Abstract
The NF-κB family member RelB has many properties not shared by other family members such as restricted subunit association and lack of regulation by the classical IκB proteins. We show that the protein level of RelB is significantly reduced in the absence of p100 and reduced even more when both p100 and p105 are absent. RelB stabilizes itself by directly interacting with p100, p105, and their processed products. However, RelB forms complexes with its partners using different interaction modes. Although the C-terminal ankyrin repeat domain of p105 is not involved in the RelB-p105 complex formation, all domains and flexible regions of each protein are engaged in the RelB-p100 complex. In several respects the RelB-p52 and RelB-p100 complexes are unique in the NF-κB family. The N-terminal domain of p100/p52 interacts with RelB but not RelA. The transcriptional activation domain of RelB, but not RelA, directly interacts with the processing region of p100. These unique protein-protein contacts explain why RelB prefers p52 as its dimeric partner for transcriptional activity and is retained in the cytoplasm as an inhibited complex by p100. This association-mediated stabilization of RelB implies a possible role for RelB in the processing of p100 into p52.
The dimeric NF-κB transcription factors are formed from five family members, p50 (NF-κB1), RelA (p65), p52 (NF-κB2), c-Rel, and RelB. These proteins share an ∼300-residue long homologous region located near the N terminus. This element, referred to as the Rel homology region (RHR),5 is responsible for DNA binding, dimerization, inhibitor binding, and nuclear localization. p50 and p52 are the processed products of precursor proteins, p105 and p100, respectively (1, 2). RelA and c-Rel homo- and heterodimers are tightly regulated by a class of inhibitor proteins known as IκB through the formation of stable IκB-NF-κB complexes that are unable to bind DNA. Activation of these dimers requires degradation of IκB. A large number of stimuli activate IκB degradation through phosphorylation of IκB by IκB kinase (IKK) leading to ubiquitination, 26 S proteasome recruitment, and degradation of IκB by the proteasome (3, 4). Signaling pathways leading to NF-κB activation through degradation of classical IκB proteins (IκBα, IκBβ, and IκBε) are classified as the canonical pathways.
RelB displays characteristics that are not shared by the other NF-κB subunits: 1) Prototypical IκB proteins do not regulate RelB-containing NF-κB dimers (5, 6). 2) The RelB homodimer does not have DNA binding activity, suggesting that unlike other members, RelB may not form a stable detectable homodimer in vivo (7). 3) RelB has a N-terminal extension, known as the leucine zipper (LZ) domain because of the presence of a leucine-rich heptad repeat that is not present in other NF-κB family members (8). 4) The x-ray crystal structure of the RelB dimerization domain (DD) revealed an intertwined domain swapped arrangement of the two monomers, suggesting that this RelB domain fold might be unstable and is stabilized by domain intertwining (9). 5) In unstimulated cells, RelB primarily associates with p100 and to a lesser extent with p50 (5). In induced cells the predominant RelB dimer is the RelB-p52 heterodimer. This stringent specificity for both the inhibited complex (RelB-p100) and the transcriptionally active complexes (RelB-p52 and RelB-p52) is unusual in the NF-κB family.
A distinct class of inducers such as LTβ, BAFF, and CD40 activate the noncanonical pathways, which result in the formation of the RelB-p52 heterodimer (10–15). The key event in the noncanonical pathways is the processing of p100 into p52 by the proteasome, which requires the activation of NF-κB-inducing kinase and IKK1 (10, 11, 16, 17). Secondary lymphoid organ development and maintenance are defective in mice deficient in RelB, p52, LTβ, IKK1, or NF-κB-inducing kinase, suggesting a close functional connection between components of the non-canonical pathway (18–23). The present study aims to investigate the structural and functional relationships between RelB and p100/p52. Understanding the reliance of RelB on specific NF-κB and IκB subunits may shed light into the mechanism of p100 processing and the formation of RelB-p52 heterodimer.
