Abstract
Arrestins are multi-functional regulators of G protein-coupled receptors. Receptor-bound arrestins interact with >30 remarkably diverse proteins and redirect the signaling to G protein-independent pathways. The functions of free arrestins are poorly understood, and the interaction sites of the non-receptor arrestin partners are largely unknown. In this study, we show that cone arrestin, the least studied member of the family, binds c-Jun N-terminal kinase (JNK3) and Mdm2 and regulates their subcellular distribution. Using arrestin mutants with increased or reduced structural flexibility, we demonstrate that arrestin in all conformations binds JNK3 comparably, whereas Mdm2 preferentially binds cone arrestin ‘frozen’ in the basal state. To localize the interaction sites, we expressed separate N- and C-domains of cone and rod arrestins and found that individual domains bind JNK3 and remove it from the nucleus as efficiently as full-length proteins. Thus, the arrestin binding site for JNK3 includes elements in both domains with the affinity of partial sites on individual domains sufficient for JNK3 relocalization. N-domain of rod arrestin binds Mdm2, which localizes its main interaction site to this region. Comparable binding of JNK3 and Mdm2 to four arrestin subtypes allowed us to identify conserved residues likely involved in these interactions.
Keywords: arrestin, cone photoreceptors, nucleo-cytoplasmic transport, protein kinases, protein-protein interactions, ubiquitin ligases
Arrestins are multi-functional regulators of G protein-coupled receptor (GPCR) signaling. Arrestins bind active phosphorylated GPCRs, thereby terminating G protein activation (Gurevich and Gurevich 2006b) and linking receptors to alternative signaling pathways (Lefkowitz and Shenoy 2005; Gurevich and Gurevich 2006a). Vertebrates have four arrestin subtypes: rod (arrestin1) and cone (arrestin4) are present in photoreceptors, whereas non-visual arrestin2 (a.k.a. β-arrestin or β-arrestin1) and arrestin3 (β-arrestin2) are ubiquitously expressed. Cone arrestin was cloned last (Murakami et al. 1993; Craft et al. 1994) and it remains the least functionally characterized member of the family. Its binding to light-activated phosphorylated cone opsins in vivo (Zhu et al. 2003) and in vivo (Sutton et al. 2005) suggests that it participates in shutting off cone opsin signaling. However, its relatively low abundance in cones (Chan et al. 2007), prominent localization to synaptic terminals (Zhu et al. 2002; Coleman and Semple-Rowland 2005), along with the ability to bind non-visual GPCRs (Sutton et al. 2005) suggest that cone arrestin may have other functions.
Receptor-bound non-visual arrestins serve as scaffolds that recruit modules of mitogen-activated protein kinase pathways to GPCRs (McDonald et al. 2000; Luttrell et al. 2001). The interaction between arrestin3 and c-Jun N-terminal kinase (JNK3) enhances JNK3 phosphorylation and keeps active JNK3 in the cytoplasm (McDonald et al. 2000). Receptorbound arrestins also act as adapters that link GPCRs to the ubiquitination machinery by interacting with E3 ubiquitin ligase Mdm2 (Shenoy et al. 2001). Arrestin affects the distribution of its interaction partners between the nucleus and cytoplasm. Transport factors that move proteins to and from the nucleus bind to specific sequences within cargo molecules: nuclear localization signals for import and nuclear export signals (NES) for export (Wen et al. 1995; Fukuda et al. 1997; Ossareh-Nazari et al. 1997). Arrestin3, which has a native NES, localizes to the cytoplasm and removes its binding partners JNK3 and Mdm2 from the nucleus (Scott et al. 2002; Wang et al. 2003a; Song et al. 2006). Other arrestin subtypes are also predominantly cytoplasmic, although they do not contain NES-like sequences identifiable by available software (http://www.cbs.dtu.dk/services/NetNES/). We have recently developed a cell-based assay to detect the interaction of arrestins with JNK3 and Mdm2 based on the ability of arrestins to remove their binding partners from the nucleus. We demonstrated that free non-visual arrestins in their basal conformation bind JNK3 and Mdm2 and that rod arrestin also binds both proteins (Song et al. 2006).
In this study, we show that cone arrestin binds JNK3 and Mdm2 and relocalizes them from the nucleus to the cytoplasm. The mutants of cone arrestin with different conformations bind JNK3 comparably, whereas Mdm2 strongly prefers the ‘constitutively inactive’ D7 mutant that mimics the conformation of free cone arrestin. Using separately expressed N- and C-domains of cone and rod arrestins, we demonstrate for the first time that both domains contain elements interacting with JNK3 and Mdm2. Comparable ability of all four vertebrate arrestins to bind JNK3 and Mdm2 identifies a limited number of conserved residues likely participating in arrestin interactions with these partners and sets the stage for targeted manipulation of these interactions to create mutant arrestins with ‘biased’ signaling capabilities.
Materials and methods
Plasmid constructs
The coding sequence of native human cone arrestin and its C-terminally tagged with Flag and green fluorescent protein (GFP) versions were subcloned into pcDNA3. Oligo 5′-AGGAGTTTACGCGGCTGGCGCTCAAAGGCGAGGAGGA-3′ was used to introduce an engineered NES (three residues Leu-Ala-Leu inserted between codons 370 and 371). ‘Constitutively active’ 3A mutant of human cone arrestin (I363A, V364A, and I365A), ‘inactive’ (D7) mutant with a seven residue deletion in the interdomain hinge (deleted residues 171, 173, 174, and 178–181), as well as truncated (Tr) mutant (1–366) were constructed by PCR-based mutagenesis. All forms of mutant cone arrestins were Flag-tagged at the C-terminus by PCR. Constructs of separate N- and C-domains of cone (residues 1–178 and 177–359) and rod (1–186 and 185–365) arrestins were made with engineered C-terminal NES and Flag tag. All constructs were verified by dideoxy-sequencing. Expression constructs for GFP-JNK3 and the human homolog of Mdm2-GFP were gifts from Drs Louis Luttrell (Medical University of South Carolina) and Gang Pei (Shanghai Institute for Biological Sciences), respectively.
Cell culture and transient transfection
Adenovirus-transformed human embryonic kidney cells (HEK293A) were routinely maintained in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen, Carlsbad, CA, USA) and penicillin and streptomycin at 37°C in a humidified incubator with 5% CO2. The cells were plated at 80–90% confluence and transfected using Lipofectamine2000 (Invitrogen) according to the manufacturers instructions. The next day, the cells were trypsinized and seeded onto Lab-Tek CC2-treated chambered slides (Campbell, CA, USA) coated with fibronectin (20 μg/mL in phosphate-buffered saline; PBS) (Sigma, St Louis, MO, USA) for immunofluoresence microscopy and onto 24-well plates coated with poly-D-lysine (15 μg/mL) for western blot analysis. The expression of cone arrestin constructs was kept in the range of 50-200 pmol/mg of total protein. This yields ~5–20 lmol/L concentration in the cytoplasm, matching physiologically relevant range estimated on the basis of recent measurements of endogenous cone arrestin in mouse retina, the proportion of cone photoreceptors in the mouse, and their size (Chan et al. 2007). Rod arrestin constructs were expressed at similar levels for comparison, even though the concentration of endogenous rod arrestin in photoreceptors is much higher, up to 1 mmol/L (Broekhuyse et al. 1985; Strissel et al. 2006; Hanson et al. 2007a,b).
