Abstract
Recently we found that visual arrestin binds microtubules and that this interaction plays an important role in arrestin localization in photoreceptor cells. Here we use site-directed mutagenesis and spin labeling to explore the molecular mechanism of this novel regulatory interaction. The microtubule binding site maps to the concave sides of the two arrestin domains, overlapping with the rhodopsin binding site, which makes arrestin interactions with rhodopsin and microtubules mutually exclusive. Arrestin interaction with microtubules is enhanced by several “activating mutations” and involves multiple positive charges and hydrophobic elements. The comparable affinity of visual arrestin for microtubules and unpolymerized tubulin (KD > 40 μM and > 65 μM, respectively) suggests that the arrestin binding site is largely localized on the individual αβ-dimer. The changes in the spin-spin interaction of a double-labeled arrestin indicate that the conformation of microtubule-bound arrestin differs from that of free arrestin in solution. In sharp contrast to rhodopsin, where tight binding requires an extended interdomain hinge, arrestin binding to microtubules is enhanced by deletions in this region, suggesting that in the process of microtubule binding the domains may move in the opposite direction. Thus, microtubule and rhodopsin binding induce different conformational changes in arrestin, suggesting that arrestin assumes three distinct conformations in the cell, likely with different functional properties.
Arrestins are soluble cytoplasmic proteins that play a critical role in the regulation of signaling by the majority of G protein-coupled receptors. Vertebrates have four different arrestin subtypes, two of which regulate rhodopsin and cone opsins in rod and cone photoreceptors, respectively, whereas two non-visual arrestins are ubiquitously expressed and regulate hundreds of different G protein-coupled receptors. All arrestins preferentially bind to the activated phosphorylated forms of their cognate receptors, shutting off G protein-mediated signaling (reviewed in Ref. 1). Non-visual arrestins also interact with numerous non-receptor binding partners, orchestrating intracellular trafficking of the arrestin-receptor complex and redirecting receptor-initiated signaling to alternative G protein-independent pathways (reviewed in Refs. 2 and 3). Recently, we identified microtubules (MTs)4 as an interaction partner of visual (rod) arrestin (4). The difference in MT binding affinity between the two splice variants of visual arrestin expressed in bovine rods (5) determines their differential subcellular localization (4). Moreover, microtubules serve as a “default” arrestin binding partner in darkadapted rod photoreceptors (6). Dynamic interactions with rhodopsin in the light and MTs in the dark underlie the massive light-dependent translocation of rod arrestin between the inner and outer segments of photoreceptor cells (6). Here we explore the molecular mechanism of visual arrestin binding to MTs and identify arrestin elements involved in this interaction. Several lines of evidence also suggest that the conformation of microtubule-bound arrestin is different from both free and receptor-bound forms.
EXPERIMENTAL PROCEDURES
Materials
[14C]Leucine and [3H]leucine were from PerkinElmer Life Sciences. All restriction enzymes were from New England Biolabs. All other chemicals were from sources previously described (7). Rabbit reticulocyte lysate was from Ambion; SP6 RNA polymerase was prepared as described (8).
In Vitro Transcription, Translation, and Evaluation of the Stability of Mutants
pGEM2-based plasmids with the arrestin coding sequence with the “idealized” 5′-UTR (8) under the control of the SP6 promoter were transcribed and translated in vitro as previously described (7). Arrestins were labeled by the incorporation of [3H]leucine and [14C]leucine with the specific activity of the mix between 1.5–3 Ci/mmol, resulting in arrestin-specific activity of 66-85 Ci/mmol (150–230 dpm/fmol). The translation of every mutant used in this study produced a single labeled protein band with the expected mobility on SDS-PAGE. The relative stability of all mutants (assessed as described in Ref. 9) exceeded 80%.
Direct Binding of Arrestin to Microtubules
Microtubules with Purified Arrestins
Purified wild type (WT) or Tr-(1–378) arrestin (4 μg) were incubated for 30 min at 30 °C in 50mM Tris-HCl, pH 7.4, 100mM NaCl, 2mM EDTA, and 1mM EGTA with 40 μg of purified tubulin (Cytoskeleton, Inc.) prepolymerized with taxol according to the manufacturer's instructions. Microtubules along with bound arrestin were pelleted by centrifugation for 10 min at 30 °C at 100,000 rpm in a TLA 120.1 rotor in a Beckman TL100 ultracentrifuge. Parallel samples with the same amount of each arrestin without microtubules served as controls. The pellet was dissolved in Laemmli's sample buffer (Sigma), and the amount of arrestin was quantified by Western blot.
Microtubules with in Vitro Translated Radiolabeled Arrestins
200 fmol of the indicated in vitro translated arrestins were incubated in 50 mM Tris-HCl, pH 7.4, 0.5 mM MgCl2, 1.5 mM dithiothreitol, 1 mM EGTA, and 50 mM potassium acetate for 20 min at 30 °C with 20 μg of prepolymerized tubulin. Microtubules along with bound arrestin were pelleted. MT-arrestin pellets were not washed because of the low affinity (i.e. high off-rate) of the interaction. The pellet was dissolved in 0.1 ml of 1% SDS, 50 mM NaOH, and bound arrestin was quantified by liquid scintillation counting. Nonspecific “binding” (arrestin pelleted without microtubules) was subtracted.
Arrestin Expression and Purification
Arrestin was expressed in Escherichia coli and purified as described (7). We constructed two cysteineless base mutants for spin labeling, CL (C63A, C128S, C143A) and CLN (C63V, C128S, C143V). The substituting residues for the native cysteines were selected to maximally preserve intramolecular interactions. Both base mutants were fully functional in terms of rhodopsin and microtubule binding and expressed in E. coli as well as WT arrestin. However, CL and CLN mutants differentially tolerated the introduction of cysteines into various parts of the arrestin molecule. To optimize expression, mutations I16C, Y58C, S60C, I72C, V74C, M75C, D82C, F85C, Q87C, Q89C, V94C, F152C, F197C, T233C, L240C, V244C, E266C, K267C, S272C, S273C, L339C, S344C, and A348C were introduced onto the CL background, whereas L77C, S78C, F79C, V139C, T157C, L173C, V376C, A381C, I16C/A381C, and V74C- (1–378) were introduced onto the CLN background.
