Abstract
Arrestins bind active phosphorylated G protein-coupled receptors, terminating G protein activation. Receptor-bound non-visual arrestins interact with numerous partners, redirecting signaling to alternative pathways. Arrestins also have nuclear localization and nuclear exclusion signals and shuttle between the nucleus and the cytoplasm. Constitutively shuttling proteins often redistribute their interaction partners between the two compartments. Here we took advantage of the nucleoplasmic shuttling of free arrestins and used a “nuclear exclusion assay” to study their interactions with two proteins involved in “life-and-death” decisions in the cell, the kinase JNK3 and the ubiquitin ligase Mdm2. In human embryonic kidney 293 cells green fluorescent protein (GFP)-JNK3 and GFP-Mdm2 predominantly localize in the nucleus, whereas visual arrestin, arrestin2(Q394L) mutant equipped with the nuclear exclusion signal, and arrestin3 localize exclusively to the cytoplasm. Coexpression of arrestins moves both GFP-JNK3 and GFP-Mdm2 to the cytoplasm. Arrestin mutants “frozen” in the basal conformation are the most efficacious. Thus, arrestins in their basal state interact with JNK3 and Mdm2, suggesting that arrestins are likely “preloaded” with their interaction partners when they bind the receptor. Robust interaction of free arrestins with JNK3 and Mdm2 and their ability to regulate subcellular localization of these proteins may play an important role in the survival of photoreceptors and other neurons, as well as in retinal and neuronal degeneration.
Arrestins specifically bind agonist-activated phosphorylated G protein-coupled receptors (GPCRs),3 terminating further G protein activation and often redirecting signaling to alternative pathways (1, 2). Visual arrestin plays a key role in the regulation of rhodopsin signaling in rod photoreceptors. Non-visual arrestins 2 and 3 are expressed in most cells and regulate the signaling of a wide variety of GPCRs. Arrestin2 is the most abundant subtype in mature neurons (3, 4). Receptor-bound non-visual arrestins link GPCRs to the activation of c-Src (5), serve as scaffolds for receptor activation-dependent phosphorylation of ERK1/2 (6) and JNK3 (7), and mobilize the E3 ubiquitin ligase Mdm2 to the arrestin-receptor complex (8), etc. Binding to the receptor is accompanied by a global conformational change in the arrestin molecule (4), which is widely believed to underlie preferential interaction of numerous nonreceptor partners with receptor-bound, rather than with free, arrestin (2, 9).
Non-visual arrestins 2 and 3 were recently shown to shuttle between the nucleus and the cytoplasm (10, 11). Arrestin3 with its native nuclear export signal (NES) removes some of its interaction partners, such as JNK3 (10) and Mdm2 (11), from the nucleus. This must be a function of free arrestin, because membrane-imbedded GPCRs are not transported through the aqueous nuclear pore. Here we used the ability of arrestin proteins to bring their binding partners out of the nucleus as a readout to study the interactions of free arrestins 2 and 3 and visual arrestin with JNK3 and Mdm2, two proteins that play a pivotal role in the regulation of cell death and survival. We found that all three arrestins interact with JNK3 and Mdm2 and dramatically change their subcellular localization. Comparison of wild-type arrestins, “constitutively active” forms, and mutants frozen in the basal state shows that both JNK3 and Mdm2 bind arrestins in their basal state and Mdm2 actually prefers the inactive conformation.
MATERIALS AND METHODS
Plasmid Constructs
The coding sequences of bovine visual arrestin (12), arrestin2 (13), and arrestin3 (14) were subcloned into pcDNA3 as described (15). NES mutants of visual arrestin L203C, L280A, and the L203C/L280A double mutant, arrestin2(Q394L) with an engineered NES, NES-less arrestin3(L394Q), and constitutively active 3A mutants of bovine visual (F375A, V376A, F377A) (16) and arrestin2 and arrestin3 (I386A, V387A, F388A in both) (17), as well as mutants with seven-residue deletions in the inter-domain hinge (deleted residues 180, 182, 183, 187–190 in visual (18), homologous residues 174, 176, 177, 181–184 in arrestin2 and 175, 177, 178, 182–185 in arrestin3), were constructed by PCR-based mutagenesis. The visual arrestin-green fluorescent protein (GFP) fusion was constructed by amplifying the entire open reading frame with HindIII at the 5′-end and ApaI at the 3′-end and subcloning it in-frame into the appropriately digested EGFP-N vector (Clontech). Arrestin2-GFP and arrestin3-GFP in pcDNA3 were gifts from Dr. J. L. Benovic (Thomas Jefferson University). Arrestin2 and arrestin3 were FLAGtagged at the C terminus by PCR. All constructs were verified by dideoxy sequencing. Expression constructs for GFP-JNK3 and the human homolog of Mdm2-GFP were gifts from Drs. Louis Luttrell (Medical University of South Carolina) and Gang Pei (Shanghai Institute for Biological Sciences), respectively.
Cell Culture and Transient Transfection
HEK-293A were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen), penicillin, and streptomycin at 37°C in a humidified incubator with 5% CO2. The cells were plated at 80–90% confluence and transfected using Lipofectamine2000 (Invitrogen). The next day, the cells were trypsinized and seeded onto Lab-Tek CC2- treated chambered slides coated with fibronectin (20 μg/ml of phosphate-buffered saline) (Sigma) for immunofluorescence microscopy and onto 24-well plates coated with poly- D-lysine (15 μg/ml) for Western blot analysis.
Animals
Animal research was conducted in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the institutional Animal Care and Use Committee. The retinas of C57 mice and the brain sections of Sprague-Dawley rats were used.
Immunohistochemistry and Immunocytochemistry
The subcellular localization of endogenous visual arrestin in dark-adapted mouse retinas was determined as described (19). Rats were anesthetized with pentobarbital (50 mg/kg intraperitoneal) and transcardially perfused with 4% paraformaldehyde. Brains were removed, postfixed in the paraformaldehyde solution overnight at 4°C, cryoprotected in 30% sucrose overnight at 4°C, frozen on dry ice, and kept at –80°C until used. 30-μm-thick free-floating sections of the brain were incubated with rabbit polyclonal antibody specific for arrestin2 (3, 4). Both primary antibodies were used in 1:500 dilution overnight at 4°C, followed by anti-rabbit biotinylated antibodies and Alexa-488-conjugated streptavidin. Sections were photographed on a Nikon EC2000 fluorescent microscope equipped with a digital camera.
