Abstract
Choline is an essential nutrient for humans, though some of the requirement can be met by endogenous synthesis catalyzed by phosphatidylethanolamine N-methyltransferase (PEMT). Premenopausal women are relatively resistant to choline deficiency compared with postmenopausal women and men. Studies in animals suggest that estrogen treatment can increase PEMT activity. In this study we investigated whether the PEMT gene is regulated by estrogen. PEMT transcription was increased in a dose-dependent manner when primary mouse and human hepatocytes were treated with 17-β-estradiol for 24 h. This increased message was associated with an increase in protein expression and enzyme activity. In addition, we report a region that contains a perfect estrogen response element (ERE) ∼7.5 kb from the transcription start site corresponding to transcript variants NM_007169 and NM-008819 of the human and murine PEMT genes, respectively, three imperfect EREs in evolutionarily conserved regions and multiple imperfect EREs in nonconserved regions in the putative promoter regions. We predict that both the mouse and human PEMT genes have three unique transcription start sites, which are indicative of either multiple promoters and/or alternative splicing. This study is the first to explore the underlying mechanism of why dietary requirements for choline vary with estrogen status in humans.—Resseguie, M., Song, J., Niculescu, M. D., da Costa, K., Randall, T. A., Zeisel, S. H. Phosphatidylethanolamine N-methyltransferase (PEMT) gene expression is induced by estrogen in human and mouse primary hepatocytes.
Keywords: choline, 17-β–estradiol, bioinformatics, ERE, evolutionarily conserved gene motifs
Choline is an essential nutrient (1); it is used to form cell membranes, it is the major source of methyl-groups in the diet, and it is a precursor for biosynthesis of the neurotransmitter acetylcholine (2). Choline is critical during fetal development, when it influences stem cell proliferation and apoptosis, thereby altering brain structure and function (3–7). Similarly, it influences neural tube development (8, 9). In later life, choline deficiency causes fatty liver, liver damage, and muscle damage (10, 11). It also reduces the capacity to handle a methionine load, resulting in elevated homocysteine (10), a risk factor for cardiovascular disease (12). Though many foods contain choline (13, 14), there is at least 2-fold variation in dietary intake in humans (9, 15). Choline can also be derived from de novo biosynthesis of phosphatidylcholine catalyzed by phosphatidylethanolamine N-methyltransferase (PEMT, EC 2.1.1.17) (16, 17). Most of this enzyme's activity is in the liver (18).
When fed a diet low in choline, premenopausal women were much less likely to develop choline-deficiency associated organ dysfunction compared to men or postmenopausal women (19). This suggests that premenopausal women might have an enhanced capacity for de novo biosynthesis of choline. Such a finding would be important during pregnancy and lactation, when the demand for choline is especially high because the transport of choline from mother to infant, via placenta or mammary gland, constitutes an extraordinary drain on maternal choline stores (20).
Studies in animal models support this hypothesis. Female rats are less sensitive to choline deficiency than are male rats (21), and female mice produce more phosphatidylcholine via the PEMT pathway than do male mice (22). Estrogen status may be important for this increased PEMT activity; estradiol treatment increased PEMT activity in pituitary of rats (23) as well as in liver of castrated-rats (24) and PEMT activity in liver was increased in diethylstilbestrol-treated roosters (25). Thus, estrogen-mediated increases in PEMT activity in humans could be an explanation for the lower dietary choline requirements of premenopausal women.
The mechanism whereby estrogen increases PEMT activity is not known. In the present study, we report that there are motifs for estrogen response elements (EREs) in the promoter region(s) of the PEMT gene.
MATERIALS AND METHODS
Materials
All reagents were obtained from Fisher Scientific (Fair Lawn, NJ, USA), unless otherwise noted.
Animals
All animal procedures were approved by the University of North Carolina Institutional Animal Care and Use Committee. Male C57BL/6J mice (5−7 wk old) were obtained from Jackson Laboratories (Bar Harbor, ME, USA) and were housed individually in cages in a climate-controlled room (24°C) exposed to a 12-hr light cycle, fed AIN76A semipurified diet (Dyets, Bethlehem, PA, USA) and water ad libitum.
Primary mouse hepatocyte isolation
Mice were anesthetized (200 mg ketamine/kg and 16 mg xylazine/kg) by subcutaneous injection, and the livers harvested. Hepatocytes were isolated from the livers by a modification of the method described previously (26). Briefly, the liver was perfused, through a needle aligned along the inferior vena cava, with buffer (pH 7.5) containing 137 mmol/L NaCl, 5.4 mmol/L KCl, 0.5 mmol/L NaH2PO4, 0.42 mmol/L Na2HPO4, 10 mmol/L HEPES, 0.5 mmol/L EGTA, 4.2 mmol/L NaHCO3, and 5 mmol/L glucose; collagenase buffer, pH 7.5, contained 137 mmol/L NaCl, 5.4 mmol/L KCl, 5 mmol/L CaCl2, 0.5 mmol/L NaH2PO4, 0.42 mmol/L Na2HPO4, 10 mmol/L HEPES, 0.15 g/L collagenase B (Boehringer Mannheim Corp, Indianapolis, IN, USA), 0.05 g/L trypsin inhibitor, 4.2 mmol/L NaHCO3, and 0.016 mmol/L phenol red. The collagenase-perfused liver was then dissected, suspended in Hanks' solution (30 ml), and filtered through cheesecloth and a 100 μm nylon membrane to remove connective tissue debris and cell clumps. Hepatocytes were subjected to centrifugation (42 g, 2 min at 4°C) and resuspended in Hanks solution; this was repeated 4×. Then hepatocytes were purified using density gradient centrifugation (45% Percoll solution, 42 g for 10 min at 4°C). Cell viability, measured by trypan blue exclusion, was more than 90%.
Preparation of estradiol-free serum for hepatocyte culture
Charcoal (Norit A, acid washed, Sigma-Aldrich, St. Louis, MO, USA) was washed twice with cold sterile water immediately before use. A 5% charcoal–0.5% dextran T70 (Pharmacia-LKB, Uppsala, Sweden) suspension was prepared, and aliquots were centrifuged at 1600 g for 10 min. Supernatants were aspirated, and fetal bovine serum (Biomeda, Foster City, CA, USA) was mixed with the charcoal pellets. This charcoal-serum mixture was kept in suspension by rolling at 4 cycles/min at 37°C for 1 h. After centrifugation at 1600 g for 20 min, the supernatant was passed through a 0.45 μm filter (Nalgene, Rochester, NY, USA). The charcoal stripped serum was then stored at −20°C until needed. More than 99% of serum sex steroids are reported to be removed by this treatment (27).
Murine hepatocyte culture
Primary mouse hepatocytes were maintained in DMEM/F12 medium (GIBCO-Life Technologies, Inc., Carlsbad, CA, USA), supplemented with 10% fetal bovine serum (Biomeda) and antibiotics (100 U/ml penicillin and 100 μg/ml of streptomycin; Sigma-Aldrich), at 37°C in humidified air containing 5% CO2. Briefly, 1.5 × 106 cells were seeded onto 60 mm culture dishes (BD Biosciences, Franklin Lakes, NJ, USA), incubated in fresh medium for 24 h and then washed with 1× phosphate buffered saline (PBS) twice before further incubation in medium that contained 90% phenol red-free DMEM/F12 supplemented with 10% charcoal stripped E2-free serum for 48 h. This medium was used to effect growth-factor deprivation (28). Cells then were treated with 0−100 nmol/L 17-β-estradiol (E2; prepared and stored in phenol red-free DMEM/F12 at 73 μmol/L per manufacturer's instructions; Sigma-Aldrich). After 24 h incubation with E2, cells were harvested for total RNA; after 48 h incubation with E2, cells were harvested for the microsomal protein fraction.