Electron Paramagnetic Resonance (EPR) Sample Preparation
Cysteine mutants were dialyzed against 50mMMOPS,100mMNaCl, pH 7.0, and labeled with a 10-fold molar excess of the sulfhydryl-specific 2,2,5,5-tetramethylpyrroline-3-yl-methanethiosulfonate spin label (MTSL, Toronto Research Chemicals) overnight at 4 °C. Excess label was removed by extensive dialysis, and arrestin was concentrated using Microcon YM-30 (Amicon). Final concentrations were determined by the BCA protein assay (Pierce) using bovine serum albumin as a standard. Polymerized MTs were incubated overnight at 25 °C with a 10-fold molar excess of a sulfhydryl blocking reagent, methylmethanethiosulfonate (MMTS, Toronto Research Chemicals). MMTS-blocked MTs were washed, concentrated by centrifugation, and resuspended in 80 mM PIPES, pH 6.9, 1mM MgCl2, 1 mM EGTA, and 20 μM taxol. Electron paramagnetic resonance (EPR) spectroscopy samples contained 10 μg of spin-labeled arrestin and 150 μg ofMTs in a final volume of 10 μl. For the spin-labeled V74C-(1–378) titration final concentrations of MTs and unpolymerized tubulin are expressed as molar concentration of the tubulin dimer,Mr = 110 kDa. Interspin distances for the double labeled mutant were analyzed using simulation software developed by Dr. C. Altenbach (10).
Electron Paramagnetic Resonance Spectroscopy
Continuous wave EPR spectroscopy was carried out at X-band on a Bruker ELEXSYS E500 fitted with a super high Q cavity. Samples were contained in a glass capillary, and spectra were recorded at room temperature over 100 G at a microwave power of 10 milliwatt with a scan time of 42 s, and typically the signal was averaged 25–36 times. Tubulin dimer binding experiments were carried out at 4 °C to prevent spontaneous polymerization using a variable Digital Temperature Controller (Bruker).
RESULTS
Direct Binding of Wild Type and Truncated Arrestin to Microtubules
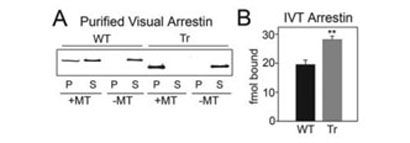
Microtubule binding plays a role in the differential localization of arrestin splice variants in bovine rods (4) and in arrestin translocation in mouse rod photoreceptors (6). To ascertain that this interaction does not require “helper” proteins, we tested the binding of purified full-length-(1–404) and truncated-(1–378) (Tr) arrestin to MTs polymerized in vitro from pure tubulin. As shown in Fig. 1A, robust binding of truncated arrestin as well as lower affinity binding of full-length arrestin was easily detectable in these experiments. These data demonstrate that arrestin binding to MTs is direct and that the behavior of the two forms of purified arrestin in this reconstituted system recapitulates their behavior in live photoreceptors.
FIGURE 1. Direct binding of WT and truncated arrestin to microtubules.
A, E. coli expressed and purified WT and truncated arrestin-(1–378)(Tr) (4 μg) were incubated in the presence or absence of 40 μg of taxol-polymerized tubulin (MT) in a volume of 50 μl. Arrestin bound to MTs (P) was separated from free arrestin (S) by centrifugation at 100,000 rpm for 10 min and quantified by Western blot with F4C1 anti-arrestin antibody (47). B, 200 fmol of in vitro translated radiolabeled WT and Tr arrestin were bound to MTs and separated as in A. Arrestin in the pellet fraction was quantified by liquid scintillation counting; nonspecific binding (in the absence of MTs) was subtracted. Means ± S.D. from three experiments are shown. **, p < 0.01, as compared with WT.
Identification of Microtubule-binding Elements in Arrestin
Next we set out to identify arrestin elements involved in microtubule binding. MT-binding domains tend to be rich in lysine and valine residues (11, 12), and several microtubule-associated proteins use positively charged elements to bind the acidic C termini of tubulin α- and β-subunits (13). Arrestin has numerous exposed positive charges (Fig. 2A) (14, 15) and binds polyanions (15–17). Therefore we performed charge-reversal and -neutralization mutagenesis of individual exposed positive charges in both arrestin domains. Because the total number of necessary mutations exceeds 40 (15), we first tested the feasibility of using a relatively high throughput direct binding assay with radiolabeled arrestins expressed in cell-free translation, similar to the assay we routinely use to measure arrestin binding to purified Gprotein-coupled receptors (18, 19). Microtubules can easily be pelleted by brief centrifugation, so we chose this method for the separation of bound and free arrestin. This assay faithfully reproduces the difference in microtubule binding between full-length and truncated arrestin (Fig. 1B) that we detected using purified proteins (Fig. 1A) and observed in vivo (4).
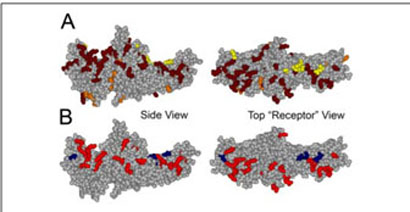
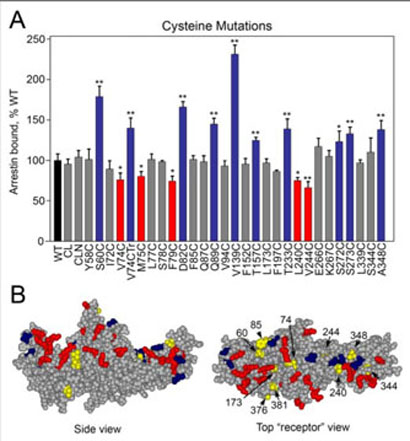
FIGURE 2. Positive charges important for MT binding map to the concave surfaces of both arrestin domains.
A, the positive charges (Arg and Lys) in arrestin are highlighted; the majority of these charges are in two “patches,” one on the concave surface of each domain. Most of the positive charges on the surface of the molecule were mutated for this study, shown in dark red, all others are in orange, extra positive charges added (see Fig. 3C) are in yellow. B, map of the charge reversal mutations in arrestin that significantly decrease (red) or enhance (blue) binding to microtubules (see Fig. 3). Left panel, side view; right panel, top view down the cavities of the two domains from the “receptor point of view.”
