Abstract
Science is beginning to understand how genetic variation and epigenetic events alter requirements for, and responses to, nutrients (nutrigenomics). At the same time, methods for profiling almost all of the products of metabolism in a single sample of blood or urine are being developed (metabolomics). Relations between diet and nutrigenomic and metabolomic profiles and between those profiles and health have become important components of research that could change clinical practice in nutrition. Most nutrition studies assume that all persons have average dietary requirements, and the studies often do not plan for a large subset of subjects who differ in requirements for a nutrient. Large variances in responses that occur when such a population exists can result in statistical analyses that argue for a null effect. If nutrition studies could better identify responders and differentiate them from nonresponders on the basis of nutrigenomic or metabolomic profiles, the sensitivity to detect differences between groups could be greatly increased, and the resulting dietary recommendations could be appropriately targeted. It is not certain that nutrition will be the clinical specialty primarily responsible for nutrigenomics or metabolomics, because other disciplines currently dominate the development of portions of these fields. However, nutrition scientists' depth of understanding of human metabolism can be used to establish a role in the research and clinical programs that will arise from nutrigenomic and metabolomic profiling. Investments made today in training programs and in research methods could ensure a new foundation for clinical nutrition in the future.
Keywords: Nutrigenomics, epigenetics, metabolomics, single-nucleotide polymorphism, clinical nutrition
INTRODUCTION
Nutrigenomics (the study of the bidirectional interactions between genes and diet) and metabolomics (the integrated study of the many small molecules produced by metabolism) are rapidly developing new bodies of knowledge that will change future research and practice in human nutrition. Just as the published human genome is an average representation of genes in humans, there is a human metabolome that is an average representation of metabolic potential for humans. However, there is significant variation from the average in both genome and metabolome in any given individual. Nutrigenomic and metabolomic profiling will help identify mechanisms that underlie individual variations in dietary requirements as well as in the capacity to respond to food-based interventions.
Although nutrition clinicians eventually may be able to provide personalized nutrition recommendations, they are most likely, in the immediate future, to use this knowledge to improve dietary recommendations for populations. Currently, estimated average requirements are used to set dietary reference intakes, because scientists cannot adequately identify subsets of the population that differ in their requirements for a nutrient. Recommended intakes must exceed the actual required intake for most of the population to ensure that those persons with the highest requirement ingest adequate amounts of the nutrient. As a result, dietary reference intakes often are set so high that diet guidelines suggest an almost unattainable intake of some foods. Once it is possible to identify common subgroups that differ in nutrient requirements by using nutrigenomic and metabolomic profiling, interventions can be targeted and recommendations can be made that avoid suggesting that the entire population needs to use almost all discretionary calories to meet nutrient requirements. Similarly, nutrigenomic and metabolomic profiling can enhance nutrition epidemiology and nutrition intervention research. When a large variance exists in response to a nutrient, statistical analyses often argue for a null effect. If responders could be differentiated from nonresponders on the basis of nutrigenomic and metabolomic profiles, this statistical noise could be eliminated and the sensitivity of nutrition research greatly increased.
It is apparent that a niche will exist for health professionals who can effectively place the new information generated by nutrigenomic and metabolomic profiling into a context that enables integration, interpretation, and, subsequently, individualized dietary recommendations. Thus, the field of nutrition would benefit by establishing itself as the predominant discipline using this knowledge in clinical and public health practice.
Concrete examples can help nutrition scientists to envision how such new knowledge will change current practice. In this report, several aspects of nutrigenomic and metabolomic profiling are considered, and concrete examples from current research are provided that foreshadow how nutrition science and practice can make use of such data.
Nutrient-gene interactions
There are 3 major conceptual groupings for thinking about nutrient-gene interactions: 1) direct interactions: nutrients, sometimes after interacting with a receptor, behave as transcription factors that can bind to DNA and acutely induce gene expression; 2) epigenetic interactions: nutrients can alter the structure of DNA (or of histone proteins in chromatin) so that gene expression is chronically altered; and 3) genetic variations: common genetic variations [single-nucleotide polymorphisms (SNPs)] can alter the expression or functionality of genes. All of these mechanisms can result in altered metabolism of and altered dietary requirements for nutrients.
Acute effects of nutrients on gene transcription
There are many examples of nutrients acting as transcription factors that modify gene expression. Vitamin A, or rather retinoid derivatives of vitamin A, interact with retinoic acid receptor proteins, and these complexes activate or repress transcription when they bind to motifs (eg, retinoic acid response elements) in gene promoter regions (1). Dietary fatty acids can interact with peroxisome proliferator–activated receptors, which then bind to DNA and modify gene expression (2). Other examples of nutrient-response element interactions that modify gene expression include the interactions of vitamin D with the vitamin D receptor, calcium with calcineurin, and zinc with metal-responsive transcription factor 1 (3). All of these examples involve a nutrient that acts as a short-term signal to acutely alter gene transcription; this effect usually stops if the exposure to the nutrient is removed. Because such examples are familiar to most nutrition scientists, they are not a focus of this discussion.
