolactin in the presence of microsomes (Figure 1C). All the reconstituted particles were able to promote translocation of preprolactin confirming that they possessed intact signal recognition and targeting activities. Importantly, the lack of elongation arrest activity reduced the translocation efficiency. RCA6-12 promoted efficient translocation.
To examine the targeting capacities of elongation-arrest-defective particles, we monitored the capacity of RCwt and RCA5 to target artificially arrested RNCs, which carry a 86 amino acid-long nascent chain of preprolactin, to microsomes (Flanagan et al., 2003). RCA5 and RCwt had comparable targeting efficiencies (Figure 1D).
Hence, the conserved amino acid residues 96-100 in h14 are essential for the elongation arrest function of SRP and for efficient translocation into microsomes. In contrast, the signal recognition and the targeting functions were not affected by the mutations.
GFP-h14 proteins are properly assembled and restore normal SRP levels
To analyze the physiological importance of elongation arrest activity in mammalian cells, we developed a complementation assay. Endogenous h14 was depleted in HEK 293T cells by expressing shRNAs (Lakkaraju et al., 2007). Endogenous h14 levels are reduced to about 50% at 120 hr and to less than 5% at 144-168 hr post transfection. As a negative control, we expressed shRNA against firefly luciferase. The h14 proteins characterized in the cell-free assay system were expressed as C-terminal fusions with GFP (G14, G14A12, G14A5 and G14A6-12) to complement cells for the absence of h14. To monitor GFP-h14 assembly into functional SRP, we fractionated cell extracts on glycerol gradients (Figure S1B). The migration of h19, another SRP subunit, in fractions 5 and 6 designated the presence of SRP. In mock-depleted cells, h14 was seen in the first six fractions consistent with the presence of an excess of h14 in human cells (Bovia et al., 1995) whereas in h14-depleted cells, the protein was hardly detectable (middle and lower panels). G14 and G14A5 proteins also migrated in fractions 5 and 6 demonstrating that both proteins were assembled into SRP. In addition, all h19 migrated with SRP in cells expressing G14A5 (lower panel) confirming that SRP assembly was normal and efficient.
7SL RNA (human SRP RNA) levels decrease rapidly upon depletion of individual SRP proteins (Lakkaraju et al., 2007). The 7SL RNA level is therefore a sensitive tool to monitor the assembly and the cellular levels of SRP. In cells depleted of h14, the levels of 7SL RNA are decreased nine-fold (Figure S1C). Significantly, the levels of 7SL RNA in cells expressing any of the four different GFP-h14 protein chimeras were restored to approximately 90% as compared to wild-type levels. These results confirmed that the GFP-h14 protein chimeras assemble into SRP and thereby restore almost normal levels of cellular SRP.
Elongation arrest-defective SRP impairs the accumulation of membrane and secreted proteins
To examine protein secretion and membrane protein accumulation in cells expressing elongation arrest-defective GFP-h14 proteins, we monitored two specific reporter and one endogenous protein: SEAP, a secreted version of alkaline phosphatase, the plasma membrane proteins VSV-G and transferrin receptor (TfnR). The secretion efficiency of SEAP was determined by measuring the enzymatic activity accumulating in the medium between 144-168 hr after transfection. VSV-G and TfnR cell surface accumulation was determined at 168 hr after the initial transfection by labeling the protein with fluorescent antibodies in the absence of permeabilizing detergents. As expected from previous results (Lakkaraju et al., 2007), depletion of h14 without complementation (shRNA(14)/GFP) resulted in a strongly reduced amount of SEAP secreted into the medium whereas the complementation with wild-type G14 restored normal levels of SEAP secretion (Figure 2A). The secretion efficiencies of cells in which h14 was replaced with G14A5 and G14A12 was reduced to about 50% of the wild-type levels. The G14A6-12 protein restored normal secretion levels.
Figure 2. G14A12 and G14A5 fail to complement h14-depleted cells for efficient protein secretion and membrane protein accumulation.
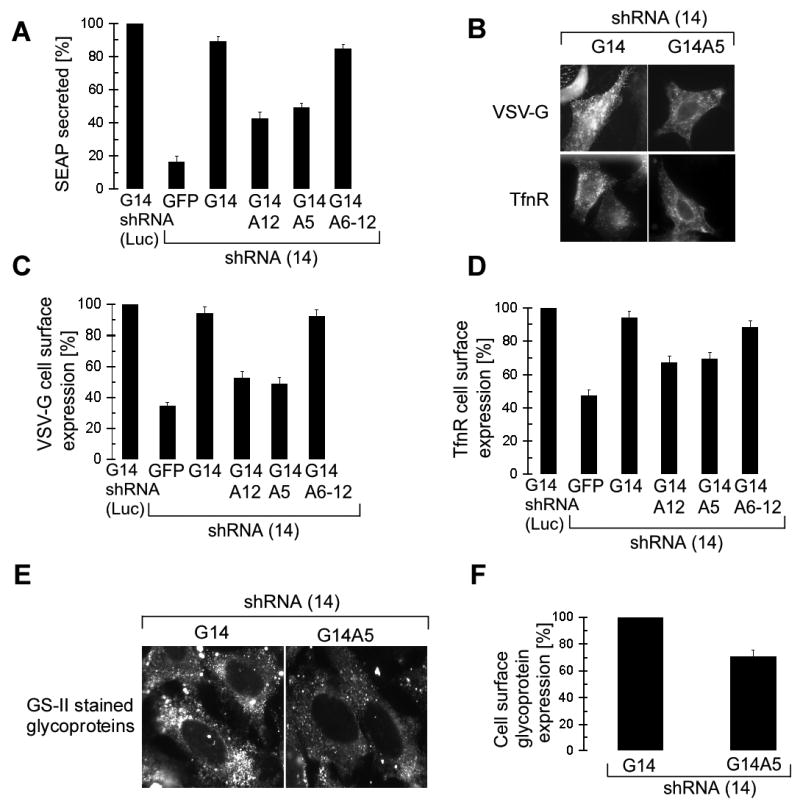
(A) SEAP activity in medium of 293T cells collected between 144-168 hr and standardized to the amount of protein present in extracts prepared from the secreting cells. shRNA(Luc)/G14: Activities were normalized to the one of mock-depleted cells expressing G14 (n=3).
