Abstract
Hedgehog (HH) signaling in the epidermis is primarily mediated by the zinc finger transcription factors GLI1 and GLI2. Exquisite regulation of HH/GLI signaling is crucial for proper specification of the epidermal lineage and development of its derivatives, whereas dysregulation of HH/GLI signaling disrupts tissue homeostasis and causes basal cell carcinoma (BCC). Similarly, bone morphogenetic proteins (BMPs) and activins have been described as key signaling factors in the complex regulation of epidermal fate decisions, although their precise interplay with HH/GLI is largely elusive. Here we show that, in human epidermal cells, expression of the activin/BMP antagonist follistatin (FST) is predominantly up-regulated by the HH effector GLI2. Consistently, we found strong FST expression in the outer root sheath of human hair follicles and BCC. Detailed promoter analysis showed that two sequences with homology to the GLI consensus binding site are required for GLI2-mediated activation. Interestingly, activation of the FST promoter is highly GLI2-specific, because neither GLI1 nor GLI3 can significantly increase FST transcription. GLI2 specificity requires the presence of a 518-bp fragment in the proximal FST promoter region. On the protein level, sequences C-terminal to the zinc finger are responsible for GLI2-specific activation of FST transcription, pointing to the existence of GLI-interacting cofactors that modulate GLI target specificity. Our results reveal a key role of GLI2 in activation of the activin/BMP antagonist FST in response to HH signaling and provide new evidence for a regulatory interaction between HH and activin/BMP signaling in hair follicle development and BCC.
The development and differentiation of mammalian epidermis and hair follicles requires precisely regulated interactions of a multitude of signals exchanged between and within the epidermis and the underlying dermis. Hedgehog (HH),3 Wnt, Notch, and transforming growth factor β signaling are key among the pathways controlling epidermal lineage and homeostasis. The well orchestrated interplay between these signals ensures temporally and spatially coordinated proliferation, specification, and differentiation.
The HH pathway has been the subject of intense investigation not only for its role in development but also in different types of cancer. The pathway is activated by binding of secreted HH ligand to the transmembrane receptor/repressor Patched (Ptch). This permits the positive mediator Smoothened to activate the transcriptional mediators of the pathway, Gli1, Gli2, and Gli3 (reviewed in Refs. 1–5). In addition, all Glis are subject to regulation by phosphorylation and proteasomal degradation (6–10). Although Gli1, Gli2, and Gli3 can bind the same consensus sequence through a highly conserved zinc finger binding domain (11), there are some well characterized differences between the three Gli proteins. Gli1 has only an activator function, is not proteolytically processed, and lacks the N-terminal repression domain present in Gli2 and in Gli3 (12, 13), the third Gli family member, which has predominantly repressive activity (14). In contrast to the latent transcription factors Gli2 and Gli3, Gli1 is not directly activated by HH signaling but is a transcriptional target gene of Gli2 and Gli3 (15, 16). Differences and overlaps in the roles of the three Glis in development are difficult to resolve in view of the complexity resulting from context-dependent differences in expression and activation state. Insight comes from phenotypes of single and compound mutations: Gli1-/- mice have no phenotype (17), while Gli2 knockout is lethal (18–20). The compound mutation Gli1-/-, Gli2+/- does have a phenotype, and the double knockout has a more severe phenotype than Gli2-/- alone pointing to an overlap of function (17). Addressing functional overlap between Gli1 and Gli2, a knock-in of Gli1 into the Gli2 locus leads to dosage-dependent, almost complete rescue, only a hair phenotype is seen postnatally (21). In addition to this in vivo demonstration of functional equivalence of Gli1 and Gli2 in mouse embryonic development, functional redundancy of activator Glis has also been shown in chick neural tube development (22). Graded Shh signaling, which is normally mediated by the Gli code represented by the activator and repressor forms of the three Glis (23), can be artificially mimicked by a gradient of different Gli3 activator and Gli3 repressor forms (22). These results emphasize the common properties of the Gli family members, but there is also considerable evidence for different biochemical and functional properties in contexts that are not related to a phenotype in embryogenesis.
The specific effects of expression of each Gli are not completely conserved between vertebrates. In neural tube development the requirements for Gli1 and Gli2 are different for Xenopus, zebrafish, and mouse (24–27). Although Gli1 acts as an activator in all three species, its inactivation has no effect in mouse but in zebrafish GLI1 is absolutely required. Gli2 has important repressor function in zebrafish but functions only as an activator in mouse neural tube development (17). Karlstrom and colleagues propose a function for Gli1 as an important amplifier of an activation signal, which may be required only in some contexts (25). Generally, the relative contribution of any Gli to a process is difficult to unravel, because not only the balance but also cooperativity and physical interaction of the Glis may play a role in establishing the Gli code regulating gene expression (27). Whether Gli1 and Gli2 are independently or cooperatively involved in HH pathway induced carcinogenesis is not known. In skin, constitutive pathway activation leads to basal cell carcinoma (BCC) and can be caused by mutational inactivation of the receptor/repressor Patched (28, 29), activation of the positive mediator Smoothened (30), and, in transgenic models, by overexpression of Shh (31) or either of the transcription factors Gli1 and Gli2 (32, 33). Although GLI1 is most consistently overexpressed in human tumors, including BCC (34, 35), Gli2 was shown to be required for BCC maintenance in a conditional expression model (36). Furthermore, in human hepatocellular carcinoma cell lines antisense knockdown experiments point to a special role of GLI2 in proliferation and target gene activation (37).
We have previously shown that the transcriptional response to overexpression of GLI1 and GLI2 in keratinocytes is only partially overlapping (38), suggesting differences in target gene specificity, which may indicate nonredundant function. Apart from moderate quantitative differences there are a number of genes expressed almost exclusively in response to one of the two transcription factors. A striking example is follistatin (FST), an activin/BMP antagonist, which binds activins and several members of the BMP family thereby blocking downstream signaling (reviewed in Refs. 39 and 40). We have investigated the differential transcriptional response in more detail to show that FST is a direct target gene preferentially activated by GLI2 rather than GLI1. Moreover we show that FST is co-expressed with GLI2 in hair follicles and BCC. The role of FST in many different developmental processes and, in particular, skin and hair follicle development was demonstrated not only by the knockout phenotype in mouse (41) but also by ectopic induction of feather buds or hair follicles by application of FST (42, 43). We identified the DNA region responsible for GLI2 specificity by deletion analysis of the FST promoter and localized the relevant region of the protein to the C-terminal part, but outside the VP16-like transactivation domain. This difference between GLI1 and GLI2 suggests a special role of GLI2 in hair follicles and might also affect BCC.
EXPERIMENTAL PROCEDURES
Cloning—For the FST promoter reporter plasmid (FSTprom) a 3088-bp MscI fragment of the human BacPac clone RP11–937I3 (obtained from Children's Hospital Oakland Research Institute) containing the putative FST promoter region was cloned into the SmaI restriction site of the luciferase reporter plasmid pGL3basic (Promega, Madison, WI). GLI binding sites were mutated at the essential positions 5, 6, and 7 to Gly by using the QuikChange site-directed mutagenesis kit (Stratagene). For FSTdelA and FST-A, FSTprom was digested with AatII/XhoI or NheI/AatII, blunted with T4 polymerase, and re-ligated. The reporter vector 6bsSV40 consists of a 6-Gli binding site cassette (15), which was cloned into the BglII site of pGL3promoter (Promega). For 6bsFST-A, FSTprom was digested with NheI/AatII, blunted with T4 DNA polymerase, and ligated with a blunted BglII fragment containing the Gli binding site cassette of 6bsSV40. GLI1, activated human GLI2 (GLI2act, N-terminally truncated), and GLI121 expression constructs have been described previously (44). Activated human GLI3 (GLI3act, amino acids 431–1580) was cloned into p3xFLAG-CMV-10 (Sigma-Aldrich). For the GLI112 expression construct the N-terminal region of GLI1, including the zinc finger (amino acids 1–379), was fused to the C-terminal region of GLI2 (amino acids 254–1258 of GLI2act). Similarly, for GLI221, the N-terminal region of GLI2, including the zinc finger (amino acids 1–253 of GLI2act), was fused to the C-terminal region of GLI1 (amino acids 380–1106). Briefly the N- and C-terminal regions of either GLI protein were amplified by PCR and cloned into the EcoRI/XbaI sites of pBluescript II KS (Stratagene). N-terminal regions were fused to the C-terminal region using a single BspEI site, which was introduced into the GLI sequences. The internal EcoRI site in GLI1 was eliminated by site-directed mutagenesis. The resulting chimeric GLI fusion constructs GLI112 and GLI221 were cloned into the EcoRI/XbaI site of the expression vector pcDNA4/TO (Invitrogen). For GLI1TA2, amino acids 1135–1258 of GLI2act were fused to amino acids 1–1017 of GLI1. The corresponding construct GLI2TA1 was constructed by fusing the C-terminal amino acids 1018–1106 of GLI1 to amino acids 1–1134 of GLI2act. For cloning, an AvrII site was introduced by silent mutagenesis. For the expression construct p4TO-FST344 a 344-amino acid-long isoform of human FST was amplified from the IMAGE clone IRAUp969F0524D (obtained from imaGenes) by PCR and cloned into the EcoRI/XhoI sites of the expression vector pcDNA/4TO (Invitrogen).