Epigenetics and nutrition
It is only relatively recently that science began to understand epigenetic mechanisms for sustained effects of nutrients on gene expression. These sustained effects are mediated by methylation of DNA or by methylation, acetylation, or biotinylation of his-tones, or by both functions (4). Such epigenetic modifications can result in changes in gene expression that can last throughout a person's life and can even persist across generations.
DNA methylation usually occurs at cytosine bases that are followed by a guanosine (5′-CpG-3′ islands) (5), and it influences gene transcription and genomic stability (6–8). In mammals, 60−90% of 5′-CpG-3′ islands are methylated (9). When this modification occurs in gene promoter regions, expression is altered (10). Increased methylation is usually associated with gene silencing or reduced gene expression (9), because methylated 5′-CpG-3′ islands attract capping proteins that hinder access to the gene for the transcription factors that normally induce gene expression (Figure 1) (11). There are some exceptions, in which gene methylation prevents the binding of inhibitory factors that results in derepression of the gene, but that is uncommon. Once 5′-CpG-3′ islands in genes are methylated, the methylation is reproduced every time the gene is copied. Thus, the effects of methylation can persist.
FIGURE 1.
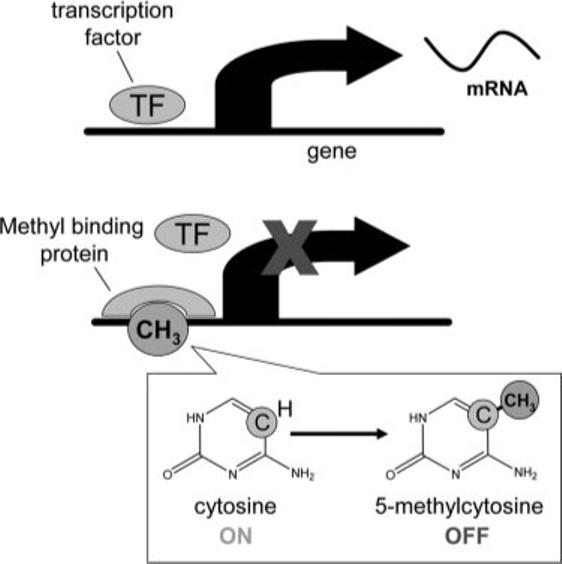
DNA methylation can silence gene expression. Methylation of cytosine located in cytosine-guanosine groupings in gene promoter regions (called 5′-CpG-3′ islands) attracts capping proteins that hinder access to the gene for the transcription factors that normally turn on gene expression and formation of messenger RNA (mRNA). When the transcription factor does not bind to the promoter area of the gene, transcription of mRNA does not occur, and the gene is silenced.
DNA is wrapped on proteins (histones) that, when packed tightly together, prevent access to the promoter sequences of genes. Methylation and acetylation [and perhaps biotinylation (12, 13)] of these histones can uncoil them, creating channels through which transcription factors can pass and activate gene promoters. These changes in histones are often triggered subsequent to methylation of 5′-CpG-3′ islands in gene DNA (4).
Changes in the dietary availability of methyl groups can induce stable changes in gene methylation, altering gene expression and the resulting phenotype (14, 15). For example, feeding pregnant pseudoagouti Avy/a mouse dams a methyl-supplemented diet altered the epigenetic regulation of agouti gene expression in their offspring, as indicated by increased agouti or black mottling of their coats (14, 16); hair color was permanently altered because the pregnant mother ate more or less of a nutrient that changed the methylation of DNA in the fetus. In a similar study, maternal dietary intake of methyl groups influenced methylation of the Axin Fused gene and determined whether offspring had permanently kinked tails (17). Feeding pregnant mice or rats more or less choline (a methyl donor) influenced the rate at which brain progenitor cells proliferated in the fetus; it also influenced the rates of apoptosis (cell suicide) in these cells (18, 19). Decreased choline availability to pregnant dams was associated with changes in DNA methylation in fetal brains that were specific to some CpG islands, and even to specific CpG sites, within genes that regulate cell cycling (20, 21). Fetuses of dams fed a low-choline diet during days 11−17 of gestation had half as much neural progenitor cell proliferation and twice as much progenitor cell apoptosis in the hippocampus (memory center) as did fetuses from mothers fed choline-adequate diets (19, 22), and the former group of fetuses also had less visuospatial and auditory memory than did those of the latter group (23). The feeding of greater amounts of choline (≈4 times dietary levels) to pregnant dams enhanced visuospatial and auditory memory in their offspring by ≤30% in the adult animals (23–29). Indeed, adult rodents lose memory function as they age, and offspring exposed to extra choline in utero did not show this “senility” (26, 28).
