Abstract
Mycophenolate mofetil (MMF), a prodrug of mycophenolic acid (MPA), is widely used as an immunosuppressive agent. MPA selectively inhibits inosine monophosphate dehydrogenase (IMPDH), a rate-limiting enzyme for the de novo synthesis of guanine nucleotides, leading to depletion of the guanine nucleotide pool. Its chemotherapeutic effects have been attributed to its ability to induce cell cycle arrest and apoptosis. MPA treatment has also been shown to induce and activate p53. However, the mechanism underlying the p53 activation pathway is still unclear. Here, we show that MPA treatment results in inhibition of pre-rRNA synthesis and disruption of the nucleolus. This treatment enhances the interaction of MDM2 with L5 and L11. Interestingly, knockdown of endogenous L5 or L11 markedly impairs the induction of p53 and G1 cell cycle arrest induced by MPA. These results suggest that MPA may trigger a nucleolar stress that induces p53 activation via inhibition of MDM2 by ribosomal proteins L5 and L11.
Inosine monophosphate dehydrogenase (IMPDH)3 is an essential, rate-limiting enzyme for the de novo synthesis of guanine nucleotides. It catalyzes the nicotinamide adenine dinucleotide (NAD)-dependent oxidation of inosine-5′-monophosphate (IMP) to xanthosine-5′-monophosphate (XMP), which is the committed step in de novo guanine nucleotide biosynthesis (1). This reaction is particularly important to B and T lymphocytes, which are singularly dependent on the de novo pathway, rather than the salvage pathway, for purine biosynthesis (2). There are two separate, but very closely related IMPDH isoenzymes, termed type I and type II, that share 84% amino acid identity (3). Expression of IMPDH, particularly the type II enzyme, is significantly up-regulated in many tumor cells, including leukemia cells (1, 4–7); thus, IMPDH is a target for cancer as well as immunosuppressive chemotherapy. Inhibitors of IMPDH such as mycophenolate mofetil (MMF, Cellcept), a prodrug of mycophenolic acid (MPA), have been used in organ and stem cell transplantation and in autoimmune diseases as highly effective immunosuppressive agents (8).
MPA, the active metabolite of MMF, is a selective inhibitor of IMPDH (8). It can effectively induce cell-cycle arrest in late G1 phase in lymphocytes (9–11), and results in differentiation (12–14) or apoptosis (15–18) in cultured cell lines depending on cell type. It has been shown that MPA treatment inhibits the induction of cyclin D3, a major component of cyclin-dependent kinase (CDK), and degradation of p27kip1, a CDK inhibitor, resulting in G1 cell cycle arrest (9). MPA causes apoptosis in interleukin-3-dependent murine hematopoietic cell lines through inhibiting both the Ras-MAPK and mTOR pathways (15). Also, the induction of apoptosis in multiple myeloma cell lines occurs through both caspase-dependent (18) and caspase-independent (19) mechanisms. However, these signaling pathways are only the potential downstream targets; the upstream mechanisms that sense the depletion of guanine nucleotide and trigger the cell cycle arrest or apoptosis are still not very clear. Interestingly, it has been shown that certain specific inhibitors of ribonucleotide biosynthesis, including MPA, cause a reversible p53-dependent G1 arrest, and p53 has been proposed to serve as a sensor of ribonucleotide pool perturbation (20), although MPA has been shown to inhibit DNA synthesis (21, 22). p53 has also been shown to mediate the cell cycle arrest and apoptosis in response to guanine nucleotide depletion in human neuroblastoma cell lines (17, 23). However, how p53 senses this nucleotide depletion-induced stress remains elusive. In this study, we show that MPA treatment results in drastic reduction of pre-rRNA synthesis and disruption of the nucleolus as evident by the massive translocation of nucleophosmin (also called B23), a nucleolar marker, from the nucleolus to the nucleoplasma. This treatment enhances the interaction of MDM2 with ribosomal proteins L5 and L11. Interestingly, knockdown endogenous L5 or L11 markedly impairs the induction of p53 and G1 cell cycle arrest induced by MPA. These results suggest that MPA may trigger a nucleolar stress, resulting in p53 activation that requires the inhibition of MDM2 activity by ribosomal proteins L5 and L11.
MATERIALS AND METHODS
Cell Lines, Reagents, and Antibodies—Human p53-proficient osteosarcoma U2OS cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 50 units/ml penicillin, and 0.1 mg/ml streptomycin at 37 °C in a 5% CO2 humidified atmosphere as previously described (24). The cells were treated with MPA (Sigma) or vehicle methanol where indicated. Anti-L5 (25), anti-L11 (26), and anti-MDM2 (2A10 and 4B11) (24, 25) antibodies have been described. Anti-p21 (NeoMarkers), anti-p53 (DO-1, Santa Cruz Biotechnology), and anti-MDM2 (SMP14, Santa Cruz Biotechnology) were purchased.