Cell Culture—HaCaT cells were cultured in Dulbecco's modified Eagle medium (high glucose, PAA Laboratories) with 10% fetal calf serum (PAA Laboratories), 100 μg/ml streptomycin, and 62.5 μg/ml penicillin (Invitrogen) at 37 °C, 5% CO2. For double stable inducible HaCaT lines expressing either human GLI1 (GLI1-HaCaT) or GLI2act (GLI2-HaCaT) (38, 45), medium was supplemented with 25 μg/ml Zeocin (Invitrogen) and 8 μg/ml blasticidin-S (ICN-Biomedica). For growth factor treatment HaCaT cells were cultured in Defined Keratinocyte-SFM medium (Invitrogen) and either treated for 24 h with recombinant human BMP4 (R&D Systems) or for 6 h with recombinant human activin A (R&D Systems). The activin/transforming growth factor-β pathway inhibitor SB431542 (Sigma-Aldrich) dissolved in DMSO (Sigma-Aldrich) was used at a final concentration of 10 μm in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum. Transgene expression was induced by adding 1 μg/ml tetracycline (Invitrogen) to the medium. Murine keratinocyte line C5N and murine BCC tumor cell lines ASZ001 and CSZ1 (46, 47) were cultured in 154-CF medium (Cascade Biologics) supplemented with 0.05 mm CaCl2, 2% fetal bovine serum (PAA Laboratories), 100 μg/ml streptomycin, and 62.5 μg/ml penicillin (Invitrogen) at 37 °C, 5% CO2.
Western Blot—Cells were lysed in 125 mm Tris (pH 6.8), 5% glycerol, 2% SDS, 1% β-mercaptoethanol, 0.006% bromphenol blue, and extracts resolved by SDS-PAGE. GLI2 and FST protein were detected using polyclonal rabbit-anti-GLI2 antibody (GLI2-H300, Santa Cruz Biotechnology), affinity-purified polyclonal goat-anti-FST antibody (R&D Systems), secondary goat-anti-rabbit horseradish peroxidase-conjugated antibody (Santa Cruz Biotechnology) and a chicken-anti-goat horseradish peroxidase-conjugated antibody (Santa Cruz Biotechnology). Proteins were visualized using the ECL detection system (Amersham Biosciences).
Immunohistochemistry—Staining for FST was performed on routinely formalin-fixed paraffin embedded tissue, using a standardized automated system (Autostainer, Dako) in combination with an Envision detection system (Dako). A mouse monoclonal antibody directed against human FST (R&D Systems, dilution, 1:100) was used. Archival formalin-fixed paraffin-embedded sections (4 μm thick) were deparaffinized with xylene, hydrated, followed by heat-induced epitope retrieval in antigen retrieval buffer, pH 9. Endogenous peroxidase blocking was carried out 10 min with 3% H2O2 in absolute methanol, and normal serum was applied. Primary antibodies were incubated at room temperature for 1 h, and after several washes detection was performed using the Envision detection system, followed by development with diaminobenzidine. Slides were counterstained with hematoxylin.
qRT-PCR Analysis—Total RNA was isolated and purified (High Pure RNA Isolation Kit, Roche Applied Science), and cDNA was synthesized from 4 μg of total RNA with Superscript II (RNase H-) reverse transcriptase (Invitrogen) using oligo(dT) primers, according to the manufacturer's instructions. qRT-PCR analysis was performed on a Rotor-Gene 3000 (Corbett Research) using iQ™ SYBR Green Supermix (Bio-Rad). Human large ribosomal protein P0 (RPLP0) or mouse acidic ribosomal phosphoprotein P0 (Arbp) was used for normalization in qRT-PCR analysis (48). For primer sequences see supplemental Table S1.
Luciferase Reporter Assay—HaCaT cells were grown in 12-well plates to 80% confluence and transfected in triplicate with the respective GLI expression constructs, pGL3 luciferase reporter plasmids as indicated in the figures and Renilla luciferase (pRL-SV40, Promega) expression plasmids for normalization. Transfection was carried out using SuperFect transfection reagent (Qiagen) according to the manufacturer's protocol. Cells were harvested 48 h after transfection, and luciferase activity was measured with a LucyII luminometer (Anthos) using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Data were normalized for Renilla luciferase activity.
Electrophoretic Mobility Shift Assay—EMSA was carried out with 5 μg of recombinant NHIS-GLI2-zinc finger protein (amino acids 409–602 of human GLI2, NP_005261), purified as described before (16), and 10 ng of [32P]dCTP (Amersham Biosciences) labeled double-stranded oligonucleotide in 30 μl of 20 mm Tris, pH 7.8, 25 mm KCl, 5 mm MgCl2, 0.5 mm dithiothreitol, 10 mm ZnSO4, 0.77 μg of poly(dI-dC) (Sigma-Aldrich), 10% glycerol. Reactions were incubated for 25 min at room temperature. Competition was carried out with 5- or 10-fold excess of specific (bs1, bs2, or bs3) or mutant unlabeled oligonucleotide (bsm) or 1.2–2.4 μg of poly(dI-dC). Samples were separated on 6% acryl amide gels. Following electrophoresis, gels were dried, exposed overnight, and scanned with a BAS-1800II (Fuji). Oligonucleotides used for EMSA are shown in supplemental Table S3).
Chromatin Immunoprecipitation—ChIP was done according to the Chromatin Immunoprecipitation Kit protocol (Upstate), with some modifications. In brief, GLI1- or GLI2-HaCaT cells were grown for 48 h in the presence of 1 μg/ml tetracycline to induce GLI expression. Cells were washed once with warm 1× phosphate-buffered saline prior to addition of Dulbecco's modified Eagle medium containing 1% formaldehyde and incubation for 10 min at room temperature. Cross-linking was stopped by adding 2.5 m glycine to a final concentration of 125 mm. Unless otherwise indicated, all following steps were either performed on ice or at 4 °C, and buffers were supplemented with protease inhibitor mixture (Sigma-Aldrich). Cross-linked cells were washed twice with ice-cold 1× phosphate-buffered saline, scraped, pelleted by centrifugation, resuspended in lysis buffer (5 mm PIPES, 85 mm KCl, 0.5% Nonidet P-40, pH 8), and Dounced to release nuclei. After centrifugation at 5000 rpm, nuclei were lysed in SDS lysis buffer and chromatin was fragmented by sonication with a Sonopuls HD2070 Sonifier (Bandelin Electronics) to an average fragment size between 100 and 1000 bp. Debris was removed by centrifugation, and the size of chromatin checked on agarose gels after de-cross-linking and proteinase digestion. Aliquots corresponding to 107 cells were diluted 1:10 with ChIP dilution buffer. Protein G-Sepharose 4 Fast Flow (Amersham Biosciences) was pre-blocked with 1 mg/ml herring sperm DNA (Sigma-Aldrich) and 5 mg/ml bovine serum albumin (Carl Roth). Each aliquot of chromatin (2 ml) was precleared with 25 μl of a 25% Sepharose slurry for 30 min. Goat polyclonal anti-GLI1 (GLI1-C18), goat polyclonal anti-GLI2 (GLI2-N20) (Santa Cruz Biotechnology), rabbit polyclonal anti acetyl histone H3 (Upstate Biochemicals), or species-matched normal IgG (Santa Cruz Biotechnology) was added, and samples were incubated with the antibodies overnight with rotation. Antibody complexes were precipitated by addition of 60 μl of pre-blocked 25% Sepharose slurry for 4h, while rotating. Supernatants were removed, from the normal IgG samples 500 μl was kept and processed as input controls, and Sepharose beads were washed twice with each ice-cold wash buffer. Protein-DNA complexes were eluted in 500 μl of elution buffer (0.2% SDS, 100 mm NaHCO3), 20 μl of 5 m NaCl was added, and samples were incubated for at least 5 h in a 65 °C water bath to open cross-links. Input samples were adjusted to 200 mm NaCl and processed as above. After reverting cross-links, eluates and input samples were treated with RNase A and proteinase K, and final DNA clean up was performed with GFX PCR and Gel band kit (Amersham Biosciences), DNA was eluted in 60 μl of RNase and DNase free sterile water, and 3.8 μl of eluates was used for PCR analysis. Input samples were further diluted 1:80, and 3.8 μl was used in PCR reactions. For ChIP-PCR, the FailSafe™ GREEN Real-Time PCR System (Epicenter Biotechnologies) was used according to the manufacturer's instructions. Primers used for PCR are shown in supplemental Table S2. PCR was performed in 10-μl reaction volume in a Rotor-Gene 3000 (Corbett Research). PCR was done in duplicates, both samples were pooled, and products were visualized on 3% MetaPhor (Cambrex Bio Science)/1× Tris-acetate-EDTA gels.