Immunohistochemistry and immunocytochemistry
Human embryonic kidney cells 293A were fixed 72 h post-transfection in 4% p-formaldehyde on ice for 15 min. The cells were rehydrated with PBS, permeabilized with 0.1% Triton in PBS, and blocked with 3% albumin in PBS for 1 h at 22°C. Untagged and Flag-tagged arrestins were visualized with F4C1 monoclonal anti-arrestin antibody (Donoso et al. 1990) or rabbit polyclonal antibody raised against F4C1 epitope (peptide VDPVDGVVLVDPDYL) and M2 anti-Flag antibody (Sigma), respectively, followed by the appropriate secondary antibodies (Molecular Probes, Eugene, OR, USA).
Animal research was conducted in compliance with the NIH Guide for the Care and Use of Laboratory Animals and approved by the institutional Animal Care and Use Committee. Salamander (Ambystoma tigrinum) was killed by cervical transection, eyes were enucleated, and eyecups were fixed overnight in 4% p-formaldehyde in PBS, cryoprotected in 30% sucrose, and frozen on dry ice. Freefloating 35-μm thick sections were incubated with monoclonal F4C1 antibody (1 : 500) overnight, followed by biotinylated anti-mouse antibody (1 : 200) (Vector Laboratories, Burlingame, CA, USA) and streptavidin-Alexa488 (1 : 200) (Invitrogen), 1 h each. The sections were mounted in 4′,6-diaminido-2-phenylindole-containing medium (Vector Laboratories). The images were acquired using Nikon EC2000 inverted fluorescent microscope (Tokyo, Japan).
Nuclear exclusion assay
Green fluorescent protein-JNK3 and the human homolog of Mdm2- GFP were transfected alone (control) or in combination with different forms of cone arrestin. Cells expressing these proteins were fixed in 4% p-formaldehyde for 15 min at 4°C. The slides were air-dried and mounted in the medium containing 4′,6-diaminido-2-phenylindole to visualize the nuclei. The GFP-JNK3 and GFP-Mdm2 were visualized using an epifluorescence microscope equipped with a CCD camera. Experiments were performed at least three times and in each experiment the distribution of JNK3, Mdm2 and/or indicated arrestin in at least 20 cells was scored (nucleus > cytoplasm; nucleus = cytoplasm; nucleus < cytoplasm), based on the proportion of the signal in the nucleus and cytoplasm. The results of three experiments were analyzed by one-way ANOVA with arrestin as a main factor (with Bonferroni correction for multiple comparisons).
Quantitative western blot
To compare the expression levels of wild-type and mutant arrestins in each experiment an aliquot of cells after each transfection was dissolved in lysis buffer (Ambion, Austin, TX, USA). Total protein was measured by Bio-Rad assay (Hercules, CA, USA). Proteins were methanol-precipitated, resolved by 10% sodium dodecyl sulfate—polyacrylamide gel electrophoresis, and transferred to polyvinylidene difluoride membrane (Millipore, Bedford, MA, USA). Protein bands were visualized with F4C1 or M2 mouse antibodies. Known aliquots of the corresponding Escherichia coli-expressed purified arrestin (Gurevich and Benovic 2000) were run alongside samples to construct the calibration curves for quantification.
Results
Subcellular localization of cone arrestin in cone photoreceptors and HEK293 cells
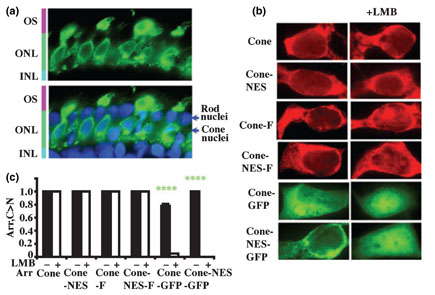
Different arrestins demonstrate distinct subcellular distribution in cells expressing them endogenously. Rod arrestin and arrestin3 are predominantly cytoplasmic, whereas the localization of arrestin2 is dramatically different in several types of neurons, from cytoplasmic to mostly nuclear (Song et al. 2006). Nucleo-cytoplasmic shuttling of cone arrestin has never been studied. To determine the subcellular localization of endogenous cone arrestin we used salamander retina, where it can be selectively detected by F4C1 monoclonal antibody that recognizes both rod and cone subtypes in other species (Smith et al. 2000). In agreement with previous studies (Zhu et al. 2002; Zhang et al. 2003), we found that in photoreceptors adapted to room light (~600 lux) cone arrestin localizes to outer segments, synaptic terminals, and cell bodies. Cone arrestin is almost exclusively cytoplasmic, with very low amounts detected in the nuclei despite fairly high concentration in perinuclear space (Fig. 1a). Untagged and Flag-tagged cone arrestin expressed in HEK293 cells at physiologically relevant levels (Chan et al. 2007) predominantly localized in the cytoplasm, faithfully reproducing the localization of the endogenous cone arrestin in photoreceptors. GFP-tagged cone arrestin (which is ~20 kDa bigger and therefore cannot diffuse via nuclear pore freely) is also mostly cytoplasmic, with somewhat greater proportion in the nucleus (Fig. 1b and c).
Fig 1.
Similar localization of endogenous cone arrestin in cone photoreceptors and HEK293 cells. (a) Endogenous cone arrestin is predominantly cytoplasmic in photoreceptor cells. Cone arrestin in salamander retina was visualized with F4C1 antibody (green). Lower panel shows overlay of cone arrestin immunostaining (green) and nuclear staining (4′,6-diaminido-2-phenylindole, blue). OS, outer segments; ONL, outer nuclear layer (photoreceptor nuclei); INL, inner nuclear layer. (b) The distribution of cone arrestin in HEK293A cells. Flag-and green fluorescent protein (GFP) -tagged cone arrestin was visualized with M2 anti-Flag antibody (red), and by native GFP fluorescence (green), respectively. Untagged and flag-tagged cone arrestin localizes almost exclusively in the cytoplasm, similar to endogenous cone arrestin in photoreceptors. GFP-tagged cone arrestin (which is ~20 kDa bigger) is also predominantly cytoplasmic, with a slight increase of the nuclear content. Exportin1 inhibitor leptomycin B (LMB) had little effect on the distribution of untagged and Flag-tagged cone arrestin, whereas the distribution of GFP-tagged protein changed dramatically from predominantly cytoplasmic to almost exclusively nuclear. An engineered nuclear export signal (NES) had minor effect on arrestin distribution with or without LMB treatment. Untagged, Flag- and GFP-tagged cone arrestin were expressed at comparable levels of 71 ± 9, 50 ± 15, and 66 ± 6 pmol/mg protein, respectively, as measured by quantitative western blot with F4C1 antibody with calibration curve with purified recombinant cone arrestin. (c) C > N: The proportion of cells showing stronger fluorescent arrestin signal in the cytoplasm than in the nucleus. Mean ± SD of three experiments are shown. ****p < 0.0001 (green), indicates the significance of LMB effect. Cone and Cone-F, untagged and Flag-tagged cone arrestin; Cone-NES and Cone-NES-F, untagged and Flag-tagged cone arrestin with an engineered NES in the C-tail; Cone-GFP, GFP-tagged cone arrestin; Cone-NES-GFP, GFP-tagged cone arrestin with an engineered NES.
