Summary
Biofilms of Bacillus subtilis consist of long chains of cells that are held together in bundles by an extracellular matrix of exopolysaccharide and the protein TasA. The exopolysaccharide is produced by enzymes encoded by the epsA-O operon and the gene encoding TasA is located in the yqxM-sipW-tasA operon. Both operons are under the control of the repressor SinR. Derepression is mediated by the antirepressor SinI, which binds to SinR with a 1:1 stoichiometry. Paradoxically, in medium promoting derepression of the matrix operons, the overall concentration of SinR in the culture greatly exceeded that of SinI. We show that under biofilm-promoting conditions sinI, which is under the control of the response regulator Spo0A, was expressed only in a small subpopulation of cells, whereas sinR was expressed in almost all cells. Activation of Spo0A is known to be subject to a bistable switch, and we infer that SinI reaches levels sufficient to trigger matrix production only in the subpopulation of cells in which Spo0A is active. Additionally, evidence suggests that sinI is expressed at intermediate, but not low or high, levels of Spo0A activity, which may explain why certain nutritional conditions are more effective in promoting biofilm formation than others.
Introduction
Most bacteria are capable of forming surface-associated, architecturally complex communities of cells, which are known as biofilms (Kolter & Greenberg, 2006, O’Toole & Kaplan, 2000, Stoodley et al., 2002). Biofilms assemble on solid surfaces or as pellicles at air/liquid interfaces. A hallmark of biofilms is the presence of an extracellular matrix that holds the cells together (Branda et al., 2005). The matrix typically consists of exopolysaccharides and proteins and sometimes nucleic acid (Sutherland, 2001, Whitchurch et al., 2002). Remarkably, the mechanisms governing the production of the matrix differ markedly from bacterium to bacterium, an observation that suggests that the capacity to assemble into communities arose independently many times in the microbial world (Branda et al., 2005, Davies et al., 1998). Here we are concerned with the mechanisms governing the production of the extracellular matrix in the spore-forming bacterium Bacillus subtilis.
Wild strains of B. subtilis, such as the 3610 strain used in the present study, form elaborate biofilms in which spore formation takes place preferentially at the tips of aerial structures that protrude from the surface of the community (Branda et al., 2001). Biofilm formation and sporulation are also connected in that both processes are dependent on Spo0A, the master regulator for entry into sporulation (Branda et al., 2001, Hamon & Lazazzera, 2001, Sonenshein, 2000). Indeed, and as we will argue here, expression of genes involved in matrix production may be a prelude to the process of spore formation. In B. subtilis, the matrix consists of an exopolysaccharide, which is produced under the direction of the 15-gene operon epsA-O (henceforth simply eps) (Branda et al., 2001) and the secreted protein TasA, which is encoded within the yqxM-sipW-tasA operon (henceforth simply yqxM) (Branda et al., 2006). Both operons are under the direct negative control of the repressor SinR (Chu et al., 2006, Kearns et al., 2005). Derepression under conditions promoting biofilm formation occurs, at least in part, through the action of SinI, an antagonist of SinR (Bai et al., 1993, Kearns et al., 2005). SinI binds to the SinR repressor in a one-to-one stoichiometry, thereby preventing the repressor from binding to DNA (Lewis et al., 1998, Lewis et al., 1996).
The starting point for this investigation was the puzzling observation that the level of expression of the sinR gene, and as a consequence, the level of accumulation of its product, is much higher than that of sinI. If SinI acts at a one-to-one stoichiometry with SinR, then how is derepression of the eps and yqxM operons achieved when the antirepressor is present at much lower concentrations than the repressor? Here we show that sinI is expressed at a high level but only in a small subpopulation of the cells, leading to the hypothesis that in these sinI-expressing cells the antirepressor reaches a threshold concentration sufficient to trigger depression. We further suggest that at the early stages of biofilm formation a subpopulation of eps- and yqxM-expressing cells supplies matrix to the entire community.
Results
Strategy
The goal of this work was to study the role of the SinR-antagonist, SinI, in the derepression of the operons (eps and yqxM) governing extracellular matrix formation. A complication in attempting to do so was that biofilms are an architecturally complex community of tightly packed cells in which it is difficult to carry out quantitative studies on gene expression. To circumvent this problem, we took advantage of the fact that transcription from the promoters for the eps and yqxM operons is strongly induced by one hour after the end of exponential phase growth under conditions in which the cells are uniformly dispersed in shaking cultures in the biofilm-promoting medium MSgg (Kearns et al., 2005, Chu et al., 2006, Branda et al., 2006). Accordingly, and for the purposes of studying the role of SinI in derepression of the eps and yqxM operons, we carried out our experiments with cells in homogeneous suspension in shaking cultures.
SinR is much more abundant than SinI
Based on the 1:1 stoichiometry of SinR and SinI in the heteromeric complex of the two proteins (Lewis et al., 1998, Lewis et al., 1996), the cellular concentration of SinI is expected to be at least as great as that of SinR under conditions in which SinR-controlled genes are derepressed. Indeed, previous Electrophoretic Mobility Shift Assays have shown that the concentration of SinI must be equal to, or in excess of, that of SinR to displace the repressor from its operator (Kearns et al., 2005, Bai et al., 1993). We were therefore puzzled to discover that the level of expression of the sinI gene, as judged by using lacZ fused to the sinI promoter (PsinI-lacZ), was approximately 15-fold lower than that of PsinR-lacZ in cells of the wild strain 3610 growing in a medium (MSgg) that promotes biofilm formation (Branda et al., 2001) (Fig. 1A).
Figure 1. SinI levels greatly exceed that of SinR.