Epigenetic effects of nutritional variation are not restricted to fetal life, nor are they seen only in the rodent. Investigators found longer survival and a 75% lower risk of diabetes mellitus in humans whose paternal grandfathers experienced food scarcity during the slow growth period just before puberty than in those whose paternal grandfathers did not (30–32). Pembrey (32) effectively argued that these effects of nutrition must occur via epigenetic imprinting of paternal genes and pointed out that the slow growth period before puberty occurs when the first viable pools of spermatocytes emerge and when reprogramming of DNA methylation imprinting begins. In a similar fashion, epi-genetic events can modulate carcinogenesis in the adult; for example, a number of single-copy genes whose expression is restricted to the testes (32) in adults become hypomethylated and are reexpressed in some cancers (33).
Technology will soon make it practical to catalog epigenetic changes in many genes at one time (34). Because a significant portion of human genes are regulated by methylation, many more examples should accrue of diet-related changes in gene methylation.
Genetic variation can influence dietary requirements
Although humans share the same genes, there are many individual variations in the codon sequences for these genes; in total, >10 million SNPs exist that occur in >1% of the population (35). Some common SNPs occur in 5% to >50% of the population. Most humans are heterozygous for ≥50 000 SNPs across their genes (36). Some fraction of these SNPs results either in alteration of gene expression or in changes in the gene product such that protein structure and function are altered. Mastering the ways in which millions of SNPs may influence nutrient requirements is daunting, but recent advances make this effort more practical. Even though the costs of genotyping have dropped markedly, the number of markers now available per gene makes the genotyping of all markers expensive. Because of this, scientists need to select for genotyping a set of markers that would eliminate the necessity of measuring all SNPs. Within short regions (ie, ≤500 kb) of a gene, it is possible to find combinations of linked SNPs that are found in multiple unrelated persons (haplotype blocks). These SNPs stay linked to one another and are inherited over many generations. An international scientific consortium has recently characterized patterns of SNP linkage in haplotype blocks (36, 37). The identification of a few alleles of a haplotype block can unambiguously identify all other polymorphic sites in its region within a group of persons of similar race and ethnicity, thereby making SNP analysis practical for the nutrition scientist or clinician.
A number of relatively common SNPs are known to influence nutrient requirements. For example, the enzyme 5,10-methylenetetrahydrofolate reductase (MTHFR) is involved in folate metabolism. The MTHFR gene has a common SNP (C677T allele) that results in reduced enzymatic activity, and homozygous persons have elevated plasma homocysteine concentrations unless they ingest high amounts of folate (38). This SNP occurs in 15−30% of the population.
Another example comes from work on SNPs that modify the risk of developing organ dysfunction or damage when humans are fed diets low in choline (39–42). Some persons developed fatty liver and liver and muscle damage, whereas others did not. Premenopausal women who were carriers of a very common SNP (MTHFD1-G1958A) were >15 times as likely as noncarriers to develop signs of choline deficiency on a low-choline diet (43). It is of interest that the risk of having a child with a neural tube defect increased in mothers with this SNP (44), and that, compared with women eating diets in the highest quartile for choline intake, women eating diets in the lowest quartile had 4 times the risk of having a baby with a neural tube defect (45). It would be interesting to determine whether those with the greatest risk have the G1958A SNP in MTHFD1.
The PEMT gene encodes for a protein responsible for endogenous formation of choline in the liver (46), and it is induced by estrogen (47). In studies of organ dysfunction after choline deficiency in humans, an SNP in the promoter region of the PEMT gene (rs12325817) was associated with greatly increased susceptibility to choline deficiency in women but not in men (42). The frequency of this variant allele was 0.74 (this common allele is called the “variant” one because wild type was determined by the codon present in lower mammals). Studies are underway to identify other genetic differences that contribute to individual variability in dietary requirements for choline.
Although all of the above examples of genetic variation that change nutrient requirements involved one-carbon metabolism, SNPs have been identified that affect other metabolic pathways. For example, an SNP in the SREBP gene affects fructose-induced hepatic lipogenesis (48). The gene for peroxisome proliferator–activated receptor-α has an SNP that has been associated with alterations in total cholesterol, LDL-cholesterol, and apolipoprotein B concentrations (49), and this SNP altered the response to the dietary intake of n–6 polyunsaturated fatty acid. In persons with the variant allele, increased n–6 polyunsaturated fatty acid intake was associated with a marked reduction in triacylglycerol concentration (49).
A database of functionally important SNPs would make it possible for a nutrition specialist—one who has an understanding of the effects of such changes on metabolism and nutrient requirements—to use SNP profiling to make practical recommendations as part of clinical practice (eg, a recommendation of higher dietary choline intake during pregnancy in women with SNPs that alter PEMT expression).