HEK 293A cells were fixed 72 h post-transfection in 100% methanol at –20°C for 5 min. The cells were rehydrated with phosphate-buffered saline and blocked with 3% bovine serum albumin in phosphate-buffered saline for 1 h at room temperature. Untagged and FLAG-tagged arrestins were visualized with F4C1 anti-arrestin (20) and M2 anti-FLAG antibody (Sigma), respectively, followed by the anti-mouse secondary antibodies (Molecular Probes, Eugene, OR).
Nuclear Exclusion Assay
GFP-JNK3 and the human homolog of Mdm2-GFP were transfected alone (control) or in combination with different arrestins. Where indicated, the cells were treated with 50 ng/ml of leptomycin B (LMB) for 14 h to inactivate exportin1. The effect of LMB after 2, 4, and 14 h was essentially the same (not shown). Cells were fixed in 4% paraformaldehyde for 15 min at 4°C. The slides were air dried and mounted with the mounting medium (Vector Laboratories) containing 4′,6-diamidino-2-phenylindole to visualize the nuclei. The GFP-JNK3 and GFP-Mdm2 were visualized using an epifluorescence microscope equipped with a CCD camera. Experiments were performed three to five times. In each experiment the distribution of JNK3, Mdm2, and/or arrestin in at least 20 cells was scored (nucleus>cytoplasm; nucleus = cytoplasm; nucleus <cytoplasm). Where appropriate, the scores were first analyzed by two-way analysis of variance with arrestin and expression level as main factors. Where the effects of both main factors and the interaction between factors were significant, post-hoc comparison of means with Bonferroni correction for multiple comparisons was used.
Quantitative Western Blot
In each experiment an aliquot of cells after each transfection was grown and dissolved in lysis solution (Ambion, Austin, TX). Total protein was measured by Bio-Rad assay. Protein was methanol precipitated, resolved by 10% SDS-PAGE, and transferred to polyvinylidene difluoride membrane (Millipore, Bedford, MA). Arrestin was visualized with F4C1 mouse monoclonal antibodies (20). Aliquots of the corresponding purified proteins were run alongside samples to construct the calibration curves for quantification.
RESULTS
Subcellular Localization of Endogenous Arrestins in Native Tissues
Subcellular distribution of endogenous arrestins in native tissues has not been studied. Therefore, we analyzed the localization of the two arrestins, visual (19) and arrestin2 (4), for which subtype-specific antibodies suitable for immunohistochemistry are available. Non-visual arrestins 2 and 3 are expressed in all brain regions (3). Arrestin2 predominates in mature neurons, where its concentration exceeds that of arrestin3 ~10–20-fold (4). Although arrestin2 is present in both the nuclei and cytoplasm in all types of neurons (Fig. 1), its relative distribution between these two compartments varies. For example, in cortical pyramidal neurons, particularly in layers III and V, the concentration of arrestin2 in the nucleus exceeds that in the cytoplasm (Fig. 1, A and B). Conversely, in striatal medium spiny neurons arrestin2 is predominantly cytoplasmic (Fig. 1, C and D). In other types of neuronal cells (the globus pallidus, nucleus of the diagonal band, and a number of other structures), arrestin2 is evenly distributed between the nucleus and the cytoplasm (not shown). The arrestin2 protein is small enough to passively move through the nuclear pore. Therefore, its preferential localization in the nucleus or the cytoplasm suggests that there are cell type-specific mechanisms that keep arrestin2 in one of these compartments.
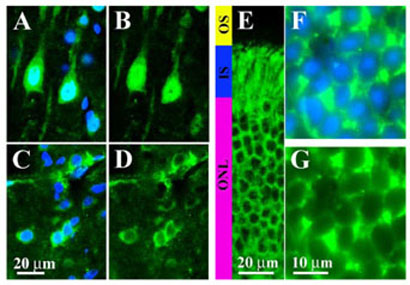
FIGURE 1. Differential subcellular distribution of endogenous arrestins in different types of neurons.
Sections of the brain (A–D) or retina (E–G) were labeled immunohistochemically with arrestin subtype-specific antibodies and counterstained with 4′,6-diamidino-2-phenylindole to visualize nuclei as described under “Materials and Methods.” A, C, and F, overlays of immunostaining for arrestins (green) and 4′,6-diamidino-2-phenylindole staining of nuclei (blue).B,D, E and G, the corresponding images of arrestin staining. A and B, high power photomicrographs of the arrestin2 subcellular distribution in the layer V pyramidal cells of the rat primary somatosensory cortex. Note the high concentration of arrestin2 in the nuclei. C and D, photomicrographs of rat striatal neurons showing mostly cytosolic localization of arrestin2. E–G, subcellular localization of visual arrestin in dark-adapted rod photoreceptors. Low power photomicrograph of the dark-adapted (overnight) retina is shown in panel E. In the dark, visual arrestin is localized to the inner segments (IS) and cell bodies of photoreceptors (outer nuclear layer, ONL), whereas outer segments (OS) of photoreceptors are devoid of arrestin. F and G, high power microphotographs of photoreceptor cells in the outer nuclear layer of the retina demonstrating that visual arrestin is completely excluded from the nuclei.
The localization of visual arrestin in rod photoreceptors is light dependent (19). In the light, it concentrates in the outer segments, whereas in the dark it is mostly sequestered in the inner segments and cell bodies (Fig. 1E). Visual arrestin is completely excluded from the nuclei even though it is clearly present in the perinuclear cytoplasm (Fig. 1, F and G). Because the size of visual arrestin is not large enough to prevent its passive entrance via the nuclear pore (21), it must be actively exported from the nucleus.
Subcellular Localization of Visual and Arrestins 2 and 3 in HEK293 Cells
Next we tested the localization of the three arrestins expressed in HEK293 cells. Wild-type (WT) arrestin3 is localized virtually exclusively in the cytoplasm, whereas visual arrestin and arrestin2 demonstrate predominantly cytoplasmic localization with a small proportion in the nucleus (Fig. 2). Thus, arrestin2 distribution in HEK293 cells is similar to its distribution in striatal neurons (Fig. 1, C and D). The localization of arrestins 2 and 3 that have a small FLAG tag (which does not considerably change their size) matches the localization of the native proteins (Fig. 2). Thus, the distribution of arrestins between the nucleus and the cytoplasm of HEK cells recapitulates their distribution in neurons that naturally express these proteins. The addition of a large (~20 kDa) GFP tag slightly increases the nuclear content of visual arrestin and arrestin3 but does not appreciably change the distribution of arrestin2 (Fig. 2).