(B) HeLa cells stained with antibodies for cell surface expression of VSV-G and TfnR in the absence of detergent.
(C) and (D) Surface staining intensities of HeLa cells expressing the GFP-h14 protein as indicated (n=2) were quantified for 200 cells each and normalized to the one of G14/shRNA(Luc).
(E) Cell surface expression of glycoproteins revealed in HeLa cells stained with GS-II lectin.
(F) Quantification of cell surface glycoprotein staining in G14A5 cells normalized to cells complemented with G14 (n=2).
VSV-G and TfnR cell surface accumulation was also reduced in G14A12 and G14A5 expressing cells (Figure 2B). The quantification (C and D) revealed similar results as observed with SEAP. Notably, cells depleted of h14 that failed to express GFP-h14 chimeras were disregarded in the quantification. The decrease in VSV-G and TfnR expression is therefore specifically due to the presence of the elongation arrest-defective SRP and not due to low levels of SRP resulting from h14 depletion. Moreover, the decrease in the cell surface expression of TfnR was not due to a recycling defect, as we could not detect an internal accumulation of TfnR in the cells expressing G14A5 and G14A12 (data not shown). Since we measured steady state levels of proteins in these experiments, and since TfnR has a low turnover rate (19±6 h, Rutledge et al., 1991), it might not be surprising that the observed effects were diminished as compared to SEAP and VSV-G.
Next, we compared the cell surface glycoprotein expression of G14 and G14A5 expressing cells. The cell surface proteins were labeled with fluorescent GS-II lectin under non permeabilizing conditions (Figure 2E). There was a difference of 30% in the surface staining intensities of glycoproteins between cells complemented with either G14A5 or G14 (Figure 2F) taking into account cells that expressed the GFP-h14 proteins. Considering that cell surface proteins have turnover rates of 30-100 h (Hare, 1990), the observed decrease in glycoprotein expression is highly significant and indicates that many proteins are affected by the absence of the elongation arrest function.
We further examined the levels of several proteins associated with ER translocation: Sec61α, β, TRAM and TRAPα as well as the chaperone BiP (Alder and Johnson, 2004; Rapoport, 2007). Of the five proteins only the level of BiP was notably decreased. The levels of three other proteins were even increased. This observation may in part be explained by experimental variations (Figure S2). Moreover, the Sec61α protein was not mislocalized. The results are consistent with a very low turnover rate of these components. If the proteins are as stable as SRP subunits, a significant decrease in their levels would only be expected to occur at 96 hr after the lack of elongation arrest activity has become effective (120 hr + 96 hr). These findings suggested that decreased levels of translocon components were not the major cause for the defects observed in the absence of the elongation arrest function. In addition, overall protein synthesis was not affected in these experiments as shown later.
Hence, replacing endogenous h14 with either of two elongation arrest-defective proteins G14A12 and G14A5 caused specific and significant defects in protein secretion and membrane protein accumulation.
The elongation arrest function is required for efficient translocation in vivo
Inefficient translocation of proteins can be revealed by monitoring the accumulation of preproteins (proteins with an uncleaved signal sequence). Bovine preprolactin, a well-studied SRP substrate in cell-free translocation systems, was chosen as a reporter and was modified with a triple flag-tag at its C-terminus (pPrl3f). The translocation efficiency was studied in pulse-labeling experiments with 293T cells grown in the presence of proteasome inhibitor (MG132). In its absence, no preprotein was detectable, presumably due to the its rapid degradation (not shown). Immunoprecipitation of prolactin and preprolactin indicated that the relative translocation efficiency was reduced to 44% in G14A5 cells (Figure 3A). Precursor accumulation was also detected by immunoblotting extracts that were prepared from cells grown in the presence of MG132 for 8 hr (Figure 3B) and the relative amount of prolactin secreted into the medium during this time period was reduced to 45% in G14A5-expressing cells. The perceptible accumulation of modified and unmodified prolactin in cells expressing G14A12 and G14A5 suggested that steps later in the secretory pathway might have slowed down as a consequence of inefficient translocation of membrane proteins and modifying enzymes.
Figure 3. Elongation arrest-defective SRP causes a defect in preprotein translocation.

(A) Preprolactin (pPL3f) and prolactin (PL3f) pulse-labeled with [35S]methionine and immunoprecipitated with anti-flag antibodies. T: translocation efficiency.
(B) Preprolactin (pPL3f) and prolactin (PL-3f and *PL3f) accumulation revealed with anti-flag antibodies. *PL: Phosphorylated prolactin in transit for secretion. *PL3f/SUP: relative levels of *PL in the medium.
(C) Same as (A) with pSEAP3f. 3fSEAP and p3fSEAP were precipitated with antibodies against SEAP (upper panel). p3fSEAP was precipitated with flag-tag antibodies (lower panel).