The challenges in applying nutrigenomic data to nutrition
The above examples show that it is now possible to think about how individual gene SNPs alter nutrient requirements and about how methylation of a particular gene is influenced by nutrition. Obviously, these concepts are just a simplified version of what nutrition science will have to deal with in the future. It is possible to detect tens of thousands of SNPs and at least the same number of epigenetic changes in genes. It is likely that these changes interact with each other in a complex way. Whereas their sequences are known, the functions of many of the genes identified in the human genome are unknown, as are the functional consequences of most of the identified SNPs in humans. Even when these functions are known, in order to integrate any differences and predict their effects on body function, scientists will need to overlay each of these changes onto the familiar map of metabolic pathways. Then algorithms will have to be developed that allow the identification of patterns in the measured changes—eg, groupings of related genes that regulate metabolic pathways and then groupings of related metabolic pathways. Although these are daunting tasks, managing gene expression data also appeared difficult 10 y ago, but current off-the-shelf bioinformatics software makes such data accessible to the interested scientist. In a short time, software will make gene expression data accessible to the clinician. Only the magnitude of the complexity involved in nutrigenomics makes it harder to develop the bioinformatics tools needed for these data to be accessible; conceptually, there is little in the way.
Nutrition and metabolomics
The integrated study of the many small molecules formed by metabolism has always been a primary domain for nutrition science. For this reason, most nutrition scientists immediately grasp the advantages gained from being able to measure many metabolites rather than a few. In the past, analytic limitations made life relatively simple, and the nutritional biochemist dealt with perhaps a half-dozen metabolites, developed an integrated theory for how they related to each other, and predicted the effects on cell function or disease. The complexity created by technology that permits the analysis of thousands of metabolites simultaneously spawned the new field of metabolomics.
The unique challenges of applying metabolomics methods to nutrition science have been reviewed elsewhere (50). A blood, saliva, or urine sample generates thousands of peaks on a mass spectrometer or nuclear magnetic resonance analysis. A standard chart of human metabolism lists ≈800 molecules, and available databases suggest that there will be 2000 molecules in the human metabolome (with thousands more if bacterial metabolites are included). Because the new instruments are so sensitive, it is likely that previously unsuspected molecules will be detected in human biofluids, which will lead to a reconsideration of the understanding of metabolism. The field is limited by the fact that the chemical identity of these peaks is not yet known, by methodologic problems in quantitation of peaks, and by a lack of sufficiently refined bioinformatics tools that permit analyses in a metabolic pathways context.
Many investigators use principal components analysis to detect relations between a given intervention and a measured metabolome. Principal components analysis reduces multidimensional datasets to lower dimensions for analysis by using the characteristics of the dataset that contribute most to its variance. Investigators can then recognize patterns of change, even though they may not be certain of the identity of the peaks that contribute to this pattern. For example, a patient with renal disease may have a pattern of peaks in a chromatogram that is characteristically different from that of a healthy person. To move beyond principal components analyses, scientists need to systematically catalog the chemical identity of the unknown peaks detected in the human metabolome. The peaks for all the known metabolites of nutrients are mixed in with peaks derived from the many non-nutrient molecules that are absorbed, metabolized, passed through the blood, and secreted in both urine and saliva. The large-bowel microflora produce many or most of these peaks that constitute the metabolome of biofluids in humans, and this contribution presents an additional challenge, because our understanding of microflora metabolism is not nearly as complete as our understanding of human metabolism. Eventually, nutrition scientists will use such measures of microflora metabolism to develop a better understanding of the role of gut microflora in human nutrition. For example, metabolomic analyses showed that the altered availability of choline caused by metabolism by gut microflora was associated with fatty liver in insulin-resistant mice (51).
In addition to peak identification, there is a need to verify that peak areas are proportional to concentrations for each metabolite. In some methods, molecules in neighboring peaks can modify detector response, thereby altering the relation of area to concentration. In addition, scientists must establish that data generated from total-metabolome assay methods correlate with data generated by using gold-standard single metabolite assays. Multidisciplinary research teams are currently working to validate methods and catalog the human metabolome, and it is reasonable to expect that this will be accomplished within 5 y. The commercial sector has already developed platforms for SNP assessment that are readily available for a reasonable price. It appears that epigenetic assessment availability is only a year or so away. Metabolomic methodology is being developed and is currently at the experimental stage in university, industry, and government laboratories. Notable among these efforts are the European Union–funded Network of Excellence (NuGo) and the Human Metabolome Project funded by Genome Canada. These efforts can be augmented if the major academic units with nutrition expertise aggregate their intellectual mass to work together to create usable analytic methods.