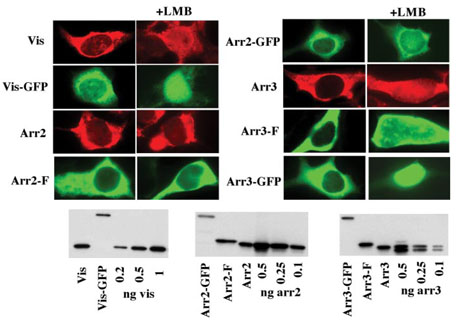
FIGURE 2. Subcellular distribution of wild-type and differentially tagged arrestins in HEK293 cells.
The indicated untagged, FLAG-tagged (F), and GFP-tagged arrestins were visualized with F4C1 pan-arrestin antibody (20), M2 anti-FLAG antibody, and native GFP fluorescence, respectively. Note the exclusive (visual and arrestin3) or preferential (arrestin2) cytoplasmic localization of untagged and FLAG-tagged arrestins. The addition of an ~20-kDa GFP tag, as well as incubation with an inhibitor of NES-dependent nuclear export, leptomycin B (LMB) (50 ng/ml), increase the proportion of visual and arrestin2 in the nucleus. Wild-type and FLAG-tagged arrestins 2, 3, and visual were expressed at comparable levels of 74 + 24, 53 + 11, and 196 + 37 pmol/mg of protein, respectively, as measured by quantitative Western blot (100 ng of total protein/lane) with the indicated amount of the corresponding purified proteins as standards. The expression levels of non-visual arrestins are higher than the levels observed in mature neurons (3, 4), whereas for visual arrestin it is lower than its expression in rod photoreceptors (44, 45). Visual arrestin-GFP was expressed at 88 + 21 pmol/mg of protein, whereas GFP fusions of both non-visual arrestins were expressed at 11 + 1 pmol/mg. Here and in the rest of the figures the images of representative cells (of 30–50 cells inspected and scored) are shown. Vis, visual (rod) arrestin; Arr2, arrestin2; Arr3, arrestin3.
A protein could be excluded from the nucleus for two reasons: it either does not enter because it is too large to diffuse via the nuclear pore and does not have a nuclear localization signal to gain entry via a controlled import mechanism, or it may enter and be actively exported if it is equipped with a nuclear export signal (NES). The size of the native and FLAG-tagged arrestins is not large enough to preclude their passive entry, whereas GFP-tagged arrestins are larger than 65 kDa, so that they can only enter and exit using the nuclear import/export machinery (21). To test the mechanisms responsible for the observed subcellular distribution of arrestin proteins, we used LMB, an inhibitor of a nuclear export receptor, exportin1 (22, 23). Preincubation of arrestin-expressing cells with 50 ng/ml of LMB notably increases the proportion of visual arrestin and arrestin3 in the nucleus, whereas no appreciable change in arrestin2 distribution (untagged or FLAG tagged) was observed (Fig. 2). The difference in the LMB-induced redistribution becomes much greater when GFP-tagged versions are used; visual arrestin and arrestin3 dramatically relocalize to the nucleus, and the nuclear content of arrestin2-GFP clearly increases (Fig. 2). These data suggest that wild-type arrestin2 is predominantly exported by an LMB-insensitive mechanism, whereas arrestin2-GFP and the other two arrestin subtypes rely on an LMB-sensitive pathway to a greater extent. However, the distribution of all three arrestins suggests that the export of even visual arrestin and arrestin3 out of the nucleus is totally dependent on LMB-sensitive processes only when these proteins are “burdened” with a large 20-kDa GFP tag.
Proteins that move out of the nucleus via the exportin1-dependent pathway carry a leucine-rich NES (23), although a broad spectrum of hydrophobic residues can form a functional NES motif (24). Only arrestin3 was shown to have a native NES motif localized in its C terminus, which can be destroyed by a single L394Q mutation (NES–) (10, 11), whereas the Q394L mutation in arrestin2 (NES+) is sufficient to create a functional NES (11). However, the effects of both mutations were only characterized using arrestin-GFP fusions (10, 11), the distribution of which differs from that of wild-type and FLAG-tagged arrestins (Fig. 2). Therefore, we tested the effects of the NES mutations in the context of WT arrestins 2 and 3 (Fig. 3). We found that engineering the NES in the C terminus of arrestin2 does not change its subcellular distribution, whereas inactivation of the native NES in arrestin3 results in arrestin2-like distribution. Both mutants are preferentially localized in the cytoplasm. The effects of LMB on their localization are as small as its effects on the parental WT proteins. Interestingly, in the presence of LMB the nuclear content of NES-less arrestin3 does not change (similar to the naturally NES-less wild-type arrestin2), whereas the nuclear content of NES+ arrestin2 increases to the same extent as that of arrestin3 that has a native NES (Figs. 2, 3). Thus, it appears that both non-visual arrestins can be exported out of the nucleus via LMB-sensitive and -insensitive pathways, suggesting that the presence or absence of an exportin1 binding NES is not a good predictor of their subcellular localization.
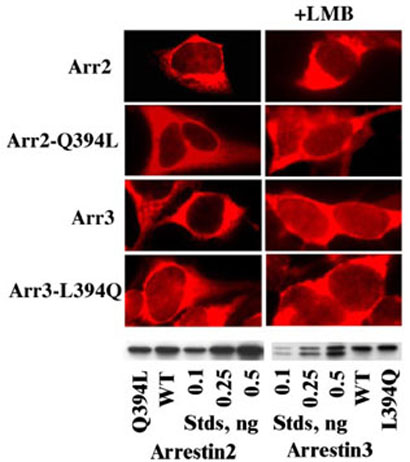
FIGURE 3. The role of the C-terminal nuclear export signal in the subcellular distribution of non-visual arrestins.
The indicated untagged arrestins were visualized in HEK293 cells with F4C1 pan-arrestin antibody (20). The presence (in arrestin3 and the arrestin2(Q394L) mutant) or absence (in arrestin2 and the arrestin3(L394Q) mutant) of the C-terminal NES does not dramatically change the subcellular distribution of untagged arrestins. Arrestin expression levels were the same as in Fig. 2.
