Abstract
The critical role of protein-protein interactions in the chemistry of polyketide synthases is well established. However, the transient and weak nature of these interactions, in particular those involving the acyl carrier protein (ACP), has hindered efforts to structurally characterize these interactions. We describe a chemo-enzymatic approach that crosslinks the active sites of ACP and their cognate ketosynthase (KS) domains, resulting in the formation of a stable covalent adduct. This process is driven by specific protein-protein interactions between KS and ACP domains. Suitable manipulation of the reaction conditions enabled complete crosslinking of a representative KS and ACP, allowing isolation of a stable, conformationally constrained adduct suitable for high-resolution structural analysis.
Keywords: acyl carrier protein, ketosynthase, crosslinking, polyketide synthase, protein-protein interaction
Since the discovery of colinearity between gene sequence and product structure of the 6-deoxyerythronolide B synthase, 1,2 there has been considerable effort towards harnessing the programmable biochemistry of multimodular polyketide synthases (PKSs). More recently, there has been a growing realization of the importance of protein-protein interactions in the successful engineering of these megasynthases. 3,4,5 A key challenge encountered in studying these protein-protein interactions is their inherent weakness (Kd > 1 µM in most cases). Here we describe the development of a new method for studying the weak but crucial non-covalent interaction between the acyl carrier protein (ACP) and the ketosynthase (KS) domains of a modular PKS.
The acyl carrier protein (ACP) is a small (~10 kDa), highly conserved domain found in PKSs as well as the related fatty acid synthases and nonribosomal peptide synthetases. Although it lacks intrinsic catalytic activity, it is involved in every step of polyketide biosynthesis, collaborating with enzymes involved in building block incorporation (the acyl transferase, AT), β-carbon processing (the ketoreductase, KR, dehydratase, DH, and enoyl reductase, ER), and finally chain termination (the thioesterase, TE). The specificity of these protein-protein interactions has been noted,6,7,8,9,10 and the high-resolution structure of a prototypical ACP domain from module 2 of the 6-deoxyerythronolide B synthase (DEBS) has also been reported.11 However, elucidating the structure of a complex between an ACP domain of a modular PKS and its partner domains has been challenging, presumably due to the exceptionally high mobility of this domain.
To directly visualize the interactions between the KS domain of a modular PKS and its cognate ACP partner, we have taken the approach of cross-linking the two domains using chemically synthesized Coenzyme A (CoA) analogues. Although 1,3-dibromopropanone has been used as a cross-linking reagent to study FASs,12 we found this approach to be unsuitable for the problem at hand due to low yields of the crosslinked product. In contrast, the mechanism-based crosslinking method of Burkart and coworkers13 yielded more promising results, as described in this report. Briefly, this method harnesses enzymes from the CoA biosynthetic pathway to yield CoA analogues that can be attached to the active site serine of an ACP using a non-specific phosphopantetheinyl transferase (PPTase). The crosslinkers of greatest interest harbor an electrophilic warhead in place of the nucleophilic cysteamine moiety, and are therefore able to react irreversibly with the active site cysteine of the KS domains. Several CoA analogues capable of crosslinking the KS and ACP proteins of the E. coli fatty acid synthase were reported by Burkart and coworkers.13,14
Given the mechanistic and structural similarity between fatty acid synthases and PKSs, we screened compounds 1–7 (Figure 1) for their ability to crosslink the didomain [KS][AT] proteins of DEBS modules 3 and 5 with their cognate stand-alone ACP domains. The X-ray crystal structures of both these didomain proteins have been solved to atomic resolution.15,16 A one-pot reaction was performed containing compounds 1–7 individually, along with three CoA biosynthetic enzymes (CoaA, CoaD and CoaE), the Sfp PPTase and either ACP3 or ACP5 in phosphate buffer. Thereafter, the [KS3][AT3] protein or the [KS5][AT5] protein was added, and the reaction products were analyzed by SDS-PAGE (Figure 2).17 In the presence of the electrophilic CoA analogues, bands with apparent molecular weights corresponding to a [KS][AT]-ACP adduct were observed with both modules. Interestingly, compound 1 showed the highest efficiency for cross-linking the DEBS proteins, whereas its geometric isomer 2 was inactive. Compound 1 was also the most effective in crosslinking the E. coli fatty acid synthase proteins.13 However, unlike the latter system, where all compounds showed reasonable crosslinking efficiency, the DEBS proteins were much more selective. Analysis of reaction mixtures incubated without [KS3][AT3] by matrix assisted laser desorption/ionization – time of flight mass spectrometry showed greater than 90% conversion of apo to crypto-ACP18 for compounds 1–7, eliminating differences in pantetheinylation as a possible cause of the observed selectivity. Structural or computational modeling studies with these alternative CoA analogues docked in the active site of the KS may offer some insight into the mechanistic origins of this discriminative power.