RESULTS
Induction of FST Expression in Response to GLI2 in Human Keratinocytes—The activin/BMP antagonist FST was identified as a putative GLI2 target gene in an array-based screen of HaCaT keratinocytes expressing GLI1 (GLI1-HaCaT) or GLI2 activator form (GLI2act) (GLI2act-HaCaT) (38) in response to tetracycline treatment. In contrast to the majority of GLI target genes identified in this screen, FST was turned on almost exclusively in response to GLI2act and not to GLI1 expression. To verify the specific activation of FST by GLI2, we analyzed FST expression at several time points after GLI2act or GLI1 induction by qRT-PCR (Fig. 1A) and Western blot (Fig. 1C). Elevated levels of FST mRNA were detected already 24 h after GLI2act induction (3-fold increase compared with uninduced samples), whereas in GLI1-HaCaT cells FST levels were unchanged. At later time points, FST mRNA increases up to 20-fold in GLI2act-induced samples compared with controls, with only a moderate increase in GLI1-induced cells (3.5-fold at 72 h). The latter increase might also be due to the indirect induction of GLI2 by GLI1 leading to delayed GLI2 expression in GLI1-HaCaT cells (45). Induction of the well characterized GLI target gene PTCH by GLI1 and GLI2act was comparable as expected (Fig. 1A, inset). To exclude that differential induction of FST is a specific property of the HaCaT cells used in these experiments, we repeated the experiments in the immortalized human keratinocyte cell line N/TERT-1 (49). N/TERT-1 keratinocytes were retrovirally transduced with either human GLI1, GLI2act, or EGFP (50) and analyzed after 60 h of transgene expression for mRNA levels of FST and PTCH (Fig. 1B). As in HaCaT cells, FST was predominantly activated by GLI2act (11.3-fold increase), whereas no induction was observed in response to GLI1. PTCH transcription was comparable in both GLI1 and GLI2act expressing N/TERT-1 keratinocytes. Induction of FST by GLI2act was also observed at the protein level as shown by Western blot analysis (Fig. 1C). The three bands may represent different FST isoforms (51) present in human keratinocytes. GLI2act protein is not detectable in uninduced samples. In summary these data suggest that in human keratinocytes FST expression is mainly activated by GLI2.
FIGURE 1.
FST expression is preferentially induced by GLI2 in human keratinocytes. A, qRT-PCR analysis of FST mRNA levels in HaCaT keratinocytes expressing either GLI1 (GLI1-HaCaT) or GLI2act (GLI2act-HaCaT) under tetracycline control for the time indicated. As a control for GLI activity the known target gene PTCH is shown (inset). B, FST and PTCH mRNA levels in human N/TERT-1 keratinocytes retrovirally transduced with GLI1 or GLI2act measured by qRT-PCR. The -fold change refers to the mRNA ratios for induced to uninduced cells (A) and cells infected with a GLI1/2 to control EGFP expressing virus (B). C, Western blot analysis of FST protein levels in GLI2act-HaCaT cells. Samples were taken from tetracycline-treated and untreated GLI2act-HaCaT cells as indicated. The upper panel shows protein expression of GLI2act transgene. FST protein was detected using a specific antibody recognizing all FST isoforms (lower panel). For control HaCaT cells were transiently transfected with an expression plasmid expressing human FST isoform 344 (p4TO-FST344) or empty expression vector (p4TO). *, unspecific signal.
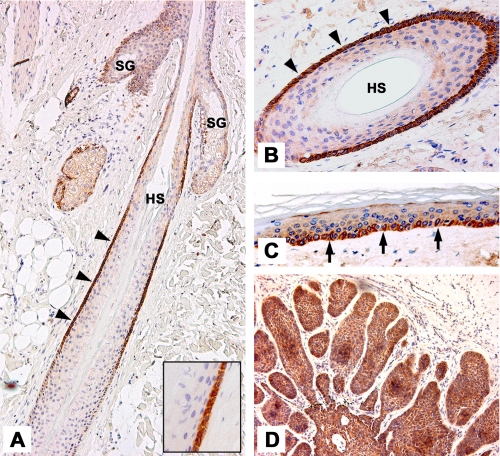
FST Is Expressed in Human Hair Follicle, Interfollicular Epidermis, and BCC—Previous studies have shown that in human skin GLI2 expression is restricted to the outermost layer of the outer root sheath (ORS) of the hair follicle and is also detectable in keratinocytes of the basal layer of the interfollicular epidermis (16). There is also increased expression of GLI2 in BCC (16, 45). As a direct target of GLI2 in hair follicle keratinocytes and human BCC, FST should be expressed in the same regions. We therefore stained sections of paraffin-embedded human scalp (Figs. 2, A–C) and samples from nodular BCCs (Fig. 2D) with an antibody recognizing all isoforms of FST. In hair follicles intense staining for FST was detected in the ORS (Fig. 2, A and B) as has been described previously (52). Notably, expression of FST was restricted to the outermost cell layer in a region below the sebaceous gland (Fig. 2A). Strong staining was also observed in basal keratinocytes of the interfollicular epidermis (Fig. 2C). The staining appears not to be homogenously distributed throughout the basal layer of the epidermis, with some cells showing a strong signal for FST, while others are weakly or not at all stained. Moreover, all BCC samples tested (n = 5) show intense staining for FST throughout the tumor island (Fig. 2D). Consistent with the in vitro data from cultured human keratinocytes and mouse BCC tumor cell lines (supplemental Fig. S3) we found FST protein in GLI2-expressing compartments of the hair follicle, interfollicular epidermis, and BCC, supporting regulation of FST expression by GLI2 in vivo.
FIGURE 2.
FST expression in normal human skin and BCC. Immunostaining of sections of human scalp and BCC with an FST specific antibody. A, FST protein localizes to the outer root sheath (ORS) (arrowheads) of human anagen hair follicles (HF). Higher magnification (inset) shows expression in the outermost layer of the ORS. B, cross-section of a human HF. C, FST protein is clearly detectable in basal keratinocytes (arrows) of the interfollicular epidermis. D, strong staining is seen in BCC tumor islands. SG, sebaceous gland; HS, hair shaft.