Arrestin-dependent Relocalization of JNK3
GFP-JNK3 expressed alone in HEK293 cells was found both in the nucleus (excluding the nucleoli) and in the cytoplasm (Fig. 4). Co-expression of wild-type arrestin3 dramatically relocalizes GFPJNK3 to the cytoplasm (Fig. 4). LMB prevents arrestin3-dependent nuclear exclusion of JNK3 (Fig. 4). Surprisingly, we found that the presumably NES-less arrestin3(L394Q) mutant also induces partial relocalization of GFPJNK3. This process is also LMB sensitive. These data suggest that either arrestin3 has additional functional NES-like motif(s) or that LMB inhibits some exportin1-independent pathways. Co-expression of wildtype arrestin2 did not affect the localization of GFP-JNK3, whereas NES+ arrestin2 excluded GFPJNK3 from the nucleus as effectively as arrestin3 (Fig. 4). This effect was completely abolished by LMB, suggesting that the complex of the NES+ arrestin2 mutant with JNK3 exits the nucleus via a different pathway than free NES+ arrestin2, which was not affected by LMB (compare Figs. 3 and 4).
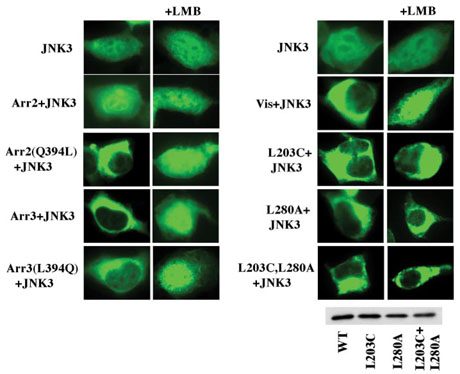
FIGURE 4. Arrestin-dependent nuclear exclusion of JNK3.
GFP-JNK3 was expressed alone or co-expressed with the indicated wild-type and mutant arrestins and visualized by GFP fluorescence. Wild-type visual arrestin, arrestin3, and NES+ arrestin2 relocalize JNK3 from the nucleus to the cytoplasm via an LMB-sensitive mechanism, whereas wild-type arrestin2 does not. Visual arrestin mutants L203C, L280A, and L203C+L280A with one or both putative NES disabled retain their ability to redistribute JNK3 but become LMB insensitive. Disabling the C-terminal NES in arrestin3 by the L394Q mutation only partially inhibits its ability to redistribute JNK3. Arrestin expression levels were the same as in Fig. 2.
Interestingly, co-expression of visual arrestin, which was not suspected of interacting with JNK3, effectively excluded GFP-JNK3 from the nucleus via an LMB-sensitive mechanism (Fig. 4). Visual arrestin does not have a conventional leucine-rich NES in its C terminus, although it has two internal NES-like sequences 203LRLAVSL209 and 278LTLVPLL284 that are exposed in the crystal structure (25). In an attempt to determine whether either of these sequences plays a role in the export of visual arrestin, we constructed two single mutants, L203C and L280A, and a combination mutant L203C/L280A and compared their ability to move JNK3 out of the nucleus to that of WT protein (Fig. 4). We found that neither mutation prevented visual arrestin-dependent nuclear exclusion of JNK3; the mutants with one or both putative NES disabled translocate GFP-JNK3 as effectively as WT. Unexpectedly, we found that in contrast to WT visual arrestin, the ability of the mutants to translocate JNK3 was insensitive to LMB (Fig. 4). Thus, WT visual arrestin (at least in complex with JNK3) is apparently exported via a mechanism involving these NES-like sequences and their elimination redirects it to an alternative export pathway. Collectively, our data demonstrate that all three arrestins interact with JNK3 and that wild-type visual arrestin and arrestin3 dramatically change the subcellular localization of this important signaling molecule.
Does JNK3 Binding Depend on Arrestin Conformation?
Arrestin3 was shown to act as a receptor-regulated scaffold for JNK3 activation (7). These data imply that receptor-bound arrestin in its “active” state interacts with JNK3 (9). To test whether JNK3 binding depends on arrestin conformation, we took advantage of the availability of functionally (16, 17) and conformationally “loose” (26) constitutively active arrestin mutants, as well as mutants with large deletions in the interdomain hinge that are frozen in the basal state and cannot bind the receptor (18, 27). The constitutively active forms of arrestins we chose have a triple alanine substitution in the C-tail (3A) that destabilizes its interaction with β-strand I and α-helix I (one of the conserved “clasps” holding arrestin in the basal conformation) (25, 28, 29). This interaction is destabilized by the receptor in the process of arrestin binding (30). The “constitutively inactive” forms of arrestins we used have a seven-residue deletion in the inter-domain hinge (D7) that leaves it only five residues long, i.e. just enough to cover the distance between the domains in the basal state (25). This deletion “freezes” arrestin in its basal conformation and blocks its ability to bind the receptor (18), precluding the conformational change involved in the process (31).
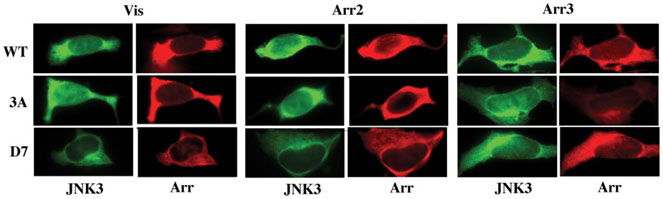
To detect even subtle differences in JNK3 interaction and score only cells that express the respective arrestins, we used FLAG-tagged versions of these mutants in all experiments and visualized arrestin with anti-FLAG primary and “red” secondary antibodies (Fig. 5). We found that both “pre-activated” 3A and constitutively inactive D7 mutants effectively relocalize JNK3. In every cell that had detectable levels of arrestin expression JNK3 was localized predominantly or exclusively in the cytoplasm (Fig. 5), whereas in a few cells that expressed GFPJNK3 but no arrestin (which can be considered internal controls) JNK3 was predominantly localized in the nucleus. If anything, the effectiveness of D7 mutants was somewhat higher than that of WT and 3A arrestins: even though the amount of JNK3 in the cytoplasm exceeded that in the nucleus in all cases, the nuclei of cells expressing D7 arrestins consistently appeared to be more “empty” (Fig. 5). Thus, JNK3 binds arrestin in its basal state, suggesting that a certain proportion of free arrestin in the cytoplasm is likely preloaded with JNK3.