Figure 1.
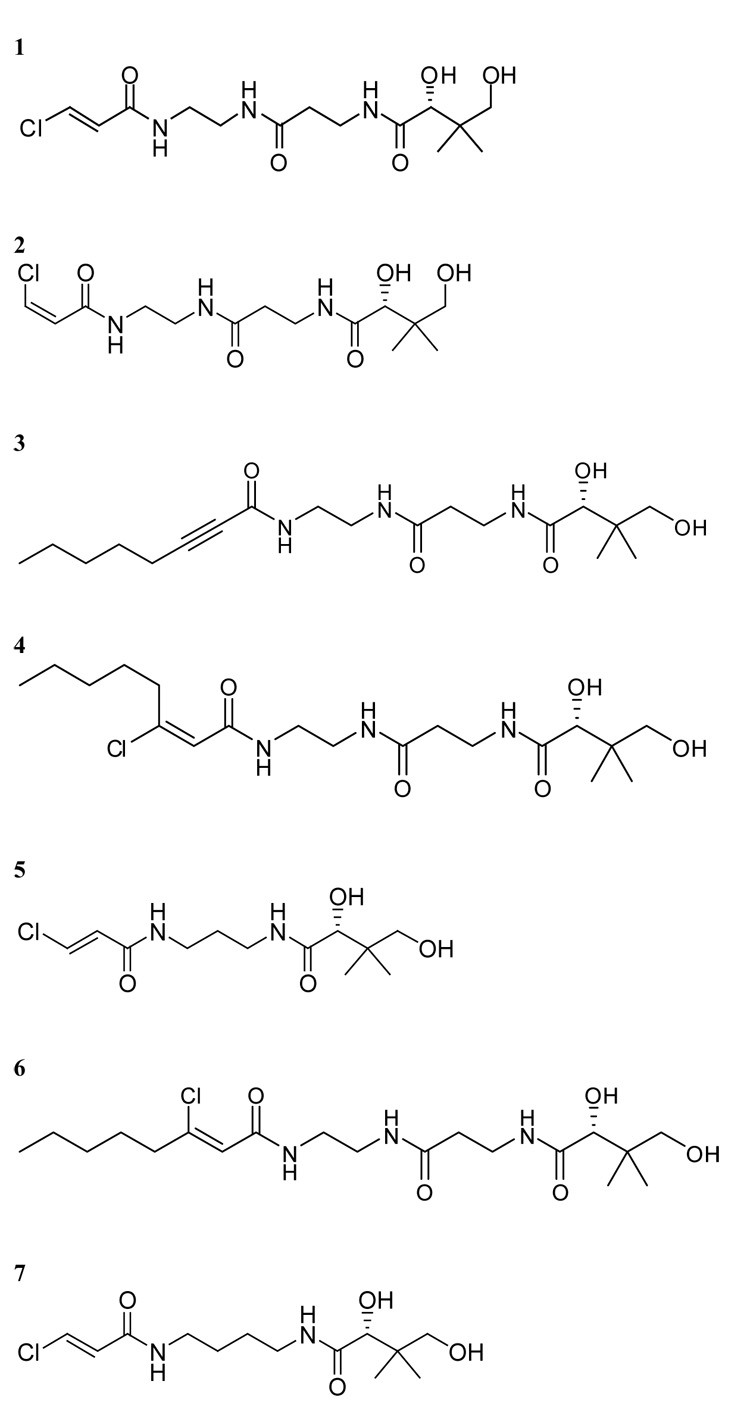
Structures of crosslinkers used in this study.