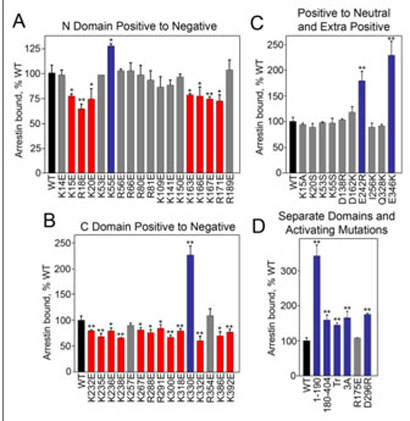
Using this assay we tested the effects of arrestin Arg/Lys to Glu mutations on binding to MTs (Fig. 3). In the N-domain, 10 positive charge reversals had no effect on arrestin binding; seven mutations reduced it by 20–35%, whereas K55E increased it by 25% (Fig. 3A). In the C-domain, only two positive charge reversals did not affect the binding, whereas 12 reduced and one (K330E) dramatically enhanced it (Fig. 3B). These data support the idea that positive charges in arrestin play a role in the MT interaction. Importantly, the dramatic position dependence of the effects of these mutations (ranging from 40% reduction to >2-fold increase in binding) strongly suggests that arrestin binding to microtubules is specific, i.e. a particular spatial configuration of charges is necessary for the interaction. Next we compared charge neutralization and reversal mutations at selected positions. In contrast to the reversal, charge neutralization at positions 15 (K15A) and 20 (K20S) does not reduce the binding, whereas charge neutralization at position 55 (K55S) does not enhance MT binding (Fig. 3C). The effects of additional positive charges are also strictly position-specific; D138R, D162K, I256K, and Q328K do not affect the binding, whereas E242R and E346K result in a dramatic increase in binding (Fig. 3C). Thus, the arrestinmicrotubule interaction appears to be dependent on the exposed charged side chains in arrestin and requires very specific positioning of these charges. Next we tested the salt sensitivity of arrestin binding and found that it is not dramatically changed between 50–1000 mM potassium acetate (data not shown). These data suggest that both hydrophobic interactions (enhanced by high salt) and ionic interactions (inhibited by high salt) participate in arrestin binding to MTs. This is reminiscent of MT interactions with various MT-associated proteins where both positive charges and hydrophobic residues often play a role (11–13).
FIGURE 3. Asubset of exposed charges in arrestin is important for microtubule binding.
The indicated in vitro translated radiolabeled arrestins (200 fmol) were incubated with 20 μg of MTs in a total volume of 100 μl for 20 min at 30 °C. The samples were centrifuged and MT-bound arrestin in the pellet was quantified as described in Fig. 1. The effects of positive charge reversal mutations in the arrestin N-domain (A) and C-domain (B) on MT binding are shown. C, charge neutralization mutations do not have the same effect as charge reversal at the same position. The addition of extra positive charges only affects binding in specific locations. D, the separately expressed N-domain-(1–190) and C-domain-(180–404) bind significantly better than full-length arrestin. Mutations that activate arrestin with regard to receptor binding also enhance MT binding, with the exception of R175E. Mutations that significantly decrease, increase, or do not change binding compared with WT are shown in red, blue, and gray, respectively. 3A (F375A, V376A, F377A), Tr (1–378). Means ± S.D. of three experiments are shown. *, p < 0.05; **, p < 0.01, as compared with WT.
The charged residues involved in MT binding are evenly distributed between the two arrestin domains (Fig. 2B). Both domains appear to be independent folding units and can be expressed separately (18). The N-domain even functions as a “miniarrestin” as far as receptor binding is concerned (16, 18–20). Therefore we tested microtubule binding of the N- and C-domains separately, and found that both domains bind microtubules substantially better than full-length arrestin (Fig. 3D). These data further support the involvement of both arrestin domains in MT binding. The enhanced binding of the separated domains suggests that each domain can “fit” its binding site on the MTs better when it is not impeded by the other, i.e. that arrestin may undergo a conformational rearrangement in the process of its binding to MTs.
Mutations That Destabilize Key Intramolecular Interactions in Arrestin Enhance Its Binding to Microtubules
The basal conformation of arrestin is stabilized by intramolecular interactions. The most important of these are the “polar core” (several interacting solvent-excluded charged residues that include Arg-175 and Asp-296) and the hydrophobic three-element interaction between the arrestin C-tail,β-strand I, and α-helix I (15, 21). The disruption of either of these interactions by mutagenesis facilitates its receptor bindinginduced conformational transition and allows arrestin to bind with high affinity not only to light-activated phosphorylated rhodopsin (P-Rh*), but also to the light-activated unphosphorylated and phosphorylated dark forms (22, 23). The deletion of the arrestin C-tail yields a similar phenotype (9, 18, 20). C-tail deletion also enhances arrestin interaction with microtubules (Figs. 1 and 3D) (4). We found that other “activating” mutations also significantly increase arrestin binding to MTs, D296R in the polar core and a triple alanine substitution in the C-tail (3A) (Fig. 3D). These results suggest that like the receptor, microtubules may destabilize the basal conformation of arrestin. One notable exception is the R175E mutation, which does not appreciably affect microtubule binding (Fig. 3D). Conceivably, glutamate in position 175 is repelled by the other negative charges in the polar core and pushed toward the cavity of the N-domain where its negative charge would interfere with the positive charges that participate inMTbinding (Figs. 2 and 3) counteracting any possible enhancement.