FIGURE 5. Arrestin interaction with JNK3 does not depend on arrestin conformation.
Three forms of each arrestin were co-expressed with GFP-JNK3: wild type (WT), C-terminal triple alanine mutants with a loose active-like conformation (3A), and the mutants frozen in the basal conformation by the deletion of seven residues in the inter-domain hinge region (D7). GFP-JNK3 was visualized by intrinsic fluorescence (green); the indicated FLAG-tagged arrestins were visualized with anti-FLAG antibody (red). Typical cells from two independent experiments (with >20 cells inspected in each) are shown. In every cell that had detectable arrestin expression all three forms of visual arrestin, arrestin2(NES+), and arrestin3 effectively relocalized JNK3 to the cytoplasm. Arrestin expression levels (in pmol/mg of protein) were: visual WT, 240 + 30; 3A, 288 + 28; D7, 55 + 15; arrestin2 WT, 65 + 26; 3A, 162 + 69; D7, 66 + 12; arrestin3 WT, 47 + 9; 3A, 2.7 + 1.8; D7, 12 + 6.
Mdm2 Preferentially Binds Arrestin in the Basal Inactive State
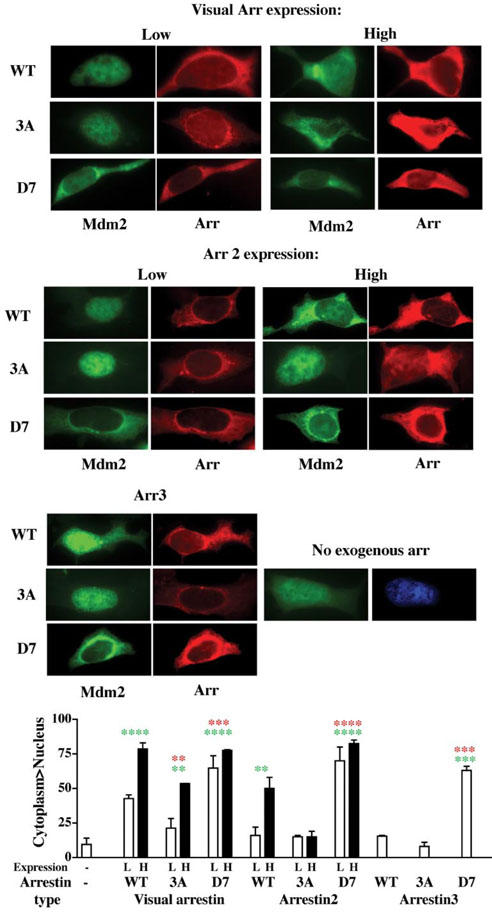
Next we used the same experimental paradigm to study the conformational dependence of arrestin interactions with the E3 ubiquitin ligase Mdm2 (Fig. 6). GFP-Mdm2 expressed alone is found almost exclusively in the nucleus. Co-expression of visual arrestin effectively excluded GFP-Mdm2 from the nucleus, indicating that visual arrestin interacts with Mdm2. As could be expected, this effect is more pronounced at higher arrestin expression. Interestingly, the 3A mutant is much less efficient, so that Mdm2 relocalization is only observed in cells expressing 3A arrestin at high level (Fig. 6). Unexpectedly, we found that the D7 mutant interacts with Mdm2 more robustly than WT, so that even at relatively low expression levels it effectively excludes Mdm2 from the nucleus. This result is remarkable because the overall expression of the visual arrestin-D7 mutant was 3–6 times lower than that of the WT and 3A forms. The relative ability of the three forms of NES+ arrestin2 to relocalize Mdm2 follows the same pattern: D7 »WT »3A (Fig. 6) (in this case all three proteins expressed at about the same level). The D7 mutant effectively moved Mdm2 to the cytoplasm even at relatively low expression, and the effect of WT arrestin2 was significant only at higher expression level, whereas 3A failed to appreciably redistribute Mdm2 (Fig. 6). Arrestin3 also tends to move Mdm2 to the cytoplasm, but redistribution of Mdm2 in the population of scored arrestin-expressing cells reached statistical significance only in the case of the D7 mutant (due to more even expression of arrestin3 we did not have high and low expressing cells to score separately). At the same expression level WT arrestin3 is less effective in redistributing Mdm2 than arrestin2 (Fig. 6), suggesting that either its affinity for Mdm2 is lower or that a higher proportion of arrestin2 present at equilibrium in the nucleus increases its chances of encountering and binding Mdm2. The expression dependence of the effects of visual arrestin and arrestin2 indicates that all forms bind Mdm2 but the affinity of the arrestin- Mdm2 interaction depends on arrestin conformation. Thus, all three arrestins interact with Mdm2, and in all cases the Mdm2 binding of mutants frozen in the basal conformation is more robust. These data suggest that free arrestin in the cytoplasm may be preloaded with Mdm2 before binding to its cognate receptor and that receptor binding is likely to significantly weaken the arrestin-Mdm2 interaction, releasing Mdm2 from the complex.
FIGURE 6. Mdm2 preferentially interacts with arrestin frozen in the basal conformation.
Three forms of visual arrestin, arrestin2(NES+), and arrestin3 were co-expressed with GFP-Mdm2: wild type (WT), C-terminal triple alanine mutant with a loose active-like conformation (3A), and the mutant frozen in the basal conformation by the deletion of seven residues in the inter-domain hinge region (D7). GFP-Mdm2 was visualized by intrinsic fluorescence (green); the indicated FLAG-tagged arrestins were visualized with anti-FLAG antibody (red). The D7 mutant effectively moves Mdm2 to the cytoplasm at relatively low (L) and high (H) expression levels; WT arrestin is less effective, whereas the effect of arrestin-3A mutants can only be observed in cells expressing them at very high levels. Typical cells are shown. Bar graph, for each protein >20 cells expressing arrestin at each (high and low) level, as judged by the intensity of FLAG signal, were scored in each experiment. For each arrestin subtype, the results of three experiments were analyzed by one-way analysis of variance with protein (none, WT, 3A, D7) as a main factor. The effect of protein was significant (p < 0.0001). The significance of the arrestin-induced changes in Mdm2 localization, as revealed by post-hoc analysis (Bonferroni) is indicated: **, p < 0.01; ***, p < 0.001; ****, p < 0.0001 (green, as compared with Mdm2 alone; red, as compared with the corresponding WT arrestin). Arrestin expression levels were the same as in Fig. 5.