(D) TfnR-GFP was revealed with anti-GFP antibodies. preTfnR-GFP most likely represents the preprotein. *TfnR-GFP: modified mature protein. The preTfnR-GFP values were standardized to actin and normalized to shRNA(Luc)/G14 (n=2).
All experiments were done in 293T cells and equal amounts of cell extracts were loaded in each lane for Western blot analysis.
We also analyzed the secretory protein SEAP which, in order to recognize the preprotein, was modified with a triple flag-tag at its very N-terminus (p3fSEAP). The relative size difference between the precursor and the mature SEAP proteins is very small and the pulse-labeled proteins immunoprecipated with SEAP antibodies therefore migrated together in SDS-PAGE. However, the specific analysis of the [35S]methionine-labeled preprotein immunoprecipitated with the flag antibodies showed its accumulation in G14A5-expressing cells during the pulse of 3 min. (Figure 3C). A similar precursor accumulation was also revealed in extracts from cells grown in the presence of MG132 for 8 h (not shown).
To examine the accumulation of preTfnR, we used a plasmid expressing the TfnR-GFP fusion protein. (Figure 3D). We observed an accumulation of unmodified TfnR-GFP, which most likely represents preTfnR-GFP, in cells with elongation arrest-defective SRP. Moreover, accumulation of mature TfnR-GFP was decreased by 40% in the cells expressing G14A12 and G14A5 consistent with the results shown previously for the endogenous TfnR (Figure 2D).
These results demonstrated that the elongation arrest function is essential for efficient translocation in vivo and its absence leads to defects in protein secretion and membrane protein accumulation.
Expression of the mutant versions of h14 significantly reduces cell growth
Next, we examined whether replacement of h14 with G14A12 and G14A5 impaired cell growth. Experiments were started with an equal number of cells and cell growth was quantified by counting manually the live cells collected every 24 hours starting at 96 hr. The cell count is expressed as fold increase in the number of cells present at the start (Figure 4A). Between 96 and 120 hr, when the endogenous h14 levels decrease from 100-50% in shRNA(14)-expressing cells (Lakkaraju et al., 2007), there is no detectable growth difference. However, after 120 hr, when cells became dependent on the GFP-h14 proteins for proper SRP assembly, G14A12 and G14A5-expressing cells displayed a significant growth defect. The doubling time of mock-depleted cells and of cells complemented with wild-type G14 was in the range of 21-23 hr. In contrast, it was 30-34 hr for cells expressing the elongation arrest-defective chimeras G14A12 and G14A5 (Figure 4B).
Figure 4. The absence of the elongation arrest function impairs cell growth.

(A) 293T cells were plated at equal densities and live cells counted at the times indicated (n=3). h14 depletion starts at 120 hr (Lakkaraju et al., 2007). No increased cell death was observed with GA12 and GA5 cells (see 0-300 AU in C).
(B) The doubling time was calculated for cell counts between 120-168 hr. Doubling times of 30-34 hr represent a 50-60% increase as compared to 21 hr of the mock treated cells.
(C) Propidium iodide-stained 293T cells were analyzed for cell cycle progression using FACSort. FI: Fluorescence intensity in arbitrary units [AU].
(D) Quantification of cells in different cell cycle phases (n=5). , The sum of cells in the three phases was set to 100%. p<0.05 for cells in G0/G1phase indicates statistical significance of the data at 95% confidence levels.
To analyze whether cell growth was delayed at a particular stage in cell cycle, we quantified the cells present in different phases of the cell cycle using propidium iodide staining followed by flow cytometric analysis (Figure 4C). The analysis revealed a specific and statistically significant increase in accumulation of the cells in G0/G1 phase in G14A5 cells as compared to the G14 cells (Figure 4D). Hence, reduced cell growth is, at least partially, explained by a delay in the G0/G1 phase of the cell cycle. After 168 hr, endogenous h14 levels start to increase again (Lakkaraju et al., 2007) and the negative effects of G14A12 and G14A5 expression disappeared rather rapidly (not shown). Our studies on cell growth were therefore limited in time. Cells expressing shRNAs stably after viral infection could not be used in the complementation assay, because they displayed higher levels of endogenous h14 than the cells in the transient transfection experiments (A.K.K.L. and K.S., unpublished results).
Slowing down nascent chain elongation with antibiotics reverts the translocation defect caused by mutant proteins
Next, we examined whether slowing down elongation of the nascent chains with low doses of protein synthesis inhibitors could rescue the mutant phenotype. We used two reversible inhibitors anisomycin and cycloheximide. Anisomycin prevents peptide bond formation, whereas cycloheximide interferes with translocation of the tRNA from the A to the P site (Gale et al., 1981) in the elongation cycle. SEAP secretion was monitored to follow the effects of the antibiotics on translocation. At 136 hr, 293T cells were treated for 2 hr with varying low doses of antibiotics. Subsequently, the medium was changed to remove already secreted SEAP and the cells were incubated for another 6 hr with the antibiotics before collecting the medium.