In this report, we show that the stability of RelB protein requires the presence of p100/p52 and p105/p50. However, RelB uses two distinct modes to interact with p100 and p105. RelB binds to p105 through the N-terminal p50 region of p105, whereas contacts between p100 and RelB are more extensive. Protein-protein contact between RelB and p52 also involves the N-terminal domains, which is not known to occur in non-RelB NF-κB dimers. The intimate association between p100/p52 and RelB explains why RelB preferentially forms a complex with p100. This association may imply their reliance for each other for their biological function.
EXPERIMENTAL PROCEDURES
Escherichia coli Expression and Protein Purification—Human p100 CTD(344–899), p100 CTDΔGRR(406–899), p100 CTDΔn(489–899), and p100 CTDΔc(406–765) were subcloned into pGEX-4T2 vector (Amersham Biosciences) with an N-terminal GST tag. Mouse RelB(1–400) was cloned into pET15b vector (Novagen) with a N-terminal His tag and a thrombin cleavage site and into pET21d vector (gift from Dr. Greg Van Duynne) with an N-terminal His tag and a TEV cleavage site. RelB(1–400) cloned into pET21D was used for refolding experiment and GST pulldowns. RelB(1–400) cloned into pET15b was used to form the RelB-p52 RHR complex used for GST pulldowns and fluorescence polarization assays. His-tagged RelB was purified the same way as RelB DD, which was described earlier (9). GST-tagged proteins were purified using glutathione-Sepharose 4B affinity column (Amersham Biosciences) followed by size exclusion chromatography. All of the constructs used in this study were verified by DNA sequencing of the entire cDNA fragments.
GST Pulldown—Equal amounts of GST-tagged protein and putative binding partner were mixed in a buffered solution (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm dithiothreitol, and 0.5% Triton X-100) and incubated for 30 min at room temperature before adding 15 μl of slurry of glutathionine-Sepharose 4B. The samples were then incubated for another 30 min at room temperature. The supernatant was removed after centrifugation followed by washing twice with dilution buffer. The samples were boiled with 1× SDS buffer, and bound proteins were separated by SDS-PAGE and visualized by IB or Coomassie staining.
Refolding Experiment—p52 and RelB were expressed and purified as described above. The heterodimer was formed by mixing a 1.1:1 molar ratio of His-RelB to p52 in a denaturation buffer (8 m urea, 250 mm NaCl, 20 mm Tris-HCl, pH 7.5, 1 mm dithiothreitol, 10% glycerol, 0.5 mm EDTA, and 0.5 mm phenylmethylsulfonyl fluoride) with a final protein concentration of 0.70 mg/ml. The heterodimer was formed by slow renaturation by dialysis using the same buffer without urea. After dialysis, the complex was purified by S-Sepharose ion exchange chromatography to remove excess RelB and separate out the heterodimer and p52 homodimer complexes. A salt gradient was run from 50 mm NaCl to 500 mm NaCl. The peak fractions, flow through, and what was loaded onto the S column were resolved by SDS-PAGE.
Fluorescence Polarizations Assay—Fluorescence polarization competition assays were done as described previously (24). Briefly, varying concentrations of p100 CTD were mixed with constant amounts of RelB-p52 heterodimer pre-equilibrated with fluorescein-labeled IgκB DNA in 20 mm Tris-HCl, pH 7.5, and 100 mm NaCl. Fraction DNA bound = (Asample - Amin)/(Amax - Amin).
Transfection—Human p100 and a series of p100 deletion mutants were generated by PCR and cloned into pEYFP-C1 vector (Clontech), which was modified by deleting yellow fluorescent protein and adding a N- or C-terminal FLAG tag. Mouse RelB was generated by PCR and cloned into pEGFP-N1 vector (Clontech) to express C-terminal GFP fusion proteins in mammalian cells. Human embryonic kidney (HEK) 293 cells were cultured with Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mm glutamine, and antibiotics. The cells were seeded in 12-well plates (for direct IB) and 6-well plates (for IP) and transfected the next day using Lipofectamine™ 2000 reagent (Invitrogen). The cell lysates were prepared 48 h post-transfection. An empty vector was transfected when needed to ensure that equal amounts of DNA were used.