Immunoblot and Co-immunoprecipitation Analyses—Cells were lysed in lysis buffer consisting of 50 mm Tris/HCl, pH 8.0, 0.5% Nonidet P-40, 1 mm EDTA, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, 1 μg/ml pepstatin A, and 1 mm leupeptin. Equal amounts of cleared cell lysates were used for immunoblot analysis as described previously (24). Co-immunoprecipitation assays were conducted as described previously (24). Bound proteins were detected by immunoblot using antibodies as indicated in the figure legends.
RNA Interference (RNAi)—RNAi-mediated knockdown of endogenous L5 and L11 was performed essentially as described (24). The target sequences for L5, L11, and the control scrambled II RNA were described (24, 25). All the siRNA duplexes with a 3′-dTdT overhang were synthesized by Dharmacon (Lafayette, CO). These siRNA duplexes (0.2 μm) were introduced into cells using SilentFect (Bio-Rad) following the manufacturer's protocol. Cells were harvested 48 h after transfection for immunoblot, reverse transcription (RT) real-time PCR, and cell cycle analyses.
Reverse Transcription and Real-time PCR Analyses—Total RNA was isolated from cells using Qiagen RNeasy Mini kits (Qiagen, Valencia, CA). Reverse transcriptions were performed as described (25). Quantitative real-time PCR was performed on an ABI 7300 real-time PCR system (Applied Biosystems) using SYBR Green Mix (Applied Biosystems) as described previously (27). All reactions were carried out in triplicate. The relative gene expression was calculated using the ΔCτ method following the manufacturer's instruction. The primers for p21, mdm2, nucleolin, and GAPDH were described (26, 27). To detect pre-rRNA, two pairs of primers were used. Primers 5′-GCTCTACCTTACCTACCTGG-3′ and 5′-TGAGCCATTCGCAGTTTCAC-3′ were used for amplifying a 112-bp pre-rRNA fragment encompassing 5′-external transcribed sequence (ETS) and 18S rRNA. Primers 5′-TGAGAAGACGGTCGAACTTG-3′ and 5′-TCCGGGCTCCGTTAATGATC-3′ were used to amplify a 96-bp pre-rRNA fragment from 18S rRNA to internal transcribed sequence (ITS)-1. The primers for amplifying 5S rRNA were 5′-GGCCATACCACCCTGAACGC-3′ and 5′-CAGCACCCGGTATTCCCAGG-3′. The primers for tRNATyr were 5′-CCTTCGATAGCTCAGCTGGTAG-3′ and 5′-GGAATCGGAACCAGCGACCTAAG-3′. The primers for acidic ribosomal phosphoprotein (ARPP) P0 were 5′-AGATCAGGGACATGTTGCTGG-3′ and 5′-AGCCTGGAAAAAGGAGGTCTTC-3′.
Immunofluorescence Staining—Cells treated with MPA or methanol control were fixed and stained with monoclonal anti-B23 antibody followed by staining with Alexa Fluor 488 (green) goat anti-mouse antibody (Molecular Probes, OR) as well as 4′,6′-diamidino-2-phenylindole (DAPI) for DNA staining. Stained cells were analyzed under a Zeiss Axiovert 25 fluorescent microscope.
Cell Cycle Analyses—U2OS cells were transfected with scrambled, L5, or L11 siRNA as indicated in figure legends. Cells were fixed and stained in 500 μl of propidium iodide (PI, Sigma) stain buffer (50 μg/ml PI, 30 μg/ml polyethylene glycol 8000, 200 μg/ml RNase A, 0.1% Triton X-100, 0.38 m NaCl, pH 7.2) at 37 °C for 30 min. The cells were then analyzed for DNA content using a Becton Dickinson FACScan flow cytometer. Data were analyzed using the CellQuest and Modfit software programs.