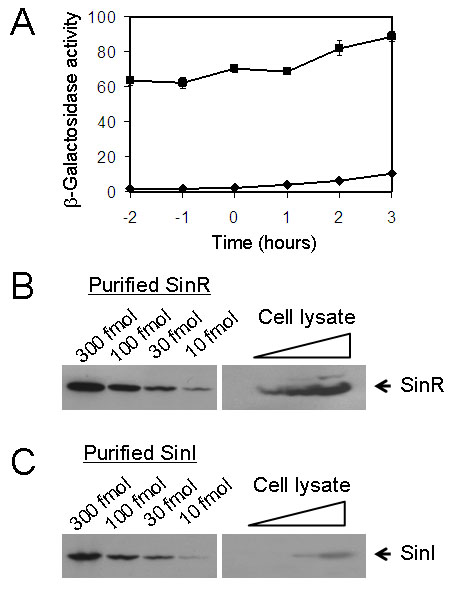
(A) Assays of β-galactosidase specific activity of cells carrying either the PsinR-lacZ (filled squares; strain YC108) or the PsinI-lacZ (filled diamonds; strain YC127) fusion at the amyE locus on the chromosome. Assays were performed for cells grown in MSgg medium and harvested at the indicated times. Time zero refers to the end of exponential phase growth. (B), (C) Quantitative immunoblots of SinR and SinI. Left-hand panels show affinity-purified, recombinant SinR and SinI proteins that were loaded at the indicated amounts. In the right-hand panels, cleared protein lysates prepared from early stationary phase cultures (one hour into stationary phase) were loaded on the same gel in a series of dilutions.
We wondered whether this difference in promoter activity was reflected in the relative cellular concentrations of the two proteins. Accordingly, we carried out quantitative immunoblot analyses with antibodies directed against SinI or SinR, using as standards purified SinR and SinI proteins that had been tagged with histidine (His6-SinI and His6-SinR). The results show that the cellular concentration of SinI (~50 molecules/cell) was eighteen-fold lower than that of SinR (~900 molecules/cell) in cells reaching early stationary phase (Fig. 1B and 1C), the time at which derepression of SinR-controlled genes commences (Kearns et al., 2005). Thus, the concentration of SinI would appear to be too low to counteract SinR effectively.
Cell population heterogeneity could explain the SinI/SinR paradox
A clue to resolving the paradox comes from the transcriptional regulation of sinR and sinI. Previous work has shown that sinR is expressed constitutively from a σA-dependent promoter whereas sinI is under the control of Spo0A, the master regulator for sporulation (Gaur et al., 1988, Shafikhani et al., 2002). Spo0A is known to be active in only a subset of cells in the population (Fujita & Losick, 2005, Chung et al., 1994, Gonzalez-Pastor et al., 2003). Thus, under conditions that activate Spo0A, the culture bifurcates into a subpopulation of Spo0A-ON cells and a subpopulation of Spo0A-OFF cells. This cell population heterogeneity is believed to be governed by a bistable switch involving the phosphorelay and positive feedback loops that control the synthesis and phosphorylation of Spo0A (Veening et al., 2006, Veening et al., 2005, Burbulys et al., 1991, Stephenson & Hoch, 2002). [Actually, this is not a simple ON/OFF switch. Rather, when Spo0A is switched ON, the level of Spo0A increases in a graded manner over time with target promoters responding rapidly or slowly depending on their affinity for Spo0A (Fujita & Losick, 2005, Jiang et al., 2000)). However, for present purposes we will simply consider Spo0A to be subject to an ON/OFF switch.] These considerations led us to hypothesize that SinR is produced more or less uniformly in all cells but that SinI is produced in only the subpopulation of Spo0A-ON cells. In other words, the protein measurements of Figure 1 reflected the relative abundance of SinI and SinR averaged over the entire population. We also hypothesize that in those cells that are producing SinI, the protein antagonist reaches a sufficient concentration to overcome SinR-mediated repression. If so, then it further follows that only some cells in the population produce the polysaccharide and protein components of the extracellular matrix for the entire community. Indeed, earlier work has shown that mutants of the eps and yqxM operons can complement each other extracellularly, a finding that demonstrates that matrix components can be shared between cells (Branda et al., 2006).
sinI is expressed in a subpopulation of cells
To test our hypothesis, we fused the promoters for sinI and sinR to the gene (gfp) for the Green Fluorescent Protein (GFP). The PsinI-gfp and PsinR-gfp fusions were then integrated into the chromosome at the amyE locus of strain 3610, a wild strain of B. subtilis that forms architecturally complex biofilms (Branda et al., 2001). Cells were grown to early stationary phase in MSgg medium and cell samples were prepared for fluorescent microscopy as described in the Experimental Procedures. The results (Fig. 2A, left-hand panel) show that PsinR-gfp was expressed in nearly all cells whereas PsinI-gfp was expressed in only a small number of cells (the fraction of PsinI-gfp-expressing cells being about 0.02; Fig. 2A, middle panel; Fig. 2C). It follows that if only about 2% of the cells are expressing sinI, then in those cells the level of SinI would exceed that of SinR; that is, the expected ratio of SinI to SinR would be roughly 2.8, which we calculate from the ratio of the concentration of SinI to SinR in the total population (1/18) divided by the fraction of cells that are producing SinI (0.02).
Figure 2. Fluorescence microscopy of cells expressing PsinR-gfp and PsinI-gfp.