Antibodies—The RelB (C-19), β-actin (C-11), and GST (sc-138), and all secondary horseradish peroxidase-conjugated antibodies were purchased from Santa Cruz. FLAG (M2), GFP (A6–455) and penta-His antibodies were from Sigma, Invitrogen, and Qiagen, respectively. p52 antibody was a gift from Dr. Nancy Rice.
Transgenic Cell Lines—All of the constructs were cloned into the pBABE vector, and transgenic cell lines were prepared as described previously (25).
Immunoblotting and Co-immunoprecipitation—Wild type and knock-out 3T3 cells (MEF cells) were grown in Dulbecco's modified Eagle's medium supplemented with 10% calf serum, 2 mm glutamine, and antibiotics. The extracts were prepared by harvesting cells followed by lysis with buffer containing 20 mm Tris-HCl, pH 7.5, 0.2 m NaCl, 1% Triton-X-100, 1 mm EDTA, 2 mm dithiothreitol, 0.1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture (Sigma). 15–30 μg of protein from the total cell extracts were separated by 10% SDS-PAGE followed by transfer to nitrocellulose membrane. Immunodetection was done using specific antibodies. Coimmunoprecipitation was done with 600 μg of identical cell extracts with 0.5 μl of anti-RelB (Santa Cruz) or 0.1 μl of anti-p52 (Dr. Nancy Rice) antibody by incubating the mixture in the lysis buffer overnight in the presence of 15 μl of protein G-Sepharose (Upstate Biotechnology Inc.). Bound complex was washed three times and separated by SDS-PAGE followed by IB.
RNase Protection Assay—MEF cells of wild type and nfκb1-/-/nfκb2-/- double knock-out were grown followed by suspension in TRIzol buffer (contains phenol and guanidine isothiocyanate) and incubated for 5 min at 30 °C. The extracts were mixed with chloroform followed by the separation of aqueous and organic phases by centrifugation. RNAs in the aqueous phase were isolated by alcohol precipitation and pelleted. Radiolabeled RNA probe specific for RelB was prepared by inserting a 300-bp segment of the p100 coding sequence into a T7-polymerase based vector followed by transcription in the presence of a radiolabeled nucleotide. The probe was hybridized with extracted RNAs followed by treatment with RNase, and the protected RNAs were separated by PAGE under denaturing conditions. The products were visualized and quantified by phosphorimaging. The bottom panel shows the mRNA levels of ribosomal protein L32 as a loading control.
Electrophoretic Mobility Shift Assay—The electrophoretic mobility shift assay of the purified recombinant p52 RHR WT and p52 RHR R54A,Y55A double mutant was carried out as described previously (26), using a 38-mer double-stranded DNA containing the human immunodeficiency virus κB site (5′-GCTACAAGGGACTTTCCGCTGGGGACTTTCCAGAGAGG-3′ and 3′-CGATGTTCCCTGAAAGGCGACCCCTGAAAGGTCTCTCC-5′) as the probe.
