Abstract
We identified a Drosophila homologue that belongs to a subfamily of the mammalian peroxiredoxin 5 genes. The Drosophila peroxiredoxin 5 (dPrx 5) gene corresponds to CG7217 (FlyBase nomenclature), and exhibits identical polarity to that of the nearby upstream CG7215 gene, for which the deduced amino-acid sequence reveals an ubiquitin-like domain at the N-terminus. The tandem pattern of the arrangement of these two genes is well-conserved among the Drosophilids. In addition to CG7215 and dPrx5 gene-specific transcripts that could be transcribed independently from two distinct promoters, transcripts spanning both coding regions have been identified, leading to the characterization of this gene cluster as a dicistronic operon. The different transcripts exhibit stage-specific accumulation patterns. While the upstream CG7215 gene can be expressed from both mono- and dicistronic mRNAs, the downstream dPrx5 gene is likely to be expressed predominantly from its monocistronic transcripts.
Keywords: Peroxiredoxin, operon, dicistronic, expression, Drosophila
INTRODUCTION
The peroxiredoxin (Prx) gene family consists of six members, all of which are potential members of the antioxidant network. They are also implicated in the regulation of signal transduction, presumably through local effects on redox state. The last discovered isoenzyme among these family members [1], peroxiredoxin 5 (Prx5), is a distinct isoform of the subfamily of 2-cys peroxiredoxins that differs from other peroxiredoxins by its structure and by the mode of resolution of its reaction intermediate. The antioxidative and anti-apoptotic functions of Prx5 as well as the ability of Prx5 to protect DNA from damage were demonstrated in cell culture experiments [2; 3; 4; 5; 6; 7; 8].
To gain more insight into biological function of Prx5 at the organismal level, we took advantage of the Drosophila model. Based upon amino acid identity and the presence of Prx5-specific conserved domains, we have identified a Drosophila homologue of the Prx5 gene (dPrx5), corresponding to CG7217 (FlyBase nomenclature). This dPrx5 gene is the down-stream member of a two gene cluster comprising an apparent dicistronic operon. The upstream member (CG7215) has no known homologue, although its deduced amino-acid sequence revealed an N-terminal ubiquitin-like domain.
While relatively uncommon, a number of operons have been found in eukaryotes, including Drosophila [9]. For the operon comprising CG7215 and dPrx5 three distinct transcript species were identified, a dicistronic species spanning both ORFs and two monocistronic ones corresponding to the two individual ORFs. It was found that transcription of the downstream dPrx5 gene could also be initiated in the small intergenic region separating the two ORFs, suggesting the presence of an internal functional promoter. Cell culture studies are consistent with the translation of dPrx derived predominantly from the mono-cistronic transcript. In general, the expression profiles of the CG7215 and dPrx transcripts derived from this cluster revealed distinct tissue- and stage-specific variations in accumulation patterns.
MATERIALS AND METHODS
Fly strains and procedures
The y w strain has been maintained in this laboratory for >14 years. A strain containing a P-element insertion in the CG7217EY021106 ORF (stock # 15852) was obtained from the Bloomington Stock Center.
Animals used in the experiments were synchronized by collecting eggs at 2-h intervals or white prepupae and subsequently allowing development to occur on media at 25°C for the desired times. Adult flies were collected within 1-2 days post-eclosion and reared at 25°C until the desired age.
5′ and 3′RACE
To identify 3′ termini of the CG7215 monocistron, reverse transcription was performed with 3′RACE Adapter Primer (Invitrogen) followed by PCR using 7215-5′RACE and Adapter primers (Fig. 1 B and Suppl. Material). 5′ end sequences for CG7215 and dPrx transcripts were determined using 5′-Full Race Core Set (Takara Bio). Reverse transcription was performed using 5′-phosphorylated primers p7215-R and pdPrx-R followed by ligation and two consecutive PCR reactions performed with A1/S1 and A2/S2 primer sets for CG7215 transcripts and A3/S3 and A4/S4 primer sets for dPrx transcripts (Fig. 1 B), as recommended in the manufacturer’s manual. The resulting PCR fragments were subsequently sequenced (Retrogen).