RESULTS
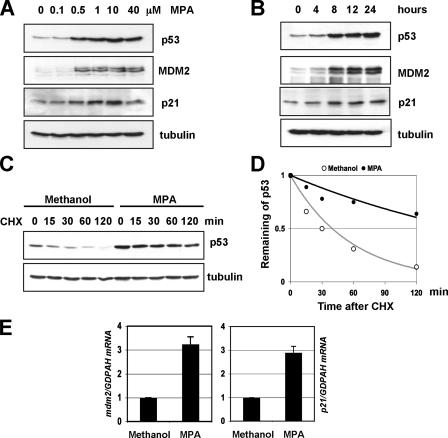
Guanine Nucleotide Depletion by MPA Stabilizes and Activates p53 and Induces G1 Cell Cycle Arrest—MPA inhibits the rate-limiting enzyme IMPDH of de novo guanine nucleotide biosynthesis, leading to depletion of intracellular guanine nucleotide, including GTPs and GDPs (28), and has been shown to activate p53 and induce p53-dependent G1 arrest in certain cell lines (14, 20). To test whether MPA activation of p53 is a general effect, we tested its effect in U2OS cells while also determining the dose and time responses. We treated U2OS cells with different doses of MPA. Cells were harvested at 12 h after the treatment for immunoblot analysis. As shown in Fig. 1A, MPA induced the levels of p53 in a dose-dependent fashion in U2OS cells at concentrations as low as 0.5 μm. Because 10 μm MPA is a clinically relevant dose (29) and also led to a peak induction of p53, we decided to use this dose for the following experiments. To determine the kinetics of MPA-induced p53 activation, we also performed time-dependent response of cells to treatment with 10 μm MPA. As shown in Fig. 1B, the induction of p53 was observed at as early as 4-h post-treatment and reached a platform from 8 to 12 h. Thus, we chose 12 h as a time point for the following experiments. To test whether the induction of p53 by MPA is due to the stabilization of p53, we performed half-life assays. U2OS cells were treated with 10 μm MPA or methanol for 12 h. The cells were then incubated with 50 μg/ml of cycloheximide and harvested at different time points for immunoblot analysis. As shown in Fig. 1, C and D, p53 was markedly stabilized by MPA treatment. The half-life of p53 was increased from about a half-hour in methanol-treated cells to more than 2 h in MPA-treated cells. These results demonstrate that MPA treatment stabilizes p53.
FIGURE 1.
MPA treatment stabilizes and activates p53. A, dose-response of p53 induction and activation by MPA. U2OS cells were treated with different doses of MPA as indicated for 12 h. Cell lysates were assayed for expression of p53, p21, and MDM2 by immunoblot analysis. B, time-dependent effect of MPA on p53 induction and activation. U2OS cells were treated with 10 μmol/liter of MPA for different time courses as indicated. Cell lysates were assayed for expression of p53, p21, and MDM2 by immunoblot analysis. C and D, MPA treatment stabilizes p53. U2OS cells were treated with 10 μmol/liter MPA for 12 h, and then 50 μg/ml cycloheximide was added to the medium. The cells were harvested at different time points as indicated and assayed for levels of p53 and tubulin by immunoblot. The bands were quantified and normalized with loading controls determined by tubulin expression and plotted in D. E, MPA treatment induces the expression of p21 and mdm2 mRNA levels. U2OS cells were treated with methanol or 10 μmol/liter MPA for 12 h. Total RNAs were prepared from cells and retrotranscribed. Real-time PCR analysis was then conducted to determine the relative expression of the p21 and mdm2 mRNA as normalized against GAPDH mRNA.
The induced p53 by MPA treatment was transcriptionally active, because the p53 target p21 and mdm2 mRNA levels were significantly induced, as determined by real-time PCR assays (Fig. 1E). Consistently, the protein levels of p21 and MDM2 were induced by MPA treatment in dose- and time-dependent manners (Fig. 1, A and B). Also, MPA treatment induced G1 arrest and resulted in a loss of the G2/M phase peak in U2OS cells (Fig. 5A). Altogether, these results suggest that guanine nucleotide depletion by MPA treatment induces and activates p53 in U2OS cells.
FIGURE 5.
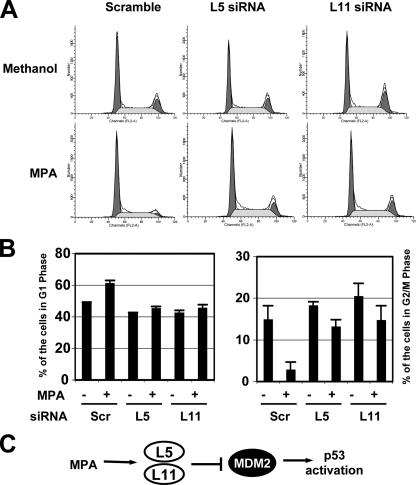
MPA treatment induces G1 cell cycle arrest that requires the ribosomal proteins L5 and L11. U2OS cells were transfected with scrambled, L5, or L11 siRNA followed by treatment with methanol or 10 μmol/liter MPA for 12 h before harvesting as indicated. The cells were then stained with PI followed by flow cytometry analysis for cell cycle profile. The histograms of PI staining from one representative experiment are shown in A. The mean percentages of cells in G1 or G2/M phase are shown in B.