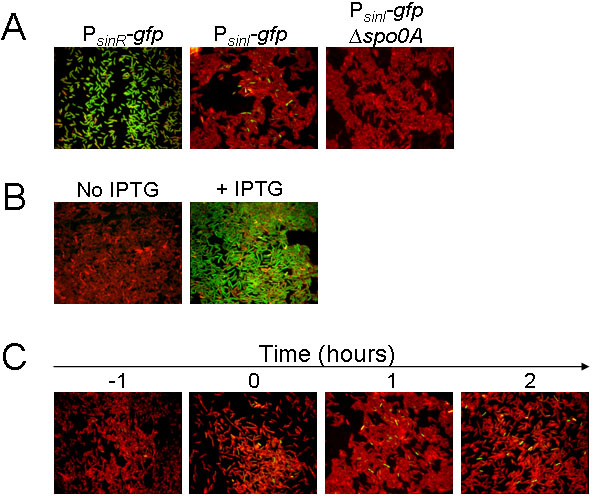
In all panels, cells were treated with the red membrane stain FM4-64. Green is fluorescence from GFP. (A) Shows cells containing PsinR-gfp (YC173) or PsinI-gfp (YC162) integrated into the chromosome at the amyE locus of strain 3610 at one hour after the end of exponential phase in MSgg medium. (B) Shows cells containing PsinI-gfp and a construct (YC170) in which the spo0A gene was replaced with the mutant allele sad67 under the control of an IPTG-inducible promoter. (C) Shows cells containing PsinI-gfp (YC162) at the indicated times before and after the end of exponential phase growth in MSgg medium.
Confirming that sinI expression is indeed under Spo0A control, the results of Fig. 2A (right-hand panel) further show that little or no fluorescence from PsinI-gfp was observed in cells of a spo0A mutant. Furthermore, the following experiment is consistent with the idea that cell population heterogeneity of Spo0A activation is the basis for the expression of sinI in only a subpopulation of cells. For this experiment, we replaced the wild-type spo0A gene with the mutant allele sad67, which encodes a truncated form of Spo0A whose activity does not depend on phosphorylation (Ireton et al., 1993). The sad67 gene was under the control of an IPTG (isopropyl β-D-1-thiogalactopyranoside)-inducible promoter and was introduced into a strain that carried PsinI-gfp at the amyE locus. Therefore, the use of this construct bypassed the normal control mechanisms that govern Spo0A synthesis and phosphorylation. The results of Fig. 2B (left-hand panel) show that when inducer was absent, little or no sinI expression was observed. However, shortly after IPTG was added to the medium (to a final concentration of 1 mM), sinI expression was observed in almost all cells (Fig. 2B, right-hand panel).
Expression of sinI varies in different media
Spo0A is known to be activated to high levels in Difco Sporulation (DS) medium (Fujita & Losick, 2005, Jiang et al., 2000). Yet, B. subtilis 3610 does not form thick pellicles in standing DS medium (data not shown) or architecturally complex colonies on solid DS medium (Fig. 3A, left-hand and middle panels). How can this medium effect be explained? The results of Fig. 3B show that the medium effect is reflected in the expression levels of the extracellular matrix operon eps (left-hand panel) and the sinI gene (right-hand panel). Thus, transcription from the eps promoter as judged by using a Peps-lacZ fusion was four-fold higher in MSgg medium than that in DS medium (Fig. 3B, left-hand panel). Likewise, transcription from the sinI promoter as measured using the PsinI-lacZ fusion described above was higher in MSgg than in DS (Fig. 3B, right-hand panel). We infer that MSgg promotes biofilm formation more robustly than does DS because sinI is expressed at a higher level in the biofilm medium than in the sporulation medium. Reinforcing this view, colonies of cells engineered to overexpress SinI (as a functional fusion with GFP) grown on DS medium partially mimicked the complex colony architecture characteristic of MSgg-grown colonies (Fig. 3A, right-hand panel).
Figure 3. Effect of medium on expression of sinI, eps, and spoIIG.
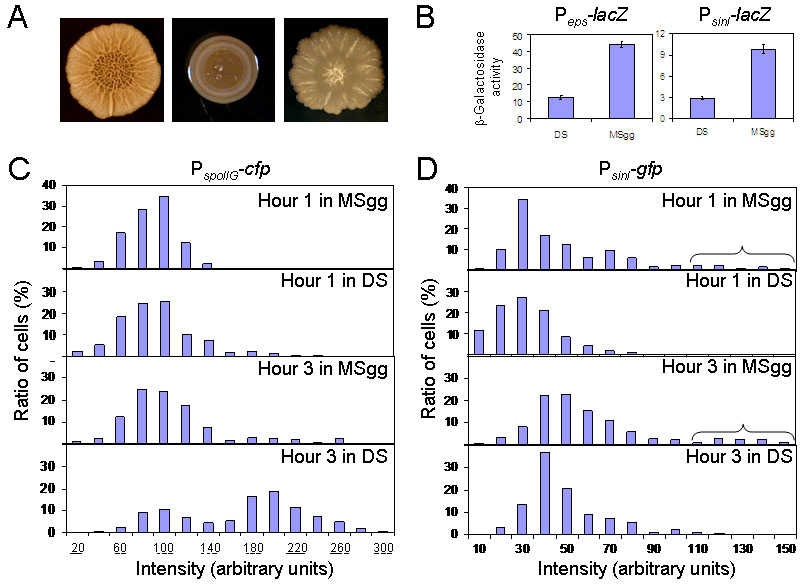
(A) Morphology of colonies of strain 3610 formed on agar plates containing MSgg (left-hand panel) or DS (middle panel) medium. The colony shown in the right-hand panel was formed on solid DS medium by a 3610 derivative overexpressing a functional fusion of SinI with GFP (YC227). (B) Assays of β-galactosidase specific activity for Peps-lacZ (left-hand panel; YC130) and PsinI-lacZ (right-hand panel; YC127) in either DS or MSgg medium. Activities of Peps-lacZ and PsinI-lacZ were measured one hour after the end of exponential phase growth. (C) and (D) Distribution of cells expressing PspoIIG-cfp (columns in 3C; FC476) or PsinI-gfp (columns in 3D; YC162) at various intensities in the indicated media and at the indicated times. Intensities for individual cells were measured using Metamorph. Each bar represents the percentage (Y axis) of cells whose fluorescence intensity was within ±5 in panel C and ±10 in panel D to the unit shown in the X axis, versus total cells in the population. The horizontal bracket in panel D identifies a subpopulation of cells that expressed PsinI-gfp highly.