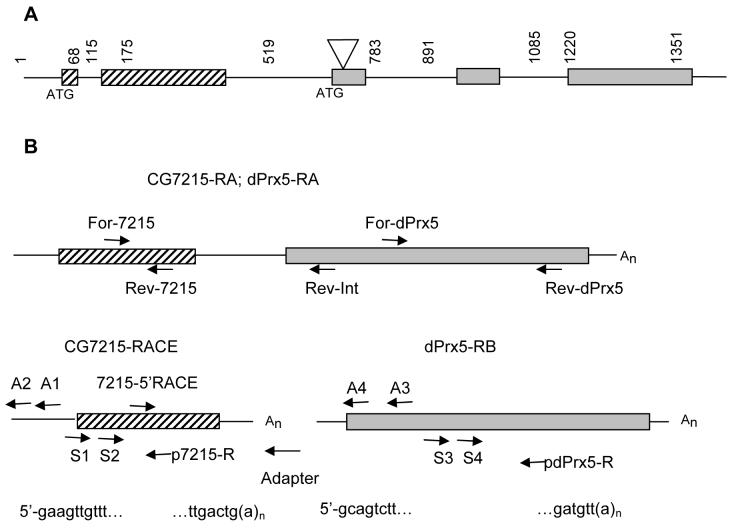
Figure 1. Schematic diagram of the structure of the CG7215 and dPrx5 genes.
A, Putative coding regions are shown as rectangles with different fills representing the CG7215 gene (stripped) and the dPrx5 gene (gray filled) and untranslated regions are shown as lines. Numbers indicated nucleotide positions. P-element insertion of the CG7217EY021106(dPrx5) mutant strain (position 853) is shown by a triangle. B, Diagrams of different mRNAs. Polycistronic transcript CG7215-RA; dPrx5-RA and a dPrx5-RB transcript are identified by sequencing of different ESTs (FlyBase) and 5′-RACE. The CG7215-RACE transcript was defined by 5′ and 3′-RACE. Partial sequences of the transcription start site are shown under a diagram for each transcript. Shown by arrows are primers used for the 5′ and 3′ RACE, RT-PCR and Northern blot analyses. Primer sequences are provided in the Supplemental Material.
Northern blot analysis and RT-PCR
Primers used for RT-PCR analysis (For-7215, Rev-7215, Rev-Int and For-dPrx5, Rev-dPrx5) and for generation of hybridization probes for CG7215 and dPrx genes (For-7215 and Rev-7215, For-dPrx5 and Rev-dPrx5) are depicted in Fig. 1 and Suppl. Material. PolyA+ RNA fractions were isolated by PolyA Track kit (Promega). Northern blot analysis using total RNA or polyA+ fractions was performed as in Radyuk et al. [10]. RT-PCR analysis was performed as described [11].
RNA interference
dsRNA synthesis and introduction into S2 cells were performed, as described in Radyuk, et al. [11]. Primers for dsRNA generation were designed to include T7 promoter sequence, see Suppl. Material.
Construction of the derivatives of pIB/V5-His vector
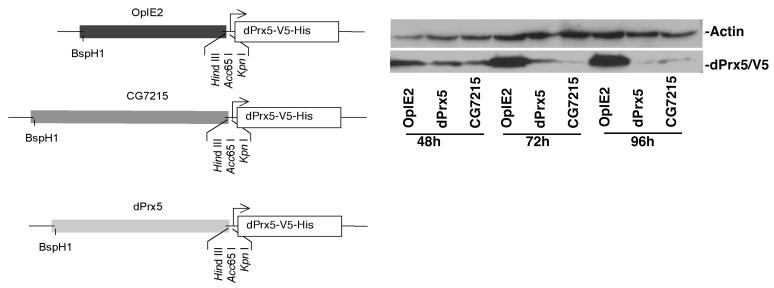
The whole OpIE2 promoter sequence of the pIB/V5-His construct that expresses V5-tagged dPrx5 protein was excised with BspH1 and HindIII endonucleases and substituted with sequences of the putative promoters that were predicted for the CG7215/dPrx operon (Fig. 4). S2 cells were then transfected and expression of the vector-encoded V5-tagged dPrx5 protein was followed by immunoblot analysis.