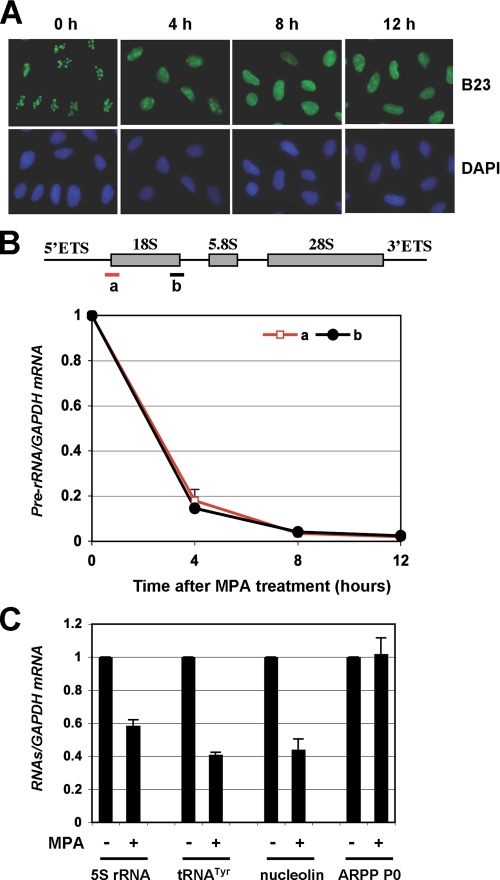
Guanine Nucleotide Depletion Mediated by MPA Treatment Inhibits Pre-rRNA Synthesis and Induces Redistribution of B23 into the Nucleoplasm—We have shown that depletion of intracellular deoxynucleotide pool by 5-fluorouracil (5-FU) resulted in a nucleolar stress-p53 activation response (27). To determine whether p53 activation mediated by MPA-induced guanine nucleotide depletion also involves nucleolar stress (also called ribosomal stress), we first examined the cellular localization of the nucleophosmin, a nucleolar marker, in response to MPA treatment. As shown in Fig. 2A, MPA treatment led to redistribution of B23 into the nucleoplasm, consistent with the previous study (30). These results suggest that MPA treatment affects the integrity of the nucleolus structure, suggesting that MPA may also trigger nucleolar stress.
FIGURE 2.
MPA treatment induces redistribution of B23 into the nucleoplasm and inhibits pre-rRNA synthesis. A, MPA treatment induces redistribution of B23 into the nucleoplasm. U2OS cells were treated with methanol or 10 μmol/liter MPA for different time points (h). The cells were immunostained with anti-B23 (green) and anti-NS (red) as well as DAPI for DNA. B, MPA treatment inhibits pre-rRNA synthesis. U2OS cells were treated with 10 μmol/liter MPA for different time points (h). Total RNAs were prepared from cells and retrotranscribed. Real-time PCR analysis was then conducted to determine the relative expression of the pre-rRNA as normalized against GAPDH mRNA. Similar results are shown using two pairs of primers amplifying a fragment between 5′-ETS and 18S rRNA (a) and a fragment between 18S rRNA and ITS-1 (b), respectively, as indicated in the diagram illustrating the pre-rRNA gene structure in the top panel. C, effect of MPA treatment on levels for 5S rRNA, tRNATyr, nucleolin, and ARPP P0 RNAs. U2OS cells were treated with 10 μmol/liter MPA for 12 h, and total RNAs were prepared from cells and retrotranscribed. Real-time PCR analysis was then conducted to determine the relative expression of the RNAs as normalized against GAPDH mRNA.
Accumulating evidence suggests that perturbation of ribosomal biogenesis mediated by the inhibition of rRNA synthesis, processing, and ribosome assembly causes ribosomal stress, leading to p53 activation (31, 32). For example, inhibition of RNA polymerase I activity by a low dose of actinomycin D (32), loss-of-function mutations of the rRNA-processing factor Bop1 (31), or serum starvation (33) induces p53. To determine whether depletion of GTP by MPA treatment also affect ribosomal biogenesis, we observed the precursor rRNA (pre-rRNA) synthesis in cells in response to MPA. As shown in Fig. 2B, RT-real-time PCR assays using primers to amplify a fragment bridging 5′-ETS and 18S rRNA (fragment a) as well as a fragment between 18S rRNA and ITS-1 (fragment b) clearly indicated that MPA treatment drastically reduced the level of pre-rRNAs. The levels of pre-rRNAs was sharply decreased as early as 4 h after MPA treatment and continuingly decreased at 12 h by more than 50-fold. These results suggest that MPA treatment drastically inhibits rRNA synthesis, resulting in nucleolar stress. We also detected other RNAs, including RNA polymerase (Pol) III-mediated transcripts such as 5S rRNA and tRNATyr, and Pol II-mediated transcripts such as nucleolin and ARPP P0. As shown in Fig. 2C, the levels of 5S rRNA, tRNATyr, and nucleolin mRNA were decreased by 40∼60% upon MPA treatment, whereas the ARPP P0 transcripts were not significantly reduced by the treatment. These results suggest that MPA-mediated guanine nucleotide depletion primarily inhibits rRNA synthesis.