We used leptomycin B (LMB), a known inhibitor of exportin1 that mediates NES-dependent export from the nucleus (Fukuda et al. 1997; Kudo et al. 1999), to test whether exportin1 affects cone arrestin localization. LMB (50 ng/mL, overnight) did not change the distribution of untagged or Flag-tagged cone arrestin, whereas the distribution of GFP-tagged protein changed dramatically, from predominantly cytoplasmic to almost exclusively nuclear (Fig. 1b and c). Cone arrestin does not have identifiable NES, and engineering of a NES in untagged and Flag-tagged cone arrestin (NES+) did not change its distribution, with or without LMB treatment (Fig. 1b and c). Thus, cone arrestin apparently enters the nucleus and exits it via exportin1-independent pathway. However, extra NES slightly reduced the proportion of GFP-tagged cone arrestin in the nucleus, consistent with LMB sensitivity of its export (Fig. 1).
Cytoplasmic localization of cone arrestin makes nuclear exclusion assay suitable to test its interactions with the binding partners that predominantly localize to the nucleus, such as protein kinase JNK3 and ubiquitin ligase Mdm2.
Cone arrestin relocalizes JNK3 and Mdm2 from the nucleus to the cytoplasm
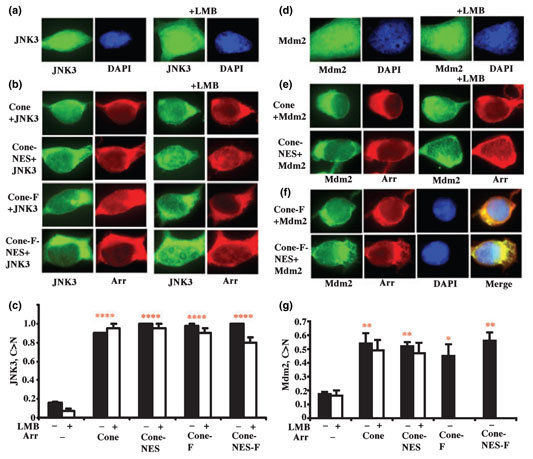
Green fluorescent protein-JNK3 expressed alone in HEK293 cells is predominantly nuclear and LMB does not affect its localization (Fig. 2a). Co-expression of wild-type cone arrestin effectively moves GFP-JNK3 to the cytoplasm; an extra NES does not change its efficiency (Fig. 2a and b). LMB treatment did not have a significant effect on JNK3 distribution in cells expressing untagged or Flag-tagged cone arrestin with or without an engineered NES (Fig. 2b and c). Thus, cone arrestin removes JNK3 from the nucleus via LMB-insensitive pathway. Moreover, enabling its interaction with exportin1 via engineered NES does not redirect it to LMB-sensitive pathway. Importantly, native and Flag-tagged cone arrestin have identical effects on JNK3 distribution (Fig. 2a–c).
Fig 2.
Cone arrestin dramatically changes the subcellular distribution of c-Jun N-terminal kinase (JNK3) and Mdm2 in HEK293 cells. (a) JNK3-green fluorescent protein (GFP) localizes mostly to the nucleus [visualized with 4′,6-diaminido-2-phenylindole (DAPI) staining, blue]. (b) Co-expression of untagged or Flag-tagged cone arrestin relocalizes JNK3-GFP (green) to the cytoplasm. Untagged and Flag-tagged cone arrestin was visualized with rabbit polyclonal antibody raised against F4C1 epitope and with M2 anti-FLAG antibody, respectively (both red). leptomycin B (LMB) did not affect JNK3-GFP distribution in cells co-expressing untagged or Flag-tagged cone arrestin. An engineered nuclear export signal (NES) did not change arrestin effect on JNK3 distribution, with or without LMB treatment. Untagged and Flag-tagged cone arrestins were expressed at comparable levels of 218 ± 5 and 152 ± 13 pmol/mg protein, respectively. (c) C > N: The proportion of cells showing stronger JNK3-GFP fluorescent signal in the cytoplasm than in the nucleus. ****p < 0.0001 (red), indicates the significance of the arrestin-induced changes in JNK3 localization when compared with JNK3 alone. (d) GFP-Mdm2 localizes mostly in the nucleus (visualized with DAPI staining, blue). Co-expression of untagged (e) and Flag-tagged (f) cone arrestin removes Mdm2 to the cytoplasm. GFP-Mdm2 was visualized by native GFP fluorescence (green), untagged and Flag-tagged cone arrestins were visualized with rabbit polyclonal antibody raised against F4C1 epitope and M2 anti-FLAG antibody, respectively (red). LMB did not reverse arrestin-induced changes in Mdm2 localization (e). An engineered NES did not change arrestin effect on Mdm2 distribution (e and f), with or without LMB treatment (e). Untagged and Flag-tagged cone arrestin was expressed at comparable levels of 176 ± 15 and 183 ± 11 pmol/mg protein, respectively. (g) C > N: The proportion of cells showing stronger GFP-Mdm2 fluorescent signal in the cytoplasm than in the nucleus. *p < 0.05 (red) and **p < 0.01 (red) indicate the significance of arrestin-induced changes in Mdm2 localization. Mean ± SD of three experiments are shown in panels (c and g) (filled and empty bars represent the data obtained without and with LMB treatment, respectively). Cone, untagged cone arrestin; Cone-NES, untagged cone with an engineered NES; Cone-F, Flag-tagged cone arrestin; Cone-NES-F, Flag-tagged cone arrestin with an engineered NES; Arr, cone arrestin.
Green fluorescent protein-Mdm2 expressed alone localizes almost exclusively in the nucleus (Fig. 2d). Co-expressed wild-type cone arrestin relocalizes GFP-Mdm2 to the cytoplasm, similar to the situation with JNK3, but with lower efficiency (Fig. 2e–g). Neither an engineered NES nor exportin1 inhibitor LMB appreciably changed the effect of untagged arrestin (Fig. 2e and g). The LMB effect on cells co-expressing Mdm2 and Flag-tagged constructs could not be tested due to severe interference of LMB with the survival of these cells.