Figure 4. Identification of putative promoters for the CG7215/dPrx5 operon.
A, Structures of the constructs. Thin lines represent parts of the vector, open bars represent a coding region of the dPrx5 gene tagged with V5 epitope and Hisx6 sequence. Promoters are shown as solid lines. The arrow indicates polarity of transcription. The pIB-dPrx5 construct was made by cloning of the whole coding region of the dPrx5 gene into Kpn1 and Xba1 sites of the pIB/V5-His vector (Invitrogen). The first construct was made by substituting the OpIE2 promoter with a putative promoter comprising a 770 bp genome fragment upstream of the ATG start codon of the CG7215 gene. The second construct was made by substituting the OpIE2 promoter with a putative promoter comprising a 689 bp genome fragment upstream of the ATG start codon of the dPrx5 gene. B, S2 cells were transfected with vectors, where expression of dPrx5/V5 recombinant protein was driven by different promoters. Proteins were extracted at different time intervals post-transfection followed by Western blot analysis with anti-V5 antibodies and anti-actin antibodies to control for loading.
Immunoblotting
Protein extraction, SDS-PAGE and immunoblot analysis were performed essentially as previously described [11]. Drosophila dPrx5 protein was detected with antibodies raised against a recombinant protein using services of the Proteintech Group, Inc. (www.ptglabs.com). To control for a loading, anti-actin antibodies were used (MP Biomedicals). Recombinant dPrx5 protein tagged with V5 epitope was detected using anti-V5 HRP conjugated antibodies (Invitrogen).
RESULTS
Identification of a Drosophila homologue of the peroxiredoxin 5 gene
Based on amino acid sequence similarity search, we have identified a Drosophila melanogaster candidate peroxiredoxin gene (CG7217, synonym CG32390) that apparently belongs to the subfamily of peroxiredoxins designated as peroxiredoxin 5 [1; 5]. The deduced amino acid sequence of the CG7217 gene shows significant homology to the known mammalian and human peroxiredoxin 5 genes, including the presence of conserved motifs, typical for this subtype of peroxiredoxin (Suppl. Material). We further designated CG7217 gene as dPrx5. This gene was cloned and expressed in E. coli and the putative dPrx5 protein was shown to have thiol-dependent peroxidase activity in vitro (data not shown).
dPrx5 is the second gene in a dicistronic operon
The analysis of genomic organization using FlyBase data showed that dPrx5 gene is the second gene in a cluster, having the characteristics of a dicistronic operon. Both genes exhibit the same transcriptional polarity and dPrx5 is separated from the first gene, CG7215, by a 268 bp intercistronic region (Fig. 1 A).
Two types of transcripts for this operon are reported in the FlyBase database. One type, CG7215-RA or CG-7217-RA, contains ORFs of both genes while the second type, CG7217-RB, contains only the dPrx5 coding region. An additional search in the GeneBank data revealed a third type of transcript, derived from the adult testis cDNA library, which contained the coding region of CG7215 gene (GeneBank # AT18329).
For a more accurate characterization of mRNAs transcribed from this operon, we performed RACE analysis of the transcripts derived from the CG7215 and dPrx5 genes as described in Materials and Methods. The 5′ RACE analysis revealed two different mRNA 5′ ends: one extending 68 bp upstream of the CG7215 start codon, and the other 59 bp stretch upstream of the dPrx5 translational start codon in the intergenic domain (Fig. 1 B). Using 3′ RACE analysis, we also established that the monocistronic CG7215 has a termination site in the intergenic region located 147 bp downstream of the stop codon of the CG7215 gene. The sequence of the 3′ end identified by RACE analysis was identical to that of the monocistronic CG7215 transcription unit as deduced from the EST clone (GeneBank # AT18329).