RESULTS
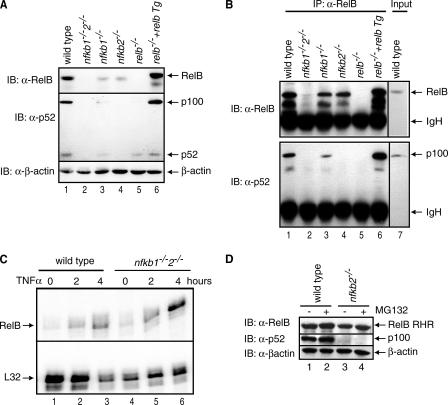
p100 Stabilizes RelB in Vivo—To understand the biochemical relationship between RelB and p100 in vivo, we compared the steady state levels of RelB in wild type (wt), nfκb1-/-, nfκb2-/-, and nfκb1-/-/nfκb2-/- mouse embryonic fibroblast (MEF) cells. Strikingly, RelB is almost absent in nfκb1-/-/nfκb2-/- MEF cells (Fig. 1A, top panel, lane 2). Immunoprecipitation (IP) experiments demonstrate the association between RelB and p100 in MEF cells (Fig. 1B). We confirmed using RNase protection assay that the level of RelB transcript in uninduced nfκb1-/-/nfκb2-/- MEF cells is not reduced when compared with uninduced wt MEF cells (Fig. 1C). Therefore, p50 and p52 do not play an important role in the basal transcription of RelB. The level of RelB is increased in tumor necrosis factor α-induced cells, which confirms prior observations that RelB expression is induced by NF-κB (Fig. 1C). However, the p100 protein level is reduced in nfkb1-/- cells, suggesting that the basal p100 expression is, at least in part, regulated by p105/p50 (Fig. 1A, middle panel, compare lanes 1 and 3). The reduction of p100/p52 protein level thus appears to be the primary reason for reduced levels of RelB. Variation in RelB protein levels with p100 was further confirmed by stably expressing transgenic RelB in relb-/- MEF cells. In these cells, levels of both RelB and p100 are higher than that in wt cells (Fig. 1A, compare lanes 1 and 6). Together these observations suggest that in addition to other modes of regulation, RelB is also regulated at the level of protein stability.
FIGURE 1.
Stability of RelB depends on the presence of p100 protein. A, extracts of wild type, nfkb1-/-, nfkb2-/-, nfkb1-/-/nfkb2-/-, relb-/-, and RelB transgene (Tg) in relb-/- MEF cells were separated by SDS-PAGE followed by immunoblot with RelB antibody (top panel), p52 antibody (middle panel), and β-actin antibody (bottom panel). B, cell extracts from the same cells as in A were used to immunoprecipitate RelB-bound proteins using RelB antibody, and the complexes were separated by SDS-PAGE followed by immunoblot with RelB antibody (top panel) and p52 antibody (bottom panel). C, RNase protection assay of RelB mRNA in wt and nfkb1-/-2-/- MEF cells in resting and tumor necrosis factor α-induced cells in the top panel and a control mRNA (ribosomal protein L32) in the bottom panel. D, extracts of wild type and nfkb2-/- MEF cells were separated by SDS-PAGE followed by immunoblot with RelB antibody (top panel), p52 antibody (middle panel), and β-actin antibody (bottom panel).
To examine whether the proteasome is involved in the regulation of RelB protein levels, we used the proteasome inhibitor MG132 and tested whether the presence of the inhibitor increased RelB protein level. Wild type and nfkb2-/- MEF cells treated with MG132 for 1 h showed a slight enhancement in the RelB protein level as compared with untreated cells (Fig. 1D). The lack of significant enhancement of RelB protein level in the inhibitor treated cells suggests that the proteasome plays some role in RelB degradation; however, additional factors may also be involved.
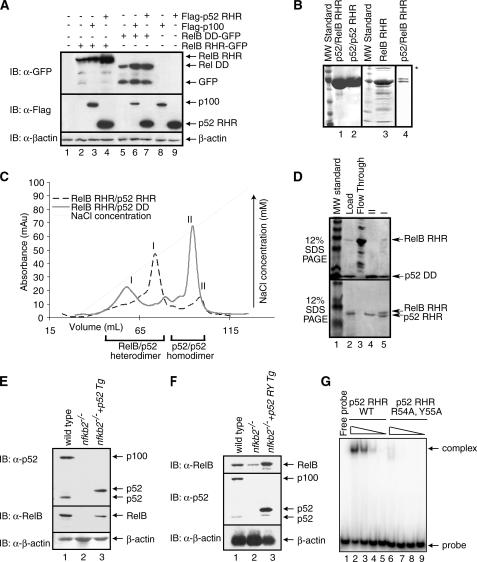
p52 Stabilizes RelB by Forming the RelB-p52 Heterodimer—In stimulated cells, RelB associates with p52, the processed product of p100. This suggests that the stabilization of RelB in induced cells should also be conferred by p52. To investigate whether and how p52 stabilizes RelB, we used a transient transfection system where the RelB RHR-GFP fusion protein was expressed alone or together with FLAG-p52 RHR or FLAG-p100. Western blot analysis reveals enhanced RelB RHR levels with concomitant decrease in the amount of degradation products when RelB is co-expressed with p52 or p100 as compared with when RelB is expressed alone (Fig. 2A, top panel, compare lanes 2–4). In contrast, p52 RHR expresses as a stable protein irrespective of the presence or absence of RelB (Fig. 2A, middle panel, compare lanes 4 and 9). We also tested the stability of RelB DD independently. As shown in Fig. 2A, RelB DD-GFP is also an unstable protein that gives rise to cleaved products. In the presence of p100 or p52, the levels of RelB DD are enhanced. These observations led to the suggestion that unstable RelB RHR can be stabilized by both p100 and p52 RHR.