In light of these findings, we decided to compare the levels of Spo0A-directed gene expression in MSgg and DS media using a cfp fusion to the promoter for the sporulation operon spoIIG (PspoIIG). PspoIIG is known to have a relatively low affinity for the response regulator and is activated only when cells have attained a high level of Spo0A activity (Fujita et al., 2005, Baldus et al., 1994). As seen in Fig. 3C, a substantial subpopulation of cells could be seen by hour three of sporulation in DS medium that were actively expressing the PspoIIG–cfp fusion. In contrast, few cells expressing PspoIIG-cfp at a high level were seen in MSgg medium at either hour one or hour three. Fig. 3D shows that sharply different results were obtained with PsinI-gfp; even at hour three, few cells expressing PsinI-gfp at high levels were seen in DS medium. However, in MSgg medium, a small proportion of cells expressed sinI at high levels at both hour one and hour three.
eps and yqxM are expressed in a subpopulation of cells
Our finding that sinI is highly expressed only in a small population of cells (Fig. 2A) predicts that transcription of the eps and yqxM operons would similarly be limited to a subpopulation of cells. In confirmation of this expectation, fluorescent microscopy experiments using cells carrying a gfp or a cfp fusion to the promoters for eps (Peps-gfp) and yqxM (PyqxM-cfp) showed that both fusions were expressed in only a subset of cells (Fig. 4A and 4B). Furthermore, when a null mutation of sinR was introduced into the fusion-bearing strains, cell population heterogeneity was eliminated and almost all cells in the population were observed to express Peps-gfp and PyqxM-cfp (Fig. 4C and 4D). In contrast, when a sinI mutation was introduced into the fusion-bearing strains, the proportion of cells expressing Peps-gfp and PyqxM-cfp was greatly reduced (Fig. 4E and 4F). We conclude that expression of the matrix operons is subject to cell population heterogeneity and that this heterogeneity is dependent on SinI-mediated relief from repression by SinR.
Figure 4. Fluorescence microscopy of cells expressing Peps-gfp or PyqxM-cfp.
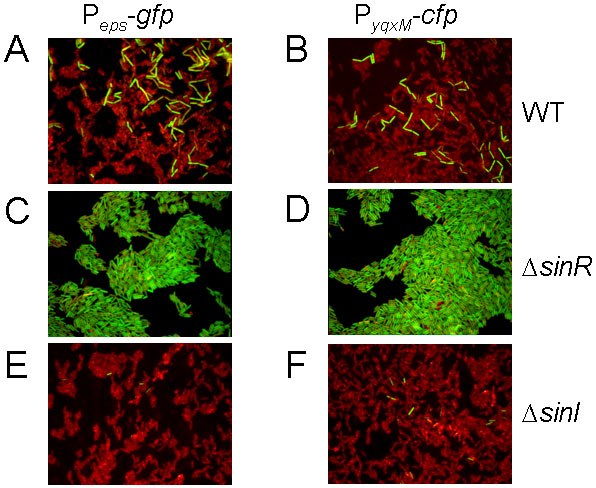
The Peps-gfp or PyqxM-cfp was in a wild background (YC164 or YC189) or in a sinR (YC167 or YC221) or sinI (YC168 or YC190) mutant background. The cells were treated with the red membrane stain FM4-64. Fluorescence from PyqxM-cfp was false-colored green.
Finally, to extend the analysis further, we asked whether in the case of yqxM, the expressing cells corresponded to the same cells that were expressing sinI. To address this question, we carried out a dual-labeling experiment using a fusion of cfp (false-colored green in Fig. 5) to the promoter for sinI and a yfp fusion (false-colored red in Fig. 5) to the promoter for yqxM. The PsinI-cfp fusion was introduced into the chromosome at the thrC locus and the PyqxM-yfp fusion at the amyE locus.
Figure 5. Fluorescence microscopy of cells harboring both PyqxM-yfp and either PsinI-cfp (YC243) or PsinR-cfp (YC244).
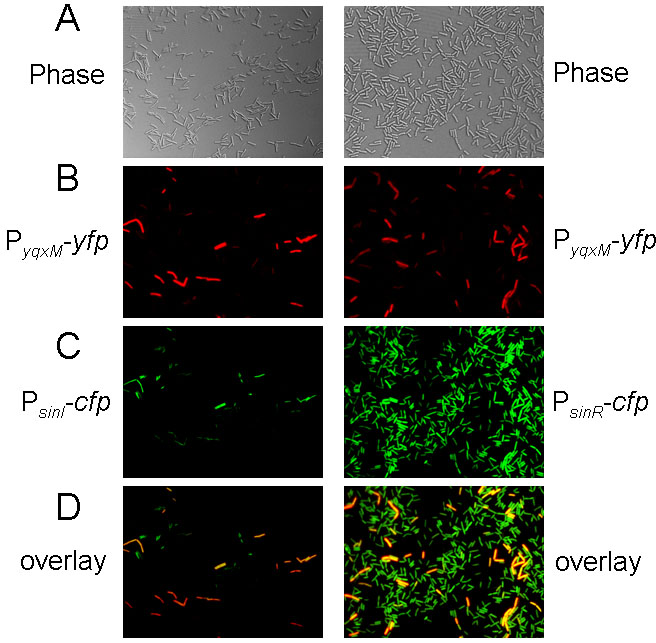
Fluorescence from PyqxM-yfp was false-colored red. Fluorescence from PsinI-cfp and PsinR-cfp was false-colored green. Images in panel D are overlays from the corresponding images in B and C.