Deletions in the Interdomain Hinge Region Increase Arrestin Binding to Microtubules
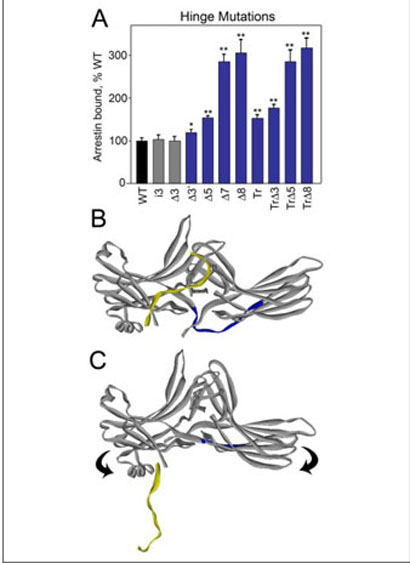
The N- and C-domains of arrestin are connected by a 12-residue-long loop termed the “hinge” region (Fig. 4B). Receptor binding is severely impeded by deletions in the hinge, suggesting that the movement of the two arrestin domains relative to each other is necessary for P-Rh* interaction (24). Because the mutations that “loosen up” the basal conformation of arrestin enhance MT binding, we tested whether interaction with MTs involves domain movement by manipulating the length of the hinge. It should be noted that the stability of the hinge (Fig. 4) and “activated” mutants (Fig. 3) was evaluated prior to binding (see “Experimental Procedures”) (9) and found to be equal or even greater than that of WT protein in all cases. We found that the addition of three extra residues to the hinge does not affect MT (Fig. 4A) or P-Rh* (24) binding. In contrast, deletions of increasing length, which progressively inhibit P-Rh* binding (24), actually enhance arrestin binding to MTs up to 3-fold (Fig. 4A). The same deletions in the context of truncated arrestin-(1–378) further enhance the binding. However, when eight residues are deleted, leaving the hinge just four residues long and likely forcing arrestin to bend “backwards” (Fig. 4C), the binding of the full-length and truncated forms is essentially the same. Conceivably, the extreme shortening of the hinge facilitates the detachment of the C-tail, so that its presence or absence no longer affects binding (Fig. 4C). The unexpected positive effect of hinge deletions on MT binding suggests that if the arrestin domains move in this process, they actually move in the direction opposite to that necessary for rhodopsin binding.
FIGURE 4. Deletions in the interdomain hinge enhance arrestin binding to MTs.
A, binding of in vitro translated arrestins with mutations in the hinge region were performed as described in Fig. 3. The mutations are designated as follows: deletions∆3,∆3′, ∆5, ∆7, and ∆8 indicate the deletion of 3 (positions 188–190), 3′ (180, 182, 183), 5 (180, 182, 188–190), 7 (180, 182, 183, 187–190), and 8 (180, 182–184, 187–190) residues, respectively; i3, insertion of three extra alanines after 180; and Tr, truncated arrestin 1–378. Mutations that significantly enhance or do not change binding are shown in blue and gray, respectively. Deletions further enhance binding in the context of Tr arrestin until the entire hinge region is removed (Tr∆8). Means ± S.D. of three experiments are shown. *, p <0.05; **, p <0.01, as compared with WT. B, crystal structure of visual arrestin in its basal conformation. The 12-residue-long hinge region between β-strands X and XI connecting the two domains is highlighted in blue; the C-tail is in yellow. C, because at least five residues in the hinge are necessary to cover the distance between the two domains, deletion of eight residues (leaving only four), would cause the molecule to bend backwards. This configuration may disrupt important interactions between the body of the molecule and the C-tail causing it to detach, making N-domain residues necessary for MT binding more accessible.
EPR Spectroscopy Corroborates the Microtubule-binding Interface
MT-binding residues on arrestin map to a well defined MT “footprint” extending through a large part of the concave sides of both domains (Fig. 2B). To corroborate these findings by an independent method, we used site-directed spin labeling (SDSL) EPR spectroscopy, which requires the introduction of a unique spin label that reports on localized regions of a protein. Thus, native cysteines must be eliminated so that the cysteine introduced in the position of interest is unique. This cysteine is then chemically modified with a sulfhydryl-specific EPR probe, which is approximately the size of a tryptophan residue (25). We constructed two cysteineless base mutants of visual arrestin (CL and CLN), and ascertained that both interact with MTs as well as WT arrestin (Fig. 5A). Next we introduced a series of unique cysteines on the background of these mutants and tested their effects on MT binding using radiolabeled translated arrestins. We found that 14 (of 28) cysteine substitutions did not affect the binding (Fig. 5A). Where the binding changed, replacement of hydrophilic residues (S60C, D82C, Q89C, T157C, T233C, S272C, S273C) enhanced binding, and replacement of hydrophobic residues (V74C, M75C, F79C, L240C, V244C) generally decreased binding, with a few exceptions (V74CTr, A348C, and V139C) (Fig. 5A). These results highlight the importance of specific non-ionic interactions in arrestin binding to MTs and further support our earlier observation that it is mediated by both ionic and hydrophobic interactions. The additional residues that participate in binding are also localized on the concave surfaces of both domains (Fig. 5B).
FIGURE 5. Residues important for MT binding map to the concave surfaces of both arrestin domains.
A, replacement of several specific hydrophilic and hydrophobic residues with cysteine significantly affects arrestin binding to MTs. Experiments were performed as described in Fig. 3. Cysteine-less arrestins used as base mutants bind MTs like WT: CL (C63A, C128S, C143A) and CLN (C63V, C128S, C143V). One cysteine mutant (V74C) was made on the background of cysteine-less Tr arrestin (V74CTr) and binds like Tr arrestin (see Figs. 1 and 3). Mutations that significantly enhance or reduce binding are shown in blue and red, respectively. Means±S.D. of three independent experiments are shown. *, p <0.05; **, p <0.01, as compared with WT. B, map of mutations in arrestin that significantly decrease (red) or increase (blue) binding to MTs (see Figs. 3 and 5). Positions where the mobility of the spin label changed upon microtubule binding (see Fig. 6) are labeled and shown in yellow. Top panel, side view; bottom panel, top view down the cavities of the two domains.
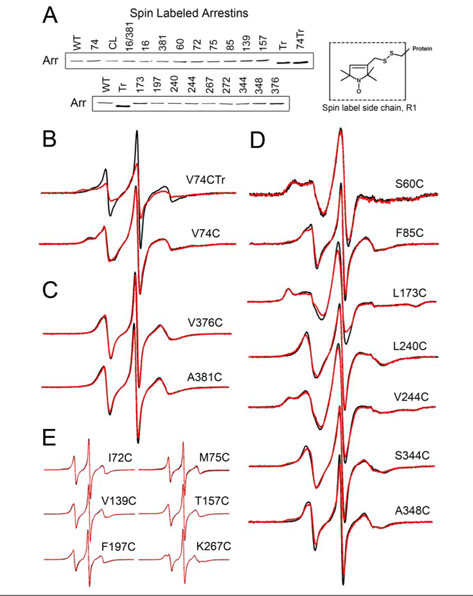
To provide adequate coverage of the molecule for EPR we expressed and purified 18 cysteine mutants, modified them with spin label (side chain R1, Fig. 6A, inset), and compared their MT binding to that ofWT full-length and truncated arrestin. We found that unlike many of their cysteine counterparts, spin label-modified arrestins bind MTs essentially like WT(Fig. 6A), likely because the size and hydrophobicity of the spin label makes it a good substitute for a hydrophobic side chain. Thus, all of these mutants can be used as EPR reporters for the arrestin-microtubule interaction.