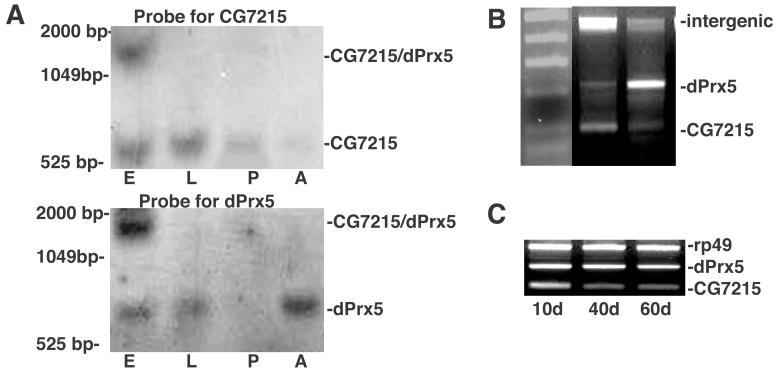
Results from Northern blot analysis, using polyA+ RNA isolated from adult flies, confirmed the existence of three distinct RNA species that could be derived from the CG7215/dPrx5 cluster. Hybridization with a probe specific for CG7215 gene detected two mRNA species, approximately 600 bp and 1500 bp long in samples isolated from embryos (Fig. 2 A). The longer transcript (∼1500 bp) was also detected when the blot was re-hybridized with a dPrx5-specific probe. In addition, hybridization with the dPrx probe picked up a distinct (∼800 bp) mRNA species, which did not cross-hybridize with the CG7215 probe. Thus, these data are consistent with two different monocistronic transcripts and a dicistronic transcript carrying the coding regions of both CG7215 and dPrx5.
Figure 2. Transcriptional profiles of the expression of CG7215 and dPrx5 genes.
A, Northern blot analysis of polyA+ RNA fraction isolated from embryos (E), 2nd-3rd instar larvae (L), 1-2 days pupae (P) and freshly eclosed adults (A). RNA molecular size markers are shown on the left. The blot was hybridized with a probe specific to the CG7215 ORF, then re-stripped and hybridized with a probe specific for the dPrx5 ORF. RNA size standards are shown on the left. B, RT-PCR analysis of transcription of CG7215/dPrx5 in embryos (E) and adult flies (A). Hi-Lo™ marker is shown on the left. Sets of primers used for the analysis are shown in Fig. 1. PCR products correspond to a fragment of the CG7215 gene obtained with For-7215/Rev-7215 primers (110 bp), a fragment of the dPrx5 gene obtained with For-dPrx5/Rev-dPrx5 primers (226 bp) and a fragment of intercistronic region obtained with For-7215/Rev-Int primers (451 bp). C, expression of CG7215 and dPrx5 genes in flies of different ages. The CG7215 gene was co-amplified with the dPrx5 gene and a house-keeping gene (rp49) as a control.
Transcription of CG7215 and dPrx5 genes
Frequently, a dicistronic gene organization in eukaryotes is the result of a duplication event, where the two genes may be functionally related and/or display similarities in gene structure. However, analysis of the nucleotide sequence of CG7215 and dPrx5 revealed no significant homology. Moreover, at the amino acid level, CG7215 exhibits none of the peroxiredoxin signatures noted in dPrx5 and in fact carries a ubiquitin-like domain. Nevertheless, given the close proximity of the two genes and the existence of dicistronic mRNA, it was possible that these two genes show patterns of co-regulation.