FIGURE 2.
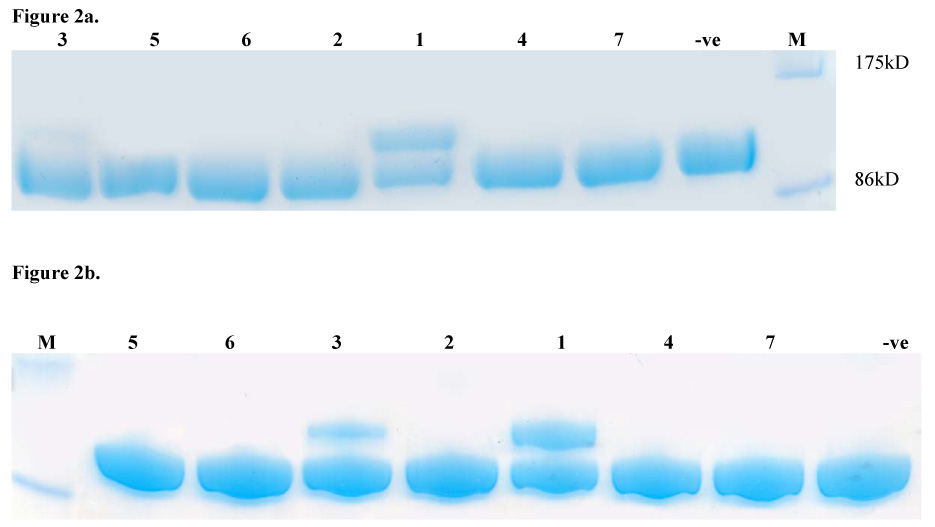
FIGURE 2a. SDS-PAGE analysis of crosslinking reaction mixtures containing compounds 1–7, [KS3][AT3] and ACP3. For compound identity refer to Figure 1.
FIGURE 2b. SDS-PAGE analysis of crosslinking reaction mixtures containing compounds 1–7, [KS5][AT5] and ACP5. For compound identity refer to Figure 1.
The ACP construct utilized in this study was engineered with the FLAG epitope19 (DYKDDDDK) the presence of which can be easily detected by Western blot analysis utilizing commercially available antibodies. SDS-PAGE followed by Western blot analysis20 (Figure 3a) revealed that the observed higher molecular weight band was indeed the expected [KS][AT]-ACP adduct.
Figure 3.
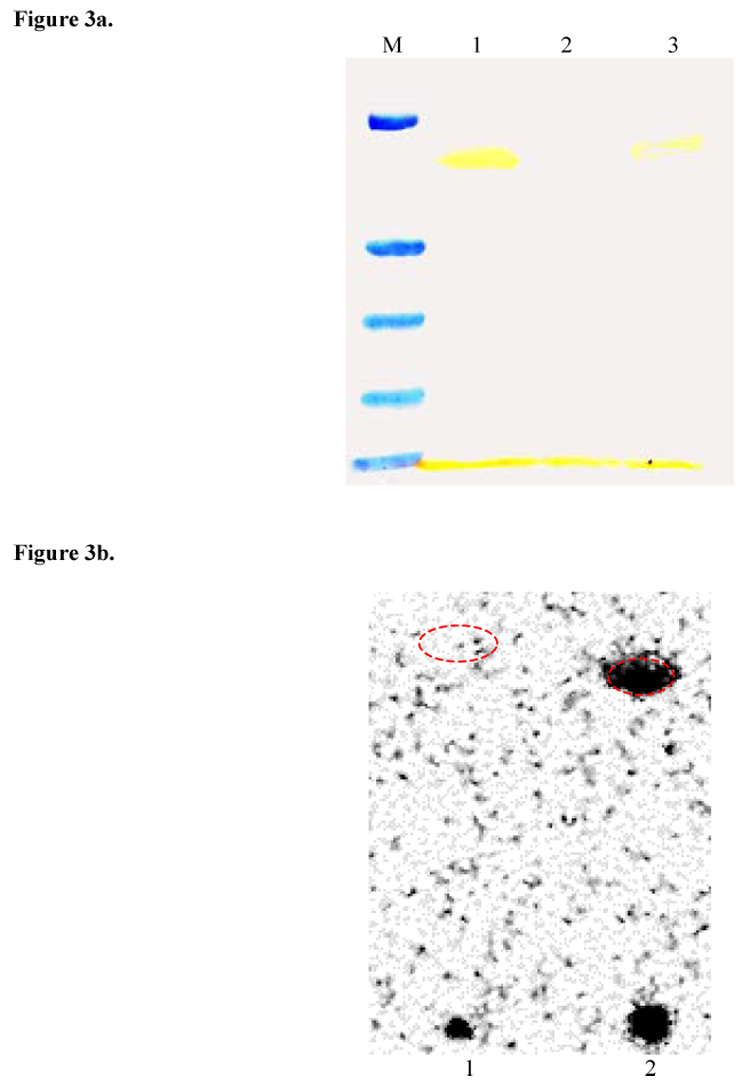
Figure 3a. Western blot analysis of lane 6 from Figure 2a with anti-FLAG M2 antibody confirming formation of [KS3][AT3]-ACP3 crosslinked adduct(lane 1).Negative control(lane 2) contained no crosslinker. Bands visible at the bottom are due to unreacted ACP3. Lane 3 is a 2-fold dilution of lane 1.
Figure 3b. Radio SDS-PAGE image of [14C](2S,3R)-2-Methyl-3-hydroxypentanoyl-N-acetylcysteamine thioester labeling of purified crosslinked adduct(lane 1) and [KS3][AT3](lane 2). Red dashed ellipses indicate the position of the bands on the SDS-PAGE gel visualized by coomassie blue staining.