Differential effects of cone arrestin conformation on JNK3 and Mdm2 binding
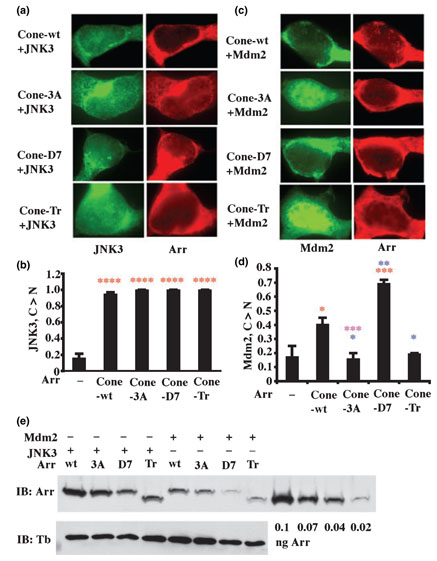
We used cone arrestin mutants to test whether the interaction with JNK3 is sensitive to arrestin conformation. JNK3 was co-expressed with Flag-tagged wild-type, 3A, D7 or truncated cone arrestin to enable simultaneous visualization of JNK3 and arrestin. We found that all forms of cone arrestin translocate JNK3 from the nucleus to the cytoplasm with similar high efficiency (Fig. 3a and b), suggesting that JNK3 binding is relatively insensitive to arrestin conformation. These data also demonstrate that cone arrestin in basal conformation binds JNK3 and that arrestin C-tail is not involved in JNK3 interaction.
Fig 3.
The effects of cone arrestin conformation on its interactions with c-Jun N-terminal kinase (JNK3) and Mdm2. (a) JNK3 binding is not significantly affected by arrestin conformation. Four forms of cone arrestin were co-expressed with green fluorescent protein (GFP)-JNK3: wild-type (Cone-wt), C-terminal triple alanine mutant with a ‘loose’ active-like conformation (Cone-3A), the mutant ‘frozen’ in the basal conformation by the deletion of seven residues in the interdomain hinge region (Cone-D7), and truncated cone arrestin(1–366) lacking the C-tail (Cone-Tr). GFP-JNK3 was visualized by intrinsic fluorescence (green); Flag-tagged arrestins were visualized with M2 anti-Flag antibody (red). Cone arrestin expression levels were: wt, 239 ± 21; 3A, 156 ± 20; D7, 73 ± 20; and Tr, 134 ± 35 pmol/mg protein. (b) C > N: The proportion of cells showing stronger JNK-GFP fluorescent signal in the cytoplasm than in the nucleus. ****p < 0.0001 (red), indicates the significance of the arrestin-induced changes in JNK3 localization. (c) Mdm2 preferentially binds cone arrestin in basal conformation. GFP-Mdm2 was visualized by intrinsic fluorescence (green); the indicated Flag-tagged arrestins were visualized with M2 anti-Flag antibody (red). Note that different forms of cone arrestin move Mdm2 from the nucleus to the cytoplasm with different efficiency in the order of: D7 > WT > 3A~Tr, which is the opposite of the order of receptor binding. Cone arrestin expression levels were: wt, 199 ± 16; 3A, 82 ± 19; D7, 55 ±0.5; and Tr, 71 ± 25 pmol/mg protein. (d) C > N: The proportion of cells showing stronger GFP-Mdm2 fluorescent signal in the cytoplasm than in the nucleus. *p < 0.05 (red); ***p < 0.001(red), indicates the significance of the arrestin-induced changes in Mdm2 localization; *p < 0.05 (blue); **p < 0.01 (blue), when compared with wild-type; ***p < 0.001 (purple), comparison between Cone-3A and Cone-D7. Mean ± SD of three experiments are shown in panels (b and d). (e) Representative quantitative western blot used to determine cone arrestin expression levels. Expressed in Escherichia coli and purified recombinant human cone arrestin was used to construct calibration curve (four lanes on the right). Lighter exposures (to ensure that the bands are not saturated) were used for protein quantification on VersaDoc with QuantityOne software (Bio-Rad). Equal amount of protein in cell lysates were loaded. Tubulin blots (lower panel) were run in parallel to test for equal loading.
Using the same experimental paradigm, we tested the conformational dependence of arrestin interactions with Mdm2 (Fig. 3c). According to the scores (Fig. 3d), the ‘constitutively inactive’ D7 mutant is much more efficient in redistributing Mdm2 from nucleus to the cytoplasm than wild-type cone arrestin. In contrast, the conformationally ‘loose’ 3A and truncated mutants do not appreciably change Mdm2 distribution in the cell (Fig. 3c and d). Thus, unlike JNK3, Mdm2 is very sensitive to arrestin conformation. Mdm2 binding is enhanced by the arrestin mutation that ‘freezes’ the molecule in the basal state that was previously considered ‘inactive.’
JNK3 and Mdm2 interaction sites on arrestin
Arrestins are elongated molecules in which several intramolecular interactions determine the relative orientation of the two domains (Hirsch et al. 1999; Han et al. 2001; Sutton et al. 2005). Arrestin domains can be expressed separately, fold independently, and retain certain functions (Gurevich and Benovic 1993; Gurevich et al. 1994, 1995; Hanson et al. 2006, 2007c; Nelson et al. 2007). In order to localize the binding sites for JNK3 and Mdm2, we expressed separate N-and C-domains of both cone and rod arrestin. As both domains are small enough to diffuse through the nuclear pore, an extra NES was added to these constructs to ensure cytoplasmic localization of arrestin domains. We found that both N- and C-domains of cone and rod arrestin move JNK3 to the cytoplasm as efficiently as full-length protein (Fig. 4a and c). These data provide the first indication that JNK3 interacts with both domains of arrestin proteins. Apparently the affinity of the partial sites on individual domains is sufficiently high to remove JNK3 from the nucleus. The N-domain of rod arrestin effectively moves Mdm2 to the cytoplasm, indicating that the main Mdm2 interaction site is localized within this element (Fig. 4b and d). In contrast, the N-domain of cone arrestin or the C-domain of either protein does not have this effect (Fig. 4b and d). The fact that full-length cone arrestin relocalizes Mdm2 (Figs 2 and 3), whereas neither domain affects its distribution (Fig. 4b and d), suggests that high affinity Mdm2 binding requires the participation of both domains.
Fig 4.