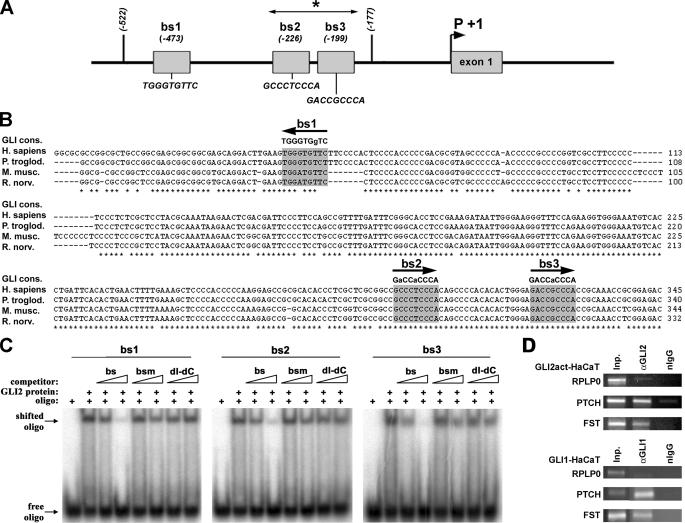
The FST Promoter Contains Three Putative GLI Binding Sites—To investigate whether FST is a direct transcriptional target of GLI2 we searched for putative GLI binding sites in a region 3 kb upstream and 0.5 kb downstream of the FST transcriptional start site (according to RefSeq NM_013409). Using the GLI consensus binding site (53) we identified three potential binding sites, two with one (bs1 and bs3) and one with two (bs2) mismatches to the GLI consensus binding sequence GACCACCCA (Fig. 3A). All three sites are located within 300 bp in close proximity to the transcriptional start site (Fig. 3A) in a region highly conserved in mammals (Fig. 3B). We tested GLI binding by EMSAs using a recombinant His-tagged GLI2 zinc finger domain protein (amino acids 409–602 of human GLI2, NP_005261). This protein specifically binds to all three sites in vitro, and binding could be competed with excess of binding sequences bs1, bs2, and bs3 (bs) but not or only much less efficiently with mutated oligonucleotide (bsm) or poly(dI-dC) (Fig. 3C). This suggests that transcriptional activation is mediated directly by GLI2. To corroborate binding of GLI to this region in vivo we performed ChIP (Fig. 3D) from GLI2act-HaCaT cells induced for 48 h. As shown in Fig. 3D, ChIP analysis confirms binding of GLI proteins to the predicted promoter region within the active FST promoter.
FIGURE 3.
The FST promoter contains three putative GLI binding sites. A, overview of the localization and sequence of three putative GLI binding sites in the human FST promoter. Numbers indicate distance from the transcription start site (according to RefSeq NM_013409). B, cross-species alignment of the region surrounding the putative GLI binding sites shows high conservation between the indicated species. Putative GLI binding sites are highlighted in gray boxes, in the first line, the consensus GLI binding site is indicated, and diverging bases are shown in lowercase. Arrows indicate the orientation of the putative binding sites. C, EMSA analysis shows specific binding of recombinant GLI2 zinc finger domain protein to all three putative GLI binding sites in vitro. Specificity of binding was tested by competing with increasing amounts of unlabeled oligonucleotide corresponding to either the wild type binding sequence (bs) or mutated binding site (bsm) sequences or unspecific competitor dI-dC. D, chromatin immunoprecipitation shows specific binding of GLI2 to the FST promoter in vivo. Chromatin isolated from induced GLI2act- or GLI1-HaCaT cells was precipitated with either specific (αGLI2 and αGLI1) or unspecific (normal IgG) antibodies. A 127-bp fragment (asterisk in A) spanning bs2 and bs3 of the FST promoter was amplified only from the specific precipitate, a 148-bp fragment from the PTCH promoter was used as positive control. No amplification was detected from the unspecific precipitates and for a 284-bp fragment from the promoter of the human acidic ribosomal protein P0 (RPLP0) (negative control). A representative of three independent experiments is shown.
Two of the Three Putative GLI Binding Sites Are Required for Activation of the FST Promoter—Having shown that GLI2 binds to sequences present in the FST promoter in vitro and in vivo, we focused on the role of the individual binding sites. We cloned a 3088-bp fragment comprising the putative GLI binding sites, the transcription start site, the first exon, and part of the first intron into the pGL3basic luciferase reporter vector (FSTprom) (Fig. 4A). As expected this FST promoter was activated by GLI2act, whereas neither GLI1 nor an activator form of GLI3 (GLI3act) were able to significantly stimulate reporter activity. The functionality of all constructs was confirmed using the well characterized promoter of the GLI target gene PTCH (PTCHprom)4 (54) (Fig. 4B). To assess the role of each putative binding site we introduced mutations at essential positions (see “Experimental Procedures”) into bs1, bs2, and bs3, and both bs1 and bs3. FST luciferase reporter containing wild-type (FSTprom) or mutated GLI binding sites (FSTprom-bs1mut, FSTprom-bs2mut, FSTprom-bs3mut, and FSTprom-bs13mut) (Fig. 4C) were co-transfected with GLI2act into HaCaT cells. Mutations in bs1 or bs3 or a combination of both almost completely abolished GLI2-dependent FST promoter activation (Fig. 4D). Interestingly, mutation of bs2 did not result in any reduction of promoter activity, although it binds the GLI2 zinc finger domain in vitro as shown by EMSA (Fig. 3C). This observation is consistent with data showing that many consensus GLI binding sequences within promoter regions are not bound and/or not functional in vivo (55).5 The data show that GLI2 and the presence of two GLI binding sites, bs1 and bs3, within the FST promoter are required for activation.
FIGURE 4.
The human FST promoter is predominantly activated by GLI2. A, luciferase reporter assay with a cloned 3088-bp fragment of the human FST promoter (FSTprom, schematic drawing). HaCaT cells were co-transfected with FSTprom reporter construct and GLI expression constructs as indicated. Only GLI2act expression resulted in strong activation of the FST promoter. B, activity of the expression constructs used in A was tested on a reporter construct (PTCHprom) containing a 1313-bp fragment of the human PTCH promoter. C, activation of the FST promoter is dependent on two GLI binding sites. A schematic view of constructs with mutated bs1, bs2, and bs3 is shown. D, luciferase reporter constructs were either co-transfected with GLI2act expression construct or empty vector as control (pc, right graph). Mutation of bs1 or bs3 or a combination of both abolished GLI2act-dependent activation of the FST promoter, whereas mutation of bs2 showed no influence on activation.
GLI2 Specificity Is Controlled by Regulatory Elements in the FST Promoter—GLI1 and GLI2 bind in vitro to the same consensus sequence (GACCACCCA) with comparable affinities (11), suggesting that differences in target gene specificity are primarily determined by interactions or combinatorial promoter binding with cofactors rather than differences in binding site affinity. We therefore set out to localize cis-regulatory sequences that may serve as binding sites for potential cofactors regulating GLI target specificity. Using luciferase reporter assays we identified a 518-bp region (fragment A) downstream of the GLI binding sites bs1 to bs3, which influences GLI2 specificity (Fig. 5A). A luciferase reporter construct (FSTdelA) containing bs1 and bs3 but lacking this 518-bp region (Fig. 5A, schematic) was co-transfected with either GLI1 or GLI2act or an empty expression vector as control (pc) into HaCaT cells. As shown in Fig. 5A, deletion of fragment A led to an increase in overall activation with concomitant loss of GLI2 specificity compared with the wild-type promoter (FSTprom) (Fig. 5A). Overall GLI1 activity when normalized to GLI2act (100%) increases from ∼5% on FSTprom to about 80% on FSTdelA, which is comparable to the normal ratio of activation by GLI1 to GLI2act on unspecific promoters such as PTCH (Fig. 4B) or the artificial GLI reporter construct 6bsSV40 (Fig. 5B, inset). The increase in overall promoter activity of FSTdelA compared with FSTprom suggests that the 518-bp fragment A contains negative regulatory sequences which specifically GLI2 but not GLI1 is able to override. In fact, fragment A by itself can mediate GLI2 specificity as was shown by luciferase reporter assays using the artificial reporter construct 6bsSV40 (Fig. 5B). When fragment A is inserted to replace the SV40 promoter between the 6xGLI binding sites sequence and the luciferase gene (6bsFST-A), GLI2 specificity is conferred upon the unspecific construct 6bsSV40 (Fig. 5B). In the absence of 6xGLI binding sites (FST-A) no luciferase activity is observed (Fig. 5B). These results suggest that the relative activities of GLI1 and GLI2 on the FST promoter are controlled by sequences other than the GLI binding sites. Although ChIP results show binding of GLI1 to upstream GLI binding sites (Fig. 3D), activation is only possible in the absence of fragment A pointing to modulation of transcriptional activity by specific cofactors.
FIGURE 5.