Murine Pemt relative real-time mRNA quantification
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Primers and probes for murine Pemt and β-actin were designed using JaMBW software (http://www.bioinformatics.org/JaMBW[b]; Table 1). The Pemt probe was labeled with a reporter dye (FAM, 6-carboxyfluorescein) at the 5′ end and a quencher dye (TAMRA, 6-carboxytetramethylrhodamine) at the 3′ end. The β-actin probe was labeled with a reporter dye (TET, tetramethylrhodamine) at the 5′ end. Quantitation of Pemt and β-actin mRNA levels was performed by a real-time RT-PCR assay using an ABI prism 7700 sequence detection system (ABI, Foster City, CA, USA). A 30 μL reaction mixture contained 500 ng of total RNA and 0.5 μmol/L of each primer. The reaction conditions were designed as follows: reverse transcription (RT) at 48°C for 30 min and initial denaturation at 95°C for 10 min followed by 40 cycles with 15 s at 95°C for denaturing and 1 min at 60°C for annealing and extension. Relative quantification of Pemt mRNA expression was calculated by the comparative threshold (Ct) method described elsewhere (29). The relative quantification value of the target gene, normalized to an endogenous control gene and relative to a calibrator, was expressed as 2−ΔΔCt (Power/folds), where ΔCt = Ct of target gene (Pemt) – Ct of endogenous control gene (β-actin), and ΔΔCt = ΔCt of samples for target gene (estrogen treatment) - ΔCt of the calibrator (without estrogen treatment, negative control) for the Pemt gene. Final results are expressed as mean ratio of Pemt gene expression for each estrogen treatment normalized to Pemt gene expression without estrogen.
TABLE 1.
Real-Time PCR primers and probes used for gene expression analysis
| Gene | GenBank Accession # | Forward Primer | Reverse Primer | Amplicon Length (bp) | Murine Gene Expression Probe |
|---|---|---|---|---|---|
| Pemt1 | NM_008819 | ACTCATGCATGCTAGTCCCA | AGCAGTGAAGGGCTCTTCAT | 178 | 5′-FAM-CGAGACAATTGCCACCAGCACGT-3′ |
| β-actin | NM_007393 | CTGCCTGACGGCCAAGTC | CAAGAAGGAAGGCTGGAAAAGA | 220 | 5′-TET-CACTATTGGCAACGAGCGGTTCCG-3′ |
| PEMT1 | NM_148172 | AAGACCCGCAAGCTGAGCA | AGTACATGGGGTTGTCCAGGA | 298 | |
| NM_007169 | |||||
| NM_148173 | |||||
| TBP | NM_003194 | TTCGGAGAGTTCTGGGATTG | TGGACTGTTCTTCACTCTTGGC | 227 | |
| PI-9 | L40378 | GTGGCAGGCCCTGCATCA | CACCCTTTATGGCGATGA | 271 |
Primers and probes for murine Pemt and β-actin were designed using JaMBW software (http://www.bioinformatics.org/JaMBW. The Pemt probe was labeled with a reporter dye (FAM, 6-carboxyfluorescein) at the 5′ end and a quencher dye (TAMRA, 6-carboxytetramethyrhodamine) at the 3′ end. The β-actin probe was labeled with a reporter dye (TET, tetramethylrhodamine) at the 5′ end. PCR primers for human PEMT, proteinase inhibitor-9 (PI-9; positive control) and TATA-box binding protein (TBP; housekeeping gene) were designed using online Gene Fisher interactive primer design software (http://bibiserv.techfak.uni-bielefeld.de/genefisher/.
Primers bind all three Pemt transcript variants.
Murine Pemt immunoblot
Protein (50 μg) was separated by SDS-polyacrylamide gel electrophoresis on a 12.5% polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Sigma-Aldrich), which was probed with anti-PEMT antibody (a kind gift from Dr. Dennis E. Vance), washed extensively, and then probed with horseradish peroxidase-conjugated goat anti-rabbit IgG (Pierce, Rockford, IL, USA). PEMT protein was visualized by a reaction with Supersignal chemiluminescent substrate (Pierce) and exposed to X-ray film (Denville Scientific, Metuchen, NJ, USA).
Human hepaocyte isolation
Primary human liver cells were provided as a gift by Admet Technologies (Durham, NC, USA). They used donated livers not suitable for orthotopic liver transplantation obtained from federally designated organ procurement organizations. Informed consent was obtained from next of kin for use of the livers for research purposes. Isolation of hepatocytes was performed by members of the Admet Technologies research team as described previously (30). These hepatocytes were transferred to us and used within 96 h of isolation.
Human hepatocyte culture
Cells were plated at a density of ∼1.8 × 106 cells per well on collagen-coated 6-well culture plates (BD Biosciences) and incubated for 6 h at 37°C in humidified air containing 5% CO2 in William's complete medium E (WCME; GIBCO-Life Technologies, Inc.) containing 10% Dextran-treated charcoal stripped fetal bovine serum (Biomeda) and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin; Sigma-Aldrich). Medium was replaced at 6 h with serum-free WCME. Following a 48 h “recovery period” in serum-free WCME, human primary cells were incubated for 24 h in WCME with 0−1000 nmol/L 17-β-estradiol (prepared and stored per manufacturer's instructions). After 24 h incubation with E2, cells were harvested for cytoplasmic mRNA; after 48 h incubation with E2, cells were harvested for total cellular protein fraction.
Human PEMT mRNA quantification
Cytoplasmic RNA from liver cells was extracted using the RNeasy mini kit (Qiagen, Valencia, CA, USA). PCR primers for human PEMT, proteinase inhibitor-9 (PI-9; positive control) and TATA-box binding protein (TBP; housekeeping gene) were designed using online Gene Fisher interactive primer design software (http://bibiserv.techfak.uni-bielefeld.de/genefisher/; Table 1) and purchased from Operon (Germantown, MD, USA). Real-time PCR reactions were performed using 100 ng RNA, 0.5 μmol of each primer, and the one-step QuantiTect SYBR Green RT-PCR kit (Qiagen) in a 40 μl reaction. The reaction conditions were designed as follows: RT at 50°C for 30 min and PCR initial activation step at 95°C for 15 min. Initial denaturation at 94°C for 15 s was followed by 40 cycles of annealing (58°C for 30 s) and extension (72°C for 30 s). Changes in PEMT mRNA levels were detected using an iCycler iQ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Relative quantification of PEMT mRNA expression was calculated by the Ct method as described earlier, and final results were expressed as a ratio change relative to untreated and normalized for TBP mRNA expression. We used expression of the PI-9 gene, known to be estrogen responsive (31), as a positive control (data not shown).
PEMT protein expression and activity assay
Mouse and human hepatocytes were harvested 48 h after 17-β-estradiol treatment with two washes of 1× PBS, scraped into medium 1 (250 mmol/L sucrose, 10 mmol/L Tris, pH 7.4, 1 mmol/L EDTA, and 1 mmol/L dithiothreitol) and homogenized with 10 complete strokes in a Teflon™-glass homogenizing vessel. For the murine cellular homogenate, an aliquot of total cellular homogenate was then centrifuged at 100,000 g for 1 h to obtain total particulate (membrane) fraction. Total murine particulate protein (50 μg) was used for PEMT activity assays as described previously using phosphatidyldimethylethanolamine (PDME; Avanti Polar Lipids, Alabaster, AL, USA) as the methyl acceptor and S-adenosyl-L-[methyl-3H] methionine (American Radiolabeled Chemicals Inc., St. Louis, MO, USA) as the methyl group donor (32).
Comparative bioinformatics promoter analysis
CAGE analysis
CAGE analysis (cap analysis gene expression) data were retrieved from the FANTOM3 website (http://fantom.gsc.riken.go.jp/). CAGE analysis (33) was used to identify the major transcription start sites (TSS) for the mouse and human PEMT gene.
EST abundance of various PEMT transcripts
The positions of expressed sequence tag (EST) sequences from the NCBI dbEST (http://www.ncbi.nlm.nih.gov/dbEST/; last accessed on 01/03/07) were displayed relative to the mouse and human PEMT gene of the Mouse and Human genome (May 2004 release) using the GenomBench tool of Vector NTI 10.1 (Invitrogen). All potentially full-length ESTs were selected and found to group into three clusters, differing in the 5′ start of the ESTs for both human and mouse.