DISCUSSION
Arrestins are ubiquitous regulators of GPCR signaling that bind the phosphorylated activated form of their cognate receptors with high affinity, blocking further G protein activation (32). In the process of receptor binding arrestin undergoes a global conformational rearrangement so that the shapes of free and receptor-bound arrestin are different (31, 33). This conformational change is believed to make the interaction sites for non-receptor binding partners more accessible, ensuring preferential binding of various signaling molecules to the receptor-arrestin complex (2, 9). Shutting down rhodopsin signaling is widely considered the only biological function of visual arrestin, whereas receptor-bound non-visual arrestins 2 and 3 also interact with clathrin (34) and AP2 (35), targeting the complex for internalization. In addition, the receptor-arrestin complex mobilizes and activates c-Src (5), scaffolds kinase cascades leading to the activation of ERK1/2 (6) and JNK3 (7), and recruits the ubiquitin ligase Mdm2 to the receptor, promoting receptor ubiquitination (8). Receptor activation-dependent JNK3 phosphorylation was described as a function of only one arrestin, arrestin3 (7). The RRSLHL motif in rat arrestin3 was tentatively identified as a JNK3-binding site (36), although it is not conserved in other mammals (25).
The information regarding the biological functions of free arrestins, other than receptor binding, is only beginning to emerge. Recently non-visual arrestins were shown to enter the nucleus (37), although no identifiable nuclear localization signal motifs were found in their sequence. Arrestin3 carries a functional NES in its C terminus that enables its shuttling between the nucleus and the cytoplasm, redistributing Mdm2 (Fig. 6) (11) and JNK3 (Figs. 4, 5) (10) in the process. Because GPCRs are not transported to the nucleus, these data suggest that free arrestin3 in its basal conformation interacts with these two partners. NES+ arrestin2 acquires the ability to remove Mdm2 (Fig. 6) (11) and JNK3 (Figs. 4, 5) from the nucleus, suggesting that it also binds both Mdm2 and JNK3 in its free (basal) state. In addition, arrestin2 was recently shown to regulate transcription of certain genes, suggesting that it has nucleus-specific functions (37).
Virtually all of the data on the subcellular localization of arrestins come from the studies of overexpressed proteins, mostly arrestin-GFP fusions (10, 11). Our analysis of the localization of native visual arrestin and arrestin2 in neurons that naturally express them shows that visual arrestin is completely excluded from the nucleus, whereas arrestin2 demonstrates variable distribution in different types of neurons (Fig. 1). Interestingly, in HEK293 cells only wild-type arrestins and arrestins that carry a small FLAG tag recapitulate this behavior, whereas the substantially larger arrestin-GFP fusions behave quite differently. The effects of LMB on the subcellular localization of visual arrestin and arrestin3 become more dramatic, and in sharp contrast to the parental protein the distribution of the arrestin2-GFP fusion becomes LMB sensitive (Fig. 2). The size of the native arrestins (44–46 kDa) suggests that arrestins are just small enough to diffuse through the nuclear pore, so that any increase in their size makes their movement to and from the nucleus subject to more strict control of the import/export machinery. In terms of LMB sensitivity arrestin-GFP fusions behave like the even larger complexes of arrestins with JNK3 and Mdm2 (Figs. 2–6).
Very modest effects of disabling mutations in the C-terminal NES of arrestin3 and both putative NES sequences in visual arrestin on the localization of these proteins suggest that either arrestins have multiple NES elements, some of which are not recognized by available software, or that they can be exported via alternative LMB-insensitive pathways. Our data indicate that all three arrestins can use both LMB-sensitive and -insensitive pathways to exit the nucleus, whereas in complex with interaction partners, arrestins 2 and 3 mostly exit via an LMB-sensitive pathway. The case of visual arrestin is most curious: the ability of wild-type protein to exclude JNK3 from the nucleus is LMB sensitive, suggesting the involvement of NES motifs in the process. However, visual arrestin with disabling mutations in one or both NES-like sequences redistributes JNK3 just as effectively as the wild-type protein but becomes insensitive to LMB. LMB covalently modifies Cys529 in exportin1 (38). Two mechanisms of LMB inhibition of exportin1 function have been proposed. The first is direct interference with the binding of the NES to exportin1 (38). Alternatively, LMB may inhibit the formation of a productive ternary complex comprising the NES-carrying cargo, exportin1, and Ran- GTP, without direct blockade of the NES-exportin1 interaction (39). The first model predicts two possible outcomes: 1) the inactivation of all NES motifs in visual arrestin should have the same effect as exportin1 inhibition, i.e. preclude arrestin-dependent JNK3 redistribution regardless of the presence of LMB; 2) if visual arrestin carries additional NES motifs that were not destroyed by the mutations, it would be expected to behave like wild type, i.e. redistribute JNK3 in an LMB-sensitive fashion. Neither prediction matches our data (Fig. 4). Thus, our results can only be rationalized in the context of the second model: conceivably, wild-type visual arrestin with two NES sequences binds exportin1 (which is crippled by LMB) and gets stuck in the nucleus along with its interaction partner, whereas NES-less mutants are not trapped in the complex with inactivated exportin1, being free to use an alternative NES-independent nuclear export pathway that is not affected by LMB (Fig. 4).
We used the ability of arrestins to relocalize their binding partners from the nucleus to the cytoplasm as a tool to study their interactions with JNK3 and Mdm2. In agreement with previous observations (11) we found that both free non-visual arrestins bind Mdm2 (Fig. 6). Unexpectedly, we found that visual arrestin interacts with Mdm2 as robustly as the non-visual subtypes (Fig. 6). Similarly, we found that all three arrestins bind JNK3 tightly enough to bring it out of the nucleus (Figs. 4, 5), suggesting that this interaction is not arrestin subtype specific, at least as far as free arrestins are concerned. Thus, two of two proteins believed to bind preferentially to the arrestin-receptor complex were found to interact with free arrestins, including visual. These data raise two biologically important questions that need to be addressed experimentally. First, do other proteins that interact with the receptor-arrestin complex also bind free arrestins, and if so, what is the functional significance of these interactions? Second, does visual arrestin interact with other known non-receptor partners of arrestins 2 and 3 and what role do these interactions play in photoreceptor cells? Comparison of the sizes of arrestin-receptor complex and numerous partners that arrestins recruit (2, 33) suggests that no more than 3–5 signaling proteins can simultaneously interact with the complex, so that non-receptor binding partners likely compete for the “parking space” on receptor-bound arrestin. In this context, the binding of signaling proteins to free arrestin may predetermine which particular signaling pathway will be activated upon arrestin recruitment to the receptor.