The results showed that in cells highly expressing PsinI-cfp, the expression of PyqxM-yfp was also generally very strong (Fig. 5, left-hand column). Nevertheless, some cells could be observed that expressed PyqxM-yfp but seemingly not PsinI-cfp. Conversely, a few cells weakly expressing PsinI-cfp were observed that were not measurably expressing PyqxM-yfp. That some cells exhibited PyqxM-yfp but not PsinI-cfp expression could indicate that transcription of sinI is transient and that transcription from the matrix operon persists after the sinI gene is no longer expressed. Conversely, the cells that were weakly expressing PsinI-cfp but not PyqxM-yfp may simply represent cells that have not yet accumulated enough SinI to derepress the yqxM operon.
As a comparison, we also created a control strain harboring the PyqxM-yfp fusion and a cfp fusion to the promoter for sinR (PsinR-cfp). The results of Fig. 5 (right-hand column) show that PsinR-cfp was expressed in more or less all cells in the population whereas PyqxM-yfp was expressed in only a subpopulation.
In toto the results of the dual-labeling experiments demonstrate that expression of the yqxM matrix operon does indeed exhibit cell population heterogeneity. Moreover, the results are consistent with the idea that this heterogeneity can be wholly, or at least partly, attributed to heterogeneity in the expression of sinI. Nevertheless, because we could not demonstrate a one to one correlation of sinI and yqxM expression in all cases, we do not rule out the possibility that additional as yet undefined levels of regulation contribute to cell population heterogeneity in matrix operon expression.
Discussion
Cell fate is generally thought of as being deterministic. That is, the fate that cells adopt is in most cases governed by the history of the cell or its proximity to inductive signals from other cells. However, an increasing number of cases are now known in which cell fate is controlled by stochastic processes. Examples of cell fate that are governed by stochastic mechanisms are entry into the persister state in E. coli (Balaban et al., 2004, Lewis, 2007), and several cases in B. subtilis (Chung et al., 1994, Dubnau & Losick, 2006, Kearns & Losick, 2005, Maamar et al., 2007), including entry into the state of genetic competence, the choice between swimming and chains of sessile states, and entry into sporulation. The decision to enter sporulation is governed by a noise-driven, bistable switch controlling the accumulation of the transcription factor Spo0A~P (Veening et al., 2005, Veening et al., 2006). A further twist to the sporulation case is that the activation of Spo0A~P not only eventually leads to spore formation but first unleashes a cannibalistic-like process in which cells that have activated Spo0A~P kill sibling cells that have not activated the transcription factor (Gonzalez-Pastor et al., 2003, Ellermeier et al., 2006). The nutrients thereby released stall sporulation. Therefore, Spo0A~P both promotes sporulation and indirectly impedes it. Cannibalism may be a mechanism to help ensure that cells do not commit to sporulation in response to what proves to be only a transient depletion of nutrients. Here we have shown that bistable control of Spo0A~P plays an additional role in the control of genes involved in production of extracellular matrix.
The principal finding of our investigation is that activation of sinI, which triggers derepression of the matrix operons eps and yqxM, occurs in only a subpopulation of the cells. In contrast, sinR, whose product is the target of SinI, is expressed more or less uniformly in the entire population. On this basis we propose that in the sinI-expressing cells, the concentration of the antirepressor reaches or exceeds that of the repressor, even though the overall level of SinI in the culture is much lower than that of SinR. It further follows from this that the matrix operons are derepressed only in a subpopulation of the cells, as confirmed by fluorescence microscopy using cells engineered to express fluorescent reporter genes joined to the promoters for the eps and yqxM operons. This implies that at the early stages of biofilm formation a subpopulation of cells are specialized for the production of matrix. Given earlier evidence indicating that matrix components can be exchanged between cells (Branda et al., 2006), it is tempting to speculate that the subpopulation of sinI-expressing cells is providing matrix components for the entire community.
The basis for this cell population heterogeneity is that sinI is under the positive control of Spo0A (Shafikhani et al., 2002, Molle et al., 2003, Gaur et al., 1988), whose synthesis and phosphorylation are known to be governed by a bistable switch (Veening et al., 2005). Consistent with this interpretation, cells engineered to produce a mutant form of Spo0A (Spo0A-Sad67) whose activity does not depend on phosphorylation (Ireton et al., 1993), expressed sinI in almost all cells in the population. Nonetheless, expression of sinI exhibited an unexpected medium dependence that seems at first glance to be at odds with the view that it is under the exclusive control of Spo0A~P. At one and three hours after the end of exponential phase growth, a subpopulation of sinI-expressing cells was readily detected in the biofilm-promoting medium MSgg. In contrast, no sinI-expressing cells were detected at hour 1 and very few at hour 3 in the sporulation medium DS. Yet, growth in DS medium was much more effective in activating Spo0A than was growth in MSgg medium as judged from the expression of the promoter for the sporulation operon spoIIG. The spoIIG operon is under the direct positive control of Spo0A~P, but like other sporulation genes and operons under Spo0A~P control spoIIG has a relatively low affinity for the response regulator, representing a so-called high-threshold target of the response regulator (Fujita et al., 2005, Fujita & Losick, 2005). Hence its expression requires relatively high levels of Spo0A~P. As seen in Fig. 3C, no spoIIG-expressing cells were observed at hour 1 in MSgg medium and relatively few at hour 1 in DS medium. However, by hour 3 in DS medium half or more of the cells were expressing spoIIG.
A possible clue to the basis for the dissimilar behaviors of sinI and spoIIG comes from an examination of the nucleotide sequence of the σA-controlled promoter governing sinI transcription (YC, unpublished analysis). Just upstream of the -35 region is a perfect match to the consensus binding site for Spo0A~P (the so-called OA box), which presumably functions as a high-affinity, activation site for the response regulator (Shafikhani et al., 2002). However, just downstream of the start site are three imperfect matches to the OA box. We speculate that these are operator sites at which Spo0A~P binding blocks the access of RNA polymerase to the promoter and thereby represses sinI but only when the response regulator reaches high enough concentrations to adhere to these postulated weak binding sites. We therefore posit that in MSgg medium Spo0A~P accumulates to intermediate levels that suffice to activate sinI but not repress it, thereby allowing for sustained synthesis of the antirepressor. In contrast, in DS medium Spo0A~P levels rapidly rise to levels that repress the biofilm-inducing gene. Our hypothesis therefore explains why MSgg medium but not DS medium promotes biofilm formation. Consistent with our hypothesis, colonies of cells engineered to overexpress sinI exhibited a rugose appearance on solid DS medium rather than the smooth-colony morphology characteristic of unengineered cells grown on the sporulation medium (Fig. 3A, middle and right-hand panels).