Identification of evolutionarily conserved regions (ECRs) and conserved transcription factor binding sites
The analysis of human and mouse syntenic relationships and conservation profiles was done through the annotation of ECRs in the alignments of genomes. We used the BLASTZ-based genome alignments generated by the ECR Browser (http://ecrbrowser.dcode.org; last accessed 01/03/07) (34). A genomic interval was annotated as an ECR if it was >100 bp and >70% identity as defined by the number of nucleotide matches in a sliding window (default settings). Prediction of potential conserved transcriptional binding sites was then done using the rVista 2.0 search tool within the ECR Browser. The Transfac V10.2 database was used to scan these ECRs using default settings.
Estrogen response element identification
EREs were identified with detection parameters set on the basis of optimized settings for the Dragon ERE Finder (http://sdmc.lit.org.sg/ERE-V2/index; last accessed 01/03/07) using the default 83% sensitivity (35). Conserved EREs were identified as elements present in both the human and mouse PEMT gene at distances comprised between −10 to + 15 kb from their respective transcription start sites (TSS A, TSS B, TSS C).
Data analysis
Prior to analysis, all data were normalized to the 0 nmol/L estrogen treatment for each individual (mouse and human). Data were expressed as mean ratio change of each estrogen treatment vs. no treatment ± se. Ratios were transformed to a log(2) scale to ensure normal distribution of the data, required by the parametric testing assumptions. Protein expression and gene expression significance of change statistical differences were assessed by one-way analysis of variance (ANOVA) followed by the Tukey-Kramer (P<0.05) multiple comparison test (JMP Version 3.2, SAS Institute Inc, Cary, NC, USA). Logistic regression analysis was performed to assess the trend significance of the dose response to estrogen (JMP Version 3.2), using ratio values. Linear Fit was used to determine the correlation between PEMT gene expression and activity, using only the points that have both gene expression and activity data.
RESULTS
Estrogen up-regulates murine Pemt gene expression, protein levels and enzyme activity
In mouse hepatocytes gene expression, protein expression and activity all were significantly increased by 17-β-estradiol treatment (Fig. 1), and a strong dose-response relationship was found for both activity (EC50 = 1 nmol/L; P<0.0001) and gene expression (EC50=5 nmol/L; P<0.0001). Additionally, a strong correlation was found between gene expression and activity (correlation coefficient=0.91).
Figure 1.
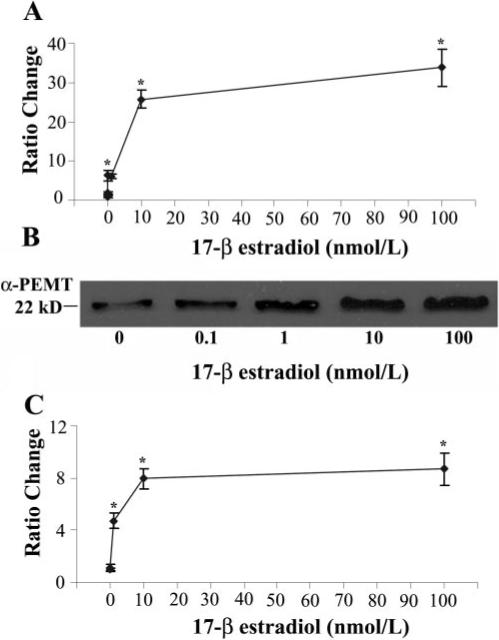
Estrogen induces Pemt gene and protein expression and increases enzyme activity in primary cultured mouse hepatocytes. Primary mouse hepatocytes were incubated with 17-β-estradiol (0−100 nmol/L) for 24 h before harvesting. A) Detection of Pemt and β-actin mRNA levels was performed by a real-time RT-PCR assay. Relative quantification of Pemt mRNA expression was calculated by the comparative threshold cycle (Ct) method as described in Experimental Procedures. Results are expressed as mean ratio change in gene expression ± sem. (n=3/point). *P < 0.05 different from no treatment. B) Total particulate protein was isolated and analyzed for PEMT protein expression by Western blotting as described in Experimental Procedures. Equal protein loading of lanes was assessed by Coomasie Blue gel staining (data not shown). C) PEMT enzyme activity was assayed by a radio-enzymatic assay as described in Experimental Procedures. Results are expressed relative to no treatment as mean ratio change ± sem (n=3/point). *P < 0.05 different from no treatment.
Estrogen up-regulates human PEMT gene expression and enzyme activity
In human hepatocytes, gene expression and activity were significantly increased by 17-β-estradiol treatment (Fig. 2) and a strong dose-response relationship was found for both activity (EC50=1 nmol/L; P<0.0006) and gene expression (EC50=10 nmol/L; P<0.0001) using logistic regression analysis. Gene expression and activity further increased at 1000 nmol/L estrogen treatment (data not shown). However, a linear fit model suggests that there is only a weak correlation between PEMT gene expression and activity (0.34) correlation coefficient.
Figure 2.
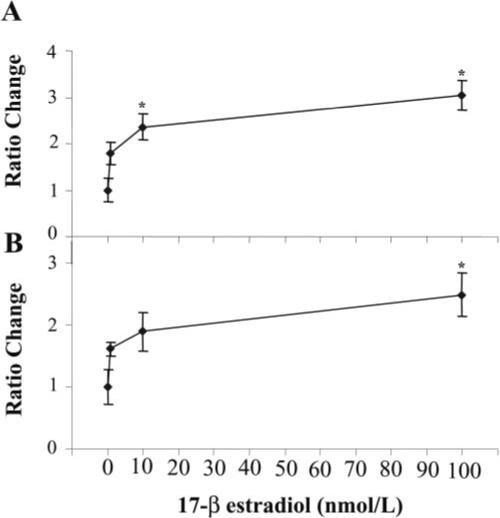
Estrogen induces PEMT gene expression and increases enzyme activity in primary cultured human hepatocytes. Primary human hepatocytes from various donors were incubated with 17-β estradiol (0−100 nmol/L) for 24 h before harvesting. A) Detection of PEMT and TATA-box binding protein (TBP) mRNA levels was performed by a real-time RT-PCR assay. Relative quantification of PEMT mRNA expression was calculated by the comparative threshold cycle (Ct) method described in Experimental Procedures. Results are expressed as ratio fold change in gene expression relative to untreated samples ± sem. (n=4−5/point). B) PEMT enzyme activity was assayed by a radio-enzymatic assay as described in Experimental Procedures. Results are expressed as ratio of change in enzyme activity (pmol/mg protein/min) relative to untreated samples ± sem. (n=4−7/point). *P < 0.05 different from no treatment.
Characterization of the major human and mouse PEMT transcripts
The human PEMT gene, which is located on chromosome 17p11.2, has 9 exons and 8 introns spanning its 86 kb length, and it encodes three variant transcripts (http://www.ensembl.org/Homo_sapiens/geneview?gene=OTTHUMG00000059290;db=vega, date last accessed 1/02/07). Each of the three transcript variants has only 7 exons and 6 introns due to splicing of the leading exons 1−3, which are alternatively spliced to a common exon 4, where translation begins (36). Three RefSeq accessions have been annotated for the human PEMT gene by NCBI, NM_148172, NM_007169, and NM_148173. Murine Pemt, located on chromosome 11, has been reported to have 7 exons and 6 introns spanning ∼75 kb and encoding one major transcript, annotated by NCBI RefSeq accession NM_008819. (http://www.ensembl.org/Mus_musculus/geneview?gene=OTTMUSG00000005808;db=vega, date last accessed 1/02/07) (37). Primer extension analysis of the murine Pemt gene demonstrated experimentally that the major Pemt TSS is located 139 bp upstream of the initiator methionine codon (37). Human and murine PEMT proteins are 80% homologous, but unlike its human counterpart, murine PEMT protein translation initiation begins in exon 2. It was previously reported that the transcript that begins with exon 2 [NM_007169] is the most abundant in human liver and that only one transcript [NM_008819] exists for the mouse Pemt gene (36, 37).