GLI2-specific activation of the FST promoter is controlled by sequences located downstream of the GLI binding sites. A, HaCaT cells were co-transfected with luciferase reporter constructs FSTprom and FSTdelA (upper panel) and GLI expression constructs as indicated. The 518bp (fragment A) deletion in construct FSTdelA resulted in reduction of GLI2 specificity associated with an overall increase in promoter activity compared with the wild type promoter FSTprom. B, luciferase assay showing that fragment A can confer GLI2 specificity in combination with GLI binding sites (6bsFST-A). The reporter constructs 6bsSV40 (unspecific), 6bsFST-A or FST-A (control) (schematic, top), were co-transfected with either GLI1 or GLI2act expression constructs or empty expression vector (pc).
Preferential Activation of FST by GLI2 Is Mediated by a Region C-terminal to the DNA Binding Domain—Having localized the DNA sequences responsible for GLI2-specific activation of FST expression we next searched for domains within GLI2 involved in specific activation of FST expression. Very roughly, GLIs can be divided into the zinc finger DNA binding domain flanked by an N- and a C-terminal part, which have been shown to contain many regulatory elements controlling stability and activity. In another example of GLI2-specific activation, Regl and colleagues recently used a hybrid GLI protein consisting of GLI2act with the zinc finger domain of GLI1 instead of GLI2 to show that the predominant activation of the human BCL2 promoter by GLI2 is dependent on the presence of the GLI2 zinc finger domain (44). Co-transfection of this GLI hybrid (GLI121) (Fig. 6A) with the FST promoter produced no increase in luciferase activity compared with wild-type GLI1 (Fig. 6B). It therefore seems likely that, in the case of FST, sequences other than the zinc finger domain are responsible for specific transcriptional activation. This is consistent with the result that GLI1 is able to activate the FST promoter provided that fragment A has been deleted (Fig. 5A).
FIGURE 6.
A region C-terminal to the zinc finger domain of GLI2 is responsible for specific activation. A, schematic drawing of chimeric GLI proteins used in the luciferase reporter assays shown in A and B. B, luciferase reporter assay showing that sequences downstream of the GLI2 zinc finger are required for FST promoter activation. Expression constructs GLI1, GLI2act, GLI121, GLI112, and GLI221 were co-transfected with FSTprom reporter into HaCaT cells. Transcription activation of the FST promoter by GLI112 is comparable to GLI2act, whereas the GLI221-activating function is reduced. GLI121 shows comparable activity to GLI1. C, luciferase assay shows that the C-terminal transactivation domain is not responsible for GLI2-specific FST activation. The overall activation potential of GLI1TA2 and GLI2TA1 is slightly reduced on the unspecific GLI reporter 6bsSV40 compared with the respective wild-type GLI proteins (inset), GLI2TA1 is a more potent activator of the FSTprom reporter construct than GLI1 or GLI1TA2.
Numerous publications have described sequences in the C-terminal region, which are important for post-translational modifications, processing, and protein stability or for interaction with other factors of the transcriptional machinery (reviewed in Refs. 56–58). The contribution of these sequences to GLI target gene specificity is unknown. We therefore exchanged the regions C-terminal to the zinc finger between GLI1 and GLI2act, thus generating the chimeric constructs GLI112 and GLI221 (Fig. 6A). GLI112 activates the FST luciferase reporter almost as efficiently as GLI2act, whereas the activity of GLI221 is strongly decreased (Fig. 6B). Both chimeric proteins activate the PTCH reporter (PTCHprom) and the 6xGLI binding site promoter (6bsSV40) at comparable levels (data not shown). These results suggest that the dramatic difference in activation potential on FST between GLI1 and GLI2 is mediated by sequences C-terminal to the zinc finger domain. The transcriptional co-activator CBP/p300, which is required for full transcriptional activity of the Drosophila GLI homologue Cubitus interruptus (59, 60) has been shown to interact with sequences in the C terminus of mammalian GLI3 (15) and GLI26 (but not GLI1 (15). We addressed a possible involvement of CBP in GLI2 activation of the FST promoter using a CBP-specific small interference RNA-mediated knockdown. RNA interference-mediated inhibition of CBP expression (supplemental Fig. S1C) did not affect the activation of the FST promoter in response to GLI2 (supplemental Fig. S1A), suggesting that CBP does not play a major role in conferring GLI2 specificity of the FST promoter. It has been shown that a transactivation domain at the C terminus of the GLI proteins is essential for activation of target gene transcription (12, 24, 61). We therefore exchanged the C-terminal 88 and 123 amino acids of GLI1 and GLI2, respectively, to generate GLI1TA2 and GLI2TA1 (Fig. 6A). Compared with the respective wild-type GLIs the overall transcriptional activation potential of the chimeric proteins was decreased on both the unspecific 6bsSV40 and the FST promoter. GLI2TA1, however, still activates the FST promoter much more efficiently than GLI1TA2 (Fig. 6C). This indicates that, even though the C-terminal transactivation domain is necessary for transcriptional activity, it does not account for the specific activation of FST transcription by GLI2. Together these results suggest that on selected promoters, GLI2 is able to interact with protein factors/DNA sequences in a way that GLI1 cannot.
GLI2 Antagonizes the Effect of activin A and BMP4 in Human Keratinocytes—Having established the activin/BMP antagonist FST as the GLI2 target gene we reasoned that GLI2 expression should antagonize BMP and activin signaling and result in repression of BMP and activin target genes, respectively. In agreement with this hypothesis, GLI2 expression down-regulated a panel of well established activin/BMP target genes (GLI2act-HaCaT, Fig. 7A) such as ID1, IVL, MXD1, CDKN1, SPRR1A, SPRR1B, and SPRR3 (62–66), which in the absence of GLI2 were induced upon BMP4 (Fig. 7B) or activin A (Fig. 7C) treatment. By contrast, GLI1 expression in keratinocytes (GLI1-HaCaT) only moderately reduced levels of activin/BMP targets, consistent with its inability to induce high levels of FST (Fig. 1A). Further, inhibition of activin signaling in HaCaT keratinocytes by treatment with the specific transforming growth factor-β/activin inhibitor SB431542 resulted in down-regulation of activin target genes (Fig. 7C) to levels comparable to those seen in GLI2- and FST-expressing cells (Fig. 7A). These data support the model that GLI2 but not GLI1 can inhibit activin/BMP signaling via activation of FST expression. Finally, it will be interesting to address the role of this antagonism in the context of hair follicle morphogenesis and skin cancer in vivo.
FIGURE 7.
GLI2 down-regulates activin A and BMP4 target genes in HaCaT keratinocytes. A, qRT-PCR analysis of mRNA levels of activin A and BMP4 target genes in HaCaT cells expressing either GLI1 (GLI1-HaCaT) or GLI2act (GLI2act-HaCaT) for 24 h. B, qRT-PCR analysis of mRNA levels of the BMP targets SPRR1A, SPRR1B, SPRR3, ID1, and CDKN1A in HaCaT cells treated with recombinant human BMP4 (400 ng/ml medium). C, mRNA expression levels of the activin A target genes IVL, MXD1, and CDKN1A in HaCaT cells treated with either recombinant human activin A (20 ng/ml medium) or with the synthetic transforming growth factor-β/activin signaling inhibitor SB431542 (10 μm) as indicated. The -fold change refers to the ratio of mRNA levels in treated to untreated cells. Negative values reflect down-regulation of mRNA levels compared with untreated controls.
DISCUSSION
Gli1, Gli2, and Gli3, the mediators of the HH signal, share a highly homologous zinc finger DNA binding domain and bind the same consensus Gli binding sequence GACCACCCA with comparable affinities (11, 53). The predominantly activating factors Gli1 and Gli2 have overlapping and distinct functions and show a high degree of tissue and context specificity in their actions (13, 24, 27, 67). It is, however, difficult to define their specific function, because of their functional redundancy during embryonic development as is evident from studies in mice (17–21) and their partly overlapping expression domains.