In the absence of antibiotics, we observed the same reduction in SEAP secretion for G14A12 and G14A5-expressing cells as already reported (Figure 2A and 5A). Adding increasing amounts of anisomycin increased the relative amounts of SEAP secreted by cells with an elongation arrest-defective SRP (G14A12 and G14A5) when compared to cells with fully functional SRP (G14). At the lowest dose of antibiotics, the relative secretion efficiency of SEAP was already improved and it became equal at a concentration of 0.03 μg/ml of anisomycin, (Figure 5A, see Figure S3 for the results with HeLa cells). The antibiotic specifically rescued the phenotype caused by G14A12 and G14A5, since it failed to restore secretion levels of cells depleted of h14 and complemented with GFP. With cycloheximide, we saw the same dose-dependent increase in secretion efficiency (Figure 5B). In contrast to anisomycin, cycloheximide was capable to compensate for low levels of SRP at higher concentrations. This is because SRP is thought to interact with the ribosome at the step in the elongation cycle, which is slowed down by cycloheximide (Ogg and Walter, 1995). By slowing down this step, the low amounts of SRP present in h14-depleted cells become sufficient for efficient ER-targeting (Lakkaraju et al., 2007).
Figure 5. The mutant phenotypes of G14A12 and G14A5 are specifically rescued by slowing down nascent chain elongation.

(A) (B) and (C) SEAP secretion from 293T cells in the presence of anisomycin (ANM), cycloheximide (CHX) or hippuristanol. For each concentration, the secretion activities were normalized to the one of shRNA(Luc)/G14 cells (n=3). The absolute activities of mock-depleted cells were decreased to 11, 16 and 15 % at 0.03 μg/ml of anisomycin, at 3 μg/ml cycloheximide and at 0.75 μM hippuristanol, respectively, as compared to control cells.
(D) Preprotein accumulation in the presence of anisomycin at 168 hr. Equal amounts of cell extracts were displayed by SDS-PAGE and pPL3f revealed with anti-flag antibodies (n=2).
(E), (F) and (G) Effects of antibiotics on the cellular protein synthesis rate in mock-depleted 293T cells. Cells were labeled for 15 min. with [35S]methionine/cysteine, labeled proteins were acid-precipitated and quantified (n=3). In parallel experiments we found that the [35S]-uptake in shRNA(Luc)-G14, shRNA(14)-G14 and shRNA(14)-G14A5 cells without antibiotics was 31'354, 33'047 and 29'149 total counts/106 cells, respectively, indicating that protein synthesis was unaffected by GFP-h14A5 expression.
Next, we monitored the relative cellular protein synthesis rate by quantifying the [35S]-uptake in the presence of the antibiotics. There was no difference in the incorporation between the mock-depleted cells and the cells complemented with h14-GFP proteins (Figure 5, legend). The cellular protein synthesis rate was reduced by 4-4.5-fold at the concentration of anisomycin or cycloheximide required to rescue the secretion defect caused by the absence of the elongation arrest function (anisomycin 0.03 μg/ml, cycloheximide at 3 μg/ml, Figure 5E and F).
To confirm that the antibiotics restored secretion by increasing the translocation efficiency, we monitored preprolactin accumulation in anisomycin-treated cells. In the presence of anisomycin, the preprotein was barely detectable in G14A5-expressing cells (Figure 5D).
The reversion of the phenotype was specific for inhibitors of translation elongation, since hippuristanol, an inhibitor of initiation (Bordeleau et al., 2006) failed to restore SEAP secretion (Figure 5C and G). At five-fold decreased protein synthesis rate, SEAP secretion was only increased to 65%. Notably, hippuristanol increased the secretion efficiency partially in a dose-dependent fashion consistent with a subsequent rate-limiting step.
These results demonstrated that a delay in elongation is essential for efficient translocation of preproteins and indicated the presence of a rate limiting step subsequent to the formation of the SRP-RNC complex.
Cellular SR levels are rate-limiting in targeting
To determine whether SR was rate-limiting in targeting, we expressed exogenous human SRα and SRβ from two separate plasmids transfected into HeLa cells at the same time as the expression plasmids for the GFP-h14 chimeras. When expressed together, the levels of SRα and SRβ were increased by 2.4 and 2.1 fold, respectively, as compared to the endogenous proteins and both proteins were found associated with the ER (Figure 6A and B). The subunits expressed individually also accumulated stably and were properly localized (Figure 6 and data not shown). As before, without the elongation arrest function, SEAP secretion was reduced to 44% as compared to control cells (Figure 6C). Expression of individual receptor subunits increased SEAP secretion only slightly. Expression of both subunits rescued secretion to 72% as compared to cells not depleted of h14. These results showed that the availability of SR is rate-limiting in targeting.
Figure 6. Increasing receptor levels compensate the effects of protein mutants.

(A) Western blot analysis from HeLa cells co-expressing either G14 or G14A5 with one or both SRα and SRβ. b/a indicates the increase in SR subunit expression standardized to β-actin.
(B) Cellular localization of SRα and SRβ. Immunofluorescent images from HeLa cells expressing G14A5 and both SR subunits labeled with anti-SRα and anti-SRβ antibodies.
(C) SEAP secretion of cells co-expressing either G14A5 or, as controls, G14 or GFP and one or both SR subunits.
In the presence of the elongation arrest function, when all nascent chains become successfully targeted to the translocon, and in the absence of exogenous h14 (at low levels of SRP) the expression of SRα and SRβ did not significantly change the secretion efficiency (Fig. 6C, G14 and GFP). Similar results were obtained in a set of experiments which were done in 293T cells with the receptors subunits expressed as fusion proteins tagged with GFP (GFP-SRα) and three flag epitopes (SRβ-3f, Figure S4).