To more fully characterize the various alternative TSS in the human and mouse PEMT orthologs, we estimated the relative abundance of the major PEMT transcripts in human and mouse liver based on the ESTs in dbEST and used FANTOM3 CAGE analysis viewer to determine the location of the various TSS (Fig. 3). Using publicly available data from dbEST, we found that the vast majority of the 5′ ends of the ESTs cluster around the start point of NM_148172 (bp 17,435,719 of ch. 17) (denoted as human TSS A), suggesting that this is the most abundant transcript in the liver for the human PEMT gene. This TSS is independently confirmed by the results of the FANTOM CAGE analysis where the representative CAGE TSS for the PEMT gene is at bp 17,435,731, 12 bp upstream of the RefSeq defined start. For NM_007169 (start defined at bp 17,426,470) (denoted as human TSS B), a representative CAGE TSS is found 40 bp upstream at bp 17,426,510. For NM_148173 (start defined at bp 17,421,504) (denoted as human TSS C), a representative CAGE start is found at bp 17,421,107 while the start based on the average of ESTs is 17,421,043; both are several hundred basepairs downstream of the RefSeq defined start.
Figure 3.
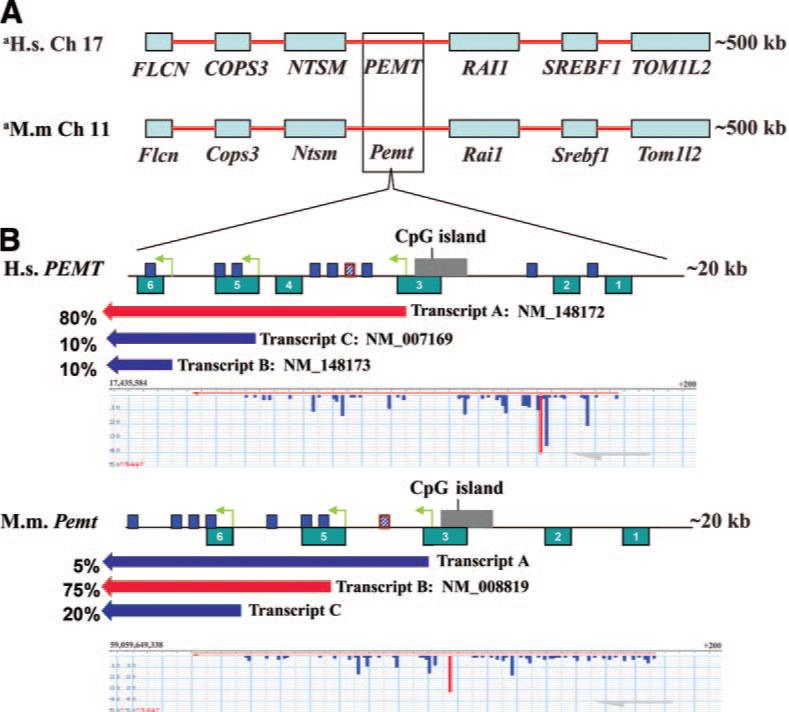
Comparative analysis of murine and human PEMT gene promoters. A) 500 kbp alignment of conserved syntenic blocks of human chromo-some 17 and mouse chromosome 11, with blue boxes showing the relationships between orthologous genes. B) A 20 kbp enlargement of the human PEMT gene encoded on the minus strand, the promoter region, and the mouse orthologous region. The PEMT gene promoter region of the human and mouse contains six distinct evolutionarily conserved regions (ECR 1-ECR 6). ECR 4 in murine Pemt has been translocated to a different chromosome (data not shown). Three conserved estrogen response elements are within ECR 5 and 6, shown as dark blue boxes; a perfect consensus ERE in a distal promoter/enhancer region ∼7.5 kb from transcription start site (TSS) B is shown as hatched blue boxes with red outline. Block arrow bars displayed beneath PEMT gene promoter regions represent the major transcripts relative to TSS A, B, C, (indicated by yellow-green arrows) with the percentages reflecting the EST abundance of each transcript in liver (purple arrows indicate minor transcripts; red indicates the major transcript. Beneath the transcript arrow bars is shown a representative CAGE analysis for the major human and mouse TSS. The highest peak, shown in red, represents the predominant TSS for the PEMT gene. Lower peaks around the major TSS (s) would represent some variation in the actual in vivo TSS.
We also suggest that the mouse Pemt gene has two previously unreported transcripts (denoted murine transcripts A and C) in addition to the previously described transcript, NM_008819 (denoted murine transcript B), which is also the most abundant transcript based on the abundance of ESTs in dbEST. The majority of the 5′ ends of the available ESTs (the EST average start; 59,649,465) cluster around the start point of NM_008819 (bp 59,649,379 of ch. 11) (denoted murine TSS B), while CAGE analysis predicts the representative TSS at bp 59,649,447, which is 68 bp upstream of the RefSeq defined start for NM_008819. Available EST data suggest two additional mouse transcripts, transcript A, initiating ∼9 kp upstream of NM_008819; and transcript C, initiating ∼5 kb downstream. A representative CAGE TSS for transcript A is at 59,659,083, denoted murine TSS A, while the EST average start suggests a start at 59,658,975, within 108 bp of the CAGE start. For transcript C a representative CAGE TSS is at 59,644,334, denoted murine TSS C, and with the EST average start position is 59,644,300. Thus, based on both EST sequences from dbEST and from CAGE analysis from the FANTOM3 database we conclude that both the mouse and human PEMT genes have three unique transcription start sites that are indicative of multiple promoters. While the identification of two additional transcripts in the mouse gene suggests conserved transcriptional regulation between species, it is interesting to speculate why transcript variant B (NM_008819) is the more abundant transcript in mice, whereas transcript variant A (NM_148173) is more abundant in humans.
Initial analysis of the human PEMT putative promoter B sequence for the exon-2 containing transcript, NM_007169, suggested that it contained transcription factor binding sites critical for SREBP and C/EBP; however, promoter B lacks core promoter elements such as a TATA or CCAAT boxes (36). We report that the region upstream of putative promoter A, exon-1 containing and the most abundant transcript variant NM_148172, contains a CpG island as well as core promoter elements including an experimentally validated 600 bp RNA polymerase II binding site (Pol 9419, P<0.00001) (Fig. 3, Table 2) (38).
TABLE 2.