The goal of the current investigation was to study early events in the derepression of the extracellular matrix operons. For this purpose, we carried out our investigation using liquid cultures under conditions of continuous shaking in order to keep the cells dispersed and under uniform nutritional and aeration conditions. Normally, of course, cultures are not shaken during biofilm formation so that the cells can assemble into pellicles at the air/liquid interface or into colonies on solid medium. In light of our finding of the bifurcation of the culture into a subpopulation of sinI-expressing cells, it will be of high interest in future work to visualize the appearance, distribution, and abundance of matrix-producing cells within the biofilm itself over the course of its formation.
Experimental Procedures
Strains and media
Bacillus subtilis strains PY79, 3610, and other derivatives were grown in Luria-Bertani (LB), Difco sporulation (DS) or MSgg (Branda et al., 2001), at 37°C or 22°C as indicated. Escherichia coli strain DH5a was used as a host for molecular cloning and was grown at 37°C in LB medium. 1.5% agar was included when making solid agar medium. When appropriate, antibiotics were included at the following concentrations: 10 μg per ml of tetracycline, 100 μg per ml of spectinomycin, 20 μg per ml of kanamycin, 5 μg per ml of chloramphenicol, and 1 μg per ml of erythromycin.
Colony morphology analysis
For colony architecture analysis on solid media, strains were grown to exponential growth phase in LB broth and 3 μl of cells were then applied to solid medium containing 1.5% Bacto agar. The plates were incubated at 22°C for three days. Images of the colonies on the plates were taken using a Nikon CoolPix 950 digital camera.
Strain construction
To construct strains with promoter-lacZ fusions integrated into the chromosome at the amyE locus, the promoter sequences of sinI, sinR, and the eps operon were amplified by Polymerase-Chain-Reaction (PCR). B. subtilis 3610 chromosomal DNA was used as the template in all PCR reactions, and oligonucleotides PsinI-F1 and PsinI-R1, PsinR-F1 and PsinR-R1, and PepsA-F1 and PepsA-R1 were used for amplifications of the promoter sequences of sinI, sinR, and the eps operon, respectively. PCR products were cloned into the plasmid pDG268 (Antoniewski et al., 1990). The resulting recombinant plasmids were then transformed into PY79 following the standard transformation protocol for B. subtilis (Gryczan et al., 1978). Transformants were selected for a double crossover recombination at the amyE locus on the chromosome of PY79. The promoter-lacZ fusions at the amyE locus were then transferred from the PY79 background into the 3610 background using SPP1-mediated transduction as described previously (Yabsin & Young, 1974, Chu et al., 2006).
To construct promoter-gfp fusions, similar procedures as described above were applied except that oligonucleotides PsinI-F2 and PsinI-R2, PsinR-F1 and PsinR-R2, and PepsA-F1 and PepsA-R2 were used for amplification of the promoter sequences of sinI, sinR, and the eps operon, respectively. The amplified PCR products were cloned into another plasmid pYC121. Plasmid pYC121 contains a promoter-less gfp gene flanked by the amyE sequences and itself was constructed as follows: the gfp gene was amplified by PCR from the vector pNGFP (Chastanet & Losick, 2007) using oligonucleotides gfp-F1 and gfp-R1, and the PCR products were cloned into the HindIII and BamHI sites of the vector pDG1662 (Guérout-Fleury et al., 1996). The sequences of all PCR products were verified by DNA sequencing at Harvard DNA Sequencing Facilities.
For the construction of dual-labeled strains, the same promoter sequences of sinI and sinR to make promoter-gfp fusions were also cloned into plasmid pYC136 to generate promoter-cfp fusions. The recombinant plasmids were then transformed into PY79 and selected for a double crossover recombination at the chromosomal thrC locus. The promoter-cfp fusions at the thrC locus were then transferred from the PY79 background into strain YC222, which contains a PyqxM-yfp fusion integrated at the chromosomal amyE locus in 3610 (The PyqxM-yfp fusion was a gift of H. Vlamakis and C. Aguilar).
All insertion deletion mutations were generated by long-flanking homology PCR that has been described previously (Wach, 1996). A detailed description of primers used in this work and all resulting strains is provided in Table S1.
β-galactosidase assays
Cells were incubated in MSgg or DS medium at 37°C in a water bath with shaking. 1 ml culture was collected at each time point. Cells were spun down and pellets were resuspended in 1 ml Z buffer (40 mM NaH2PO4, 60 mM Na2HPO4, 1 mM MgSO4, 10 mM KCl, and 38 mM β-mercaptoethanol) supplemented with 200 μg ml−1 freshly made lysozyme. Resuspensions were incubated at 30°C for 15 min. Reactions were started by adding 200 μl of 4 mg ml−1 ONPG (2-nitrophenyl β-D-galactopyranoside) and stopped by adding 500 μl of 1 M Na2CO3. Samples were briefly spun down. The soluble fractions were transferred to cuvettes (VWR), and OD420 values of the samples were recorded using a Pharmacia ultraspectrometer 2000. The β-galactosidase specific activity was calculated according to the equation [OD420/time × OD600] × dilution factor × 1000. Assays were conducted at least in duplicate.