FIGURE 6. The changes in spin label mobility inducedbyvisual arrestin interaction with microtubules.
A, purified mutants with unique cysteines modified with spin label at the indicated position were tested for their ability to bind MTs as described in Fig. 1. Arrestin in the pellet fraction was quantified by Western blot. Full-length spin-labeled mutants bind comparably to WT, whereas cysteineless Tr arrestin with spin label at position 74 (spin-labeled V74CTr) binds comparably to Tr arrestin. Inset, the R1 side chain generated by reacting the cysteine mutants with the methanethiosulfonate nitroxide reagent. B-E, for each spin-labeled arrestin, the overlay of the EPR spectra in the absence (black) or presence (red) of MTs are shown. EPR samples contained 10 μg of spin-labeled visual arrestin and 150 μg of MTs in a final volume of 10 μl.
The shape of the EPR spectrum of a spin label reflects its mobility. A highly mobile spin label on the surface of a protein has a characteristic narrow spectrum with sharp peaks, whereas the reduction in its mobility because of intramolecular constraints or occlusion by being “trapped” in a protein-protein interaction interface is reflected in the broadening of the spectrum and/or the appearance of an additional peak in the low field region (26, 27). We recorded the EPR spectra of purified full-length arrestins individually spin-labeled at 16 different positions and of truncated arrestin (Tr)-(1–378) labeled at position 74 in the absence and presence of 135 μM polymerized tubulin (Fig. 6, B–E). Arrestin association with MTs notably decreases the mobility of the spin label at position 74 in the truncated form of arrestin, whereas the spin label mobility slightly increases at this position in full-length arrestin (Fig. 6B). The mobility of the spin label side chain also slightly increases at sites 376 and 381 in the C-tail (Fig. 6C). The majority of the sites studied showed a small decrease in mobility, as seen at positions 60, 85, 173, 240, 244, 344, and 348 (Fig. 6D), whereas essentially no changes were observed at positions 72, 75, 139, 157, 197, and 267 (Fig. 6E). Consistent and reproducible mobility changes in these positions identify essentially the same MT-binding interface as the direct binding assay: the concave sides of both arrestin domains (Fig. 5B).
Arrestin Affinity for Microtubules and theαβ-Tubulin Dimer
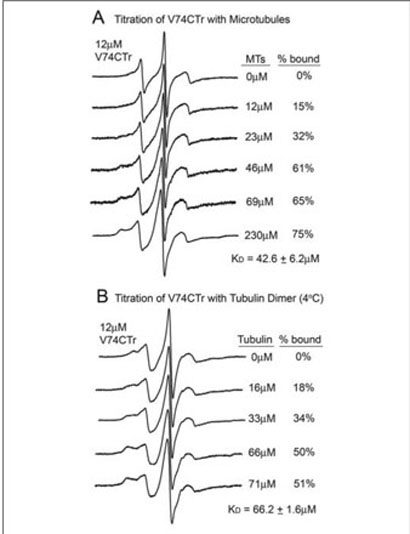
Most of the changes in the EPR spectra are not as dramatic as one would expect for a tight protein-protein interaction, suggesting that arrestin affinity for MTs is fairly low. This is consistent with the fact that in vivo MT-bound arrestin is readily released as soon as a more high affinity binding partner, such as P-Rh*, emerges (4, 6). To test this idea we compared MT-induced changes in the spectrum of the label at position 74 in the context of full-length (V74C) and truncated arrestin (V74CTr) (Fig. 6B). Spin label in free V74CTr is more mobile than in the full-length form, supporting the notion that arrestin truncation loosens up its basal conformation. In sharp contrast to V74C, MT binding of V74CTr results in a dramatic decrease in the mobility of the label (Fig. 6B). This change is large enough to calculate the affinity of the interaction by performing a “titration” of spin labeled V74CTr with MTs to determine the extent of the spectral change as a function of MT concentration (Fig. 7A). These data yielded a Kd for the V74CTr-MT interaction of 43 ± μM. Thus, even the affinity of truncated arrestin is fairly low, suggesting that the Kd of full-length arrestin is likely 50–100 μM. At equilibrium, Kd=Kd/Ka; because the diffusion-limited Ka of a protein the size of arrestin at normal viscosity is in the range of 104-105 M-1 s1, a midmicromolar Kd suggests the dissociation rate of the complex is very fast (t1/2 <1 s). This is consistent with microtubules serving as high capacity, very low affinity “sinks” for visual arrestin in dark-adapted photoreceptors, releasing bound arrestin fast enough to make its rapid translocation to the outer segment of the photoreceptor possible (6).
FIGURE 7. The affinity of truncated arrestin for microtubules and tubulin dimer.
The spectra of 12 μM truncated arrestin spin-labeled at V74CTr was measured in the presence of increasing concentrations of microtubules (A) or tubulin dimer (B). Final concentrations of tubulin are expressed as molar concentration of the tubulin dimer (Mr = 110 kDa). The percent bound value for each spectrum was quantified by spectral subtraction of the corresponding free arrestin spectrum containing no tubulin from each of the composite spectra in the titration series. With the unbound arrestin spectrum subtracted out, each remaining spectrum included only the bound fraction. The percent arrestin bound values were calculated by comparison of the integrated intensities of each resulting bound spectrum with the corresponding composite spectrum. EPR spectroscopy of samples containing tubulin dimer was carried out at 4 °C to prevent spontaneous polymerization. The mobility of the spin label side chain is decreased due to the decrease in temperature, thus the spectrum of V74CTr free in solution in (B) differs from that recorded at room temperature (A).
Tubulin in the cell exists in dynamic equilibrium between αβ-dimer and MTs(13, 28). If the arrestin binding site on microtubules is confined within a single αβ-dimer, arrestin should also bind unpolymerized tubulin. Alternatively, if the arrestin interaction site spans more than one αβ-dimer, it should not interact appreciably with unpolymerized tubulin. To address this issue, a similar titration experiment of spinlabeled V74CTr with unpolymerized tubulin was performed. We found that truncated arrestin bound the αβ-tubulin dimer with an affinity only slightly lower than for MTs (Kd of 66 ± 2 μM) (Fig. 7B), suggesting that most (if not all) of the arrestin interaction surface on MTs is confined to the individual αβ-dimer.