Discussion
The results of these studies demonstrate for the first time that elongation arrest activity has a fundamental role in maintaining the structure and function of mammalian cells. Its absence leads to the depletion of proteins in the endogenous membrane system and to the reduction of protein secretion with profound consequences on cell growth. SRP lacking elongation arrest activity causes defects in protein translocation that are reversed when the overall rate of nascent chain elongation is decreased by four-fold. This result demonstrates that when the cellular translation elongation rate of signal sequence-containing proteins exceeds the rate of targeting those proteins to the ER membrane, translocation into the ER does not proceed efficiently. The rate-limiting factor in targeting in vivo is the SRP receptor: if its concentration is insufficient, delays in the targeting of RNCs to the ER occur because of a shortage of operational targeting sites. As a consequence, not all nascent chains will reach the translocon in time for successful engagement into translocation. But importantly, this effect may also serve a regulatory purpose. Since SRP will dissociate preferentially from SRP-RNC complexes with low affinity while awaiting a free SR, nascent chains will be targeted and processed more frequently at the ER if their SRP-signal sequence affinity is high rather than low.
In pancreatic microsomes, SR is about two-fold less abundant than active translocons (Guth et al., 2004), consistent with a rate-limiting function in secretion. Moreover, the translocon might become rate-limiting at more than two-fold increased SR levels providing an explanation for the incomplete rescue of translocation in these experiments. Exogenous SR expression in wild-type cells did not improve secretion efficiency suggesting that subsequent steps in protein translocation and secretion become limiting. SR expression also failed to improve secretion at low SRP levels consistent with the interpretation that SRP is rate-limiting under these conditions. Very little is known about the relative abundance of SRP and the membrane components in different cell types or tissues, and this issue needs to be explored more thoroughly to understand whether elongation arrest activity has the same fundamental importance for all cell types as suggested by our experiments and whether it might also be used for regulatory purposes. S. cerevisiae failed to reveal growth or translocation defects under normal growth conditions in a strain expressing an elongation arrest-defective version of the Srp14p subunit. It has a temperature-sensitive growth defect similar to the one observed for strains lacking Srp14p altogether (Mason et al., 2000). The difference may be explained by increased cellular levels of SR and by a higher capacity of S. cerevisiae to translocate proteins post-translationally.
A kinetic study in COS-1 cells revealed that the average targeting time for a reporter protein was very short (about 5 s) (Goder et al., 2000). During this time period, nascent chains only elongate by 40 amino acid residues (with an average elongation rate of 8 aa/s), making the elongation arrest function presumably unnecessary. As already discussed in (Goder et al., 2000), this time period represents the average time for all ribosomes and it is conceivable that the targeting time for the first ribosome is considerably longer than that of the subsequent ribosomes. One factor which may shorten the average targeting time is SRP-mediated ribosome stacking, which was shown in cell-free translation systems (Wolin and Walter, 1988,Wolin and Walter, 1989).
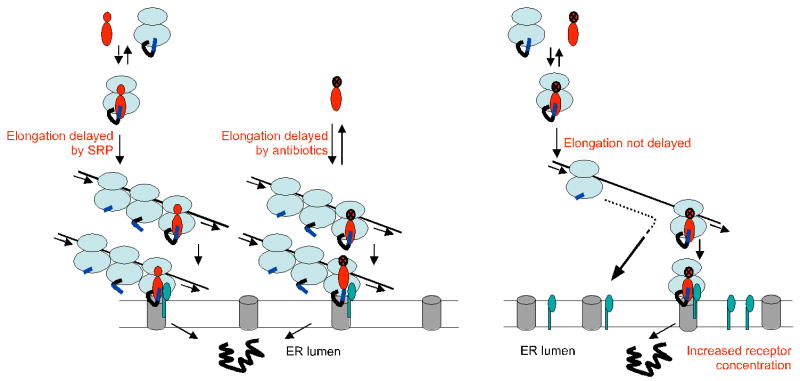
Based on these observations, our results may be explained with the following model (Figure 7). At limiting SR concentrations, the first targeting event is critical because in the absence of a delay mediated by SRP or antibiotics, nascent chains extend beyond a critical length which renders them translocation-incompetent. With SRP and antibiotics, the average length of nascent chains is decreased during the time window required for targeting, and ribosomes stack along the mRNA by switching the mode of translation from initiation-limited (Sonenberg et al., 2000) to elongation-limited. The first ribosome will be arrested by signal sequence binding of SRP, whereas the following ribosome stacks behind the first one without the need for SRP. This effect may reduce the number of SRP bound to polysomes, consistent with the 20-50-fold lower abundance of SRP as compared to ribosomes (Bovia et al., 1995; Raue et al., 2007). With antibiotics, the ribosomes will stack close to the translation start site. SRP will bind to the leading ribosome as soon as the signal sequence emerges. In both cases, targeting of the leading ribosome will increase the concentration of short nascent chains in close proximity to the membrane-bound SR. Although each nascent chain may still need to be targeted individually (Rapoport et al., 1987), the process may be accelerated and thereby decreases the average targeting time. Both antibiotics have a compensatory effect indicating that the stage at which the ribosome is arrested in the elongation cycle is apparently not critical for restoring efficient translocation.
Figure 7. A model for SRP-mediated targeting.
The elongation arrest function of SRP reduces nascent chain length and induces ribosome stacking during the targeting process to ensure highest efficacy in protein translocation at rate-limiting SR levels. At increased SR levels, more nascent chains are target successfully even in the absence of a delay in elongation (see also text). SRP: red; SRP with black cross: SRP without the elongation arrest function; SR: green.
At higher levels of membrane-bound SR, the targeting frequency of SRP-RNCs is increased resulting in the successful targeting of more nascent chains, even in the absence of a delay in elongation and subsequent ribosome stacking. The average occupancy of mRNAs is in the range of one ribosome every 80-100 nucleotides (Sonenberg et al., 2000).