The active site cysteine of a KS domain can be selectively acylated by a suitably loaded N-acetylcysteamine thioester which functions as a surrogate for the phosphopantetheinyl arm of the ACP.3,9 When incubated with [14C](2S,3R)-2-Methyl-3-hydroxypentanoyl-N-acetylcysteamine thioester, the crosslinked [KS][AT]-ACP adduct could not be labeled, whereas the control [KS][AT] protein was readily labeled (Figure 3b).21 This observation confirmed that the active site cysteine of the KS domain was the site of crosslinking between the [KS][AT] and the ACP.
Earlier studies have demonstrated that KS-ACP interactions in DEBS are specific.9 To investigate whether this differential specificity could affect KS-ACP crosslinking, [KS3][AT3] was crosslinked with ACP3 and ACP5 utilizing 1 as the linker. The KS demonstrated a clear preference for its cognate ACP, as evidenced by the higher intensity of cross-linked band (Figure 5). A similar experiment performed with [KS5][AT5] using linkers 1 and 3 confirmed the hypothesis that protein-protein interactions drive cross-linking (Figure 5).
Figure 5.
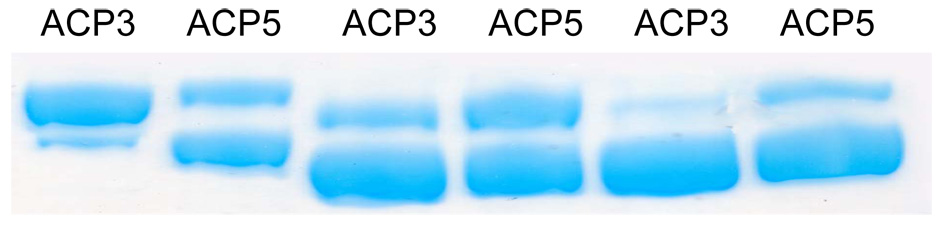
SDS-PAGE analysis of reaction mixtures containing [KS3][AT3](lanes 1 and 2) or [KS5][AT5](lanes 3–6) and alternative ACP domains. Crosslinker 1 was used in lanes 1–4, whereas compound 3 was used in lanes 5 and 6. Reaction conditions were identical to those described for lane 3 in Figure 4.