To address this issue, we carried out a preliminary study of transcriptional profiles. An abbreviated developmental Northern (Fig. 2 A) involving embryos, 2nd/3rd stage larvae, 1-2 da old pupae and newly eclosed adults shows no strong co-expression pattern. Thus CG7215-specific transcripts tend to be more abundant than dPrx5-specific transcripts during development while the opposite is true in adults. Levels of the dicistronic mRNA species are barely detectable except in the case of embryonic tissue. The differences in the proportions of monocistronic and dicistronic mRNAs in embryos and adults are more clearly demonstrated by RT-PCR analysis (Fig. 2 B), where different sets of primers were used to simultaneously amplify the intergenic region (dicistronic-specific), and regions specific for each individual gene. RT-PCR analysis was also used to obtain transcription profiles of CG7215 and dPrx5 in adult flies of different ages and in response to paraquat administration or UV exposure. Under all conditions, accumulation of the dicistronic species in adults was almost undetectable (data not shown). In older flies (40 and 60 da), both genes showed a slight decline in levels, which was more pronounced for CG7215 gene (Fig. 2 C). Paraquat administration (at a dosage of 10 mM that gives a median survivorship in the range of 4 days) or exposure of 24 h pupae to UV (50 mJ) had no impact whatsoever on either CG7215 or dPrx mRNA levels (data not shown). Thus, while there appear to be some commonalities in the expression patterns of CG7215 and dPrx, the tight co-overexpression that might be expected for members of a dicistron are clearly not evident.
Expression of CG7215 and dPrx
In eukaryotes, translation of a dicistronic mRNA is normally initiated at the first AUG codon, while translation of the down-stream gene may be initiated at a distinct internal ribosomal entry site (IRES) or perhaps re-initiated by transiting ribosomes after translation termination of the first gene, as reviewed in [12]. Alternatively, polycistronic pre-mRNA may be processed by splicing or endonuclease action to form two mature polyadenylated mRNAs that are individually translated.
As regards the CG7215/dPrx5 gene cluster, we have determined that the monocistronic mRNAs have distinct 5′ and 3′ UTRs, and are therefore not likely to be the result of alternative splicing. Furthermore, no evidence was found for the presence of an IRES in the intergenic domain, based on sequence analysis (UTRScan tool - http://www.ba.itb.cnr.it/UTR/), while in silico analysis did identify a predicted transcription start site. Finally, we found two additional in-frame AUG codons in the upstream CG7215 gene that precede the downstream ORF of the dPrx5 gene. The presence of such upstream in-frame AUG codons or ORFs usually attenuates translation of a second cistron [13]. Taken altogether, the computer analysis predicts that dPrx5 is expressed predominantly from its monocistronic mRNA, while CG7215 may be expressed from both monocistronic and dicistronic transcripts.
Individual gene silencing was used to confirm the results of in silico analysis. To silence the upstream CG7215 gene, S2 cells were treated with double-stranded RNA complementary to the CG7215 coding region. Previous analysis of S2 cells revealed that amounts of dicistronic mRNA were comparable to monocistronic species (data not shown). As expected, RNAi resulted in a severe knock-down of both RNA species (monocistronic and dicistronic) containing CG7215 (Fig. 3 A and data not shown). In contrast, there was no discernable effect on RNA levels of dPrx5, suggesting that expression of the downstream dPrx5 gene depends predominantly on the monocistronic species.
Figure 3. Analysis of CG7215 and dPrx5 expression.