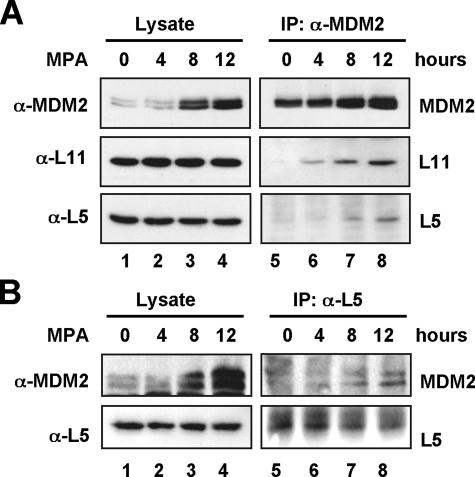
Guanine Nucleotide Depletion Mediated by MPA Treatment Enhances the Interaction of MDM2 with Ribosomal Proteins L5 and L11—Our recent studies as well as studies by others have shown that several ribosomal proteins, including L5, L11, and L23, target the MDM2-p53 feedback loop in response to nucleolar or ribosomal stress (24, 25, 33–37). These ribosomal proteins directly bind to MDM2 and inhibit MDM2-mediated ubiquitylation of p53, thus stabilizing and activating p53 (24, 25, 33–37). As noted above, MPA treatment inhibits rRNA synthesis and results in disruption of the nucleolus, we hypothesized that MPA treatment might also induce the interaction of MDM2 with ribosomal proteins. Indeed, MPA treatment drastically enhanced the interaction of MDM2 with the ribosomal proteins L5 and L11 when anti-MDM2 antibodies were used for immunoprecipitation (Fig. 3A). The increased binding of MDM2 to L5 was true in a reciprocal co-immunoprecipitation using anti-L5 antibodies (Fig. 3B). Because all the L11 antibodies tested were not suitable for co-immunoprecipitation with endogenous proteins (data not shown), reciprocal immunoprecipitation could not be done with the currently available anti-L11 antibodies. These results indicate that MPA-induced p53 activation involves suppression of MDM2 activity by ribosomal proteins, further supporting a nucleolar stress-p53 response in MPA-treated cells.
FIGURE 3.
MPA treatment enhances the interaction of MDM2 with ribosomal proteins L5 and L11. A, U2OS cells were treated with 10 μmol/liter MPA for different time courses as indicated. Cell lysates were immunoprecipitated with anti-MDM2 (4B11 and SMP14) antibodies followed by immunoblot using anti-L5, anti-L11, or anti-MDM2 (2A10) antibodies (lanes 5–8). The lysates were also directly loaded onto an SDS gel for immunoblot analysis with the above antibodies (lanes 1–4). B, U2OS cell lysates prepared as in A were used for immunoprecipitation with anti-L5 antibodies followed by immunoblot using anti-L5 and anti-MDM2 (SMP14) antibodies (lanes 5–8). The lysates were also directly loaded onto an SDS gel for immunoblot analysis with above antibodies (lanes 1–4).
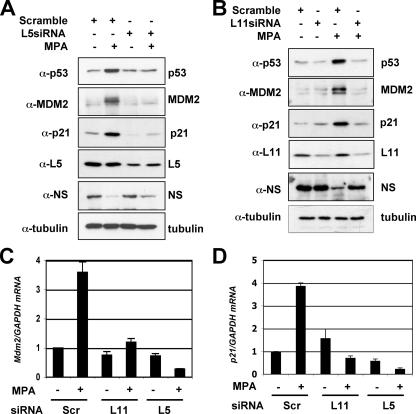
Reduction of Endogenous L5 or L11 by siRNA Alleviates MPA-induced p53 Activation and Cell Cycle Arrest—To validate the requirement of L5 and L11 for MPA-induced p53 activation, we performed siRNA-mediated ablation experiments. Indeed, reduction of either L5 (Fig. 4A) or L11 (Fig. 4B) levels by siRNA markedly inhibited the MPA-induced level of p53 compared with that in scrambled RNA-transfected cells. Consistently, knocking down either L5 or L11 abrogated MPA-induced p21 and MDM2 protein levels (Fig. 4, A and B) as well as their mRNA levels as measured by real-time RT-PCR assays (Fig. 4, C and D). Consistently knocking down either L5 or L11 significantly reduced the MPA-induced G1 cell cycle arrest and rescued the G2/M phase peak (Fig. 5, A and B). These results demonstrate that L5 and L11 are required for MPA-mediated induction of p53 activation and G1 arrest. We did not analyze whether knocking down L23 also inhibits MPA-mediated p53 activation, because knockdown of L23 itself can cause induction and activation of p53 (24, 36). However, it is possible that L23 may also play a role in MPA activation of p53.
FIGURE 4.