Because arrestin in rod photoreceptors is present at high micromolar concentrations (29), this finding prompted us to test whether arrestin affects tubulin polymerization.Wefound that purified arrestin (up to 90 μM) has no effect on tubulin polymerization (in sharp contrast to 3 μM purified MAP2, which served as a positive control) (data not shown). Thus, the arrestin binding site on the αβ-dimer does not include or affect the surfaces involved in microtubule formation.
Microtubule Binding Causes Release of the Arrestin C-tail
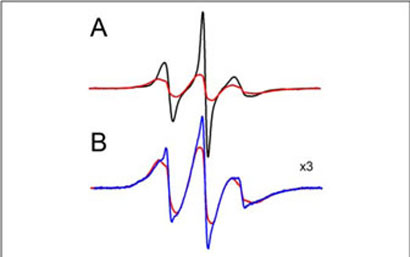
The N-domain part of the MT-binding surface is partially “shielded” by the C-tail (15). This explains the enhanced binding of the C-terminally truncated mutant in vitro (Figs. 1 and 3C, the p44 splice variant in vivo (4), and of the N-domain when expressed separately (Fig. 3D). These data suggest that microtubules may “push” the arrestin C-tail out of the way in the process of binding. This idea is also supported by an increase in mobility of the spin label in the two C-tail positions tested, 376 and 381, in sharp contrast to the decrease in mobility in all the other positions where changes were observed (Fig. 6). To test this hypothesis, we compared the spectra of an arrestin mutant spin-labeled at positions 16 (β-strand I) and 381 (C-tail) with and without MTs. A strong spinspin interaction is observed without MTs (Fig. 8A). Deconvolution of the spectrum shows that the distance between these two spin labels is 11–19 Å in solution, in good agreement with the crystallographic distance of 12 Å (15). In the presence of MTs (Fig. 8B) 16% of the labels move farther than 20 Å apart, reporting the release of the C-tail. Thus, the MTs apparently push the C-tail of bound arrestin out of its basal position, suggesting that the MT-bound conformation is distinct from the arrestin conformation in solution.
FIGURE 8. MT binding causes the release of the arrestin C-tail.
The EPR spectra are shown normalized to the same number of spins for (A) the spin labeled double mutant 16/381 (red) overlaid with the equal sum of the two singly labeled mutants 16 and 381 (black) free in solution. B, the spectra of the spin labeled double mutant 16/381 (25 μM) in the absence (red) and presence (blue) of 230 μM MTs are overlaid and magnified to illustrate the partial loss of the spin-spin interaction upon binding to MTs.
DISCUSSION
Arrestin binding to microtubules has recently been implicated in the light-dependent redistribution of visual arrestin in rod photoreceptor cells (6). Here we describe the structural basis of this interaction. Most MT binding sites on microtubule-associated proteins are rich in positive charges (11–13). Examination of the positive charges on the arrestin surface reveals that most are on the concave sides of the two domains (Fig. 2A); therefore we focused our search for MT-binding residues on this surface. Using two independent methods, site-directed mutagenesis of charges and hydrophobic residues and site-directed spin labeling, we found that several residues on the concave sides of the two domains are involved in MT binding (Figs. 2 and 5B). Arrestin has very few exposed positive charges on the “back” of the molecule (Fig. 2A), and although none of the few residues that we tested affected the MT binding (Fig. 3), we cannot exclude the participation of this surface in theMT interaction. However, like the effects of the charge mutations, the changes of spin label mobility revealed by EPR identify a very specific microtubule interaction surface on the concave surface of the arrestin molecule. For example, the mobility at the two C-tail positions (376, 381) in full-length arrestin increases upon MT binding, whereas the mobility at seven different positions (60, 85, 173, 240, 244, 344, and 348) on the body of the molecule decreases. In contrast, the mobility at six other positions (72, 75, 139, 157, 197, and 267) does not change (Fig. 6). Because we observed both increases and decreases in spin label mobility induced by MT binding, these changes cannot be explained simply by the reduction of the rotational mobility of arrestin in the complex, which appears too small to be detected, as evidenced by the absence of observed changes in label mobility at six positions (Fig. 6E). The direction and magnitude of the observed changes clearly do not depend on the mobility at a particular position in free arrestin.
Receptor-binding elements of arrestin identified by several laboratories using a wide variety of methods (Refs. 30–32 and references therein) invariably map to the same concave surface of the molecule. Our data suggest that the significant overlap between MT and rhodopsin sites is the structural basis for the mutual exclusivity of arrestin binding to these two partners (6). Thus, rhodopsin and MTs apparently compete for visual arrestin in rod photoreceptors.
The relatively low affinity of arrestin for MTs (Fig. 7) along with its nanomolar affinity for light-activated rhodopsin (33, 34) make MTs a perfect sink for arrestin in dark-adapted photoreceptors. The tubulin concentration in several cell types has been shown to be ~20–40 μM (35, 36). The tubulin content in brain tissue is much higher and reaches up to 25% of the soluble protein in the cell (36), bringing its concentration to~150 μM. As a result MTs present an enormous surface area for binding (up to 1000 μm2/cell), which is almost as large as the entire surface of the plasma membrane (28). The affinity (Kd) of microtubule-associated proteins as well as other proteins such as kinesin and GRK2 for MTs ranges from ~0.1 to 20 μM (35, 37–39). These proteins can exert their effects on MTs even though they are expressed at substantially lower concentrations than tubulin itself. The most conservative estimate of visual arrestin concentration in rod photoreceptors is~150 μM (29). Rod cells are neurons with a very high microtubule content (40). Calculations based on the law of mass action show that at 150 μM arrestin in the presence of 150 μM microtubules the proportion of MT-bound arrestin in the dark would be 45–50% at a Kd of 75–100 μM (our lowest affinity estimate for full-length arrestin). Recent measurements suggest that the arrestin concentration in rods may bemuch higher (41), which favors arrestin interaction with MTs even more. Moreover, MTs in rods are highly enriched in the inner segment (40) where arrestin is also concentrated in dark-adapted photoreceptors. Thus visual arrestin binding to MTs has particular physiological relevance in rods and demonstrates how microtubules serve as a default interaction partner in dark-adapted photoreceptors as we have shown recently (6). Our experiments in transgenic mice (6) demonstrate that even a mild increase in arrestin affinity for MTs (to 42 μM as measured for truncated arrestin, Fig. 7) significantly slows its light-dependent movement to the outer segment. Therefore the relatively low affinity of arrestin for microtubules is important for its timely redistribution in photoreceptors, which is one of the mechanisms of light adaptation (42).