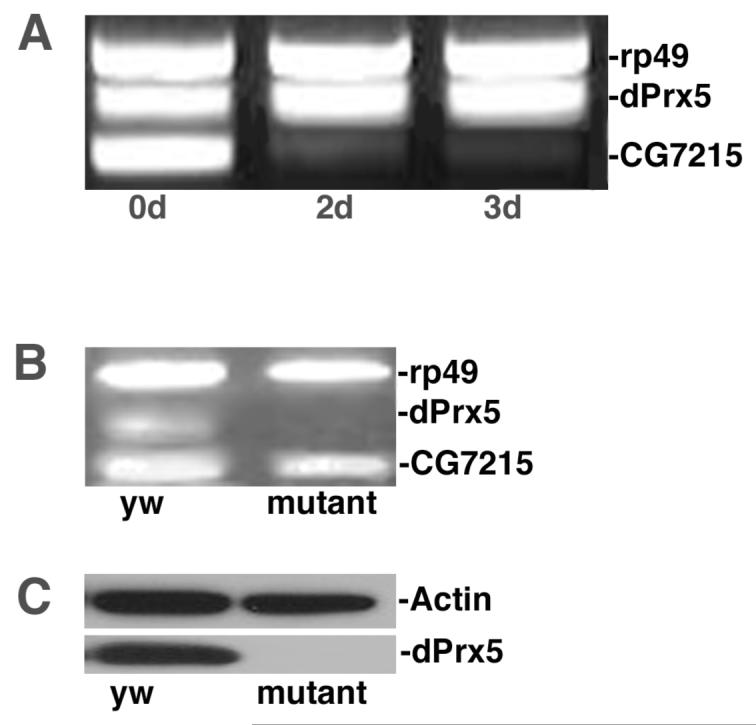
A, S2 cells were transfected with in vitro generated dsRNA, complementary to a coding region and part of the 5′UTR of the CG7215 gene. RNA samples were isolated after indicated periods of time followed by RT-PCR analysis with three different sets of primers that amplify the house-keeping rp49 gene (control), as well as the CG7215 and dPrx5 genes. B, RT-PCR analysis of CG7215 and dPrx5 gene expression in the y w strain and a mutant strain, containing a P-element insertion in the coding region of dPrx5. C, immunoblot analysis of dPrx5 protein expression in y w and P-element mutant strains using antibodies developed to dPrx5 recombinant protein. Anti-actin antibodies were used to control for loading.
The effect of dPrx5 abrogation on CG7215 RNA levels was examined in adult flies using a mutant fly strain, in which the P element is inserted in the first exon of the dPrx5 coding region (Fig. 1 A and Material and Methods). As shown by both RT-PCR and immunoblot analyses (Fig. 3 B and C), there is no discernable dPrx5 product, while CG7215 transcripts were detectable in both mutant and control.
These data support a model in which the upstream CG7215 gene could be translated from both the CG7215-specific mRNA as well as the dicistronic mRNA. Furthermore the accumulation of dPrx5-specific transcripts is not likely a result of processing of dicistronic RNA precursors but rather an independent event, where RNA polymerase transcribes monocistronic dPrx5 from its own promoter. Indeed, a candidate dPrx5 promoter domain encompassing 689 base pairs that span the intergenic region as well as some 421 base pairs of the 3′ end of the CG7215 coding region has been identified. As depicted in Fig. 4 A, two candidate promoter domains, one upstream of CG7215 and one upstream of dPrx5, were used to replace an OpIE2 promoter residing in a pIB/V5-His recombinant construct containing the dPrx5 coding region fused to the V5 epitope. S2 cells were transfected with both constructs and proteins were extracted after 48h, 72h and 96 h and subjected to immunoblot analysis. As determined by the detection of dPrx5/V5 protein both candidate promoter domains were shown to be functional, although expression levels were lower than that driven by the constitutive high-level OpIE2 promoter (Fig. 4 B).
Discussion
In an effort to facilitate functional studies at the organismal level, we have begun characterization of the Drosophila homolgue, dPrx5. This led us to identify dPrx5 as the downstream member of a two genes cluster that meets the criteria of a dicistronic operon [9; 12]. Although this type of genomic organization is unusual for eukaryotes, 31 gene pairs in the Annotated Drosophila genome have been denoted as dicistronic and 17 pairs as putative dicistronic operons [9]. For many of the predicted Drosophila dicistronic genes (31/48), there was some evidence supporting alternative monocistronic transcript(s) for either the upstream or downstream coding domain, or for both [14; 15]. The dPrx5 coding sequence resides in both dicistronic and monocistronic transcripts but is translated predominantly, if not exclusively, from the monocisrtonic species. This conclusion is based on RNAi knock-down studies targeting the dicistronic and upstream monocistronic species, which had no discernable effect on the accumulation of dPrx5 transcript. Additional support for this conclusion is based on in silico analysis, which revealed no IRES in the dicistronic transcript but did reveal two in frame AUG codons preceeding the dPrx5 ORF, an occurrence, which typically dampens downstream translation initiation [13]. Finally, it is notable that a distinct functional promoter domain associated with the expression of dPrx5 monocistronic transcripts has been identified and is immediately upstream of the dPrx5 ORF. Certainly the existence of two independent promoter domains could underlie the observed differences in transcriptional profiles.