MPA-induced p53 activation requires the ribosomal proteins L5 and L11. A, ablation of endogenous L5 by siRNA inhibits MPA-induced p53. U2OS cells were transfected with scrambled or L5 siRNA for 48 h as indicated. Twelve hours before harvesting, the cells were treated with methanol (lanes 1 and 3) or 10 μmol/liter MPA (lanes 2 and 4). Cell lysates were assayed for expression of p53, p21, and MDM2 by immunoblotting with specific antibodies. B, ablation of endogenous L11 by siRNA inhibits MPA-induced p53. U2OS cells were transfected with scrambled or L11 siRNA for 48 h as indicated. Twelve hours before harvesting, the cells were treated with methanol (lanes 1 and 2) or 10 μmol/liter MPA (lanes 3 and 4). Cell lysates were assayed for expression of p53, p21, and MDM2 by immunoblot with specific antibodies. C and D, ablation of endogenous L5 or L11 by siRNA inhibits the levels of p21 and mdm2 mRNA induced by MPA. Total RNAs were prepared from cells transfected with scrambled, L5, or L11 siRNA followed by treatment with methanol or MPA as above (A or B) and retrotranscribed. Real-time PCR analysis was then conducted to determine the expression of the mdm2 (C) and p21 (D) mRNA levels. The expression of GAPDH mRNA was used as control.
DISCUSSION
It has been shown that guanine nucleotide depletion mediated by MPA causes a reversible p53-dependent G1 arrest (20) and induces p53 in human cell lines (17, 23). However, the mechanism underlying this p53 activation pathway remains undetermined. In this study, we found that guanine nucleotide depletion by MPA treatment drastically inhibits pre-rRNA synthesis (Fig. 2B), disrupts the nucleolus resulting in massive translocation of the nucleolar protein B23 into the nucleoplasm (Fig. 2A), and induces p53 activity (Fig. 1). These results suggest that MPA activation of p53 may resemble p53 activation induced by the treatment of cells with a low dose of actinomycin D (Act D), which specifically inhibits RNA polymerase I activity (24). It has been shown that MPA treatment results in decrease of intracellular guanine nucleotide levels by 40–60% depending on dosage and cell type (17, 38). This reduction of guanine nucleotides would presumably result in global reduction of RNA synthesis. Indeed, we have detected the reduction of the levels of several RNA species, including Pol III-mediated transcripts 5S rRNA and tRNATyr and Pol II-mediated transcripts nucleolin mRNA by 40–60% (Fig. 2C). However, the levels in p21 and mdm2 mRNAs, whose genes are transcription targets of p53, were still significantly elevated in response to MPA treatment (Fig. 1E). Likewise, the level of one tested transcript, ARPP P0 mRNA, was not changed significantly (Fig. 2C). Therefore, under the limited supply of intracellular guanine nucleotides, gene transcription is still highly regulated in cells. It is likely that those genes important for cell growth and proliferation would be shut down, while others critical for negating cell growth would be turned on, ensuring the tight coordination of cell growth with cell metabolism. In contrast, the synthesis of pre-rRNA was drastically decreased by MPA treatment at as early as 4 h. These results suggest that guanine nucleotide depletion induced by MPA primarily and efficiently inhibits rRNA synthesis. This might be due to the high demand of nucleotides in rRNA synthesis, as rRNA accounts for up to 80% of total RNA and Pol I-mediated transcription of rRNA represents nearly 60% of the total transcription in the cell (39). It is also possible that guanine nucleotide depletion by MPA may regulate nucleolar trafficking of Pol I transcriptional machineries such as transcription initiation factor TIF-1A (40), thus directly inhibiting Pol I activity. Although it remains to be determined whether MPA inhibition of pre-rRNA synthesis is due to its effect on Pol I activity or a direct consequence of guanine nucleotide depletion, our results suggest that MPA treatment may also trigger nucleolar stress as in the case of Act D treatment.
Nucleolar stress can be triggered by external or internal stimuli leading to perturbation of the ribosomal biogenesis (31, 32). For example, inhibition of RNA polymerase (Pol) I activity by a low dose of Act D (32) and genetic disruption of the Pol I transcription initiation factor TIF-IA (40), inhibition of rRNA processing by loss-of-function mutations of the rRNA processing factor Bop1 (31), or treatment of cells with 5-FU (27), as well as inhibition of overall ribosomal biogenesis by serum starvation (33) or genetic inactivation of ribosomal protein S6 (41) can all induce and activate p53. Recent studies have shown that ribosomal proteins L5, L11, and L23 may play an important role in mediating p53 activation via binding to and inhibiting MDM2 E3 ligase activity toward p53 in response to nucleolar stress (24, 25, 33–37). It is proposed that these ribosomal proteins may be released from the intact ribosomes as ribosome-free ribosomal proteins that target MDM2 in response to nucleolar stress (27, 33). Thus, these ribosomal proteins may also play a role in MPA-mediated p53 activation.
Indeed, knocking down either L5 or L11 by siRNA drastically suppressed the MPA-mediated p53 activation and cell cycle arrest (Figs. 4 and 5). Also, the interaction of MDM2 with L5 and L11 was markedly enhanced by the treatment with MPA (Fig. 3). Therefore, as in the case of p53 activation by Act D and 5-FU, ribosomal proteins L5 and L11 are essential for efficient induction and activation of p53 by MPA treatment. Because knocking down L23 itself can cause induction and activation of p53 (24, 36), it is somewhat difficult to determine whether L23 is also essential for MPA-mediated p53 activation.