It should be noted that visual arrestin self-associates at physiological concentrations (43–45) with equilibrium constants in the midmicromolar range. It is possible that the visual arrestin oligomer also binds MTs and that the binding surface may ormay not be the same as for the monomer. All of our binding of cell-free translated arrestin was done at nanomolar concentrations where only monomer is present. EPR studies were performed at low micromolar concentrations of arrestin where the monomer also predominates. Thus, our binding data and determination of the MT binding site reflect that of the arrestin monomer.
Arrestin binding to MTs and receptors has a common feature: truncated and other “constitutively active” arrestin mutants in which the intramolecular “clasps” holding the two domains in their basal orientation are destabilized (reviewed in Ref. 14) bind the receptor and microtubules with higher affinity (Fig. 3D). The main reason for this phenomenon in receptor binding is that in the process arrestin undergoes a substantial conformational rearrangement (34) likely involving the movement of the two domains in the direction of their concave sides (“forward”) (24). In the basal state of arrestin only five residues are necessary to cover the distance between the two domains, but the inter-domain hinge is 12 residues long, providing sufficient “slack” to allow domain movement. Deletions in the hinge impede the forward domain movement that is required for P-Rh* binding (24). However, the highest binding of full-length arrestin to MTs was observed with a mutant that has only four hinge residues, after deletion of eight (Fig. 4). This indicates that if the two domains move upon MT binding, they move in the opposite direction (bending backwards) (Fig. 4C), suggesting that the MT-bound conformation is different from the basal state. This idea is supported by the substantially higher binding of the N- and C-domains expressed separately (Fig. 3D), which apparently impede each others' binding in the basal conformation of full-length arrestin. The observed MTbinding-induced increase in C-tail mobility (positions 376 and 381, Fig. 6C), and clear changes in spin-spin interaction between the reporters in the C-tail and β-strand I (Fig. 8) provide direct evidence that the arrestin conformation actually changes in this process.
Until now arrestin was widely believed to exist in two conformations, free and receptor-bound. In these two structural states arrestin interacts with different signaling molecules (3, 46). Our data demonstrate that arrestin exists in a third distinct conformation. Apparently microtubules do not simply serve as a sink for visual arrestin in the dark but convert it into a structural state different from free and receptor-bound arrestin. It is tempting to speculate that the functional properties of the novel MT-bound conformation of arrestin described here are unique. MT-bound arrestin may recruit new binding partners and/or organize signaling complexes that can be shuttled along the microtubules to other parts of the cell.
Acknowledgments
We thank Dr. T. Shinohara and L. A. Donoso for bovine visual arrestin cDNA and F4C1 antibody, respectively.
Footnotes
This work was supported in part by National Institutes of Health Grants EY11500 and GM63097 (to V. V. G.) and AI58024 and GM70642 (to C. S. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: MT, microtubule; UTR, untranslated region; WT, wild type; MOPS, 4-morpholinepropanesulfonic acid; PIPES, 1,4-piperazinediethanesulfonic acid; EPR, electron paramagnetic resonance; P-Rh*, light-activated phosphorylated rhodopsin.
REFERENCES
- 1.Carman CV, Benovic JL. Curr. Opin. Neurobiol. 1998;8:335–344. doi: 10.1016/s0959-4388(98)80058-5. [DOI] [PubMed] [Google Scholar]
- 2.Marchese A, Chen C, Kim YM, Benovic JL. Trends Biochem. Sci. 2003;28:369–376. doi: 10.1016/S0968-0004(03)00134-8. [DOI] [PubMed] [Google Scholar]
- 3.Lefkowitz RJ, Shenoy SK. Science. 2005;308:512–517. doi: 10.1126/science.1109237. [DOI] [PubMed] [Google Scholar]
- 4.Nair KS, Hanson SM, Kennedy MJ, Hurley JB, Vsevolod GV, Slepak VZ. J. Biol. Chem. 2004;279:41240–41248. doi: 10.1074/jbc.M406768200. [DOI] [PubMed] [Google Scholar]
- 5.Smith WC, Milam AH, Dugger D, Arendt A, Hargrave PA, Palczewski K. J. Biol. Chem. 1994;269:15407–15410. [PubMed] [Google Scholar]
- 6.Nair KS, Hanson SM, Mendez A, Gurevich EV, Kennedy MJ, Shestopalov VI, Vishnivetskiy SA, Chen J, Hurley JB, Gurevich VV, Slepak VZ. Neuron. 2005;46:555–567. doi: 10.1016/j.neuron.2005.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gurevich VV, Benovic JL. Methods Enzymol. 2000;315:422–437. doi: 10.1016/s0076-6879(00)15859-8. [DOI] [PubMed] [Google Scholar]
- 8.Gurevich VV. Methods Enzymol. 1996;275:382–397. doi: 10.1016/s0076-6879(96)75023-1. [DOI] [PubMed] [Google Scholar]
- 9.Gurevich VV. J. Biol. Chem. 1998;273:15501–15506. doi: 10.1074/jbc.273.25.15501. [DOI] [PubMed] [Google Scholar]
- 10.Altenbach C, Oh KJ, Trabanino RJ, Hideg K, Hubbell WL. Biochemistry. 2001;40:15471–15482. doi: 10.1021/bi011544w. [DOI] [PubMed] [Google Scholar]