Purification of His6-SinR and His6-SinI proteins
E. coli strains RL4219 and RL4220 (Kearns et al., 2005) were used for the production of His6-SinR and His6-SinI fusion proteins, respectively. 500-ml cultures were grown in LB broth supplemented with 25 μg ml−1 kanamycin and 50 μg ml−1 chloramphenicol at 30°C to an OD600 of 0.5. IPTG was then added to a final concentration of 1 mM and cultures were incubated at 30°C for two more hours. Cells were harvested and washed once with 50 ml cold phosphate buffer (20 mM sodium phosphate, 200 mM NaCl, 10% glycerol, 1 mM PMSF, pH 7.4). Cell pellets were suspended in 5 ml of cold phosphate buffer supplemented with 200 μg ml−1 of freshly made lysozyme solution and incubated on ice for 30 min. Lysed cells were further disrupted on ice using sonication. Cell lysates were centrifuged at 5000 rpm for 5 min to remove cell debris and were further ultracentrifuged at 35,000 rpm for 30 min at 4°C. Soluble fractions were transferred to clean cold tubes.
1 ml of Ni-NTA agarose beads (Qiagen) was added to the cleared lysate and samples were gently rotated for 2 h at 4°C. The lysate/bead mixture was then loaded onto a column and washed five times, each time with two bed volumes of wash buffer (20 mM sodium phosphate, 300 mM NaCl, 10% glycerol, 20 mM imidazole, pH 8.5). The column was eluted with 5 bed volumes of elution buffer (20 mM sodium phosphate, 300 mM NaCl, 10% glycerol, 300 mM imidazole, pH 8.5). Collected samples were dialysed against 20 mM sodium phosphate, 300 mM NaCl, 0.3 mM DTT, 10% glycerol, pH 7.4 and were quantified using a BCA Protein Assay Kit by Pierce (IL, USA). Proteins were aliquoted and stored in 50% glycerol at −80°C.
Quantitative immunoblots
Protein lysates were diluted, mixed 2:1 with protein lysis buffer (Bio-Rad) and were size-fractionated by 15% SDS-PAGE. In parallel, affinity-purified His6-SinR and His6-SinI proteins were diluted in the same way and loaded on the same SDS-PAGE. After size-fractionation, proteins were transferred to an immunoblot-specific PVMF membrane (Bio-Rad) at 250 mA for 2 hours using a Bio-Rad mini-PROTEIN II Cell. After completion of protein transfer, the membrane was briefly washed with TBS buffer (20 mM Tris-Cl, 200 mM NaCl, 0.1% Tween 20, pH 7.5) and incubated in 30 ml TBS buffer supplemented with 5% skim milk. The membrane was then transferred to a clean tip box and incubated with the affinity-purified antiserum against either SinI or SinR at a dilution of 1:10,000 in 15 ml TBS buffer. After 3 h incubation at room temperature with gentle shaking, the membrane was then washed three times, each time with 30 ml TBS buffer. A second goat-anti-rabbit antibody (Bio-Rad) was added to the membrane with a dilution of 1:6,000 in 15-ml TBS buffer and incubated at room temperature for 1 h. The membrane was again washed three times with TBS buffer. Finally, the membrane was transferred to a clean tip box, mixed with 7-ml each of Solutions A and B in the SuperSignal West Pico Chemiluminescent Kit (Pierce), and incubated for 5 min at room temperature. An X-ray film was exposed to the membrane and then developed using an Eastman Kodak RP X-OMAT processor.
To quantify the number of SinI molecules per cell, we estimated that the pixel density from the lane containing 10 fmol of purified His6-SinI was roughly equal to that from the middle lane on the right panel (Fig. 2C), which contained protein prepared from a total of 1.2 × 1011 cells. It was thus calculated that for each cell, there are about 50 molecules of SinI. The number of SinR molecules per cell was quantified in a similar way. We estimated that the pixel density from the middle lane on the right panel (Fig. 2B), which contained protein prepared from a total of 6 × 1010 cells, would have been equal to 90 fmol of purified His6-SinR. We thus calculated that there are about 900 SinR molecules per cell.
Fluorescence microscopy
Cells were grown in MSgg broth to early stationary phase. 1 ml of the culture was harvested and centrifuged. Cells were washed with PBS buffer twice (PBS buffer was autoclaved and filtered through a 0.25 μM syringe filter prior to use), and resuspended in 50 μl PBS buffer. 3 μl of resuspended cells were dropped on the center of an agar-coated microscopy slide (VWR, catalog number 48311-702), and covered by a 0.15 mm microscopy cover slide (VWR, catalog number 48366-045). Cover slides were pre-treated with polyllysine. Samples were examined using an Olympus workstation BX61. Images were taken and analyzed using an automated software program MetaMorph (Universal Imaging Corporation).
Supplementary Material
Acknowledgments
We thank members of the Losick and Kolter labs for helpful discussions, especially Drs. A. Chastenet, C. Aguilar and H. Vlamakis. We also thank D. Kearns (Indiana University) for providing affinity-purified SinR and SinI antibodies. This work was supported by NIH grant GM18568 to RL and GM58213 to RK. YC is a postdoctoral fellow of the Jane Coffin Childs Memorial Fund.
Contributor Information
Yunrong Chai, Department of Molecular and Cellular Biology, The Biological Laboratories, Harvard University, Cambridge, Massachusetts 02138.
Frances Chu, Department of Molecular and Cellular Biology, The Biological Laboratories, Harvard University, Cambridge, Massachusetts 02138.
Roberto Kolter, Department of Microbiology and Molecular Genetics, Harvard Medical School, Boston, Massachusetts 02115.
Richard Losick, Department of Molecular and Cellular Biology, The Biological Laboratories, Harvard University, Cambridge, Massachusetts 02138.