In summary, this study not only reveals a mechanistic insight into the MPA activation of p53, but also provides another example of p53 activation of cells in response to nucleolar stress via the ribosomal protein-MDM2 interaction mechanism, further emphasizing the critical role of the ribosomal proteins in mediating p53 checkpoint in response to nucleolar stress, thus ensuing the fine coordination of cell cycle progression with ribosomal biogenesis. Because siRNA knockdown experiments have shown that ribosomal proteins L5, L11, and L23 are required for p53 activation by multiple stimuli (24, 25, 27, 33, 36, 37), it is conceivable that these ribosomal proteins, or perhaps others such as S7 (42) are required for a common nucleolar stress-p53 activation pathway under growth inhibitory conditions. Further experiments on other ribosomal proteins in response to a panel of different internal or external stimuli would be necessary for testing this hypothesis. Also, it remains to be determined how exactly the ribosomal proteins are released from the nucleolus in response to nucleolar stress and target MDM2.
Acknowledgments
We thank the members of the Lu laboratory for active discussion.
This work was supported in part by NCI, National Institutes of Health Grants CA095441, CA93614, CA079721, and CA127724 (to H. L.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: IMPDH, inosine monophosphate dehydrogenase; RT, reverse transcription; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MMF, mycophenolate mofetil; MPA, mycophenolic acid; siRNA, small interfering RNA; 5-FU, 5-fluorouracil; PI, propidium iodide; Act D, actinomycin D; ETS, external transcribed sequence; DAPI, 4′,6-diamidino-2-phenylindole.
References
- 1.Jackson, R. C., Weber, G., and Morris, H. P. (1975) Nature 256 331-333 [DOI] [PubMed] [Google Scholar]
- 2.Allison, A. C., Hovi, T., Watts, R. W., and Webster, A. D. (1977) Ciba Found Symp. 48 207-224 [DOI] [PubMed] [Google Scholar]
- 3.Natsumeda, Y., Ohno, S., Kawasaki, H., Konno, Y., Weber, G., and Suzuki, K. (1990) J. Biol. Chem. 265 5292-5295 [PubMed] [Google Scholar]
- 4.Carr, S. F., Papp, E., Wu, J. C., and Natsumeda, Y. (1993) J. Biol. Chem. 268 27286-27290 [PubMed] [Google Scholar]
- 5.Nagai, M., Natsumeda, Y., Konno, Y., Hoffman, R., Irino, S., and Weber, G. (1991) Cancer Res. 51 3886-3890 [PubMed] [Google Scholar]
- 6.Nagai, M., Natsumeda, Y., and Weber, G. (1992) Cancer Res. 52 258-261 [PubMed] [Google Scholar]
- 7.Gharehbaghi, K., Burgess, G. S., Collart, F. R., Litz-Jackson, S., Huberman, E., Jayaram, H. N., and Boswell, H. S. (1994) Leukemia 8 1257-1263 [PubMed] [Google Scholar]
- 8.Allison, A. C., and Eugui, E. M. (2005) Transplantation 80 S181-S190 [DOI] [PubMed] [Google Scholar]
- 9.Laliberte, J., Yee, A., Xiong, Y., and Mitchell, B. S. (1998) Blood 91 2896-2904 [PubMed] [Google Scholar]
- 10.Heinschink, A., Raab, M., Daxecker, H., Griesmacher, A., and Muller, M. M. (2000) Clin. Chim. Acta 300 23-28 [DOI] [PubMed] [Google Scholar]
- 11.Quemeneur, L., Gerland, L. M., Flacher, M., Ffrench, M., Revillard, J. P., and Genestier, L. (2003) J. Immunol. 170 4986-4995 [DOI] [PubMed] [Google Scholar]
- 12.Inai, K., Tsutani, H., Yamauchi, T., Fukushima, T., Iwasaki, H., Imamura, S., Wano, Y., Nemoto, Y., Naiki, H., and Ueda, T. (2000) Leuk. Res. 24 761-768 [DOI] [PubMed] [Google Scholar]
- 13.Inai, K., Tsutani, H., Yamauchi, T., Nakamura, T., and Ueda, T. (1998) Adv. Exp. Med. Biol. 431 549-553 [DOI] [PubMed] [Google Scholar]
- 14.Messina, E., Micheli, V., and Giacomello, A. (2005) Neurosci. Lett. 375 97-100 [DOI] [PubMed] [Google Scholar]
- 15.Gu, J. J., Gathy, K., Santiago, L., Chen, E., Huang, M., Graves, L. M., and Mitchell, B. S. (2003) Blood 101 4958-4965 [DOI] [PubMed] [Google Scholar]