FIGURE 2.
RelB RHR stabilization by p52 in vitro. A, Western blot analysis of the steady state levels of RelB RHR and RelB DD proteins in HEK 293 cells transfected with FLAG-p100, FLAG-p52, and RelB-GFP in different combinations. In the presence of p100 and p52, RelB protein levels are enhanced. B, Coomassie-stained SDS-PAGE showing purity of p52-RelB heterodimer, p52 homodimer, and RelB RHR homodimer. A lower concentration of p52-RelB heterodimer is shown in lane 4 to visualize the separation between the two proteins. RelB continuously degrades during purification, and the degradation products are seen in the gel (lane 4). Aggregated species of RelB in lane 3 is marked by an asterisk. C, co-refolded mixtures of RelB RHR and p52 RHR (black), and RelB RHR and p52 DD (gray) were separated by cation exchange (S-Sepharose column) chromatography. D, Coomassie-stained SDS-PAGE of samples from the S column chromatography of the RelB RHR-p52 DD (top panel) and RelB RHR-p52 RHR (bottom panel). The load appears to contain excess p52, which masks the RelB RHR. E, Western blot analysis of the steady state levels of p100/p52 (top panel) and RelB (middle panel) in wt, nfkb2-/- MEF and p52-reconstituted nfkb2-/- cells. Reconstituted p52 migrates higher due to the exact site of processing of p100 is unknown. F, DNA binding-defective p52 mutant stabilizes RelB. Western blot analysis of the steady state levels of p100/p52 (top panel) and RelB (middle panel) in wt, nfkb2-/- MEF, and p52-reconstituted nfkb2-/- cells. Reconstituted p52 migrates higher due to the exact site of processing of p100 is unknown. G, electrophoretic mobility shift assay analysis of wild type p52 and p52 R54A,Y55A double mutant. Lanes 2 and 6, 3 and 7, 4 and 8, and 5 and 9 contain 1100, 250, 25, and 2.5 nm wild type and double mutant p52, respectively.
We next carried out a series of in vitro experiments to examine the relationship between RelB and p52. In contrast to other NF-κB subunits, RelB RHR is difficult to purify from an E. coli expression system because it continuously degrades during purification (Fig. 2B, lane 3). The most likely explanation is that the folding stability of RelB RHR is weak, allowing E. coli proteases to cleave the unfolded regions of RelB RHR. When RelB RHR is co-folded with p52 RHR, a stable RelB-p52 RHR heterodimer is formed that can be further purified to homogeneity using an ion exchange chromatography step (Fig. 2, B and C). Intriguingly, our attempts to generate a heterodimer between RelB RHR and p52 DD were not successful. Most of the RelB RHR remained free of p52 DD after a refolding step. The free RelB RHR does not bind to the ion exchange column and therefore comes out in the flow through of the column (Fig. 2D, top panel, lane 3). Only a small amount of the complex could be generated, which remained susceptible to degradation (Fig. 2D, top panel, lane 5). This suggests that the entire RHR of p52 is important for the stability of the RelB-p52 heterodimer and that the NTD of p52 plays a role in stabilizing the RelB-p52 heterodimer. Similar to the RelB-p52 heterodimer, we also found that the formation of the RelB-p50 heterodimer required the NTD of p50 (data not shown). In contrast, both the RelA-p50 RHR heterodimer and the RelA DD-p50 RHR heterodimer can be formed with equivalent efficiencies (24). Therefore, the requirement of the NTD of p52 and p50 for the stability of RelB appears to be unique to RelB.