The availability of constitutively active arrestin mutants (26) and mutants frozen in their basal state (18) allowed us to explore the conformational dependence of arrestin interactions with JNK3 and Mdm2. It is important to note that the 3A mutation used here releases the arrestin C-tail by destabilizing its interactions with the elements of the N-domain, in essence performing some of the “activating” functions of receptor-attached phosphates (30). Thus, the 3A mutation likely allows free arrestin to assume a conformation similar to that of arrestin bound to the inactive phosphoreceptor, which is different from the ultimate active conformation of arrestin bound to the activated phosphoreceptor (40). Although activating mutations do not force arrestin into the “true” active state identical to that of receptor-bound arrestin (28), they significantly expand the conformational space the molecule can “sample” in the direction of the active-like state (26). Interestingly, JNK3 does not demonstrate a dramatic preference for one state of arrestin over another (Fig. 5), whereas Mdm2 clearly prefers the basal arrestin conformation (Fig. 6). Because arrestin transition into its active state involves the movement of the two arrestin domains relative to each other (18, 31), an attractively simple mechanistic interpretation of this difference between the two binding partners is that the JNK3-binding site is localized on one of the arrestin domains whereas the Mdm2 interaction site involves both, so that the relative orientation of the two domains in the basal state ensures a perfect fit, which is destroyed by their movement in the process of arrestin activation. Preferential binding of Mdm2 to arrestins in their basal conformation suggests that arrestins likely come to the receptor preloaded with it. Mdm2 mobilized by arrestin was shown to ubiquitinate the receptor (8). In view of an apparent decrease of arrestin affinity for Mdm2 when it assumes a more active-like conformation (Fig. 6), it is tempting to speculate that Mdm2 brought by arrestin to the receptor has just enough time to attach one ubiquitin moiety before being released due to arrestin transition into active receptor-bound state, so that this mechanism limits the extent of arrestin-assisted receptor ubiquitination.
Subcellular localization of signaling molecules is vital for their biological function. The distribution between the nucleus and the cytoplasm is particularly important for proteins that directly or indirectly regulate transcription. This includes JNK3, which phosphorylates and activates the transcription factor c-Jun, a component of the pro-apoptotic transcription complex AP-1 in photoreceptors (41), and Mdm2, which ubiquitinates p53 and targets it for degradation, thereby suppressing p53-mediated apoptosis (42). Thus, the ability of visual arrestin expressed at high micromolar concentrations in rod photoreceptors (43) and arrestins2 and 3 expressed at relatively high concentration in certain brain regions (4) to dramatically change the distribution of JNK3 and Mdm2 between the nucleus and the cytoplasm may play a role in neuronal survival. Our data suggest that the variation of arrestin2 distribution between the nucleus and cytoplasm in different neuronal subtypes likely results in differential localization of signaling molecules with which it interacts.
Acknowledgments
We thank Drs. Jeffrey L. Benovic, Louis M. Luttrell, and G. Pei for arrestin-GFP, GFP-JNK3, and GFP-Mdm2 constructs and Dr. Susan M. Hanson for critical reading of the manuscript.
This work was supported by National Institutes of Health Grants EY11500 (to V. V. G.) and NS45117 (to E. V. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GPCR, G protein-coupled receptor; ERK, extracellular signal-regulated kinase; E3, ubiquitin-protein isopeptide ligase; NES, nuclear export signal; GFP, green fluorescent protein; HEK, human embryonic kidney; LMB, leptomycin B; WT, wild type; JNK, c-Jun N-terminal kinase.
REFERENCES
- 1.Carman CV, Benovic JL. Curr. Opin. Neurobiol. 1998;8:335–344. doi: 10.1016/s0959-4388(98)80058-5. [DOI] [PubMed] [Google Scholar]
- 2.Lefkowitz RJ, Shenoy SK. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 3.Gurevich EV, Benovic JL, Gurevich VV. Neuroscience. 2002;109:421–436. doi: 10.1016/s0306-4522(01)00511-5. [DOI] [PubMed] [Google Scholar]
- 4.Gurevich EV, Benovic JL, Gurevich VV. J. Neurochem. 2004;91:1404–1416. doi: 10.1111/j.1471-4159.2004.02830.x. [DOI] [PubMed] [Google Scholar]
- 5.Luttrell LM, Ferguson SS, Daaka Y, Miller WE, Maudsley S, Della Rocca GJ, Lin F, Kawakatsu H, Owada K, Luttrell DK, Caron MG, Lefkowitz RJ. Science. 1999;283:655–661. doi: 10.1126/science.283.5402.655. [DOI] [PubMed] [Google Scholar]
- 6.Luttrell LM, Roudabush FL, Choy EW, Miller WE, Field ME, Pierce KL, Lefkowitz RJ. Proc. Natl. Acad. Sci. U. S. A. 2001;98:2449–2454. doi: 10.1073/pnas.041604898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald PH, Chow CW, Miller WE, Laporte SA, Field ME, Lin FT, Davis RJ, Lefkowitz RJ. Science. 2000;290:1515–1518. doi: 10.1126/science.290.5496.1574. [DOI] [PubMed] [Google Scholar]
- 8.Shenoy SK, McDonald PH, Kohout TA, Lefkowitz RJ. Science. 2001;294:1307–1313. doi: 10.1126/science.1063866. [DOI] [PubMed] [Google Scholar]
- 9.Gurevich VV, Gurevich EV. Structure. 2003;11:1037–1042. doi: 10.1016/s0969-2126(03)00184-9. [DOI] [PubMed] [Google Scholar]