The interaction of individual cone and rod arrestin domains with c-Jun N-terminal kinase (JNK3) and Mdm2. (a and b) N- and C-domains of cone and rod arrestin were co-expressed with JNK3-green fluorescent protein (GFP) (a) and GFP-Mdm2 (b). The indicated Flagtagged arrestins were visualized with M2 anti-Flag antibody (red), JNK3-GFP and GFP-Mdm2 were visualized by intrinsic fluorescence (green). N- and C-domains of both cone and rod arrestin redistribute JNK3 to the cytoplasm, whereas only N-domain of rod arrestin removes Mdm2 from the nucleus. Arrestin expression levels (in pmol/mg protein) were: Cone-N, 159 ± 43; Cone-C, 129 ± 46 (co-expressed with JNK3); Cone-N, 127 ± 5.6; Cone-C, 176 ± 12 (co-expressed with Mdm2); Rod-N, 116 ± 15; Rod-C, 126 ± 13 (co-expressed with JNK3); Rod-N, 139 ± 10; Rod-C, 147 ± 5.1 (co-expressed with Mdm2). Protein expression was determined by quantitative western blot of cell lysates, as described in the legend to Fig. 3. (c and d) C > N: The proportion of cells showing stronger GFP-JNK3 (c) or GFP-Mdm2 (d) fluorescent signal in the cytoplasm than in the nucleus. **p < 0.01 (red); ***p < 0.001 (red); ****p < 0.0001 (red) indicate the significance of the arrestin-induced changes in JNK3 (c) and Mdm2 (d) localization, when compared with GFP-JNK3 and GFP-Mdm2 expressed alone; additional comparisons in panel (d): ***p < 0.001 (green); ****p < 0.0001 (green), comparison between an individual domain and corresponding full-length arrestin; ****p < 0.0001 (blue), comparison between the N- and C-domain. Mean ± SD of three experiments are shown. Cone-N: N-domain of cone arrestin (residues 1–178); Cone-C: C domain of cone arrestin (177–359); Rod-N: N-domain of rod arrestin (1–186); Rod-C: C domain of rod arrestin (185–365).
Discussion
Receptor-bound non-visual arrestins redirect GPCR signaling to mitogen-activated protein kinase cascades activating JNK3, ERK1/2, and facilitate GPCR internalization by binding to clathrin and AP2 (reviewed in Gurevich and Gurevich 2003; Lefkowitz and Shenoy 2005). They also act as adapters for E3 ubiquitin ligase Mdm2 (Shenoy et al. 2001; Shenoy and Lefkowitz 2003; Girnita et al. 2005). In contrast, little is known about the interactions of free arrestins with non-receptor partners. Demonstrated ability of non-visual arrestins to remove JNK3 and Mdm2 from the nucleus was the first indication that free arrestins bind these proteins (Scott et al. 2002; Wang et al. 2003a; Song et al. 2006). Interaction with non-receptor signaling proteins was considered a specialized function of non-visual arrestins until we demonstrated that rod arrestin binds JNK3 and Mdm2 (Song et al. 2006). Remarkable similarity of crystal structures of cone (Sutton et al. 2005), rod (Hirsch et al. 1999), and arrestin2 (Han et al. 2001) suggests that some signaling proteins may also interact with cone arrestin.
The cytoplasmic localization of endogenous cone arrestin in photoreceptors is faithfully reproduced in HEK293 cells (Fig. 1) indicating that intracellular distribution of ectopically expressed protein is controlled by physiologically relevant mechanisms. The size of arrestin (about 40 kDa) is small enough to diffuse via nuclear pore. As diffusion could only equalize protein concentrations in the two compartments, its localization suggests that cone arrestin is actively exported from the nucleus. Transport through the nuclear pore is mediated by specific soluble factors, importins and exportins (Weis 2003). Their binding to cargo is regulated by a small GTPase Ran required for the directionality of transport (Askjaer et al. 1998). Cone arrestin has neither conventional nuclear localization signals nor NES, and the distribution of untagged or Flag-tagged protein was not affected by the inhibition of exportin1, demonstrating that cytosolic localization of cone arrestin is achieved via LMB-insensitive mechanisms. It is worth noting that LMB dramatically changed the distribution of GFP-tagged cone arrestin from mostly cytoplasmic to nuclear. This result illustrates the advantage of small tags over large fluorescent protein tags. The latter often are used to visualize arrestins (Barak et al. 1997), but they can significantly change the behavior of the protein (Fig. 1).
We found that cone arrestin binds both JNK3 and Mdm2 and relocalizes them from the nucleus to the cytoplasm. These data indicate that, although cone arrestin is predominantly cytoplasmic, it actually shuttles between the two compartments. We found that all forms of cone arrestin remove JNK3 from the nucleus with comparable efficiency (Fig. 3a and b). In contrast, Mdm2 is highly sensitive to arrestin conformation, with the order of potency D7 > WT > 3A~Tr (Fig. 3c and d), i.e. the opposite compared with receptor binding. These data demonstrate that in case of cone arrestin the conformational preferences of JNK3 and Mdm2 are similar to those for other arrestin subtypes. Arrestins recruited to activated GPCRs may bring to the receptor signaling proteins that bind free arrestin. Our data suggest that JNK3 would likely remain bound to arrestins when they interact with receptors as a part of the ‘signalosome’ complex (McDonald et al. 2000). In contrast, Mdm2 would be released soon after arrestin binding to the receptor, as it has lower affinity for the receptor-bound arrestin conformation.
Arrestins preferentially bind active phosphorylated form of their cognate receptors (Gurevich and Gurevich 2006b). Cone arrestin binds light-activated phosphorylated cone pigments (Zhu et al. 2003; Sutton et al. 2005) and translocates to the outer segment of cone photoreceptors in the light (Zhu et al. 2002; Zhang et al. 2003; Coleman and Semple-Rowland 2005), similar to its rod counterpart (Broekhuyse et al. 1985; Philp et al. 1987; Elias et al. 2004; Nair et al. 2005; Strissel et al. 2006), suggesting that it participates in shutting off phototransduction in cones. The expression of cone arrestin per mouse cone photoreceptor cell was recently estimated to be about 1/400th of rod arrestin expression per rod (Chan et al. 2007). Endogenous rod arrestin expression is extremely high, translating into >1 mmol/L concentration in photoreceptors (Hanson et al. 2007b). Taking into account that cone photoreceptors are substantially smaller than rods, the physiologically relevant range of cone arrestin concentrations in the cytoplasm can be estimated at 5–20 μmol/L. This level is lower than the expression of cone pigments, in contrast to almost equimolar expression of rod arrestin and rhodopsin in rods (Broekhuyse et al. 1985; Strissel et al. 2006; Hanson et al. 2007a). Cones also co-express rod arrestin (Zhu et al. 2005) that is perfectly capable to quench cone opsins (Kefalov et al. 2003). In contrast, cone arrestin cannot take over the function of its rod counterpart to ensure rapid turnoff of rhodopsin signaling (Chan et al. 2007). A significant proportion of cone arrestin is consistently found in the synaptic terminals in vivo (Fig. 1a) (Zhu et al. 2002; Zhang et al. 2003), and in vitro it demonstrates binding to m2 muscarinic cholinergic receptor which is much closer to that of non-visual arrestins than to rod arrestin (Sutton et al. 2005). Our finding that cone arrestin binds JNK3 and Mdm2 suggests that in cones it may participate in G protein-independent signaling of cone opsins and/or non-visual GPCRs.