- 11.Goode BL, Denis PE, Panda D, Radeke MJ, Miller HP, Wilson L, Feinstein SC. Mol. Biol. Cell. 1997;8:353–365. doi: 10.1091/mbc.8.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noble M, Lewis SA, Cowan NJ. J. Cell Biol. 1989;109:3367–3376. doi: 10.1083/jcb.109.6.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amos LA. Curr. Opin. Struct. Biol. 2000;10:236–241. doi: 10.1016/s0959-440x(00)00070-1. [DOI] [PubMed] [Google Scholar]
- 14.Gurevich VV, Gurevich EV. Trends Pharmacol. Sci. 2004;25:105–111. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Hirsch JA, Schubert C, Gurevich VV, Sigler PB. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- 16.Gurevich VV, Chen C-Y, Kim CM, Benovic JL. J. Biol. Chem. 1994;269:8721–8727. [PubMed] [Google Scholar]
- 17.Palczewski K, Pulvermuller A, Buczylko J, Hofmann KP. J. Biol. Chem. 1991;266:18649–18654. [PubMed] [Google Scholar]
- 18.Gurevich VV, Benovic JL. J. Biol. Chem. 1992;267:21919–21923. [PubMed] [Google Scholar]
- 19.Gurevich VV, Dion SB, Onorato JJ, Ptasienski J, Kim CM, Sterne-Marr R, Hosey MM, Benovic JL. J. Biol. Chem. 1995;270:720–731. doi: 10.1074/jbc.270.2.720. [DOI] [PubMed] [Google Scholar]
- 20.Gurevich VV, Benovic JL. J. Biol. Chem. 1993;268:11628–11638. [PubMed] [Google Scholar]
- 21.Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Structure. 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 22.Vishnivetskiy SA, Paz CL, Schubert C, Hirsch JA, Sigler PB, Gurevich VV. J. Biol. Chem. 1999;274:11451–11454. doi: 10.1074/jbc.274.17.11451. [DOI] [PubMed] [Google Scholar]
- 23.Vishnivetskiy SA, Schubert C, Climaco GC, Gurevich YV, Velez M-G, Gurevich VV. J. Biol. Chem. 2000;275:41049–41057. doi: 10.1074/jbc.M007159200. [DOI] [PubMed] [Google Scholar]
- 24.Vishnivetskiy SA, Hirsch JA, Velez M-G, Gurevich YV, Gurevich VV. J. Biol. Chem. 2002;277:43961–43968. doi: 10.1074/jbc.M206951200. [DOI] [PubMed] [Google Scholar]
- 25.Hubbell WL, Gross A, Langen R, Lietzow MA. Curr. Opin. Struct. Biol. 1998;8:649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 26.Klug CS, Feix JB. In: Biological Magnetic Resonance, Volume 24: Biomedical EPR-Part B: Methodology an Instrumentation. Eaton SS, Eaton GR, Berliner LJ, editors. Kluwer Academic/Plenum Publishers; New York, NY: 2005. pp. 269–308. [Google Scholar]
- 27.Hubbell WL, Cafiso DS, Altenbach C. Nat. Struct. Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 28.Gundersen GG, Cook TA. Curr. Opin. Cell Biol. 2000;11:81–94. doi: 10.1016/s0955-0674(99)80010-6. [DOI] [PubMed] [Google Scholar]
- 29.Hamm HE, Bownds MD. Biochemistry. 1986;25:4512–4523. doi: 10.1021/bi00364a010. [DOI] [PubMed] [Google Scholar]
- 30.Hanson SM, Gurevich VV. J. Biol. Chem. 2006;281:3458–3462. doi: 10.1074/jbc.M512148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vishnivetskiy SA, Hosey MM, Benovic JL, Gurevich VV. J. Biol. Chem. 2004;279:1262–1268. doi: 10.1074/jbc.M308834200. [DOI] [PubMed] [Google Scholar]
- 32.Gurevich VV, Gurevich EV. Pharmacol. Ther. 2006 in press. [Google Scholar]
- 33.Osawa S, Raman D, Weiss ER. Methods Enzymol. 2000;315:411–422. doi: 10.1016/s0076-6879(00)15858-6. [DOI] [PubMed] [Google Scholar]
- 34.Schleicher A, Kuhn H, Hofmann KP. Biochemistry. 1989;28:1770–1775. doi: 10.1021/bi00430a052. [DOI] [PubMed] [Google Scholar]
- 35.Ackmann M, Wiech H, Mandelkow E. J. Biol. Chem. 2000;275:30335–30343. doi: 10.1074/jbc.M002590200. [DOI] [PubMed] [Google Scholar]
- 36.Hiller G, Weber K. Cell. 1978;14:795–804. doi: 10.1016/0092-8674(78)90335-5. [DOI] [PubMed] [Google Scholar]
- 37.Wallis KT, Azhar S, Rho MB, Lewis SA, Cowan NJ, Murphy DB. J. Biol. Chem. 1993;268:15158–15167. [PubMed] [Google Scholar]
- 38.Haga K, Ogawa H, Haga T, Murofushi H. Eur. J. Biochem. 1998;255:363–368. doi: 10.1046/j.1432-1327.1998.2550363.x. [DOI] [PubMed] [Google Scholar]
- 39.Seitz A, Kojima H, Oiwa K, Mandelkow EM, Song YH, Mandelkow E. EMBO J. 2002;21:4896–4905. doi: 10.1093/emboj/cdf503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eckmiller MS. Vis. Neurosci. 2000;17:711–722. doi: 10.1017/s0952523800175054. [DOI] [PubMed] [Google Scholar]
- 41.Strissel KJ, Sokolov M, Trieu LH, Arshavsky VY. J. Neurosci. 2006;26:1146–1153. doi: 10.1523/JNEUROSCI.4289-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arshavsky VY. Science's STKE 2003. 2003:PE43. doi: 10.1126/stke.2003.204.pe43. [DOI] [PubMed] [Google Scholar]
- 43.Schubert C, Hirsch JA, Gurevich VV, Engelman DM, Sigler PB, Fleming KG. J. Biol. Chem. 1999;274:21186–21190. doi: 10.1074/jbc.274.30.21186. [DOI] [PubMed] [Google Scholar]
- 44.Shilton BH, McDowell JH, Smith WC, Hargrave PA. Eur. J. Biochem. 2002;269:3801–3809. doi: 10.1046/j.1432-1033.2002.03071.x. [DOI] [PubMed] [Google Scholar]
- 45.Imamoto Y, Tamura C, Kamikubo H, Kataoka M. Biophys. J. 2003;85:1186–1195. doi: 10.1016/S0006-3495(03)74554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gurevich VV, Gurevich EV. Structure. 2003;11:1037–1042. doi: 10.1016/s0969-2126(03)00184-9. [DOI] [PubMed] [Google Scholar]
- 47.Donoso LA, Gregerson DS, Smith L, Robertson S, Knospe V, Vrabec T, Kalsow CM. Curr. Eye Res. 1990;9:343–355. doi: 10.3109/02713689008999622. [DOI] [PubMed] [Google Scholar]