- 16.Li, G., Segu, V. B., Rabaglia, M. E., Luo, R. H., Kowluru, A., and Metz, S. A. (1998) Endocrinology 139 3752-3762 [DOI] [PubMed] [Google Scholar]
- 17.Messina, E., Gazzaniga, P., Micheli, V., Guaglianone, M. R., Barbato, S., Morrone, S., Frati, L., Agliano, A. M., and Giacomello, A. (2004) Int. J. Cancer 108 812-817 [DOI] [PubMed] [Google Scholar]
- 18.Takebe, N., Cheng, X., Fandy, T. E., Srivastava, R. K., Wu, S., Shankar, S., Bauer, K., Shaughnessy, J., and Tricot, G. (2006) Mol. Cancer. Ther. 5 457-466 [DOI] [PubMed] [Google Scholar]
- 19.Ishitsuka, K., Hideshima, T., Hamasaki, M., Raje, N., Kumar, S., Podar, K., Le Gouill, S., Shiraishi, N., Yasui, H., Roccaro, A. M., Tai, Y. Z., Chauhan, D., Fram, R., Tamura, K., Jain, J., and Anderson, K. C. (2005) Oncogene 24 5888-5896 [DOI] [PubMed] [Google Scholar]
- 20.Linke, S. P., Clarkin, K. C., Di Leonardo, A., Tsou, A., and Wahl, G. M. (1996) Genes Dev. 10 934-947 [DOI] [PubMed] [Google Scholar]
- 21.Duan, D. S., and Sadee, W. (1988) Biochem. J. 255 1045-1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen, B. T., and Sadee, W. (1986) Biochem. J. 234 263-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Messina, E., Barile, L., Lupi, F., and Giacomello, A. (2004) Nucleosides Nucleotides Nucleic Acids 23 1545-1549 [DOI] [PubMed] [Google Scholar]
- 24.Dai, M. S., Zeng, S. X., Jin, Y., Sun, X. X., David, L., and Lu, H. (2004) Mol. Cell. Biol. 24 7654-7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai, M. S., and Lu, H. (2004) J. Biol. Chem. 279 44475-44482 [DOI] [PubMed] [Google Scholar]
- 26.Dai, M. S., Arnold, H., Sun, X. X., Sears, R., and Lu, H. (2007) EMBO J. 26 3332-3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun, X. X., Dai, M. S., and Lu, H. (2007) J. Biol. Chem. 282 8052-8059 [DOI] [PubMed] [Google Scholar]
- 28.Allison, A. C. (2005) Lupus 14 Suppl. 1, s2-s8 [DOI] [PubMed] [Google Scholar]
- 29.Weigel, G., Griesmacher, A., Zuckermann, A. O., Laufer, G., and Mueller, M. M. (2001) Clin. Pharmacol. Therap. 69 137-144 [DOI] [PubMed] [Google Scholar]
- 30.Tsai, R. Y., and McKay, R. D. (2005) J. Cell Biol. 168 179-184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pestov, D. G., Strezoska, Z., and Lau, L. F. (2001) Mol. Cell. Biol. 21 4246-4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashcroft, M., Taya, Y., and Vousden, K. H. (2000) Mol. Cell. Biol. 20 3224-3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhat, K. P., Itahana, K., Jin, A., and Zhang, Y. (2004) EMBO J. 23 2402-2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai, M. S., Shi, D., Jin, Y., Sun, X. X., Zhang, Y., Grossman, S. R., and Lu, H. (2006) J. Biol. Chem. 281 24304-24313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lohrum, M. A., Ludwig, R. L., Kubbutat, M. H., Hanlon, M., and Vousden, K. H. (2003) Cancer Cell 3 577-587 [DOI] [PubMed] [Google Scholar]
- 36.Jin, A., Itahana, K., O'Keefe, K., and Zhang, Y. (2004) Mol. Cell. Biol. 24 7669-7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, Y., Wolf, G. W., Bhat, K., Jin, A., Allio, T., Burkhart, W. A., and Xiong, Y. (2003) Mol. Cell. Biol. 23 8902-8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson, B. S., Deanin, G. G., Standefer, J. C., Vanderjagt, D., and Oliver, J. M. (1989) J. Immunol. 143 259-265 [PubMed] [Google Scholar]
- 39.Warner, J. R. (1999) Trends Biochem. Sci. 24 437-440 [DOI] [PubMed] [Google Scholar]
- 40.Yuan, X., Zhou, Y., Casanova, E., Chai, M., Kiss, E., Grone, H. J., Schutz, G., and Grummt, I. (2005) Mol. Cell 19 77-87 [DOI] [PubMed] [Google Scholar]
- 41.Panic, L., Montagne, J., Cokaric, M., and Volarevic, S. (2007) Cell Cycle 6 20-24 [DOI] [PubMed] [Google Scholar]
- 42.Chen, D., Zhang, Z., Li, M., Wang, W., Li, Y., Rayburn, E. R., Hill, D. L., Wang, H., and Zhang, R. (2007) Oncogene 26 5029-5037 [DOI] [PubMed] [Google Scholar]