- 10.Scott MG, Le Rouzic E, Perianin A, Pierotti V, Enslen H, Benichou S, Marullo S, Benmerah A. J. Biol. Chem. 2002;277:37693–37701. doi: 10.1074/jbc.M207552200. [DOI] [PubMed] [Google Scholar]
- 11.Wang P, Wu Y, Ge X, Ma L, Pei G. J. Biol. Chem. 2003;278:11648–11653. doi: 10.1074/jbc.M208109200. [DOI] [PubMed] [Google Scholar]
- 12.Shinohara T, Dietzschold B, Craft CM, Wistow G, Early JJ, Donoso LA, Horwitz J, Tao R. Proc. Natl. Acad. Sci. U. S. A. 1987;84:6975–6979. doi: 10.1073/pnas.84.20.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lohse MJ, Benovic JL, Codina J, Caron MG, Lefkowitz RJ. Science. 1990;248:1547–1550. doi: 10.1126/science.2163110. [DOI] [PubMed] [Google Scholar]
- 14.Sterne-Marr R, Gurevich VV, Goldsmith P, Bodine RC, Sanders C, Donoso LA, Benovic JL. J. Biol. Chem. 1993;268:15640–15648. [PubMed] [Google Scholar]
- 15.Pan L, Gurevich EV, Gurevich VV. J. Biol. Chem. 2003;278:11623–11632. doi: 10.1074/jbc.M209532200. [DOI] [PubMed] [Google Scholar]
- 16.Gurevich VV. J. Biol. Chem. 1998;273:15501–15506. doi: 10.1074/jbc.273.25.15501. [DOI] [PubMed] [Google Scholar]
- 17.Celver J, Vishnivetskiy SA, Chavkin C, Gurevich VV. J. Biol. Chem. 2002;277:9043–9048. doi: 10.1074/jbc.M107400200. [DOI] [PubMed] [Google Scholar]
- 18.Vishnivetskiy SA, Hirsch JA, Velez M-G, Gurevich YV, Gurevich VV. J. Biol. Chem. 2002;277:43961–43968. doi: 10.1074/jbc.M206951200. [DOI] [PubMed] [Google Scholar]
- 19.Nair KS, Hanson SM, Mendez A, Gurevich EV, Kennedy MJ, Shestopalov VI, Vishnivetskiy SA, Chen J, Hurley JB, Gurevich VV, Slepak VZ. Neuron. 2005;46 doi: 10.1016/j.neuron.2005.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donoso LA, Gregerson DS, Smith L, Robertson S, Knospe V, Vrabec T, Kalsow CM. Curr. Eye Res. 1990;9:343–355. doi: 10.3109/02713689008999622. [DOI] [PubMed] [Google Scholar]
- 21.Weis K. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 22.Ossareh-Nazari B, Bachelerie F, Dargemont C. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 23.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 24.Wen W, Meinkoth JL, Tsien RY, Taylor SS. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 25.Hirsch JA, Schubert C, Gurevich VV, Sigler PB. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- 26.Carter JM, Gurevich VV, Prossnitz ER, Engen JR. J. Mol. Biol. 2005;351:865–878. doi: 10.1016/j.jmb.2005.06.048. [DOI] [PubMed] [Google Scholar]
- 27.Hanson SM, Francis DJ, Vishnivetskiy SA, Klug CS, Gurevich VV. J. Biol. Chem. 2006;281:9765–9772. doi: 10.1074/jbc.M510738200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Structure. 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 29.Sutton RB, Vishnivetskiy SA, Robert J, Hanson SM, Raman D, Knox BE, Kono M, Navarro J, Gurevich VV. J. Mol. Biol. 2005;354:1069–1080. doi: 10.1016/j.jmb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 30.Vishnivetskiy SA, Schubert C, Climaco GC, Gurevich YV, Velez M-G, Gurevich VV. J. Biol. Chem. 2000;275:41049–41057. doi: 10.1074/jbc.M007159200. [DOI] [PubMed] [Google Scholar]
- 31.Gurevich VV, Gurevich EV. Trends Pharmacol. Sci. 2004;25:59–112. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 32.Krupnick JG, Gurevich VV, Benovic JL. J. Biol. Chem. 1997;272:18125–18131. doi: 10.1074/jbc.272.29.18125. [DOI] [PubMed] [Google Scholar]
- 33.Gurevich VV, Gurevich EV. Pharmacol. Ther. 2006;110:465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodman OB, Jr., Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, Keen JH, Benovic JL. Nature. 1996;383:447–450. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- 35.Laporte SA, Oakley RH, Zhang J, Holt JA, Ferguson SS, Caron MG, Barak LS. Proc. Natl. Acad. Sci. U. S. A. 1999;96:3712–3717. doi: 10.1073/pnas.96.7.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller WE, McDonald PH, Cai SF, Field ME, Davis RJ, Lefkowitz RJ. J. Biol. Chem. 2001;276:27770–27777. doi: 10.1074/jbc.M102264200. [DOI] [PubMed] [Google Scholar]
- 37.Kang J, Shi Y, Xiang B, Qu B, Su W, Zhu M, Zhang M, Bao G, Wang F, Zhang X, Yang R, Fan F, Chen X, Pei G, Ma L. Cell. 2005;123:833–847. doi: 10.1016/j.cell.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 38.Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Askjaer P, Jensen TH, Nilsson J, Englmeier L, Kjems J. J. Biol. Chem. 1998;273:33414–33422. doi: 10.1074/jbc.273.50.33414. [DOI] [PubMed] [Google Scholar]
- 40.Hanson SM, Francis DJ, Vishnivetskiy SA, Kolobova EA, Hubbell WL, Klug CS, Gurevich VV. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4900–4905. doi: 10.1073/pnas.0600733103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hao W, Wenzel A, Obin MS, Chen CK, Brill E, Krasnoperova NV, Eversole-Cire P, Kleyner Y, Taylor A, Simon MI, Grimm C, Reme CE, Lem J. Nat. Genet. 2002;32:254–260. doi: 10.1038/ng984. [DOI] [PubMed] [Google Scholar]
- 42.Evans SC, Viswanathan M, Grier JD, Narayana M, El-Naggar AK, Lozano G. Oncogene. 2001;20:4041–4049. doi: 10.1038/sj.onc.1204533. [DOI] [PubMed] [Google Scholar]
- 43.Strissel KJ, Sokolov M, Trieu LH, Arshavsky VY. J. Neurosci. 2006;26:1146–1153. doi: 10.1523/JNEUROSCI.4289-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamm HE, Bownds MD. Biochemistry. 1986;25:4512–4523. doi: 10.1021/bi00364a010. [DOI] [PubMed] [Google Scholar]
- 45.Kawamura S. In: Neurobiology and Clinical Aspects of the Outer Retina. Djamgoz MBA, Archer SN, Vallerga S, editors. Chapman and Hall, London: 1995. pp. 105–105. [Google Scholar]