Human and mouse conserved promoter motifs
| Human Transcript | Evolutionarily Conserved Region | Motifs1 | Position | Sequence |
|---|---|---|---|---|
| ECR 1 | PPARA | −13233 | TGATCT | |
| [−13399−−13206] | SP1 | −13118 | CCACAGCCCC | |
| ERE | −11388 | TT-GGTCA-GGC-TGGTC-TT | ||
| ECR 2 | MYB | −10839 | GGCCAGTTC | |
| [−10919−−10748] | ERE | −10266 | TT-AGTCA-CGC-TGGTC-TC | |
| Pol II binding site | −9038 | (bp 17,435,552−17,436,153) | ||
| ECR 3 | CpG island | −8774 | CpG Repeat | |
| [−9246−−9038] | Exon 1 | −9151 | ||
| Transcript A: NM_148172 | TSS A | −9205 | TTCCGGGGG (bp 17,435,719) | |
| ERE | −7647 | GG-GGTCA-TGA-TCACC-TC | ||
| Consensus | ERE | −7401 | CG-GGTCA-GGG-TGACC-CT | |
| ERE | −6707 | GA-GACCA-GCC-TGACC-AA | ||
| ERE | −5879 | CG-GGGTA-TCT-TGACT-GC | ||
| FOXA1 | −3309 | AAGTTGTTTCCATT | ||
| ECR 4 | MYB | −3264 | AAACTGCCA | |
| [−3324−−2994] | SOX6 | −3190 | TGCATTGTTATCA | |
| GATA | −3186 | TGTTATCATT | ||
| SP1 | −862 | GTGGCGTGAT | ||
| AP1 | −386 | CTGACTCCT | ||
| AP1 | −206 | CCCGAGTCAGC | ||
| Transcript B: NM_007169 | TSS+1 | +1 | TTGTCCATG (bp 17,426,514) | |
| TSS B | +40 | GACCACAA (bp 17,426,470) | ||
| ECR 5 | Exon 2 | +50 | ||
| [−35−+1073] | C/EBP | +175 | AAATTACCA | |
| AP1 | +631 | CATTAGTCATT | ||
| ERE | +725 | TG-AGACA-GGC-TGACC-TG | ||
| ERE | +741 | GA-GGCCA-TTG-GGACC-TG | ||
| Transcript C: NM_148173 | ECR 6 | TSS C | +5010 | TGTGGGCGA (bp 17,421,504) |
| [+5192−+5579] | GATA | +5284 | TTTTATCTTC | |
| ERE | +5476 | TG-GGCTA-CGT-GGACC-CC |
| Mouse | Evolutionarily Conserved Region | Motifs1 | Position | Sequence |
|---|---|---|---|---|
| ECR 1 | Ppar-α | −12980 | AGATCA | |
| [−12997−−12808] | Sp1 | −12884 | CCCACC | |
| ECR 2 | Ppar-α | −11788 | AGGTCA | |
| [−11874−−11709 | TSS A | −9602 | CATCAGATA (bp 59,659,083) | |
| ECR 3 | CpG island | −9622−−9169 | CpG repeats | |
| [−9592−−9382] | ERE | −7919 | TA-GGTCA-GGA-TGACC-TT | |
| Consensus | ||||
| Transcript A | ||||
| ECR 5 | TSS B (+1) | +1 | CCCAGTGTG (bp 59,649,481) | |
| (−190−+884) | ||||
| Transcript B: NM_008819 | TSS B | +110 | TTCCTTCTG (bp 59,649,379) | |
| Ap1 | +429 | CGTTAGTCACT | ||
| ERE | +790 | TG-AGGCA-GGC-TGACC-AG | ||
| ERE | +813 | CA-GGGCA-CGG-GGACC-TG | ||
| ERE | +5150 | TT-AGTCA-TGT-TGGCT-GC | ||
| ECR 6 | ||||
| (+4848−+5243) | Gata | +5441 | CTTTTATCTTC | |
| Transcript C | TSS C | +5147 | GCTGATCTC (bp 59,644,334) | |
| ERE | +5192 | TG-GGTTA-CAT-GGACC-CC | ||
| ERE | +5780 | GG-TGTCA-AGG-TGACC-TA | ||
| ERE | +5860 | AA-GACCA-CTG-TGACC-TC | ||
| ERE | +7961 | CA-GGTGG-GCC-TGACC-CT |
For both human and mouse PEMT genes motif chromosomal position is displayed relative to transcription start site B (+1) based on the literature defined major TSS for each (36, 37); +/− indicates upstream or downstream orientation, respectively. For the human +1 site, this corresponds to bp 17,426,514 of ch17. For mouse this corresponds to bp 59,649,481 on ch11 (UCSC May 2004 Release of the Mus musculus genome, mm5), which corresponds to bp 59,853,136 in the current release of the M. musculus genome (mm8). The mouse and human PEMT gene promoters are highly conserved in six distinct evolutionarily conserved regions (ECR 1-ECR 6). ECR 4 in mouse Pemt has been translocated to a different chromosome (data not shown). There are conserved estrogen response elements and transcription factor binding sites within ECR 5 and 6 in mouse and human proximal promoter regions. The human and murine PEMT gene contains additional EREs, including a consensus ERE in a distal promoter/enhancer region approximately 7.5kb from TSS B. The human promoter A region contains a CpG island and an experimentally validated 600 bp RNA polymerase II binding site (Pol 9419, P<0.00001) (Figure 3, Table 2) (38). ECR browser, rVista TFBS search engine, Dragon ERE Finder, and TRANSFAC were used to identify ECRs, EREs, and TFBS, respectively. Underline indicates nucleotides that differ from the estrogen response element consensus sequence. Italicized letters indicate transcription start sites for each proximal promoter region denoted A, B, C.
Motif Abbrevations for gene names are from LocusLink (www.ncbi.nlm.nih.gov/LocustLink/list.cgi; last accessed 01/02/07).
Identification of an evolutionarily shared promoter framework in the PEMT gene
In orthologous promoters, elements important for transcriptional regulation may be expected to be conserved during evolution (39). Evolutionarily conserved regions (ECRs) were identified from two species (Homo sapiens and Mus musculus) in the PEMT promoter(s) by using the comparative genomics tool ECR Browser (as described in Experimental Procedures). We determined that the PEMT gene promoters contain six evolutionarily conserved regions within the region encompassing the three TSS predicted for the PEMT gene in each species (Fig. 3, Table 2). The region containing this cluster of ECRs is ∼18 kb for each organism with ECR 6 overlapping the TSS of the shortest predicted transcript for each and ECR 1 being 4 kb upstream of NM_148172 (human) and 9.7 kb of transcript A (mouse). No other ECRs were found in the 15 kb upstream of ECR 1 in the mouse or within 29 kb of the human ECR 1. Within these conserved regions, predicted transcription factor binding sites are likely to be conserved (Table 2).
EREs previously identified in human genes often have counterparts at similar positions in their mouse orthologs. Because conservation of newly identified EREs between the two species may indicate a functional role of these elements, we searched for the presence of conserved EREs with less than 9 kb difference in distance from their respective transcriptional start sites. For estrogen binding-site predictions we used the previously described ERE model (35) and AP1 and SP1 binding-site position weight matrices from the TRANSFAC database (40). We identified one perfect consensus estrogen response element ∼7.5 kb from transcription start site B, TSS (+1), in both the human and murine PEMT promoters. In the murine Pemt gene promoter region we identified seven imperfect EREs differing from the consensus by 1−3 nucleotides, three of which are located in ECRs in close proximity to TSS B and TSS C (Fig. 3, Table 2). The human PEMT promoter region contains eight imperfect estrogen response motifs, three of which occur in an evolutionarily conserved region in close proximity to Promoter B (TSS B) and Promoter C (TSS C) (Fig. 3, Table 2). We also found several AP1, SP1 and FOXA1 sites in these evolutionarily conserved regions.
DISCUSSION
Choline is derived not only from the diet, but as well from de novo synthesis of phosphatidylcholine catalyzed by PEMT (2). We have previously reported that when deprived of dietary choline, men and postmenopausal women are more likely to develop fatty liver or muscle damage compared to premenopausal women (19). In the present study, we observed that estrogen, at doses bracketing physiological concentrations in humans (0−100 nmol/L), caused a marked up-regulation in PEMT mRNA expression and enzyme activity, and that the PEMT gene has motifs (41) that may act as EREs in its promoter regions. This observation may help to explain why premenopausal women usually do not develop organ dysfunction when fed a diet low in choline (19); they have estrogen-induced increased capacity for endogenous biosynthesis of the choline moiety.
Pregnancy and lactation are times when demand for choline is especially high. Indeed, transport of choline from mother to fetus (42, 43) depletes maternal plasma choline in humans (44). Thus, despite an apparent enhanced capacity to synthesize choline, the demand for this nutrient is so high that stores are depleted (20). Because milk contains a great deal of choline, lactation further increases maternal demand for choline, resulting in further depletion of tissue stores (20, 45). Pemt −/− mice abort pregnancies around 9−10 days gestation unless fed supplemental choline (personal observation). Women in the United States vary enough in dietary choline intake (from <300 mg/d to >500 mg/d) to influence the risk that they will have a baby with a birth defect; low dietary intake of choline during pregnancy was associated with a 4-fold increased risk of giving birth to an infant with a NTD (9) and 1.5-fold increased risk for orofacial clefts (46). Choline nutriture during pregnancy is especially important because it influences brain development in the fetus (3–5, 47–56). These observations suggest that women depend on high rates of endogenous biosynthesis of choline induced by estrogen, as well as on dietary intake of choline to sustain normal pregnancy. It is biologically plausible that, during evolution, appropriate mechanisms were developed to ensure that young women are less susceptible to dietary choline deficiency and have adequate stores of choline prior to, and during pregnancy.