Making nutrigenomics and metabolomics accessible to the nutrition clinician
At present, the data-mining and analytic software available is divided into areas according to the kind of data being analyzed. Gene expression data are mined with different software than are epigenetic data, and metabolomic data are analyzed in completely different ways. Each piece of software requires training in its use, and each has its special eccentricities. Eventually, a suite of software needs to be developed that can manage these data in an integrated platform. Changes in genes, gene methylation, and metabolism are all related and can be understood only when these disparate elements are brought together. A reasonable first step would be to adopt the common metabolic map that nutritional and biochemical science has developed over the past 6 decades. After statistical analyses (eg, significance, variance, and false discovery filters), data would be expressed in relation to the specific reactions in the metabolic pathways (Figure 2). For example, in a study in which data on genes and metabolites of glucose metabolism are collected, one would use existing information about metabolism to group the observed changes and create a graphic presentation that is immediately understandable. Good software could permit examination of the data by zooming from a micro level (pathways of gluconeogenesis) to the level of glucose metabolism as a whole or to the even larger scale of liver or whole-body metabolism. Eventually, computer algorithms would identify patterns in these changes and help the user to develop hypotheses that explain the observation and to make recommendations for diet change. This process is much more complex than but not very different conceptually from what a clinician does now: measures glucose concentration and insulin activity, uses an algorithm to decide if insulin insensitivity is present, and then intervenes as appropriate.
FIGURE 2.
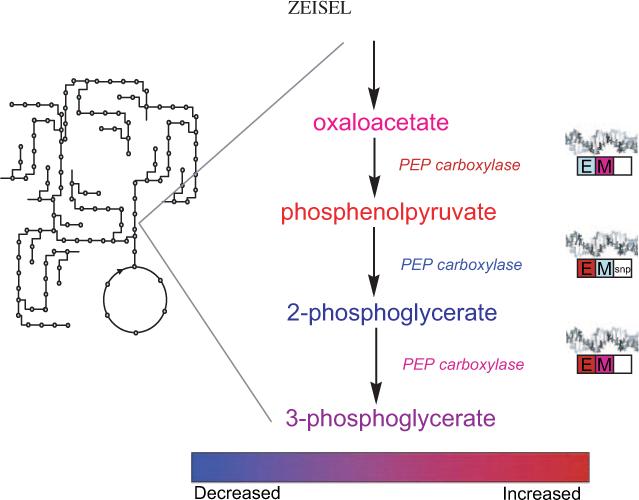
A theoretical example of what the informatics tools may look like that will make nutrigenomics and metabolomics accessible to nutrition scientists and clinicians. After appropriate statistical tests and filtering, data from nutrigenomic, epigenetic, and metabolomic platforms can be overlaid on the metabolic map that we are familiar with. By selecting an area of the metabolic map, the user can zoom in on the region and see a presentation of changes in metabolite concentrations, enzyme activities, and perhaps flux rates with color coding to indicate direction of change. Information on changes in DNA expression (E), gene methylation (M), and gene SNPs also can be presented by using similar color coding. By selecting the metabolite, enzyme, or gene box, an investigator would be able to examine the exact values and variances and perhaps even to examine the raw data. Another level of the informatics program would summarize all of the related changes by pathway and present information on false discovery and significant difference statistics. Finally, a summary of appropriate diet recommendations would be presented.
The tools currently available to nutritionists for analysis of metabolomic data are sparse, difficult to use, or beyond the financial means of individual scientists. However, given the demand for such tools, it is reasonable to expect that they will be refined and will become available within 5 y and that they will make metabolomic and nutrigenomic data accessible first to nutrition scientists and then to nutrition clinicians.
Planning for the future
Although the analytic and informatics capacities to effectively use metabolomics and nutrigenomics are a few years away, nutrition scientist and educators need to act soon to set the stage appropriately. A decade from now, science will have identified complex associations between diet and the expression of thousands of genes with simultaneous changes in thousands of metabolites. The integration of nutrigenomics and metabolomics with the metabolic map will be incredibly complex, but there are manageable pieces of this effort that could be addressed by those who have spent a lifetime developing expertise in nutrition and metabolism. The ultimate challenge will be in making sense of what will be a huge, perhaps overwhelming amount of data so that nutrition clinicians can use those data to make practical recommendations. On a smaller scale, nutrition scientists, with their integrative knowledge of metabolism, do this already. These scientists should assist with the development of informatics tools to facilitate the interpretation of exponentially more complex sets of data. The analytic and informatics tools to be developed will be of limited utility for nutrition scientists and clinicians if nutrition science does not play a substantial role in driving their creation. As a start, nutrition scientists need to give engineers and software writers a list of the highest-priority metabolites—those that we think will be most useful to nutrition science.