Structural studies have verified that the active [KS][AT] didomain protein exists as a homodimer with two active sites per complex.15,16 Thus, it can be deduced that an incompletely crosslinked [KS][AT] protein exists in solution as three distinct species with 0, 1 or 2 ACP domains covalently attached to the [KS][AT] homodimer. Although the presence of orthogonal purification tags (FLAG on ACP and hexa-His on [KS][AT]) can be exploited to purify the crosslinked species from the two starting materials via affinity chromatography, incompletely crosslinked homodimers cannot be separated from 2:2 complexes in a straightforward fashion. This represents a major roadblock for obtaining a homogeneous preparation of the desired species for X-ray crystallography. To minimize this potential problem, we optimized the crosslinking reaction so that it would proceed to completion. A number of variables including temperature, incubation time, and relative and absolute concentrations of reactants and buffer conditions were evaluated (Figure 4). The temperature and absolute concentration of reactants (particularly [KS][AT] and ACP) were the most critical variables. It should be noted that optimal crosslinking of [KS5][AT5] with ACP5 occurred using different conditions compared to the optimal crosslinking of [KS3][AT3] with ACP3 (data not shown), suggesting that each protein pair must be optimized individually.
Figure 4.

SDS-PAGE gel showing variability of [KS3][AT3] crosslinking to ACP3 as a function of alternative reaction conditions(Numbers refer to Table below). Lower temperatures allowed access to longer incubation times without inducing protein precipitation observed at higher temperatures.
In summary, we have demonstrated the utility of a chemo-enzymatic methodology to crosslink ACP and KS domains from two DEBS modules in a site specific manner, exploiting the native reactivity of these domains. Further experiments established the role of protein-protein interaction in this process and suitable manipulation of the reaction conditions allowed us to obtain homogenous crosslinked protein. Our results demonstrate the value of this approach as a route to engineer a functionally relevant, relatively rigid KS-ACP complex for high-resolution structural studies. They also highlight the value of this approach for studying other transient interactions involving ACP domains in multimodular PKSs.
Acknowledgements
This work was supported by grants from the NIH (CA 66736 to C.K., GM 22172 to D.E.C. and GM 75797 to M.D.B.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.Cortes J, Haydock SF, Roberts GA, Bevitt DJ, Leadlay PF. Nature. 1990;348:176. doi: 10.1038/348176a0. [DOI] [PubMed] [Google Scholar]
- 2.Donadio S, Staver MJ, Mcalpine JB, Swanson SJ, Katz L. Science. 1991;252:675. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 3.Gokhale RS, Tsuji SY, Cane DE, Khosla C. Science. 1999;284:482. doi: 10.1126/science.284.5413.482. [DOI] [PubMed] [Google Scholar]
- 4.Tsuji SY, Wu N, Khosla C. Biochemistry. 2001;40:2317. doi: 10.1021/bi002462v. [DOI] [PubMed] [Google Scholar]
- 5.Khosla C, Tang Y, Chen AY, Schnarr NA, Cane DE. Annu. Rev. Biochem. 2007;76:195. doi: 10.1146/annurev.biochem.76.053105.093515. [DOI] [PubMed] [Google Scholar]
- 6.Tsuji SY, Wu N, Khosla C. Biochemistry. 2001;40:2326. doi: 10.1021/bi002463n. [DOI] [PubMed] [Google Scholar]
- 7.Wu N, Cane DE, Khosla C. Biochemistry. 2002;41:5056. doi: 10.1021/bi012086u. [DOI] [PubMed] [Google Scholar]