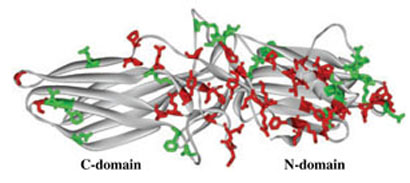
Arrestin elements on the concave sides of both domains (Fig. 5) involved in receptor binding were mapped using a wide variety of methods (reviewed in Gurevich and Gurevich 2006b). In contrast, the binding sites for relatively few of other arrestin binding partners have been identified: clathrin (Goodman et al. 1996; Kim and Benovic 2002), AP2 (Laporte et al. 1999; Kim and Benovic 2002), microtubules (Hanson et al. 2006, 2007c), and calmodulin (Wu et al. 2006). The results of previous attempts to identify JNK3 binding elements in arrestin are controversial. JNK3 interaction site was originally mapped to the C-domain (McDonald et al. 2000) and subsequently narrowed down to a short sequence RRSLHL in the loop between β-strands XI and XII on the distal tip of the C-domain of rat arrestin3 (Miller et al. 2001). However, this identification was based on the difference in apparent JNK3 binding between arrestin2 and arrestin3, which was not confirmed in subsequent studies (Wang et al. 2003a; Song et al. 2006). Moreover, RRSLHL sequence is not conserved in arrestin3 homologues in other mammalian species, making it an unlikely candidate (Gurevich and Gurevich 2006a, b).
Fig 5.
Conserved residues on the non-receptor-binding side of arrestin molecule. Comparable binding of all four vertebrate arrestins to c-Jun N-terminal kinase and Mdm2 suggests that the residues on the non-receptor-binding side that are either conserved throughout the arrestin family (side chains high-lighted in red) or conservatively substituted in different arrestin subtypes (side chains highlighted in green) mediate these interactions. These residues form three ‘patches’: two relatively large ones on the distal N-domain and near the center on the molecule and a small one on the distal C-domain. Note that a considerable portion of the C-domain does not carry conserved residues, which makes this area a likely candidate for the binding of subtype-specific arrestin partners.
To localize the binding sites for JNK3 and Mdm2, we expressed individual domains of cone and rod arrestins. Both domains move JNK3 as efficiently as full-length proteins (Fig. 4a and c). Thus, JNK3 binding site includes elements in both domains and the affinity of N- and C-domain parts of the site is relatively high. These data explain why JNK3 interaction with arrestin is conformation-independent in the nuclear exclusion assay (Fig. 3a and b): the strongest conceivable effect of an ‘unfavorable’ arrestin conformation would be the disruption of the interaction with one of the domains, which apparently does not prevent JNK3 binding (Fig. 4a and c). The presence of a strong JNK3-binding site on each domain of arrestin raises several biologically important questions. Do the elements in both domains constitute parts of an extensive high affinity site that spans the elongated arrestin molecule, or do these ‘alternative’ sites mediate arrestin-JNK3 interaction under different circumstances? Does JNK3 bind to both domains simultaneously or to one of the ‘partial’ sites when arrestin scaffolds ASK1-MKK4-JNK3 cascade? Does JNK3 bind to free and receptor-bound arrestin via the same site(s)? Targeted disruption of individual ‘parts’ of the interaction site (Fig. 5) will be necessary to answer these questions.
Mdm2 was shown to interact with the N-domain of arrestin3 (Wang et al. 2003b). Our finding that the N-domain of rod arrestin alone relocalizes Mdm2 as efficiently as full-length protein, whereas the C-domain does not (Fig. 4b and d), supports the conclusion that the N-domain contains the main Mdm2 binding site. However, neither domain of cone arrestin affects the localization of Mdm2 (Fig. 4b and d), whereas full-length protein does (Figs 2 and 3). Thus, Mdm2 also interacts with both arrestin domains, although the N-domain plays a major role. Apparent involvement of both arrestin domains in Mdm2 binding and the insufficient affinity of the ‘partial’ sites on each domain explain the high sensitivity of this interaction to conformational changes that involve domain movement (Vishnivetskiy et al. 2002). Mdm2 preference for arrestin ’frozen’ in the basal state (Fig. 3c and d) suggests that the distance between the ‘partial’ sites in this conformation provides a perfect fit for Mdm2. Receptor binding-induced conformational rearrangement likely facilitates the release of arrestin-associated Mdm2, which may explain limited modification of receptor-bound arrestin by this ubiquitin ligase (Shenoy et al. 2001; Shenoy and Lefkowitz 2003).
The data allow us to narrow down the number of arrestin residues that can be involved in JNK3 and Mdm2 interactions. Comparable binding of all four vertebrate arrestins to JNK3 and Mdm2 (Figs 2 and 3; Wang et al. 2003a; Song et al. 2006) suggests that these interactions largely involve residues that are conserved among the four vertebrate arrestins. Whereas the two non-visual arrestins are highly homologous (Attramadal et al. 1992; Sterne-Marr et al. 1993), they have diverged very far from their rod and cone ‘cousins’ (Gurevich and Gurevich 2006a), leaving a limited number of possible ‘suspects’ (Fig. 5). In addition, our data show that the least conserved arrestin element, the C-tail, is unlikely to be involved (Fig. 3). Moreover, the interaction of JNK3 and Mdm2 with the arrestin-receptor complex (McDonald et al. 2000; Shenoy et al. 2001; Shenoy and Lefkowitz 2003) excludes residues in the receptor-binding elements. This analysis identifies 21 and 12 exposed conserved residues in the N- and C-domain, respectively (red in Fig. 5) along with 13 and 8 positions with conservative substitutions (green in Fig. 5), setting the stage for more precise identification of JNK3 and Mdm2 binding sites by site-directed mutagenesis.
Acknowledgements
The authors thank Drs L. A. Donoso, George Inana, Louis M. Luttrell, and G. Pei for F4C1 antibody, human cone arrestin cDNA, GFP-JNK3, and GFP-Mdm2 constructs, respectively, and Dr Whitney M. Cleghorn for critical reading of the manuscript. This study is supported by NIH RO1 grants EY11500 (VVG), NS45117 (EVG), Vanderbilt University Discovery Grant 1040659012 (VVG), and P30 core grant in vision research EY008126 (to Vanderbilt University).