Our observation that PEMT gene expression in mice and humans is induced by physiological concentrations of estrogen in humans (0−100 nmol/L) (57) correlates with the previous finding that estrogen increases PEMT activity in liver of the bird (25, 58, 59), the rat liver (24), and the rat pituitary (23, 60). The classic actions of estrogen occur through its receptors ERα and ERβ, which bind as homodimers or heterodimers to EREs in the promoters of many estrogen-responsive genes (41). The consensus ERE (PuGGTCAnnnTGACCPy) is an inverted palindromic sequence separated by three intervening nucleotides (41). This motif is usually surrounded by 50 nucleotide-flanking regions that contain other transcription factor binding sites (61). Imperfect ERE half-site motifs (ERE1/2) also bind with ERα and ERβ (62–64). It is noteworthy that the majority of known estrogen responsive genes contain imperfect EREs that differ from the consensus sequence (65). EREs are usually found within −10 to + 5 kb of transcriptional start sites, and ∼1% of elements appear to be conserved in the flanking regions of orthologous human and mouse genes (66). Functional evolutionarily conserved EREs are most abundant in the 0 to + 1 kb region around transcriptional start sites (66). Estrogen also can mediate its effect on proximal gene promoters from distances up to 10 kb by acting as an enhancer element (66). This seems to be especially true when estrogen response motifs are located in the proximal gene region. EREs that are located far away from the promoter of a single gene can interact synergistically with EREs in the proximal promoter by DNA looping (67).
In mice, we found three conserved EREs at position + 790 bp and + 813 bp downstream of the TSS B and + 66 bp upstream of TSS C. In humans, we found three conserved EREs at positions + 725 bp and + 741 bp downstream of TSS B and + 466 bp of TSS C (Table 2). One of the ERE1/2 we found was embedded in an Alu repeat sequence. Alu elements, the most abundant interspersed repeats in the human genome, have repeatedly been found to be involved in gene rearrangements in humans (68). Ubiquitous presence of Alu repeats and their specific properties suggest a number of functions for the Alu elements, one of which is the introduction of functional estrogen response elements into gene promoters (68). Finally, not all genes that are regulated by estrogen contain an ERE. Estrogen may regulate these target genes through interactions with other transcription factors such as activator protein 1, AP1 (25, 58, 59), NF-κB (60), or specific factor, Sp1 (41), and FOXA1 (69, 70). We found several AP1, FOXA1, and SP1 sites in both conserved and nonconserved promoter regions.
It was previously reported that the murine Pemt gene encodes one major transcript and that transcription was initiated from a single promoter (37). Both EST and CAGE analysis suggests the existence of three transcripts for the murine Pemt gene and for its human ortholog. A cluster of ECRs within the putative promoter region contains the multiple TSS for each species and many conserved predicted binding sites for transcription factors within these ECRs and throughout the sequence analyzed above. Based on multiple similarities in gene and protein structure, it is reasonable to predict that the murine and human PEMT genes are regulated in a similar manner.
This report is the first to identify a possible mechanism through which estrogen induces PEMT expression and may explain the observation that premenopausal women are relatively resistant to choline deficiency. Future studies may include measuring the abundance of the three PEMT gene transcripts in human and murine hepatocytes cultured in the presence and absence of 17-β-estradiol in order to determine whether estrogen treatment causes alternative promoter use and/or splicing. We are currently determining whether recently identified single nucleotide polymorphisms that increase women's dietary requirement for choline (71) work by altering estrogen-mediated induction of the PEMT gene.
Acknowledgments
The authors thank Drs. Jianbei Deng, Yiwei Rong, Shuli Wang, and Xiaonan Zhu for their technical assistance. We thank Admet Technologies for their generous gift of human hepatocytes. This work was supported by NIH grants to SZ (DK55865, AG09525). Support for this work was also provided by grants from the NIH to the UNC Clinical Nutrition Research Unit (DK56350) and the Center for Environmental Health and Susceptibility (ES10126). None of the authors have conflicts of interest.
REFERENCES
- 1.Institute of Medicine, and National Academy of Sciences USA . Dietary Reference Intakes for Folate, Thiamin, Riboflavin, Niacin, Vitamin B12, Panthothenic Acid, Biotin, and Choline. Vol. 1. National Academy Press; Washington D.C., USA: 1998. Choline. pp. 390–422. [PubMed] [Google Scholar]
- 2.Zeisel SH. Choline: critical role during fetal development and dietary requirements in adults. Annu. Rev. Nutr. 2006;26:229–250. doi: 10.1146/annurev.nutr.26.061505.111156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albright CD, Tsai AY, Friedrich CB, Mar MH, Zeisel SH. Choline availability alters embryonic development of the hippocampus and septum in the rat. Brain. Res. Dev. Brain. Res. 1999;113:13–20. doi: 10.1016/s0165-3806(98)00183-7. [DOI] [PubMed] [Google Scholar]
- 4.Craciunescu CN, Albright CD, Mar MH, Song J, Zeisel SH. Choline availability during embryonic development alters progenitor cell mitosis in developing mouse hippocampus. J. Nutr. 2003;133:3614–3618. doi: 10.1093/jn/133.11.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mellott TJ, Williams CL, Meck WH, Blusztajn JK. Prenatal choline supplementation advances hippocampal development and enhances MAPK and CREB activation. FASEB J. 2004;18:545–547. doi: 10.1096/fj.03-0877fje. [DOI] [PubMed] [Google Scholar]
- 6.Loy R, Heyer D, Williams CL, Meck WH. Choline-induced spatial memory facilitation correlates with altered distribution and morphology of septal neurons. Adv. Exp. Med. Biol. 1991;295:373–382. doi: 10.1007/978-1-4757-0145-6_21. [DOI] [PubMed] [Google Scholar]
- 7.Meck WH, Williams CL. Metabolic imprinting of choline by its availability during gestation: Implications for memory and attentional processing across the lifespan. Neurosci. Biobehav. Rev. 2003;27:385–399. doi: 10.1016/s0149-7634(03)00069-1. [DOI] [PubMed] [Google Scholar]
- 8.Fisher MC, Zeisel SH, Mar MH, Sadler TW. Inhibitors of choline uptake and metabolism cause developmental abnormalities in neurulating mouse embryos. Teratology. 2001;64:114–122. doi: 10.1002/tera.1053. [DOI] [PubMed] [Google Scholar]
- 9.Shaw GM, Carmichael SL, Yang W, Selvin S, Schaffer DM. Periconceptional dietary intake of cho-line and betaine and neural tube defects in offspring. Am. J. Epidemiol. 2004;160:102–109. doi: 10.1093/aje/kwh187. [DOI] [PubMed] [Google Scholar]
- 10.da Costa KA, Gaffney CE, Fischer LM, Zeisel SH. Choline deficiency in mice and humans is associated with increased plasma homocysteine concentration after a methionine load. Am. J. Clin. Nutr. 2005;81:440–444. doi: 10.1093/ajcn.81.2.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.da Costa KA, Badea M, Fischer LM, Zeisel SH. Elevated serum creatine phosphokinase in cholinedeficient humans: mechanistic studies in C2C12 mouse myo-blasts. Am. J. Clin. Nutr. 2004;80:163–170. doi: 10.1093/ajcn/80.1.163. [DOI] [PubMed] [Google Scholar]