Nutrition experts need to contribute to the science but also to the training of health professionals, so as to provide them with the skill sets essential for using nutrigenomic and metabolomic data sets. The clinical nutrition specialist will need training in nutrigenomics and metabolomics. The intricacies of the biotechnology involved in measurements may concern only research scientists, and the catalog of important gene and metabolite changes will be growing too fast for clinicians to be expected to master all of the content. However, there is a critical thinking process that weights information according to the known weaknesses and strengths of the methods used to obtain the information and then places it into a metabolic context. The clinician will have to be trained to weigh metabolomic and nutrigenomic evidence and convert it into recommendations. To do this, an understanding of metabolism that is perhaps more extensive than that which current training provides will be needed. Finally, clinical nutrition must develop the clinical infrastructure needed to efficiently perform nutrigenomic and metabolomic profiling. It may be sufficient to perform SNP and epigenetic profiling once in a lifetime, but metabolomic measures would be made acutely at the time of evaluation, and it is likely that multiple measures made before and after an intervention will be used to assess metabolite flux. Investments made today in research methods and in training programs can ensure a new foundation for clinical nutrition and public health practice in the future.
It is not certain that nutrition clinicians will occupy this important clinical niche, because geneticists, chemists, engineers, and pharmacologists currently dominate the development of portions of nutrigenomics and metabolomics. However, before these new technologies can effectively penetrate into clinical practice, the information generated by these methods must be overlaid onto the integrated metabolic pathways matrix that health professionals already understand. The depth of understanding of human metabolism that nutrition scientists have is a critical asset needed to advance this area of study. This asset could be used to solidify a place for nutrition clinicians in the future of nutrigenomics and metabolomics.
Acknowledgments
SHZ serves on scientific advisory boards for Hershey Foods, Solae, Dupont, MetaboAge, and PogoHealth and has received grant funding from Mead Johnson Nutritionals and the Gerber Foundation. None of these activities represents a personal or financial conflict of interest with respect to this commentary.
Footnotes
Supported by grants no. DK55865, AG09525, and ES012997 from the National Institutes of Health; by a grant from the Gerber Foundation; and by grants from the National Institutes of Health to the Clinical Nutrition Research Unit (DK56350), Clinical Research Center (RR00046), and the Center for Environmental Health and Susceptibility (ES10126) at the University of North Carolina.
REFERENCES
- 1.Lane MA, Bailey SJ. Role of retinoid signalling in the adult brain. Prog Neurobiol. 2005;75:275–93. doi: 10.1016/j.pneurobio.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Hsu SC, Huang CJ. Reduced fat mass in rats fed a high oleic acid-rich safflower oil diet is associated with changes in expression of hepatic PPARα and adipose SREBP-1c-regulated genes. J Nutr. 2006;136:1779–85. doi: 10.1093/jn/136.7.1779. [DOI] [PubMed] [Google Scholar]
- 3.Muller M, Kersten S. Nutrigenomics: goals and strategies. Nat Rev Genet. 2003;4:315–22. doi: 10.1038/nrg1047. [DOI] [PubMed] [Google Scholar]
- 4.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005;15:490–5. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 5.Holliday R, Grigg GW. DNA methylation and mutation. Mutat Res. 1993;285:61–7. doi: 10.1016/0027-5107(93)90052-h. [DOI] [PubMed] [Google Scholar]
- 6.Jaenisch R. DNA methylation and imprinting: why bother? Trends Genet. 1997;13:323–9. doi: 10.1016/s0168-9525(97)01180-3. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA, Gonzalgo ML. Altered DNA methylation and genome instability: a new pathway to cancer? Proc Natl Acad Sci U S A. 1997;94:2103–5. doi: 10.1073/pnas.94.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet. 2000;1:11–9. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- 9.Jeltsch A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem. 2002;3:274–93. doi: 10.1002/1439-7633(20020402)3:4<274::AID-CBIC274>3.0.CO;2-S. (Published erratum appears in Chembiochem 2002;3:382.) [DOI] [PubMed] [Google Scholar]