Nascent chains may loose competence for translocation because translation has already terminated or nascent chains have elongated beyond a critical length after which they cannot be targeted (Siegel and Walter, 1985; Wiedmann et al., 1987; Flanagan et al., 2003). Why nascent chain length interferes with targeting still remains to be elucidated. It was first suggested that SRP no longer recognizes the signal sequence in longer nascent chains because it becomes sequestered (Siegel and Walter, 1985). However, more recent experiments demonstrated that SRP binds with the same affinity to RNCs with preprolactin nascent chains of different length (Flanagan et al., 2003). It is conceivable that longer nascent chains may interfere sterically with SR and/or translocon recognition.
The cellular generation time is increased by 50% in the absence of the elongation arrest function. Plausibly, the delay in growth is most prominent in the G1 phase of the cell cycle when cells depend on rapid synthesis of membrane and secretory proteins. We expect the absence of elongation arrest activity to result in cell death, since essential membrane components will eventually become limiting causing severe defects in cell structure and function.
The essential motif of only five amino acid residues in the C-terminal region of SRP14 is highly conserved. The dramatic effect of the mutation suggests that defined and limited interactions, most likely with the ribosome, cause the arrest in elongation. Based on available atomic structure data (Weichenrieder et al., 2000) combined with cryo-electron microscopy densities (Halic et al., 2004), it is not yet possible to assign a contact site in the ribosome for the SRP14 motif. A similar small basic peptide in the ER membrane protein Erj1p inhibits translation initiation (Dudek et al., 2005). In contrast to the Alu domain, the protein was found to bind at the nascent chain exit site (Blau et al., 2005).
An Alu domain-like RNA structure is present in most eukaryal and archaeal SRPs, and also in a few eubacterial SRPs. However, SRP14 proteins have only been identified in eukaryal species. Organisms lacking SRP14 may have developed other mechanisms to interfere with elongation. SRP in trypanosomes comprises a small tRNA-like RNA (sRNA-85) instead of SRP9/14 (Lustig et al., 2005) and sRNA-85 may well replace the function of SRP14 (Liu et al., 2003).
The described complementation assay may serve as a tool to study the physiological relevance of mutations in other SRP subunits and may prove useful in exploring the functions of other ribonucleoprotein particles.
Experimental Procedures
Plasmid construction and SRP assembly in vivo
Please refer to the supplementary Experimental Procedures for a detailed description.
Western blotting and antibodies
7SL RNA analysis and Western blotting with most antibodies are described in Lakkaraju et al., 2007. Additional antibodies: Anti-flag M2 (1:10'000, Sigma), anti-SRα (1:2'000, Abcam), anti-SRβ (1:500, kind gift of Dr. P. Walter (UCSF, California).
Cell culture
Human HeLa (CCL-2) and HEK 293T cells were grown at 37°C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (Sigma). Equal numbers of cells were initially transfected with shRNA(14) and, for mock depletion, with shRNA(Luc). After 24 hr, cells were selected for 24 hr in puromycin (3 μg/ml). At 72 hr, cells were plated at equal numbers into 6 cm dishes and transfected with GFP-h14-expressing plasmids (or GFP as a control) and, if applicable, with a plasmid expressing a reporter protein. Antibiotics were added initially for 2 hr at 136 hr, then the medium was changed and the cells were incubated for another 6 hr with antibiotics before collecting the medium and lysing the cells (Lakkaraju et al., 2007). The cell number was quantified manually by counting the number of live cells. Growth rates were calculated for every 24 hr. Cell cycle assays are described in supplemental Experimental Procedures.
SEAP assay, glycoprotein, VSV-G, and TfnR-GFP cell surface quantification and microscopy
Methods are described in Lakkaraju et al., 2007. Cell surface glycoprotein staining is described in supplemental Experimental Procedures.
Pulse labeling
At 168 h after initial transfection, MG132 (25 μM) was added and after 45 min. 293T cells were labeled with 200 μCi/ml of [35S]methionine/cysteine mix (Hartmann Analytic) for 3 min. Cells were lysed with 200 μl of 1% SDS in 0.1M Tris, pH 8 directly in the plate. The lysate was transferred into a microtube and heated to 70°C with occasional vortexing until the lysate became less viscous. The sample was diluted (3-fold) in the IP buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS 50mM Tris-Hcl pH 8, 1mM EDTA and 1× protease inhibitor cocktail) and the labeled proteins incubated overnight with the antibodies. The antibodies were immobilized on protein G-Sepharose (40 μl) for 4 hr. The precipitates were washed 3× with IP buffer for 15 min. and the bound proteins displayed by 12% SDS-PAGE. Visualization and quantification was done with the Bio-Rad phosphorimager.
Protein and RNA expression, purification, reconstitution of SRP and activity assays
Proteins and synthetic SRP RNA were expressed and purified as described in supplemental Experimental procedures and in (Huck et al., 2004) and (Terzi et al., 2004). The targeting assay was done as described in (Flanagan et al., 2003). Cycloheximide, SRP and EKRM were used at concentrations of 500 μM, 200 nM and 1 eq/reaction, respectively. EKRM membranes were pre-saturated as described in (Schaletzky and Rapoport, 2006). The protein contents of the supernatants and pellets were analyzed on a 15% Tricine gel and the radioactivity was detected and quantified with a Bio-Rad phosphorimager.