Here we provide evidence for distinct transcriptional activator function of GLI1 and GLI2. We show that the expression of FST, an antagonist of activin/BMP signaling, is predominantly up-regulated by GLI2 in human keratinocytes. We have found that only GLI2 but not GLI1 nor GLI3 can activate FST transcription from its promoter uncovering clear differences in target gene specificity of GLI proteins. In the 5′ regulatory region of the human FST gene there are three highly conserved GLI binding sites, of which only two are functional. Interestingly, mutation of either site completely abolishes GLI2-mediated activation, which may indicate binding of multiple GLI proteins. Co-occurrence of two or more binding sequences has been observed in other GLI-responsive promoters (44, 50) and is integrated into a new bioinformatic approach for the identification of Gli target genes (11). Indeed, Nguyen and colleagues (27) recently reported interactions of Gli proteins mediated by their zinc finger domains.
The human BCL2 promoter is preferentially activated by GLI2, and it is the DNA binding domain of GLI2 that is mainly contributing to this specificity (44). GLI2-specific activation of FST transcription is likely to be mediated by a different mechanism as we found no significant effect of the GLI2 zinc finger domain on FST activation. Of note, GLI2-specific activation depends not only on the presence of GLI binding sites but also on sequences in a 518-bp fragment downstream of those sites. Upon deletion of this region, GLI2 specificity is lost, and the promoter becomes more similar to the PTCH promoter, which can be activated by all GLI proteins to comparable levels. These sequences also have the potential to transfer GLI2 specificity to an artificial GLI-responsive promoter. In addition, we observed a significant increase in overall promoter activity upon deletion of the 518-bp region, which suggests the existence of negative regulatory elements that can be de-repressed by GLI2 only, not by the other GLI proteins. Although preliminary at this stage, it is noteworthy that an in silico search for potential binding sites of transcriptional activators and repressors within fragment A (supplemental Fig. S2) identified a cluster of four potential binding sites for ZFP161(ZF5), a zinc finger transcription factor known to repress c-myc expression (68). Further, a potential binding site for YY1 was found. YY1 is a prominent repressor of transcription, also in epidermal cells (69, 70). Neither repressor has yet been connected to HH/GLI signaling, and additional studies are necessary to address their potential involvement in the repression of FST expression. A common mechanism mediating gene repression is DNA methylation in combination with co-repressor proteins (reviewed in Ref. 71). Recently, it was shown that in human adrenocortical cells the FST promoter is methylated (72). Transcriptional repression is often accompanied by a deacetylation of histone H3 and H4, characterizing a less active transcriptional status. Chromatin immunoprecipitation at the FST transcription start site did not detect a significant difference between GLI1- and GLI2-expressing keratinocytes for acetylated histone H3 (supplemental Fig. S1D). Whether DNA methylation and/or chromatin remodeling proteins are involved in mediating GLI2-specific transcriptional activation will be an interesting question to address in future studies.
Although the importance of context-dependent regulation of Gli function is evident, not much is known about target gene specificity of the different Gli proteins, and only a limited number of interacting proteins directly involved in transcriptional activation has been identified (15, 73–77). From our results with different chimeric GLI proteins we conclude that a region C-terminal to the DNA binding domain is the main contributor to GLI2-specific activation of the FST promoter. The amino acid sequence of the C-terminal part of all Gli proteins is only moderately conserved. In addition to several phosphorylation target sites and motives involved in protein degradation, an acidic transactivation domain at the very end of the C terminus shows homology to the TAFII31 binding domain of VP16 and is common to all three Glis (61). Deletion of this region (12, 13, 61) leads to dramatic loss of transactivation function of all three Gli proteins in reporter gene assays. However, our data suggest that this domain is not responsible for GLI2-specific activation of FST, although it seems necessary for full GLI2 activity, pointing to a more complex mechanism of FST activation by GLI2.
It is known that the Drosophila GLI homologue, Cubitus interruptus (Ci) requires interaction with the transcriptional co-activator CBP/p300 for full transcriptional activity (59, 60). In mammals such an interaction has been described for GLI3, while GLI1 does not contain a potential CBP binding motif and has been shown not to interact with CBP (15). In contrast, GLI2 harbors a putative CBP interaction motif, which contributes to activation of selected target gene promoters.6 However, for the activation of the FST promoter by GLI2, our results using CBP knockdown by RNA interference suggest an ancillary role only for CBP. This is further supported by the fact that GLI3, although it has been shown to bind CBP, also lacks the potential to activate FST transcription.
In human skin FST expression broadly overlaps the GLI2 expression domain previously shown by Ikram and colleagues (16). In particular, strong FST expression was detected in the ORS, most intense in a region referred to as putative stem cell niche of human hair follicles (reviewed in Refs. 78–80). This also agrees with a recent report (52) showing that FST is a potential marker for epidermal stem cells. It has been amply demonstrated that HH/GLI signaling is essential for proliferation and downward growth of placode cells (reviewed in Ref. 81). Furthermore, GLI2 was shown to promote proliferation of epidermal cells (36, 82) and to be indispensable for hair follicle development downstream of HH (83), whereas activation of BMP signaling inhibits hair follicle morphogenesis. Consistent with a proposed role of GLI2 in antagonizing activin/BMP signaling via up-regulation of FST, we have shown that GLI2 down-regulates a number of established BMP and activin target genes in human keratinocytes. Several studies have shown that balancing of BMP signaling is crucial for maintaining the integrity of the ORS and for proper differentiation of cells in adjacent layers of the hair follicle thus underscoring the importance of BMP antagonists in this process (reviewed in Ref. 40). Recently it has been suggested that BMP antagonists may also play an important role in expansion of BCC and that BMP proteins inhibit proliferation and promote cell differentiation of BCC tumor cells ex vivo. Accordingly, the BMP antagonist Gremlin 1 (GREM1) can reverse the effect of BMPs on BCC cells, thereby promoting growth of tumor cells (62). We found FST expression in human BCC tumor islands and at highly elevated levels in mouse BCC cell lines derived from Ptch-deficient mice. It is therefore tempting to speculate that up-regulation of the activin/BMP antagonist FST by GLI2 may represent a novel mechanism of how aberrant HH signaling initiates and/or maintains BCC growth.
In summary, we identified GLI2 as a specific activator of FST in human epidermal cells. Its specificity depends on protein sequences downstream of the zinc finger domain and on DNA elements in the FST promoter pointing to a novel mechanism of differential target gene regulation. It is likely to rely on selective interaction of GLI proteins with unknown cofactors rather than on selective DNA binding alone. Given the biological function of FST in epidermal development, regulation of FST by GLI2 reveals a new regulatory interaction of HH/GLI and the activin/BMP signaling pathways by which HH signaling may control proliferation and differentiation in healthy and diseased epidermal tissue.
Acknowledgments
We thank Gerhard Regl for critical reading of the manuscript.
This work was supported by the Austrian Genome Project GENAU Ultrasensitive Proteomics and Genomics II (to A. M. F.), FWF Project 16518-B14 (to F. A.), and the University of Salzburg priority program Biosciences and Health. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3, Tables S1–S3, and additional references.
Footnotes
The abbreviations used are: HH, Hedgehog; BCC, basal cell carcinoma; FST, follistatin; BMP, bone morphogenetic protein; qRT, quantitative reverse transcription; EMSA, electrophoretic mobility shift assay; ChIP, chromatin immunoprecipitation; ORS, outer root sheath; CBP, CREB-binding protein; CREB, cAMP-response element-binding protein; FSTprom, FST promoter reporter plasmid; PTCHprom, PTCH reporter.
C. Schmid, unpublished data.
A.-M. Frischauf, unpublished observations.