- 12.Guba S, Fink L, Fonseca V. Hyperhomocysteinemia. An emerging and important risk factor for thromboembolic and cardiovascular disease. Am. J. Clin. Pathol. 1996;106:709–722. doi: 10.1093/ajcp/106.6.709. [DOI] [PubMed] [Google Scholar]
- 13.Zeisel SH, Mar MH, Howe JC, Holden JM. Concentrations of choline-containing compounds and betaine in common foods. J. Nutr. 2003;133:1302–1307. doi: 10.1093/jn/133.5.1302. [DOI] [PubMed] [Google Scholar]
- 14.Zeisel SH, Mar M-H, Howe JC, Holden JM. Erratum: Concentrations of choline-containing compounds and betaine in common foods. J. Nutr. 2003;133:1302–1307. doi: 10.1093/jn/133.5.1302. [DOI] [PubMed] [Google Scholar]; J. Nutr. 133:2918–2919. [Google Scholar]
- 15.Fischer LM, Scearce JA, Mar MH, Patel JR, Blanchard RT, Macintosh BA, Busby MG, Zeisel SH. Ad libitum choline intake in healthy individuals meets or exceeds the proposed adequate intake level. J. Nutr. 2005;135:826–829. doi: 10.1093/jn/135.4.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vance DE, Walkey CJ, Agellon LB. Why has phosphatidylethanolamine N-methyltransferase survived in evolution? Biochem. Soc. Trans. 1998;26:337–340. doi: 10.1042/bst0260337. [DOI] [PubMed] [Google Scholar]
- 17.Bremer J, Greenberg D. Methyl transferring enzyme system of microsomes in the biosynthesis of lecithin (phosphatidylcholine). Biochim. Biophys. Acta. 1961;46:205–216. [Google Scholar]
- 18.Vance DE, Walkey CJ, Cui Z. Phosphatidylethanolamine N-methyltransferase from liver. Biochim. Biophys. Acta. 1997;1348:142–150. doi: 10.1016/s0005-2760(97)00108-2. [DOI] [PubMed] [Google Scholar]
- 19.Fischer L, daCosta K, Kwock L, Stewart P, Lu T-S, Stabler S, Allen R, Zeisel S. Gender and menopausal status influence human dietary requirements for the nutrient choline. Am. J. Clin. Nutr. 2007 doi: 10.1093/ajcn/85.5.1275. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeisel SH, Mar M-H, Zhou Z-W, da Costa K-A. Pregnancy and lactation are associated with diminished concentrations of choline and its metabolites in rat liver. J. Nutr. 1995;125:3049–3054. doi: 10.1093/jn/125.12.3049. [DOI] [PubMed] [Google Scholar]
- 21.Tessitore L, Sesca E, Greco M, Pani P, Dianzani M. Sexually differentiated response to choline in choline deficiency and ethionine intoxication. Int. J. Exper. Path. 1995;76:125–129. [PMC free article] [PubMed] [Google Scholar]
- 22.Noga AA, Vance DE. A gender-specific role for phosphatidylethanolamine N-methyltransferase-derived phosphatidylcholine in the regulation of plasma high density and very low density lipoproteins in mice. J. Biol. Chem. 2003;278:21851–21859. doi: 10.1074/jbc.M301982200. [DOI] [PubMed] [Google Scholar]
- 23.Drouva SV, LaPlante E, Leblanc P, Bechet JJ, Clauser H, Kordon C. Estradiol activates methylating enzyme(s) involved in the conversion of phosphatidylethanolamine to phosphatidylcholine in rat pituitary membranes. Endocrinology. 1986;119:2611–2622. doi: 10.1210/endo-119-6-2611. [DOI] [PubMed] [Google Scholar]
- 24.Young DL. Estradiol- and testosterone-induced alterations in phosphatidylcholine and triglyceride synthesis in hepatic endoplasmic reticulum. J. Lipid Res. 1971;12:590–595. [PubMed] [Google Scholar]
- 25.Vigo C, Vance DE. Effect of diethylstilboestrol on phosphatidylcholine biosynthesis in the liver of roosters. Biochem. Soc. Trans. 1981;9:98–99. doi: 10.1042/bst0090098. [DOI] [PubMed] [Google Scholar]
- 26.Morita M, Watanabe Y, Akaike T. Protective effect of hepatocyte growth factor on interferon-gamma-induced cytotoxicity in mouse hepatocytes. Hepatology. 1995;21:1585–1593. [PubMed] [Google Scholar]
- 27.Soto AM, Sonnenschein C. The role of estrogens on the proliferation of human breast tumor cells (MCF-7). J. Steroid. Biochem. 1985;23:87–94. doi: 10.1016/0022-4731(85)90265-1. [DOI] [PubMed] [Google Scholar]
- 28.Berthois Y, Katzenellenbogen JA, Katzenellenbogen BS. Phenol red in tissue culture media is a weak estrogen: implications concerning the study of estrogen-responsive cells in culture. Proc. Natl. Acad. Sci. U. S. A. 1986;83:2496–2500. doi: 10.1073/pnas.83.8.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.LeCluyse EL, Alexandre E, Hamilton GA, Viollon-Abadie C, Coon DJ, Jolley S, Richert L. Isolation and culture of primary human hepatocytes. Methods Mol. Biol. 2005;290:207–229. doi: 10.1385/1-59259-838-2:207. [DOI] [PubMed] [Google Scholar]
- 31.Kanamori H, Krieg S, Mao C, Di Pippo VA, Wang S, Zajchowski DA, Shapiro DJ. Proteinase inhibitor 9, an inhibitor of granzyme B-mediated apoptosis, is a primary estrogen-inducible gene in human liver cells. J. Biol. Chem. 2000;275:5867–5873. doi: 10.1074/jbc.275.8.5867. [DOI] [PubMed] [Google Scholar]
- 32.Audubert F, Vance DE. Pitfalls and problems in studies on the methylation of phosphatidylethanolamine. J. Biol. Chem. 1983;258:10695–10701. [PubMed] [Google Scholar]
- 33.Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T, Fukuda S, Sasaki D, Podhajska A, Harbers M, Kawai J, Carninci P, Hayashizaki Y. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bajic VB, Tan SL, Chong A, Tang S, Strom A, Gustafsson JA, Lin CY, Liu ET. Dragon ERE Finder version 2: A tool for accurate detection and analysis of estrogen response elements in vertebrate genomes. Nucleic Acids Res. 2003;31:3605–3607. doi: 10.1093/nar/gkg517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shields DJ, Agellon LB, Vance DE. Structure, expression profile and alternative processing of the human phosphatidylethanolamine N-methyltransferase (PEMT) gene. Biochim. Biophys. Acta. 2001;1532:105–114. doi: 10.1016/s1388-1981(01)00122-6. [DOI] [PubMed] [Google Scholar]
- 37.Walkey CJ, Cui Z, Agellon LB, Vance DE. Characterization of the murine phosphatidylethanolamine N-methyltransferase-2 gene. J. Lipid Res. 1996;37:2341–2350. [PubMed] [Google Scholar]
- 38.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 39.Elnitski L, Hardison RC, Li J, Yang S, Kolbe D, Eswara P, O'Connor MJ, Schwartz S, Miller W, Chiaromonte F. Distinguishing regulatory DNA from neutral sites. Genome. Res. 2003;13:64–72. doi: 10.1101/gr.817703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heinemeyer T, Chen X, Karas H, Kel AE, Kel OV, Liebich I, Meinhardt T, Reuter I, Schacherer F, Wingender E. Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res. 1999;27:318–322. doi: 10.1093/nar/27.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walter P, Green S, Greene G, Krust A, Bornert JM, Jeltsch JM, Staub A, Jensen E, Scrace G, Waterfield M, et al. Cloning of the human estrogen receptor cDNA. Proc. Natl. Acad. Sci. U. S. A. 1985;82:7889–7893. doi: 10.1073/pnas.82.23.7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sweiry JH, Yudilevich DL. Characterization of choline transport at maternal and fetal interfaces of the perfused guinea-pig placenta. J. Physiol. 1985;366:251–266. doi: 10.1113/jphysiol.1985.sp015795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sweiry JH, Page KR, Dacke CG, Abramovich DR, Yudilevich DL. Evidence of saturable uptake mechanisms at maternal and fetal sides of the perfused human placenta by rapid paired-tracer dilution: studies with calcium and choline. J. Devel. Physiol. 1986;8:435–445. [PubMed] [Google Scholar]