References
- Antoniewski C, Savelli B, Stragier P. The spoIIJ gene, which regulates early developmental steps in Bacillus subtilis, belongs to a class of environmentally responsive genes. J Bacteriol. 1990;172:86–93. doi: 10.1128/jb.172.1.86-93.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai U, Mandic-Mulec I, Smith I. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev. 1993;7:139–148. doi: 10.1101/gad.7.1.139. [DOI] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Baldus JM, Green BD, Youngman P, Moran CP., Jr Phosphorylation of Bacillus subtilis transcription factor Spo0A stimulates transcription from the spoIIG promoter by enhancing binding to weak 0A boxes. J Bacteriol. 1994;176:296–306. doi: 10.1128/jb.176.2.296-306.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006;59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci USA. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Vik A, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Burbulys D, Trach KA, Hoch JA. Initiation of sporulation in Bacillus subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- Chastanet A, Losick R. Engulfment during sporulation in Bacillus subtilis is governed by a multi-protein complex containing tandemly acting autolysins. Mol Microbiol. 2007;64:139–152. doi: 10.1111/j.1365-2958.2007.05652.x. [DOI] [PubMed] [Google Scholar]
- Chu F, Kearns DB, Branda SS, Kolter R, Losick R. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol. 2006;59:1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x. [DOI] [PubMed] [Google Scholar]
- Chung JD, Stephanopoulos G, Ireton K, Grossman AD. Gene expression in single cells of Bacillus subtilis: evidence that a threshold mechanism controls the initiation of sporulation. J Bacteriol. 1994;176:1977–1984. doi: 10.1128/jb.176.7.1977-1984.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- Dubnau D, Losick R. Bistability in bacteria. Mol Microbiol. 2006;61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- Ellermeier CD, Hobbs EC, Gonzalez-Pastor JE, Losick R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell. 2006;124:549–559. doi: 10.1016/j.cell.2005.11.041. [DOI] [PubMed] [Google Scholar]
- Fujita M, Gonzalez-Pastor JE, Losick R. High- and Low-threshold genes in the Spo0A regulon of Bacillus subtilis. J Bacteriol. 2005;187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Losick R. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 2005;19:2236–2244. doi: 10.1101/gad.1335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur NK, Cabane K, Smith I. Structure and expression of the Bacillus subtilis sin operon. J Bacteriol. 1988;170:1046–1053. doi: 10.1128/jb.170.3.1046-1053.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- Gryczan TJ, Contente S, Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978;134:318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérout-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Hamon MA, Lazazzera BA. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol Microbiol. 2001;42:1199–1209. doi: 10.1046/j.1365-2958.2001.02709.x. [DOI] [PubMed] [Google Scholar]
- Ireton K, Rudner DZ, Siranosian KJ, Grossman AD. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 1993;7:283–294. doi: 10.1101/gad.7.2.283. [DOI] [PubMed] [Google Scholar]
- Jiang M, Shao W, Perego M, Hoch JA. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol Microbiol. 2000;38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Losick R. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 2005;19:3083–3094. doi: 10.1101/gad.1373905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R, Greenberg EP. Microbial sciences: The superficial life of microbes. Nature. 2006;441:300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Micro. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- Lewis RJ, Brannigan JA, Offen WA, Smith I, Wilkinson AJ. An evolutionary link between sporulation and prophage induction in the structure of a repressor:anti-repressor complex. J Mol Biol. 1998;283:907–912. doi: 10.1006/jmbi.1998.2163. [DOI] [PubMed] [Google Scholar]
- Lewis RJ, Brannigana JA, Smith I, Wilkinson AJ. Crystallisation of the Bacillus subtilis sporulation inhibitor SinR, complexed with its antagonist, Sinl. FEBS Letters. 1996;378:98–100. doi: 10.1016/0014-5793(95)01432-2. [DOI] [PubMed] [Google Scholar]
- Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–529. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle V, Fujita M, Jensen ST, Eichenberger P, Gonzalez-Pastor JE, Liu JS, Losick R. The Spo0A regulon of Bacillus subtilis. Mol Microbiol. 2003;50:1683–1701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- O’Toole G, Kaplan HB. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54:49–80. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- Shafikhani SH, Mandic-Mulec I, Strauch MA, Smith I, Leighton T. Postexponential regulation of sin operon expression in Bacillus subtilis. J Bacteriol. 2002;184:564–571. doi: 10.1128/JB.184.2.564-571.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshein AL. Control of sporulation initiation in Bacillus subtilis. Curr Opin Microbiol. 2000;3:561–566. doi: 10.1016/s1369-5274(00)00141-7. [DOI] [PubMed] [Google Scholar]
- Stephenson K, Hoch JA. Evolution of signalling in the sporulation phosphorelay. Mol Microbiol. 2002;46:297–304. doi: 10.1046/j.1365-2958.2002.03186.x. [DOI] [PubMed] [Google Scholar]
- Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communites. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- Sutherland IW. The biofilm matrix - an immobilized but dynamic microbial environment. Trends Microbiol. 2001;9:222–227. doi: 10.1016/s0966-842x(01)02012-1. [DOI] [PubMed] [Google Scholar]
- Veening J-W, Hamoen LW, Kuipers OP. Phosphatases modulate the bistable sporulation gene expression pattern in Bacillus subtilis. Mol Microbiol. 2005;56:1481–1494. doi: 10.1111/j.1365-2958.2005.04659.x. [DOI] [PubMed] [Google Scholar]
- Veening JW, Smits WK, Hamoen LW, Kuipers OP. Single cell analysis of gene expression patterns of competence development and initiation of sporulation in Bacillus subtilis grown on chemically defined media. J Appl Microbiol. 2006;101:531–541. doi: 10.1111/j.1365-2672.2006.02911.x. [DOI] [PubMed] [Google Scholar]
- Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene sidruptions in Saccharomyces cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- Yabsin RE, Young EF. Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol. 1974;14:1343–1348. doi: 10.1128/jvi.14.6.1343-1348.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