F. Aberger, unpublished results.
References
- 1.Pasca di Magliano, M., and Hebrok, M. (2003) Nat. Rev. Cancer 3 903-911 [DOI] [PubMed] [Google Scholar]
- 2.Lum, L., and Beachy, P. A. (2004) Science 304 1755-1759 [DOI] [PubMed] [Google Scholar]
- 3.Ruiz i Altaba, A., Sanchez, P., and Dahmane, N. (2002) Nat. Rev. Cancer 2 361-372 [DOI] [PubMed] [Google Scholar]
- 4.Ingham, P. W., and McMahon, A. P. (2001) Genes Dev. 15 3059-3087 [DOI] [PubMed] [Google Scholar]
- 5.Hooper, J. E., and Scott, M. P. (2005) Nat. Rev. Mol. Cell. Biol. 6 306-317 [DOI] [PubMed] [Google Scholar]
- 6.Riobo, N. A., Haines, G. M., and Emerson, C. P., Jr. (2006) Cancer Res. 66 839-845 [DOI] [PubMed] [Google Scholar]
- 7.Huntzicker, E. G., Estay, I. S., Zhen, H., Lokteva, L. A., Jackson, P. K., and Oro, A. E. (2006) Genes Dev. 20 276-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan, Y., and Wang, B. (2007) J. Biol. Chem. 282 10846-10852 [DOI] [PubMed] [Google Scholar]
- 9.Wang, B., and Li, Y. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 33-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan, Y., Bai, C. B., Joyner, A. L., and Wang, B. (2006) Mol. Cell Biol. 26 3365-3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hallikas, O., Palin, K., Sinjushina, N., Rautiainen, R., Partanen, J., Ukkonen, E., and Taipale, J. (2006) Cell 124 47-59 [DOI] [PubMed] [Google Scholar]
- 12.Sasaki, H., Nishizaki, Y., Hui, C., Nakafuku, M., and Kondoh, H. (1999) Development 126 3915-3924 [DOI] [PubMed] [Google Scholar]
- 13.Ruiz i Altaba, A. (1999) Development 126 3205-3216 [DOI] [PubMed] [Google Scholar]
- 14.Wang, B., Fallon, J. F., and Beachy, P. A. (2000) Cell 100 423-434 [DOI] [PubMed] [Google Scholar]
- 15.Dai, P., Akimaru, H., Tanaka, Y., Maekawa, T., Nakafuku, M., and Ishii, S. (1999) J. Biol. Chem. 274 8143-8152 [DOI] [PubMed] [Google Scholar]
- 16.Ikram, M. S., Neill, G. W., Regl, G., Eichberger, T., Frischauf, A. M., Aberger, F., Quinn, A., and Philpott, M. (2004) J. Invest. Dermatol. 122 1503-1509 [DOI] [PubMed] [Google Scholar]
- 17.Park, H. L., Bai, C., Platt, K. A., Matise, M. P., Beeghly, A., Hui, C. C., Nakashima, M., and Joyner, A. L. (2000) Development 127 1593-1605 [DOI] [PubMed] [Google Scholar]
- 18.Ding, Q., Motoyama, J., Gasca, S., Mo, R., Sasaki, H., Rossant, J., and Hui, C. C. (1998) Development 125 2533-2543 [DOI] [PubMed] [Google Scholar]
- 19.Matise, M. P., Epstein, D. J., Park, H. L., Platt, K. A., and Joyner, A. L. (1998) Development 125 2759-2770 [DOI] [PubMed] [Google Scholar]
- 20.Mo, R., Freer, A. M., Zinyk, D. L., Crackower, M. A., Michaud, J., Heng, H. H., Chik, K. W., Shi, X. M., Tsui, L. C., Cheng, S. H., Joyner, A. L., and Hui, C. (1997) Development 124 113-123 [DOI] [PubMed] [Google Scholar]
- 21.Bai, C. B., and Joyner, A. L. (2001) Development 128 5161-5172 [DOI] [PubMed] [Google Scholar]
- 22.Stamataki, D., Ulloa, F., Tsoni, S. V., Mynett, A., and Briscoe, J. (2005) Genes Dev. 19 626-641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruiz i Altaba, A., Nguyen, V., and Palma, V. (2003) Curr. Opin. Genet. Dev. 13 513-521 [DOI] [PubMed] [Google Scholar]
- 24.Ruiz i Altaba, A. (1998) Development 125 2203-2212 [DOI] [PubMed] [Google Scholar]
- 25.Karlstrom, R. O., Tyurina, O. V., Kawakami, A., Nishioka, N., Talbot, W. S., Sasaki, H., and Schier, A. F. (2003) Development 130 1549-1564 [DOI] [PubMed] [Google Scholar]
- 26.Tyurina, O. V., Guner, B., Popova, E., Feng, J., Schier, A. F., Kohtz, J. D., and Karlstrom, R. O. (2005) Dev. Biol. 277 537-556 [DOI] [PubMed] [Google Scholar]
- 27.Nguyen, V., Chokas, A. L., Stecca, B., and Altaba, A. R. (2005) Development 132 3267-3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahn, H., Wicking, C., Zaphiropoulous, P. G., Gailani, M. R., Shanley, S., Chidambaram, A., Vorechovsky, I., Holmberg, E., Unden, A. B., Gillies, S., Negus, K., Smyth, I., Pressman, C., Leffell, D. J., Gerrard, B., Goldstein, A. M., Dean, M., Toftgard, R., Chenevix-Trench, G., Wainwright, B., and Bale, A. E. (1996) Cell 85 841-851 [DOI] [PubMed] [Google Scholar]
- 29.Johnson, R. L., Rothman, A. L., Xie, J., Goodrich, L. V., Bare, J. W., Bonifas, J. M., Quinn, A. G., Myers, R. M., Cox, D. R., Epstein, E. H., Jr., and Scott, M. P. (1996) Science 272 1668-1671 [DOI] [PubMed] [Google Scholar]
- 30.Xie, J., Murone, M., Luoh, S. M., Ryan, A., Gu, Q., Zhang, C., Bonifas, J. M., Lam, C. W., Hynes, M., Goddard, A., Rosenthal, A., Epstein, E. H., Jr., and de Sauvage, F. J. (1998) Nature 391 90-92 [DOI] [PubMed] [Google Scholar]
- 31.Oro, A. E., Higgins, K. M., Hu, Z., Bonifas, J. M., Epstein, E. H., Jr., and Scott, M. P. (1997) Science 276 817-821 [DOI] [PubMed] [Google Scholar]
- 32.Grachtchouk, M., Mo, R., Yu, S., Zhang, X., Sasaki, H., Hui, C. C., and Dlugosz, A. A. (2000) Nat. Genet. 24 216-217 [DOI] [PubMed] [Google Scholar]
- 33.Nilsson, M., Unden, A. B., Krause, D., Malmqwist, U., Raza, K., Zaphiropoulos, P. G., and Toftgard, R. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 3438-3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dahmane, N., Lee, J., Robins, P., Heller, P., and Ruiz i Altaba, A. (1997) Nature 389 876-881 [DOI] [PubMed] [Google Scholar]
- 35.Ghali, L., Wong, S. T., Green, J., Tidman, N., and Quinn, A. G. (1999) J. Invest. Dermatol. 113 595-599 [DOI] [PubMed] [Google Scholar]
- 36.Hutchin, M. E., Kariapper, M. S., Grachtchouk, M., Wang, A., Wei, L., Cummings, D., Liu, J., Michael, L. E., Glick, A., and Dlugosz, A. A. (2005) Genes Dev. 19 214-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, Y., Yoon, J. W., Xiao, X., Dean, N. M., Monia, B. P., and Marcusson, E. G. (2007) Cancer Res. 67 3583-3593 [DOI] [PubMed] [Google Scholar]
- 38.Eichberger, T., Sander, V., Schnidar, H., Regl, G., Kasper, M., Schmid, C., Plamberger, S., Kaser, A., Aberger, F., and Frischauf, A. M. (2006) Genomics 87 616-632 [DOI] [PubMed] [Google Scholar]
- 39.Harrison, C. A., Gray, P. C., Vale, W. W., and Robertson, D. M. (2005) Trends Endocrinol. Metab. 16 73-78 [DOI] [PubMed] [Google Scholar]
- 40.Botchkarev, V. A., and Sharov, A. A. (2004) Differentiation 72 512-526 [DOI] [PubMed] [Google Scholar]
- 41.Matzuk, M. M., Lu, N., Vogel, H., Sellheyer, K., Roop, D. R., and Bradley, A. (1995) Nature 374 360-363 [DOI] [PubMed] [Google Scholar]
- 42.Patel, K., Makarenkova, H., and Jung, H. S. (1999) Mech. Dev. 86 51-62 [DOI] [PubMed] [Google Scholar]
- 43.Nakamura, M., Matzuk, M. M., Gerstmayer, B., Bosio, A., Lauster, R., Miyachi, Y., Werner, S., and Paus, R. (2003) FASEB J. 17 497-499 [DOI] [PubMed] [Google Scholar]