- 44.McMahon KE, Farrell PM. Measurement of free choline concentrations in maternal and neonatal blood by micropyrolysis gas chromatography. Clin. Chim. Acta. 1985;149:1–12. doi: 10.1016/0009-8981(85)90267-0. [DOI] [PubMed] [Google Scholar]
- 45.Holmes-McNary M, Cheng WL, Mar MH, Fussell S, Zeisel SH. Choline and choline esters in human and rat milk and infant formulas. Am. J. Clin. Nutr. 1996;64:572–576. doi: 10.1093/ajcn/64.4.572. [DOI] [PubMed] [Google Scholar]
- 46.Shaw GM, Carmichael SL, Laurent C, Rasmussen SA. Maternal nutrient intakes and risk of orofacial clefts. Epidemiology. 2006;17:285–291. doi: 10.1097/01.ede.0000208348.30012.35. [DOI] [PubMed] [Google Scholar]
- 47.Albright CD, Friedrich CB, Brown EC, Mar MH, Zeisel SH. Maternal dietary choline availability alters mitosis, apoptosis and the localization of TOAD-64 protein in the developing fetal rat septum. Brain. Res. Dev. Brain Res. 1999;115:123–129. doi: 10.1016/s0165-3806(99)00057-7. [DOI] [PubMed] [Google Scholar]
- 48.Albright CD, Mar MH, Craciunescu CN, Song J, Zeisel SH. Maternal dietary choline availability alters the balance of netrin-1 and DCC neuronal migration proteins in fetal mouse brain hippocampus. Brain Res. Dev. Brain Res. 2005;159:149–154. doi: 10.1016/j.devbrainres.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Albright CD, Mar MH, Friedrich CB, Brown EC, Zeisel SH. Maternal choline availability alters the localization of p15Ink4B and p27Kip1 cyclin-dependent kinase inhibitors in the developing fetal rat brain hippocampus. Dev. Neurosci. 2001;23:100–106. doi: 10.1159/000048701. [DOI] [PubMed] [Google Scholar]
- 50.Albright CD, Siwek DF, Craciunescu CN, Mar MH, Kowall NW, Williams CL, Zeisel SH. Choline availability during embryonic development alters the localization of calretinin in developing and aging mouse hippocampus. Nutr. Neurosci. 2003;6:129–134. doi: 10.1080/1028415031000084418. [DOI] [PubMed] [Google Scholar]
- 51.Albright CD, Tsai AY, Mar M-H, Zeisel SH. Choline availability modulates the expression of TGFβ1 and cytoskeletal proteins in the hippocampus of developing rat brain. Neurochem. Res. 1998;23:751–758. doi: 10.1023/a:1022411510636. [DOI] [PubMed] [Google Scholar]
- 52.Niculescu MD, Craciunescu CN, Zeisel SH. Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. FASEB J. 2006;20:43–49. doi: 10.1096/fj.05-4707com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pyapali G, Turner D, Williams C, Meck W, Swartzwelder HS. Prenatal choline supplementation decreases the threshold for induction of long-term potentiation in young adult rats. J. Neurophysiol. 1998;79:1790–1796. doi: 10.1152/jn.1998.79.4.1790. [DOI] [PubMed] [Google Scholar]
- 54.Meck W, Williams C. Perinatal choline supplementation increases the threshold for chunking in spatial memory. Neuroreport. 1997;8:3053–3059. doi: 10.1097/00001756-199709290-00010. [DOI] [PubMed] [Google Scholar]
- 55.Meck WH, Smith RA, Williams CL. Pre- and postnatal choline supplementation produces long-term facilitation of spatial memory. Dev. Psychobiol. 1988;21:339–353. doi: 10.1002/dev.420210405. [DOI] [PubMed] [Google Scholar]
- 56.Meck WH, Williams CL. Metabolic imprinting of choline by its availability during gestation: implications for memory and attentional processing across the lifespan. Neurosci. Biobehav. Rev. 2003;27:385–399. doi: 10.1016/s0149-7634(03)00069-1. [DOI] [PubMed] [Google Scholar]
- 57.Peck JD, Hulka BS, Poole C, Savitz DA, Baird D, Richardson BE. Steroid hormone levels during pregnancy and incidence of maternal breast cancer. Cancer Epidemiol. Biomarkers. Prev. 2002;11:361–368. [PubMed] [Google Scholar]
- 58.Vigo C, Vance DE. Effect of diethylstilboestrol on phosphatidylcholine biosynthesis and choline metabolism in the liver of roosters. Bioche. J. 1981;200:321–326. doi: 10.1042/bj2000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vigo C, Paddon HB, Millard FC, Pritchard PH, Vance DE. Diethylstilbestrol treatment modulates the enzymatic activities of phosphatidylcholine biosynthesis in rooster liver. Biochimica. et Biophysica. Acta. 1981;665:546–550. doi: 10.1016/0005-2760(81)90269-1. [DOI] [PubMed] [Google Scholar]
- 60.Drouva SV, Rerat E, Leblanc P, Laplante E, Kordon C. Variations of phospholipid methyltransferase(s) activity in the rat pituitary: estrous cycle and sex differences. Endocrinology. 1987;121:569–574. doi: 10.1210/endo-121-2-569. [DOI] [PubMed] [Google Scholar]
- 61.Tang S, Tan SL, Ramadoss SK, Kumar AP, Tang MH, Bajic VB. Computational method for discovery of estrogen responsive genes. Nucleic Acids Res. 2004;32:6212–6217. doi: 10.1093/nar/gkh943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lopez D, Sanchez MD, Shea-Eaton W, McLean MP. Estrogen activates the high-density lipoprotein receptor gene via binding to estrogen response elements and interaction with sterol regulatory element binding protein-1A. Endocrinology. 2002;143:2155–2168. doi: 10.1210/endo.143.6.8855. [DOI] [PubMed] [Google Scholar]
- 63.Agarwal A, Yeung WS, Lee KF. Cloning and characterization of the human oviduct-specific glycoprotein (HuOGP) gene promoter. Mol. Hum. Reprod. 2002;8:167–175. doi: 10.1093/molehr/8.2.167. [DOI] [PubMed] [Google Scholar]
- 64.Xie T, Ho SL, Ramsden D. Characterization and implications of estrogenic down-regulation of human catechol-O-methyltransferase gene transcription. Mol. Pharmacol. 1999;56:31–38. doi: 10.1124/mol.56.1.31. [DOI] [PubMed] [Google Scholar]
- 65.Driscoll MD, Sathya G, Muyan M, Klinge CM, Hilf R, Bambara RA. Sequence requirements for estrogen receptor binding to estrogen response elements. J. Biol. Chem. 1998;273:29321–29330. doi: 10.1074/jbc.273.45.29321. [DOI] [PubMed] [Google Scholar]
- 66.Bourdeau V, Deschenes J, Metivier R, Nagai Y, Nguyen D, Bretschneider N, Gannon F, White JH, Mader S. Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol. Endocrinol. 2004;18:1411–1427. doi: 10.1210/me.2003-0441. [DOI] [PubMed] [Google Scholar]
- 67.Sathya G, Li W, Klinge CM, Anolik JH, Hilf R, Bambara RA. Effects of multiple estrogen responsive elements, their spacing, and location on estrogen response of reporter genes. Mol. Endocrinol. 1997;11:1994–2003. doi: 10.1210/mend.11.13.0039. [DOI] [PubMed] [Google Scholar]
- 68.Norris J, Fan D, Aleman C, Marks JR, Futreal PA, Wiseman RW, Iglehart JD, Deininger PL, McDonnell DP. Identification of a new subclass of Alu DNA repeats which can function as estrogen receptor-dependent transcriptional enhancers. J. Biol. Chem. 1995;270:22777–22782. doi: 10.1074/jbc.270.39.22777. [DOI] [PubMed] [Google Scholar]
- 69.Laganiere J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguere V. From the Cover: Location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11651–11656. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. Chromo-some-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 71.Da Costa KA, Kozyreva OG, Song J, Galanko JA, Fischer LM, Zeisel SH. Common genetic polymorphisms affect the human requirement for the nutrient choline. FASEB J. 2006;20:1336–1344. doi: 10.1096/fj.06-5734com. [DOI] [PMC free article] [PubMed] [Google Scholar]