Supplementary Material
Acknowledgments
We thank A. Chassot, Y. Miao, and Y. Shao for technical assistance, Drs J. Pelletier, R.S. Hegde and P. Walter for providing reagents and Dr. D. Picard for suggestions on the manuscript. This work was supported by grants from the Swiss National Science Foundation, the Canton of Geneva and the MEDIC Foundation. Support from NIH grants R01GM26494 and the Robert A. Welch Foundation (Chair grant BE-0017) are also acknowledged (A.E.J.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Asvin K.K. Lakkaraju, Département de biologie cellulaire, Université de Genève, Sciences III, 1211 Geneva, Switzerland.
Camille Mary, Département de biologie cellulaire, Université de Genève, Sciences III, 1211 Geneva, Switzerland.
Anne Scherrer, Département de biologie cellulaire, Université de Genève, Sciences III, 1211 Geneva, Switzerland.
Arthur E. Johnson, Department of Molecular and Cellular Medicine, Texas A&M University System Health Science Center, College Station, TX 77843-1114, USA
Katharina Strub, Département de biologie cellulaire, Université de Genève, Sciences III, 1211 Geneva, Switzerland.
References
- Alder NN, Johnson AE. Cotranslational membrane protein biogenesis at the endoplasmic reticulum. J Biol Chem. 2004;279:22787–22790. doi: 10.1074/jbc.R400002200. [DOI] [PubMed] [Google Scholar]
- Blau M, Mullapudi S, Becker T, Dudek J, Zimmermann R, Penczek PA, Beckmann R. ERj1p uses a universal ribosomal adaptor site to coordinate the 80S ribosome at the membrane. Nat Struct Mol Biol. 2005;12:1015–1016. doi: 10.1038/nsmb998. [DOI] [PubMed] [Google Scholar]
- Bordeleau ME, Mori A, Oberer M, Lindqvist L, Chard LS, Higa T, Belsham GJ, Wagner G, Tanaka J, Pelletier J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat Chem Biol. 2006;2:213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- Bovia F, Fornallaz M, Leffers H, Strub K. The SRP9/14 subunit of the signal recognition particle (SRP) is present in more than 20-fold excess over SRP in primate cells and exists primarily free but also in complex with small cytoplasmic Alu RNAs. Mol Biol Cell. 1995;6:471–484. doi: 10.1091/mbc.6.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovia F, Wolff N, Ryser S, Strub K. The SRP9/14 subunit of the human signal recognition particle binds to a variety of Alu-like RNAs and with higher affinity than its mouse homolog. Nucleic Acids Res. 1997;25:318–326. doi: 10.1093/nar/25.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly T, Gilmore R. Formation of a functional ribosome-membrane junction during translocation requires the participation of a GTP-binding protein. J Cell Biol. 1986;103:2253–2261. doi: 10.1083/jcb.103.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly T, Rapiejko PJ, Gilmore R. Requirement of GTP hydrolysis for dissociation of the signal recognition particle from its receptor. Science. 1991;252:1171–1173. doi: 10.1126/science.252.5009.1171. [DOI] [PubMed] [Google Scholar]
- Dudek J, Greiner M, Muller A, Hendershot LM, Kopsch K, Nastainczyk W, Zimmermann R. ERj1p has a basic role in protein biogenesis at the endoplasmic reticulum. Nat Struct Mol Biol. 2005;12:1008–1014. doi: 10.1038/nsmb1007. [DOI] [PubMed] [Google Scholar]
- Flanagan JJ, Chen JC, Miao Y, Shao Y, Lin J, Bock PE, Johnson AE. Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. J Biol Chem. 2003;278:18628–18637. doi: 10.1074/jbc.M300173200. [DOI] [PubMed] [Google Scholar]
- Gilmore R, Blobel G, Walter P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J Cell Biol. 1982;95:463–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goder V, Crottet P, Spiess M. In vivo kinetics of protein targeting to the endoplasmic reticulum determined by site-specific phosphorylation. EMBO J. 2000;19:6704–6712. doi: 10.1093/emboj/19.24.6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guth S, Volzing C, Muller A, Jung M, Zimmermann R. Protein transport into canine pancreatic microsomes: a quantitative approach. Eur J Biochem. 2004;271:3200–3207. doi: 10.1111/j.1432-1033.2004.04252.x. [DOI] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CM, Grassucci RA, Frank J, Beckmann R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- Hare JF. Mechanisms of membrane protein turnover. Biochim Biophys Acta. 1990;1031:71–90. doi: 10.1016/0304-4157(90)90003-u. [DOI] [PubMed] [Google Scholar]
- Hauser S, Bacher G, Dobberstein B, Lutcke H. A complex of the signal sequence binding protein and the SRP RNA promotes translocation of nascent proteins. EMBO J. 1995;14:5485–5493. doi: 10.1002/j.1460-2075.1995.tb00235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck L, Scherrer A, Terzi L, Johnson AE, Bernstein HD, Cusack S, Weichenrieder O, Strub K. Conserved tertiary base pairing ensures proper RNA folding and efficient assembly of the signal recognition particle Alu domain. Nucleic Acids Res. 2004;32:4915–4924. doi: 10.1093/nar/gkh837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsson N, Varshavsky A. Split ubiquitin as a sensor of protein interactions in vivo. Proc Natl Acad Sci USA. 1994;91:10340–10344. doi: 10.1073/pnas.91.22.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- Lakkaraju AK, Luyet PP, Parone P, Falguieres T, Strub K. Inefficient targeting to the endoplasmic reticulum by the signal recognition particle elicits selective defects in post-ER membrane trafficking. Exp Cell Res. 2007;313:834–847. doi: 10.1016/j.yexcr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Lipp J, Dobberstein B, Haeuptle MT. Signal recognition particle arrests elongation of nascent secretory and membrane proteins at multiple sites in a transient manner. J Biol Chem. 1987;262:1680–1684. [PubMed] [Google Scholar]