- 44.Regl, G., Kasper, M., Schnidar, H., Eichberger, T., Neill, G. W., Philpott, M. P., Esterbauer, H., Hauser-Kronberger, C., Frischauf, A. M., and Aberger, F. (2004) Cancer Res. 64 7724-7731 [DOI] [PubMed] [Google Scholar]
- 45.Regl, G., Neill, G. W., Eichberger, T., Kasper, M., Ikram, M. S., Koller, J., Hintner, H., Quinn, A. G., Frischauf, A. M., and Aberger, F. (2002) Oncogene 21 5529-5539 [DOI] [PubMed] [Google Scholar]
- 46.Xie, J., Aszterbaum, M., Zhang, X., Bonifas, J. M., Zachary, C., Epstein, E., and McCormick, F. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 9255-9259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.So, P. L., Langston, A. W., Daniallinia, N., Hebert, J. L., Fujimoto, M. A., Khaimskiy, Y., Aszterbaum, M., and Epstein, E. H., Jr. (2006) Exp. Dermatol. 15 742-750 [DOI] [PubMed] [Google Scholar]
- 48.Martin, K. J., Graner, E., Li, Y., Price, L. M., Kritzman, B. M., Fournier, M. V., Rhei, E., and Pardee, A. B. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 2646-2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dickson, M. A., Hahn, W. C., Ino, Y., Ronfard, V., Wu, J. Y., Weinberg, R. A., Louis, D. N., Li, F. P., and Rheinwald, J. G. (2000) Mol. Cell Biol. 20 1436-1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kasper, M., Schnidar, H., Neill, G. W., Hanneder, M., Klingler, S., Blaas, L., Schmid, C., Hauser-Kronberger, C., Regl, G., Philpott, M. P., and Aberger, F. (2006) Mol. Cell Biol. 26 6283-6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sugino, K., Kurosawa, N., Nakamura, T., Takio, K., Shimasaki, S., Ling, N., Titani, K., and Sugino, H. (1993) J. Biol. Chem. 268 15579-15587 [PubMed] [Google Scholar]
- 52.Ohyama, M., Terunuma, A., Tock, C. L., Radonovich, M. F., Pise-Masison, C. A., Hopping, S. B., Brady, J. N., Udey, M. C., and Vogel, J. C. (2006) J. Clin. Invest. 116 249-260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kinzler, K. W., and Vogelstein, B. (1990) Mol. Cell Biol. 10 634-642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Agren, M., Kogerman, P., Kleman, M. I., Wessling, M., and Toftgard, R. (2004) Gene (Amst.) 330 101-114 [DOI] [PubMed] [Google Scholar]
- 55.Vokes, S. A., Ji, H., McCuine, S., Tenzen, T., Giles, S., Zhong, S., Longabaugh, W. J., Davidson, E. H., Wong, W. H., and McMahon, A. P. (2007) Development 134 1977-1989 [DOI] [PubMed] [Google Scholar]
- 56.Jiang, J. (2006) Cell Cycle 5 2457-2463 [DOI] [PubMed] [Google Scholar]
- 57.Kasper, M., Regl, G., Frischauf, A. M., and Aberger, F. (2006) Eur. J. Cancer 42 437-445 [DOI] [PubMed] [Google Scholar]
- 58.Wang, Y., McMahon, A. P., and Allen, B. L. (2007) Curr. Opin. Cell Biol. 19 159-165 [DOI] [PubMed] [Google Scholar]
- 59.Akimaru, H., Chen, Y., Dai, P., Hou, D. X., Nonaka, M., Smolik, S. M., Armstrong, S., Goodman, R. H., and Ishii, S. (1997) Nature 386 735-738 [DOI] [PubMed] [Google Scholar]
- 60.Chen, Y., Goodman, R. H., and Smolik, S. M. (2000) Mol. Cell Biol. 20 1616-1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoon, J. W., Liu, C. Z., Yang, J. T., Swart, R., Iannaccone, P., and Walterhouse, D. (1998) J. Biol. Chem. 273 3496-3501 [DOI] [PubMed] [Google Scholar]
- 62.Sneddon, J. B., Zhen, H. H., Montgomery, K., van de Rijn, M., Tward, A. D., West, R., Gladstone, H., Chang, H. Y., Morganroth, G. S., Oro, A. E., and Brown, P. O. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 14842-14847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seishima, M., Nojiri, M., Esaki, C., Yoneda, K., Eto, Y., and Kitajima, Y. (1999) J. Invest. Dermatol. 112 432-436 [DOI] [PubMed] [Google Scholar]
- 64.Pardali, K., Kowanetz, M., Heldin, C. H., and Moustakas, A. (2005) J. Cell Physiol. 204 260-272 [DOI] [PubMed] [Google Scholar]
- 65.Gomes, W. A., and Kessler, J. A. (2001) Dev. Biol. 237 212-221 [DOI] [PubMed] [Google Scholar]
- 66.Werner, S., Beer, H. D., Mauch, C., Luscher, B., and Werner, S. (2001) Oncogene 20 7494-7504 [DOI] [PubMed] [Google Scholar]
- 67.Lipinski, R. J., Gipp, J. J., Zhang, J., Doles, J. D., and Bushman, W. (2006) Exp. Cell Res. 312 1925-1938 [DOI] [PubMed] [Google Scholar]
- 68.Numoto, M., Niwa, O., Kaplan, J., Wong, K. K., Merrell, K., Kamiya, K., Yanagihara, K., and Calame, K. (1993) Nucleic Acids Res. 21 3767-3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu, X., Kawachi, Y., Nakamura, Y., Sakurai, H., Hirota, A., Banno, T., Takahashi, T., Roop, D. R., and Otsuka, F. (2004) J. Invest. Dermatol. 123 1120-1126 [DOI] [PubMed] [Google Scholar]
- 70.Alvarez-Salas, L. M., Benitez-Hess, M. L., and Dipaolo, J. A. (2005) Int. J. Oncol. 26 259-266 [DOI] [PubMed] [Google Scholar]
- 71.Klose, R. J., and Bird, A. P. (2006) Trends Biochem. Sci. 31 89-97 [DOI] [PubMed] [Google Scholar]
- 72.Utriainen, P., Liu, J., Kuulasmaa, T., and Voutilainen, R. (2006) J. Endocrinol. 188 305-310 [DOI] [PubMed] [Google Scholar]
- 73.Mao, J., Maye, P., Kogerman, P., Tejedor, F. J., Toftgard, R., Xie, W., Wu, G., and Wu, D. (2002) J. Biol. Chem. 277 35156-35161 [DOI] [PubMed] [Google Scholar]
- 74.Morita, K., Lo Celso, C., Spencer-Dene, B., Zouboulis, C. C., and Watt, F. M. (2006) J. Dermatol. Sci. 44 11-20 [DOI] [PubMed] [Google Scholar]
- 75.Callahan, C. A., Ofstad, T., Horng, L., Wang, J. K., Zhen, H. H., Coulombe, P. A., and Oro, A. E. (2004) Genes Dev. 18 2724-2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kogerman, P., Grimm, T., Kogerman, L., Krause, D., Unden, A. B., Sandstedt, B., Toftgard, R., and Zaphiropoulos, P. G. (1999) Nat. Cell Biol. 1 312-319 [DOI] [PubMed] [Google Scholar]
- 77.Barnfield, P. C., Zhang, X., Thanabalasingham, V., Yoshida, M., and Hui, C. C. (2005) Differentiation 73 397-405 [DOI] [PubMed] [Google Scholar]
- 78.Fuchs, E. (2007) Nature 445 834-842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cotsarelis, G. (2006) J. Invest. Dermatol. 126 1459-1468 [DOI] [PubMed] [Google Scholar]
- 80.Ohyama, M. (2007) J. Dermatol. Sci. 46 81-89 [DOI] [PubMed] [Google Scholar]
- 81.Callahan, C. A., and Oro, A. E. (2001) Curr. Opin. Genet. Dev. 11 541-546 [DOI] [PubMed] [Google Scholar]
- 82.Regl, G., Kasper, M., Schnidar, H., Eichberger, T., Neill, G. W., Ikram, M. S., Quinn, A. G., Philpott, M. P., Frischauf, A. M., and Aberger, F. (2004) Oncogene 23 1263-1274 [DOI] [PubMed] [Google Scholar]
- 83.Mill, P., Mo, R., Fu, H., Grachtchouk, M., Kim, P. C., Dlugosz, A. A., and Hui, C. C. (2003) Genes Dev. 17 282-294 [DOI] [PMC free article] [PubMed] [Google Scholar]