- 10.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–13. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 11.Fan G, Hutnick L. Methyl-CpG binding proteins in the nervous system. Cell Res. 2005;15:255–61. doi: 10.1038/sj.cr.7290294. [DOI] [PubMed] [Google Scholar]
- 12.Oommen AM, Griffin JB, Sarath G, Zempleni J. Roles for nutrients in epigenetic events. J Nutr Biochem. 2005;16:74–7. doi: 10.1016/j.jnutbio.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Kothapalli N, Sarath G, Zempleni J. Biotinylation of K12 in histone H4 decreases in response to DNA double-strand breaks in human JAr choriocarcinoma cells. J Nutr. 2005;135:2337–42. doi: 10.1093/jn/135.10.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr. 2002;132(suppl):2393S–400S. doi: 10.1093/jn/132.8.2393S. [DOI] [PubMed] [Google Scholar]
- 15.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12:949–57. [PubMed] [Google Scholar]
- 17.Waterland RA, Dolinoy DC, Lin JR, Smith CA, Shi X, Tahiliani KG. Maternal methyl supplements increase offspring DNA methylation at Axin fused. Genesis. 2006;44:401–6. doi: 10.1002/dvg.20230. [DOI] [PubMed] [Google Scholar]
- 18.Craciunescu CN, Albright CD, Mar MH, Song J, Zeisel SH. Choline availability during embryonic development alters progenitor cell mitosis in developing mouse hippocampus. J Nutr. 2003;133:3614–8. doi: 10.1093/jn/133.11.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albright CD, Tsai AY, Friedrich CB, Mar MH, Zeisel SH. Choline availability alters embryonic development of the hippocampus and septum in the rat. Brain Res Dev Brain Res. 1999;113:13–20. doi: 10.1016/s0165-3806(98)00183-7. [DOI] [PubMed] [Google Scholar]
- 20.Niculescu MD, Craciunescu CN, Zeisel SH. Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. FASEB J. 2006;20:43–9. doi: 10.1096/fj.05-4707com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niculescu MD, Yamamuro Y, Zeisel SH. Choline availability modulates human neuroblastoma cell proliferation and alters the methylation of the promoter region of the cyclin-dependent kinase inhibitor 3 gene. J Neurochem. 2004;89:1252–9. doi: 10.1111/j.1471-4159.2004.02414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albright CD, Friedrich CB, Brown EC, Mar MH, Zeisel SH. Maternal dietary choline availability alters mitosis, apoptosis and the localization of TOAD-64 protein in the developing fetal rat septum. Brain Res Dev Brain Res. 1999;115:123–9. doi: 10.1016/s0165-3806(99)00057-7. [DOI] [PubMed] [Google Scholar]
- 23.Meck WH, Williams CL. Choline supplementation during prenatal development reduces proactive interference in spatial memory. Brain Res Dev Brain Res. 1999;118:51–9. doi: 10.1016/s0165-3806(99)00105-4. [DOI] [PubMed] [Google Scholar]
- 24.Meck W, Williams C. Perinatal choline supplementation increases the threshold for chunking in spatial memory. Neuroreport. 1997;8:3053–9. doi: 10.1097/00001756-199709290-00010. [DOI] [PubMed] [Google Scholar]
- 25.Meck W, Williams C. Characterization of the facilitative effects of perinatal choline supplementation on timing and temporal memory. Neuroreport. 1997;8:2831–5. doi: 10.1097/00001756-199709080-00005. [DOI] [PubMed] [Google Scholar]
- 26.Meck W, Williams C. Simultaneous temporal processing is sensitive to prenatal choline availability in mature and aged rats. Neuroreport. 1997;8:3045–51. doi: 10.1097/00001756-199709290-00009. [DOI] [PubMed] [Google Scholar]
- 27.Meck WH, Smith RA, Williams CL. Pre- and postnatal choline supplementation produces long-term facilitation of spatial memory. Dev Psychobiol. 1988;21:339–53. doi: 10.1002/dev.420210405. [DOI] [PubMed] [Google Scholar]
- 28.Meck WH, Williams CL. Metabolic imprinting of choline by its availability during gestation: implications for memory and attentional processing across the lifespan. Neurosci Biobehav Rev. 2003;27:385–99. doi: 10.1016/s0149-7634(03)00069-1. [DOI] [PubMed] [Google Scholar]
- 29.Williams CL, Meck WH, Heyer DD, Loy R. Hypertrophy of basal forebrain neurons and enhanced visuospatial memory in perinatally choline-supplemented rats. Brain Res. 1998;794:225–38. doi: 10.1016/s0006-8993(98)00229-7. [DOI] [PubMed] [Google Scholar]
- 30.Kaati G, Bygren LO, Edvinsson S. Cardiovascular and diabetes mortality determined by nutrition during parents' and grandparents' slow growth period. Eur J Hum Genet. 2002;10:682–8. doi: 10.1038/sj.ejhg.5200859. [DOI] [PubMed] [Google Scholar]
- 31.Bygren LO, Kaati G, Edvinsson S. Longevity determined by paternal ancestors' nutrition during their slow growth period. Acta Biotheor. 2001;49:53–9. doi: 10.1023/a:1010241825519. [DOI] [PubMed] [Google Scholar]
- 32.Pembrey ME. Time to take epigenetic inheritance seriously. Eur J Hum Genet. 2002;10:669–71. doi: 10.1038/sj.ejhg.5200901. [DOI] [PubMed] [Google Scholar]