- 8.Wu N, Tsuji SY, Cane DE, Khosla C. J. Am. Chem. Soc. 2001;123:6465. doi: 10.1021/ja010219t. [DOI] [PubMed] [Google Scholar]
- 9.Chen AY, Schnarr NA, Kim CY, Cane DE, Khosla C. J. Am. Chem. Soc. 2006;128:3067. doi: 10.1021/ja058093d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen AY, Cane DE, Khosla C. Chem Biol. 2007;14:784. doi: 10.1016/j.chembiol.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alekseyev V, Liu C, Puglisi J, Khosla C. Protein Sci. 2007;16:2093. doi: 10.1110/ps.073011407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoops JK, Henry SJ, Wakil SJ. J. Biol. Chem. 1983;258:12482. [PubMed] [Google Scholar]
- 13.Worthington AS, Rivera H, Torpey JW, Alexander MD, Burkart MD. ACS Chem. Biol. 2006;1:687. doi: 10.1021/cb6003965. [DOI] [PubMed] [Google Scholar]
- 14.Worthington AS, Hur GH, Meier JL, Burkart MD. manuscript in preparation. [Google Scholar]
- 15.Tang Y, Kim CY, Mathews II, Cane DE, Khosla C. Proc. Nat.l Acad. Sci. U S A. 2006;103:11124. doi: 10.1073/pnas.0601924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Y, Chen AY, Kim CY, Cane DE, Khosla C. Chem Biol. 2007;14:931. doi: 10.1016/j.chembiol.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.A typical 10 µL reaction mixture was set up as follows: To a 100 mM potassium phosphate (pH 7.2), buffer with 12.5 mM MgCl2 and 8 mM ATP were added ACP3 (5 µg, 25µM, 5 eq.), CoaA (1.5 µM), CoaD (1.5 µM), CoaE (1.5 µM), B. subtilis Sfp (20 µM) and crosslinker (200 µM, 40 eq). The mixture was incubated at 37 °C for 1.5 hrs. [KS3][AT3] (9.1 µg, 5 µM, 1 eq.) was then added and the mixture was further incubated at 30°C for 2 hrs. Incubation for longer periods of time resulted in precipitation of [KS][AT]. All gels were run under the following conditions: 7.5% Tris-HCl, 10 well, 30uL, Bio-Rad, Ready gels. 200 V, 150 mA, 100W.
- 18.Data obtained from MALDI-TOF mass analysis of apo-ACP and crypto-ACP domains is as follows:(Figures in parentheses refer to calculated molecular weights for [M + H]+ - methionine. For compound identity refer to Figure 1). Apo-ACP: 12891.6(12884.2). 1-Crypto-ACP: 13304.15(13295). 2-Crypto-ACP: 13303.78(13295). 3-Crypto-ACP: 13345.35(13328.67). 4-Crypto-ACP: 13382.18(13365.13). 5-Crypto-ACP: 13254.26(13237.95). 6-Crypto-ACP: 13383.04(13365.13). 7-Crypto-ACP: 13270.63(13251.98).
- 19.For the expression of ACP3 with an internal FLAG epitope, pYTT01 was constructed as follows. The DNA sequence encoding DEBS ACP3 was amplified by PCR as an NdeI-EcoRI fragment using primers 5′-CATATGGACTACAAGGACGACGACGACAAGCGGCTCGCGGGGCTTTCCCCGGACGAGCAG-3′ and 5′-CGCGGCGAATTCTTAGGCGTCACCGACGAGCCGGGC-3′. This NdeI-EcoRI fragment was cloned into pET28a expression vector to yield plasmid pYTT01, encoding DEBS ACP3 with N- terminal His6-tag and an internal FLAG tag.
- 20.Western blot with ANTI-FLAG M2 monoclonal antibody(Sigma) was performed as per the manufacturers instructions using nitrocellulose membrane.
- 21.Purified crosslinked adduct (HiTrap-Q anion-exchange column, GE Healthcare) or [KS3][AT3] (10 µM, in 100 mM phosphate at pH 7.2) was incubated with 50 µM [14C](2S,3R)-2-Methyl-3-hydroxypentanoyl-N-acetylcysteamine thioester for 2 h at room temperature. Samples were quenched with SDS-PAGE loading buffer lacking any reducing agents and loaded directly onto a SDS-PAGE gel. The gel was dried using a BioRad gel-drying system and analyzed using a phosphorimager.