The rationale for the inclusion of dPrx5 in a dicistronic arrangement is not evident. The lack of any significant sequence similarities between the two genes indicates that it may not have come about due to a simple duplication event. Sequence analysis of the CG7215 gene revealed no close homologues in any non-Drosophila species, including mosquitos, silkworms and bees. For instance, while BLAST search produced sequences with 70-95% amino acid identities to CG7215 within Drosophilidae, only N-termini domain of CG7215 showed some degree of homology in other organisms, with identity percentage running from 49% in Aedes aegypti to <36% in other Dipterans. While the CG7215 and dPrx5 genes are maintained within all Drosophila species, their homologues are in close proximity only in the melanogaster subgroup, which includes D. melanogater, D. simulans, D. sechellia, D. yakuba and D. erecta (Suppl. Material), with the intergenic distance ranged from 261 to 295 bp. Furthermore, alignment analysis of intergenic sequences from the melanogaster subgroup revealed 89-93% nucleotide identity. In more distantly related species, the intergenic distances tend to increase, ranging from 376 bp (in D. ananassae) to 537 bp (in D. grimshawi) with no substantial homology to the intergenic sequence of D. melanogaster. Thus, it would appear that this gene cluster arose specifically in the Drosophila lineage, with selection on the intergenic region being maintained in the melanogaster subgroup.
Although the dicistronic context of the Drosophila dPrx5 gene appears to be unique, the Prx5 gene in other eukaryotic organisms is often associated, with longer alternative transcripts. Thus, in Apis mellifera, there is a predicted transcript with a long 5′UTR (GeneBank # XM_624803) containing three apparently untranslated ORFs (4-5 amino acids in length). An alternative Prx5 mRNA species with a long 5′ UTR containing several short ORFs was also found in mice [16]. Since the alternative transcripts produced by the Prx5 gene were localized to different mouse tissues, the authors surmised that this could provide an additional tissue-specific level of control for Prx5 gene expression, and it is plausible that a similar phenomenon might be operational in Drosophila.
Supplementary Material
Acknowledgements
This work was supported by grants R21 AG025096 from the National Institute on Aging/National Institutes of Health (to Dr. Radyuk) and RO1 AG20715 (to Dr. Orr) from the NIA/NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Knoops B, Clippe A, Bogard C, Arsalane K, Wattiez R, Hermans C, Duconseille E, Falmagne P, Bernard A. Cloning and characterization of AOEB166, a novel mammalian antioxidant enzyme of the peroxiredoxin family. J Biol Chem. 1999;274:30451–8. doi: 10.1074/jbc.274.43.30451. [DOI] [PubMed] [Google Scholar]
- [2].Zhou Y, Kok KH, Chun AC, Wong CM, Wu HW, Lin MC, Fung PC, Kung H, Jin DY. Mouse peroxiredoxin V is a thioredoxin peroxidase that inhibits p53-induced apoptosis. Biochem Biophys Res Commun. 2000;268:921–7. doi: 10.1006/bbrc.2000.2231. [DOI] [PubMed] [Google Scholar]
- [3].Banmeyer I, Marchand C, Clippe A, Knoops B. Human mitochondrial peroxiredoxin 5 protects from mitochondrial DNA damages induced by hydrogen peroxide. FEBS Lett. 2005;579:2327–33. doi: 10.1016/j.febslet.2005.03.027. [DOI] [PubMed] [Google Scholar]
- [4].Banmeyer I, Marchand C, Verhaeghe C, Vucic B, Rees JF, Knoops B. Overexpression of human peroxiredoxin 5 in subcellular compartments of Chinese hamster ovary cells: effects on cytotoxicity and DNA damage caused by peroxides. Free Radic Biol Med. 2004;36:65–77. doi: 10.1016/j.freeradbiomed.2003.10.019. [DOI] [PubMed] [Google Scholar]