- 33.Hoffmann MJ, Muller M, Engers R, Schulz WA. Epigenetic control of CTCFL/BORIS and OCT4 expression in urogenital malignancies. Biochem Pharmacol. 2006;72:1577–88. doi: 10.1016/j.bcp.2006.06.020. (Epub 2006 Jul 18) [DOI] [PubMed] [Google Scholar]
- 34.Wojdacz TK, Hansen LL. Techniques used in studies of age-related DNA methylation changes. Ann N Y Acad Sci. 2006;1067:479–87. doi: 10.1196/annals.1354.069. [DOI] [PubMed] [Google Scholar]
- 35.McVean G, Spencer CC, Chaix R. Perspectives on human genetic variation from the HapMap Project. PLoS Genet. 2005;1:e54. doi: 10.1371/journal.pgen.0010054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinds DA, Stuve LL, Nilsen GB, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–9. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 37.Crawford DC, Nickerson DA. Definition and clinical importance of haplotypes. Annu Rev Med. 2005;56:303–20. doi: 10.1146/annurev.med.56.082103.104540. [DOI] [PubMed] [Google Scholar]
- 38.Gibney MJ, Gibney ER. Diet, genes and disease: implications for nutrition policy. Proc Nutr Soc. 2004;63:491–500. doi: 10.1079/pns2004369. [DOI] [PubMed] [Google Scholar]
- 39.Busby MG, Fischer L, da Costa KA, Thompson D, Mar MH, Zeisel SH. Choline- and betaine-defined diets for use in clinical research and for the management of trimethylaminuria. J Am Diet Assoc. 2004;104:1836–45. doi: 10.1016/j.jada.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 40.da Costa KA, Badea M, Fischer LM, Zeisel SH. Elevated serum creatine phosphokinase in choline-deficient humans: mechanistic studies in C2C12 mouse myoblasts. Am J Clin Nutr. 2004;80:163–70. doi: 10.1093/ajcn/80.1.163. [DOI] [PubMed] [Google Scholar]
- 41.da Costa KA, Gaffney CE, Fischer LM, Zeisel SH. Choline deficiency in mice and humans is associated with increased plasma homocysteine concentration after a methionine load. Am J Clin Nutr. 2005;81:440–4. doi: 10.1093/ajcn.81.2.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.da Costa K, Kozyreva OG, Song J, Galanko JA, Fischer LM, Zeisel SH. Common genetic polymorphisms have major effects on the human requirement for the nutrient choline. FASEB J. 2006;20:1336–44. doi: 10.1096/fj.06-5734com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kohlmeier M, da Costa KA, Fischer LM, Zeisel SH. Genetic variation of folate-mediated one-carbon transfer pathway predicts susceptibility to choline deficiency in humans. Proc Natl Acad Sci U S A. 2005;102:16025–30. doi: 10.1073/pnas.0504285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brody LC, Conley M, Cox C, et al. A polymorphism, R653Q, in the trifunctional enzyme methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase/formyltetrahydrofolate synthetase is a maternal genetic risk factor for neural tube defects: report of the Birth Defects Research Group. Am J Hum Genet. 2002;71:1207–15. doi: 10.1086/344213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaw GM, Carmichael SL, Yang W, Selvin S, Schaffer DM. Periconceptional dietary intake of choline and betaine and neural tube defects in offspring. Am J Epidemiol. 2004;160:102–9. doi: 10.1093/aje/kwh187. [DOI] [PubMed] [Google Scholar]
- 46.Vance DE, Walkey CJ, Cui Z. Phosphatidylethanolamine N-methyltransferase from liver. Biochim Biophys Acta. 1997;1348:142–50. doi: 10.1016/s0005-2760(97)00108-2. [DOI] [PubMed] [Google Scholar]
- 47.Resseguie M, Song J, Niculescu M, da Costa K, Randall T, Zeisel S. Phosphatidylethanolamine n-methyltransferase (PEMT) gene expression is induced by estrogen in human and mouse primary hepatocytes. FASEB J. doi: 10.1096/fj.07-8227com. (in press) (Epub 2007 Apr 24) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagata R, Nishio Y, Sekine O, et al. Single nucleotide polymorphism (–468 Gly to A) at the promoter region of SREBP-1c associates with genetic defect of fructose-induced hepatic lipogenesis [corrected]. J Biol Chem. 2004;279:29031–42. doi: 10.1074/jbc.M309449200. [DOI] [PubMed] [Google Scholar]
- 49.Ordovas JM. Genetic interactions with diet influence the risk of cardiovascular disease. Am J Clin Nutr. 2006;83(suppl):443S–6S. doi: 10.1093/ajcn/83.2.443S. [DOI] [PubMed] [Google Scholar]
- 50.Gibney MJ, Walsh M, Brennan L, Roche HM, German B, van Ommen B. Metabolomics in human nutrition: opportunities and challenges. Am J Clin Nutr. 2005;82:497–503. doi: 10.1093/ajcn.82.3.497. [DOI] [PubMed] [Google Scholar]
- 51.Dumas ME, Barton RH, Toye A, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci U S A. 2006;103:12511–6. doi: 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]