- Liu L, Ben-Shlomo H, Xu YX, Stern MZ, Goncharov I, Zhang Y, Michaeli S. The trypanosomatid signal recognition particle consists of two RNA molecules, a 7SL RNA homologue and a novel tRNA-like molecule. J Biol Chem. 2003;278:18271–18280. doi: 10.1074/jbc.M209215200. [DOI] [PubMed] [Google Scholar]
- Lustig Y, Goldshmidt H, Uliel S, Michaeli S. The Trypanosoma brucei signal recognition particle lacks the Alu-domain-binding proteins: purification and functional analysis of its binding proteins by RNAi. J Cell Sci. 2005;118:4551–4562. doi: 10.1242/jcs.02578. [DOI] [PubMed] [Google Scholar]
- Mason N, Ciufo LF, Brown JD. Elongation arrest is a physiologically important function of signal recognition particle. EMBO J. 2000;19:4164–4174. doi: 10.1093/emboj/19.15.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer DI, Krause E, Dobberstein B. Secretory protein translocation across membranes-the role of the “docking protein”. Nature. 1982;297:647–650. doi: 10.1038/297647a0. [DOI] [PubMed] [Google Scholar]
- Ng DT, Brown JD, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg SC, Walter P. SRP samples nascent chains for the presence of signal sequences by interacting with ribosomes at a discrete step during translation elongation. Cell. 1995;81:1075–1084. doi: 10.1016/s0092-8674(05)80012-1. [DOI] [PubMed] [Google Scholar]
- Okun MM, Shields D. Translocation of preproinsulin across the endoplasmic reticulum membrane. The relationship between nascent polypeptide size and extent of signal recognition particle-mediated inhibition of protein synthesis. J Biol Chem. 1992;267:11476–11482. [PubMed] [Google Scholar]
- Pool MR. Signal recognition particles in chloroplasts, bacteria, yeast and mammals (review) Mol Membr Biol. 2005;22:3–15. doi: 10.1080/09687860400026348. [DOI] [PubMed] [Google Scholar]
- Powers T, Walter P. The nascent polypeptide-associated complex modulates interactions between the signal recognition particle and the ribosome. Curr Biol. 1996;6:331–338. doi: 10.1016/s0960-9822(02)00484-0. [DOI] [PubMed] [Google Scholar]
- Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- Rapoport TA, Heinrich R, Walter P, Schulmeister T. Mathematical modeling of the effects of the signal recognition particle on translation and translocation of proteins across the endoplasmic reticulum membrane. J Mol Biol. 1987;195:621–636. doi: 10.1016/0022-2836(87)90186-0. [DOI] [PubMed] [Google Scholar]
- Raue U, Oellerer S, Rospert S. Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J Biol Chem. 2007;282:7809–7816. doi: 10.1074/jbc.M611436200. [DOI] [PubMed] [Google Scholar]
- Rutledge EA, Mikoryak CA, Draper RK. Turnover of the transferrin receptor is not influenced by removing most of the extracellular domain. J Biol Chem. 1991;266:21125–21130. [PubMed] [Google Scholar]
- Schaletzky J, Rapoport TA. Ribosome binding to and dissociation from translocation sites of the endoplasmic reticulum membrane. Mol Biol Cell. 2006;17:3860–3869. doi: 10.1091/mbc.E06-05-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan SO, Stroud RM, Walter P. Mechanism of association and reciprocal activation of two GTPases. PLoS Biol. 2004;2:e320. doi: 10.1371/journal.pbio.0020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V, Walter P. Elongation arrest is not a prerequisite for secretory protein translocation across the microsomal membrane. J Cell Biol. 1985;100:1913–1921. doi: 10.1083/jcb.100.6.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V, Walter P. Removal of the Alu structural domain from signal recognition particle leaves its protein translocation activity intact. Nature. 1986;320:81–84. doi: 10.1038/320081a0. [DOI] [PubMed] [Google Scholar]
- Terzi L, Pool MR, Dobberstein B, Strub K. Signal recognition particle Alu domain occupies a defined site at the ribosomal subunit interface upon signal sequence recognition. Biochem. 2004;43:107–117. doi: 10.1021/bi0353777. [DOI] [PubMed] [Google Scholar]
- Thomas Y, Bui N, Strub K. A truncation in the 14 kDa protein of the signal recognition particle leads to tertiary structure changes in the RNA and abolishes the elongation arrest activity of the particle. Nucleic Acids Res. 1997;25:1920–1929. doi: 10.1093/nar/25.10.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol. 1981;91:557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichenrieder O, Wild K, Strub K, Cusack S. Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature. 2000;408:167–173. doi: 10.1038/35041507. [DOI] [PubMed] [Google Scholar]
- Wiedmann M, Kurzchalia TV, Bielka H, Rapoport TA. Direct probing of the interaction between the signal sequence of nascent preprolactin and the signal recognition particle by specific cross-linking. J Cell Biol. 1987;104:201–208. doi: 10.1083/jcb.104.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988;7:3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Walter P. Signal recognition particle mediates a transient elongation arrest of preprolactin in reticulocyte lysate. J Cell Biol. 1989;109:2617–2622. doi: 10.1083/jcb.109.6.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Walter P. Discrete nascent chain lengths are required for the insertion of presecretory proteins into microsomal membranes. J Cell Biol. 1993;121:1211–1219. doi: 10.1083/jcb.121.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.