- [5].Seo MS, Kang SW, Kim K, Baines IC, Lee TH, Rhee SG. Identification of a new type of mammalian peroxiredoxin that forms an intramolecular disulfide as a reaction intermediate. J Biol Chem. 2000;275:20346–54. doi: 10.1074/jbc.M001943200. [DOI] [PubMed] [Google Scholar]
- [6].Tien Nguyen-nhu N, Knoops B. Mitochondrial and cytosolic expression of human peroxiredoxin 5 in Saccharomyces cerevisiae protect yeast cells from oxidative stress induced by paraquat. FEBS Lett. 2003;544:148–52. doi: 10.1016/s0014-5793(03)00493-9. [DOI] [PubMed] [Google Scholar]
- [7].Kropotov A, Serikov V, Suh J, Smirnova A, Bashkirov V, Zhivotovsky B, Tomilin N. Constitutive expression of the human peroxiredoxin V gene contributes to protection of the genome from oxidative DNA lesions and to suppression of transcription of noncoding DNA. Febs J. 2006;273:2607–17. doi: 10.1111/j.1742-4658.2006.05265.x. [DOI] [PubMed] [Google Scholar]
- [8].Kropotov AV, Grudinkin PS, Pleskach NM, Gavrilov BA, Tomilin NV, Zhivotovsky B. Downregulation of peroxiredoxin V stimulates formation of etoposide-induced double-strand DNA breaks. FEBS Lett. 2004;572:75–9. doi: 10.1016/j.febslet.2004.07.011. [DOI] [PubMed] [Google Scholar]
- [9].Misra S, Crosby MA, Mungall CJ, Matthews BB, Campbell KS, Hradecky P, Huang Y, Kaminker JS, Millburn GH, Prochnik SE, Smith CD, Tupy JL, Whitfied EJ, Bayraktaroglu L, Berman BP, Bettencourt BR, Celniker SE, de Grey AD, Drysdale RA, Harris NL, Richter J, Russo S, Schroeder AJ, Shu SQ, Stapleton M, Yamada C, Ashburner M, Gelbart WM, Rubin GM, Lewis SE. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0083. RESEARCH0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Radyuk SN, Klichko VI, Spinola B, Sohal RS, Orr WC. The peroxiredoxin gene family in Drosophila melanogaster. Free Radic Biol Med. 2001;31:1090–100. doi: 10.1016/s0891-5849(01)00692-x. [DOI] [PubMed] [Google Scholar]
- [11].Radyuk SN, Sohal RS, Orr WC. Thioredoxin peroxidases can foster cytoprotection or cell death in response to different stressors: over- and under-expression of thioredoxin peroxidase in Drosophila cells. Biochem J. 2003;371:743–52. doi: 10.1042/BJ20021522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Blumenthal T. Operons in eukaryotes. Brief Funct Genomic Proteomic. 2004;3:199–211. doi: 10.1093/bfgp/3.3.199. [DOI] [PubMed] [Google Scholar]
- [13].Wall AA, Phillips AM, Kelly LE. Effective translation of the second cistron in two Drosophila dicistronic transcripts is determined by the absence of in-frame AUG codons in the first cistron. J Biol Chem. 2005;280:27670–8. doi: 10.1074/jbc.M500255200. [DOI] [PubMed] [Google Scholar]
- [14].Brogna S, Ashburner M. The Adh-related gene of Drosophila melanogaster is expressed as a functional dicistronic messenger RNA: multigenic transcription in higher organisms. Embo J. 1997;16:2023–31. doi: 10.1093/emboj/16.8.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gray TA, Nicholls RD. Diverse splicing mechanisms fuse the evolutionarily conserved bicistronic MOCS1A and MOCS1B open reading frames. Rna. 2000;6:928–36. doi: 10.1017/s1355838200000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lee TH, Kim SJ, Kang SW, Lee KK, Rhee SG, Yu DY. Molecular cloning and characterization of the mouse peroxiredoxin V gene. Biochem Biophys Res Commun. 2000;270:356–62. doi: 10.1006/bbrc.2000.2430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.