Abbreviations
- DAPI
4′,6-diaminido-2-phenylindole
- GFP
green fluorescent protein
- GPCR
G protein-coupled receptor
- HEK
human embryonic kidney cells
- JNK3
c-Jun N-terminal kinase
- LMB
leptomycin B
- NES
nuclear export signal
- PBS
phosphate-buffered saline
References
- Askjaer P, Jensen TH, Nilsson J, Englmeier L, Kjems J. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by Ran GTP. J. Biol. Chem. 1998;273:33414–33422. doi: 10.1074/jbc.273.50.33414. [DOI] [PubMed] [Google Scholar]
- Attramadal H, Arriza JL, Aoki C, Dawson TM, Codina J, Kwatra MM, Snyder SH, Caron MG, Lefkowitz RJ. Beta-arrestin2, a novel member of the arrestin/beta-arrestin gene family. J. Biol. Chem. 1992;267:17882–17890. [PubMed] [Google Scholar]
- Barak LS, Ferguson SS, Zhang J, Caron MG. A betaarrestin/green fluorescent protein biosensor for detecting G proteincoupled receptor activation. J. Biol. Chem. 1997;272:27497–27500. doi: 10.1074/jbc.272.44.27497. [DOI] [PubMed] [Google Scholar]
- Broekhuyse RM, Tolhuizen EF, Janssen AP, Winkens HJ. Light induced shift and binding of S-antigen in retinal rods. Curr. Eye Res. 1985;4:613–618. doi: 10.3109/02713688508999993. [DOI] [PubMed] [Google Scholar]
- Chan S, Rubin WW, Mendez A, Liu X, Song X, Hanson SM, Craft CM, Gurevich VV, Burns ME, Chen J. Functional comparisons of visual arrestins in rod photoreceptors of transgenic mice. Invest. Ophthalmol. Vis. Sci. 2007;48:1968–1975. doi: 10.1167/iovs.06-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JE, Semple-Rowland SL. GC1 deletion prevents light-dependent arrestin translocation in mouse cone photoreceptor cells. Invest. Ophthalmol. Vis. Sci. 2005;46:12–16. doi: 10.1167/iovs.04-0691. [DOI] [PubMed] [Google Scholar]
- Craft CM, Whitmore DH, Wiechmann AF. Cone arrestin identified by targeting expression of a functional family. J. Biol. Chem. 1994;269:4613–4619. [PubMed] [Google Scholar]
- Donoso LA, Gregerson DS, Smith L, Robertson S, Knospe V, Vrabec T, Kalsow CM. S-antigen: preparation and characterization of site-specific monoclonal antibodies. Curr. Eye Res. 1990;9:343–355. doi: 10.3109/02713689008999622. [DOI] [PubMed] [Google Scholar]
- Elias RV, Sezate SS, Cao W, McGinnis JF. Temporal kinetics of the light/dark translocation and compartmentation of arrestin and alpha-transducin in mouse photoreceptor cells. Mol. Vis. 2004;10:672–681. [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- Girnita L, Shenoy SK, Sehat B, Vasilcanu R, Girnita A, Lefkowitz RJ, Larsson O. b-Arrestin is crucial for ubiquitination and down-regulation of the insulin-like growth factor-1 receptor by acting as adaptor for the MDM2 E3 ligase. J. Biol. Chem. 2005;280:24412–24419. doi: 10.1074/jbc.M501129200. [DOI] [PubMed] [Google Scholar]
- Goodman OB, Jr, Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, Keen JH, Benovic JL. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2- adrenergic receptor. Nature. 1996;383:447–450. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Benovic JL. Visual arrestin interaction with rhodopsin: sequential multisite binding ensures strict selectivity towards light-activated phosphorylated rhodopsin. J. Biol. Chem. 1993;268:11628–11638. [PubMed] [Google Scholar]
- Gurevich VV, Benovic JL. Arrestin: mutagenesis, expression, purification, and functional characterization. Methods Enzymol. 2000;315:422–437. doi: 10.1016/s0076-6879(00)15859-8. [DOI] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. The new face of active receptor bound arrestin attracts new partners. Structure. 2003;11:1037–1042. doi: 10.1016/s0969-2126(03)00184-9. [DOI] [PubMed] [Google Scholar]
- Gurevich EV, Gurevich VV. Arrestins are ubiquitous regulators of cellular signaling pathways. Genome Biol. 2006a;7:236. doi: 10.1186/gb-2006-7-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV, Gurevich EV. The structural basis of arrestin-mediated regulation of G protein-coupled receptors. Pharm. Ther. 2006b;110:465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich VV, Chen C-Y, Kim CM, Benovic JL. Visual arrestin binding to rhodopsin: intramolecular interaction between the basic N-terminus and acidic C-terminus of arrestin may regulate binding selectivity. J. Biol. Chem. 1994;269:8721–8727. [PubMed] [Google Scholar]
- Gurevich VV, Dion SB, Onorato JJ, Ptasienski J, Kim CM, Sterne-Marr R, Hosey MM, Benovic JL. Arrestin interaction with G protein-coupled receptors. Direct binding studies of wild type and mutant arrestins with rhodopsin, b2-adrenergic, and m2 muscarinic cholinergic receptors. J. Biol. Chem. 1995;270:720–731. doi: 10.1074/jbc.270.2.720. [DOI] [PubMed] [Google Scholar]
- Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Crystal structure of beta-arrestin at 1.9 Å: possible mechanism of receptor binding and membrane translocation. Structure. 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- Hanson SM, Francis DJ, Vishnivetskiy SA, Klug CS, Gurevich VV. Visual arrestin binding to microtubules in-volves a distinct conformational change. J. Biol. Chem. 2006;281:9765–9772. doi: 10.1074/jbc.M510738200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SM, Gurevich EV, Vishnivetskiy SA, Ahmed MR, Song X, Gurevich VV. Each rhodopsin molecule binds its own arrestin. Proc. Natl Acad. Sci. USA. 2007a;104:3125–3128. doi: 10.1073/pnas.0610886104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SM, Van Eps N, Francis DJ, Altenbach C, Vishnivetskiy SA, Klug CS, Hubbell WL, Gurevich VV. Structure and function of the visual arrestin oligomer. EMBO J. 2007b;26:1726–1736. doi: 10.1038/sj.emboj.7601614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson SM, Cleghorn WM, Francis DJ, Vishnivetskiy SA, Raman D, Song S, Nair KS, Slepak VZ, Klug CS, Gurevich VV. Arrestin mobilizes signaling proteins to the cytoskeleton and redirects their activity. J. Mol. Biol. 2007c;368:375–387. doi: 10.1016/j.jmb.2007.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch JA, Schubert C, Gurevich VV, Sigler PB. The 2.8 Å crystal structure of visual arrestin: a model for arrestin's regulation. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- Kefalov V, Fu Y, Marsh-Armstrong N, Yau KW. Role of visual pigment properties in rod and cone phototransduction. Nature. 2003;425:526–531. doi: 10.1038/nature01992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Benovic JL. Differential roles of arrestin-2 interaction with clathrin and adaptor protein 2 in G protein-coupled receptor trafficking. J. Biol. Chem. 2002;277:30760–30768. doi: 10.1074/jbc.M204528200. [DOI] [PubMed] [Google Scholar]
- Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl Acad. Sci. USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte SA, Oakley RH, Zhang J, Holt JA, Fergusons SG, Caron MG, Barak LS. The 2-adrenergic receptor/arrestin complex recruits the clathrin adaptor AP-2 during endocytosis. Proc. Natl Acad. Sci. USA. 1999;96:3712–3717. doi: 10.1073/pnas.96.7.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- Luttrell LM, Roudabush FL, Choy EW, Miller WE, Field ME, Pierce KL, Lefkowitz RJ. Activation and targeting of extracellular signal-regulated kinases by beta-arrestin scaffolds. Proc. Natl Acad. Sci. USA. 2001;98:2449–2454. doi: 10.1073/pnas.041604898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald PH, Chow CW, Miller WE, Laporte SA, Field ME, Lin FT, Davis RJ, Lefkowitz RJ. Beta-arrestin 2: a receptor-regulated MAPK scaffold for the activation of JNK3. Science. 2000;290:1515–1518. doi: 10.1126/science.290.5496.1574. [DOI] [PubMed] [Google Scholar]
- Miller WE, McDonald PH, Cai SF, Field ME, Davis RJ, Lefkowitz RJ. Identification of a motif in the carboxyl terminus of beta-arrestin2 responsible for activation of JNK3. J. Biol. Chem. 2001;276:27770–27777. doi: 10.1074/jbc.M102264200. [DOI] [PubMed] [Google Scholar]
- Murakami A, Yajima T, Sakuma H, McLaren MJ, Inana G. X-arrestin: a new retinal arrestin mapping to the X chromosome. FEBS Lett. 1993;334:203–209. doi: 10.1016/0014-5793(93)81712-9. [DOI] [PubMed] [Google Scholar]
- Nair KS, Hanson SM, Mendez A, et al. Light-dependent redistribution of arrestin in vertebrate rods is an energy-independent process governed by protein-protein interactions. Neuron. 2005;46:555–567. doi: 10.1016/j.neuron.2005.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CD, Perry SJ, Regier DS, Prescott SM, Topham MK, Lefkowitz RJ. Targeting of diacylglycerol degradation to M1 muscarinic receptors by beta-arrestins. Science. 2007;315:663–666. doi: 10.1126/science.1134562. [DOI] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- Philp NJ, Chang W, Long K. Light-stimulated protein movement in rod photoreceptor cells of the rat retina. FEBS Lett. 1987;225:127–132. doi: 10.1016/0014-5793(87)81144-4. [DOI] [PubMed] [Google Scholar]
- Scott MG, Le Rouzic E, Perianin A, Pierotti V, Enslen H, Benichou S, Marullo S, Benmerah A. Differential nucleocyto plasmic shuttling of beta-arrestins. Characterization of a leucinerich nuclear export signal in beta-arrestin2. J. Biol. Chem. 2002;277:37693–37701. doi: 10.1074/jbc.M207552200. [DOI] [PubMed] [Google Scholar]
- Shenoy SK, Lefkowitz RJ. Trafficking patterns of beta-arrestin and G protein-coupled receptors determined by the kinetics of beta-arrestin deubiquitination. J. Biol. Chem. 2003;278:14498–14506. doi: 10.1074/jbc.M209626200. [DOI] [PubMed] [Google Scholar]
- Shenoy SK, McDonald PH, Kohout TA, Lefkowitz RJ. Regulation of receptor fate by ubiquitination of activated beta 2-adrenergic receptor and beta-arrestin. Science. 2001;294:1307–1313. doi: 10.1126/science.1063866. [DOI] [PubMed] [Google Scholar]
- Smith WC, Gurevich EV, Dugger DR, Shelamer CL, Vishnivetskiy SA, McDowell H, Gurevich VV. Cloning and functional characterization of salamander rod and cone arrestins. Invest. Ophthalmol. Vis. Sci. 2000;41:2445–2455. [PubMed] [Google Scholar]
- Song X, Raman D, Gurevich EV, Vishnivetskiy SA, Gurevich VV. Visual and both non-visual arrestins in their ‘inactive’ conformation bind JNK3 and Mdm2 and relocalize them from the nucleus to the cytoplasm. J. Biol. Chem. 2006;281:21491–21499. doi: 10.1074/jbc.M603659200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterne-Marr R, Gurevich VV, Goldsmith P, Bodine RC, Sanders C, Donoso LA, Benovic JL. Polypeptide variants of beta-arrestin and arrestin3. J. Biol. Chem. 1993;268:15640–15648. [PubMed] [Google Scholar]
- Strissel KJ, Sokolov M, Trieu LH, Arshavsky VY. Arrestin translocation is induced at a critical threshold of visual signaling and is superstoichiometric to bleached rhodopsin. J. Neurosci. 2006;26:1146–1153. doi: 10.1523/JNEUROSCI.4289-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Vishnivetskiy SA, Robert J, Hanson SM, Raman D, Knox BE, Kono M, Navarro J, Gurevich VV. Crystal structure of cone arrestin at 2.3 Å: evolution of receptor specificity. J. Mol. Biol. 2005;354:1069–1080. doi: 10.1016/j.jmb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Vishnivetskiy SA, Hirsch JA, Velez M-G, Gurevich YV, Gurevich VV. Transition of arrestin in the active receptorbinding state requires an extended interdomain hinge. J. Biol. Chem. 2002;277:43961–43968. doi: 10.1074/jbc.M206951200. [DOI] [PubMed] [Google Scholar]
- Wang P, Wu Y, Ge X, Ma L, Pei G. Subcellular localization of beta-arrestins is determined by their intact N domain and the nuclear export signal at the C terminus. J. Biol. Chem. 2003a;278:11648–11653. doi: 10.1074/jbc.M208109200. [DOI] [PubMed] [Google Scholar]
- Wang P, Gao H, Ni Y, Wang B, Wu Y, Ji L, Qin L, Ma L, Pei G. Beta-arrestin 2 functions as a G-protein-coupled receptoractivated regulator of oncoprotein Mdm2. J. Biol. Chem. 2003b;278:6363–6370. doi: 10.1074/jbc.M210350200. [DOI] [PubMed] [Google Scholar]
- Weis K. Regulating access to the genome: nucleoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- Wu N, Hanson SM, Francis DJ, Vishnivetskiy SA, Thibonnier M, Klug CS, Shoham M, Gurevich VV. Arrestin binding to calmodulin: a direct interaction between two ubiquitous signaling proteins. J. Mol. Biol. 2006;364:955–963. doi: 10.1016/j.jmb.2006.09.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Huang W, Zhu X, Craft CM, Baehr W, Chen CK. Light-dependent redistribution of visual arrestins and transducin subunits in mice with defective phototransduction. Mol. Vis. 2003;9:231–237. [PubMed] [Google Scholar]
- Zhu X, Li A, Brown B, Weiss ER, Osawa S, Craft CM. Mouse cone arrestin expression pattern: light induced translocation in cone photoreceptors. Mol. Vis. 2002;8:462–471. [PubMed] [Google Scholar]
- Zhu X, Brown B, Li A, Mears AJ, Swaroop A, Craft CM. GRK1-dependent phosphorylation of S and M opsins and their binding to cone arrestin during cone phototransduction in the mouse retina. J. Neurosci. 2003;23:6152–6160. doi: 10.1523/JNEUROSCI.23-14-06152.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Wu K, Rife L, Brown B, Craft CM. Rod arrestin expression and function in cone photoreceptors. Invest. Ophthalmol. Vis. Sci. 2005;46:1179. [Google Scholar]