To further confirm the stabilization role of p52 for RelB, we have reconstituted p52 in nfκb2-/- MEF cells. In these cells, RelB protein level is significantly higher than the RelB protein level in cells deficient in p100/p52 (Fig. 2E, middle panel, compare lanes 2 and 3). These experiments demonstrate that RelB can be stabilized by p52 and p50 and that RelB has a unique interaction with p52 and p50. It has been recently shown that the constitutive processing of truncated p100 into p52 takes place in the nucleus on DNA. Therefore, it is possible that the stabilization of RelB by p52 may require p52, which is competent in DNA binding. To test this we have generated stable cells expressing the p52 R54A,Y55A double mutant. These two residues are invariant across the NF-κB family and have been shown to contact DNA in all NF-κB-DNA complexes. Similar mutants in RelA and p50 significantly affect their DNA binding activities both in vitro and in cells (27). As expected, the double mutant is highly defective in κB DNA binding (Fig. 2G). Our results show that this DNA binding-defective mutant of p52 is fully capable of stabilizing RelB in nfkb2-/- cells (Fig. 2F). Taken together these results demonstrate that both p100 and p52 stabilize RelB.
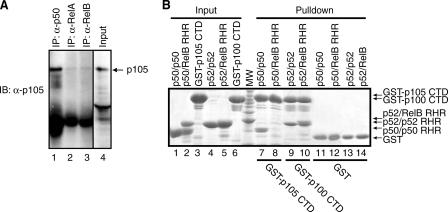
p50/p105 Contributes to RelB Stabilization—As mentioned earlier, previous transfection-based experiments demonstrated RelB interaction specificity for both p100 and p105. We wished to examine whether endogenous p105 and RelB also interact. Indeed IP results show that p105 interacts with RelB to a similar extent as RelA (Fig. 3A). Altogether, these observations suggest that RelB interacts with both the precursor/product pairs, p100/p52 and p105/p50. These results also provide an explanation for the partial stabilization of RelB protein in the absence of p100/p52.
FIGURE 3.
p105 interacts with RelB in a distinct mode. A, cell extract from HeLa cells were used to immunoprecipitate RelB, p50, and RelA-bound p105 using RelB, p50, and RelA antibodies. The complexes were separated by SDS-PAGE followed by immunoblot with p105 antibody. B, in vitro GST pulldown experiments demonstrating the difference in the binding interaction between p100 CTD (IκBδ) and p105 CTD (IκBγ) with p52-RelB. Lane 8 demonstrates the lack of interaction of p105 CTD with p50/RelB versus the stable interaction between p100 CTD and p52-RelB (lane 10). Lanes 7 and 9 are positive controls of p105 CTD interacting with p50 and p100 CTD with p52, respectively. Lanes 1–6 show the inputs, and lanes 11–14 show the controls.
It has been shown that unlike p100, p105 was unable to retain RelB in the cytoplasm (5). We reasoned that despite strong sequence and structural homology, p105 and p100 might bind RelB differently. To test this, we expressed both p100 and p105 CTDs as GST fusion proteins and carried out in vitro GST pulldown experiments. Surprisingly, the CTD of p105 does not bind the RelB-p50 heterodimer, although it binds the p50 homodimer complex strongly (Fig. 3B, compare lanes 7 and 8). In contrast, the CTD of p100 binds both RelB-p52 heterodimer and p52 homodimer. It appears that the RelB-p105 complex is unique where RelB represses the ability of p50 to interact with the CTD of p105. The differential modes of complex formation may explain differences in the nucleocytoplasmic distribution of the RelB-p100 and RelB-p105 complexes.
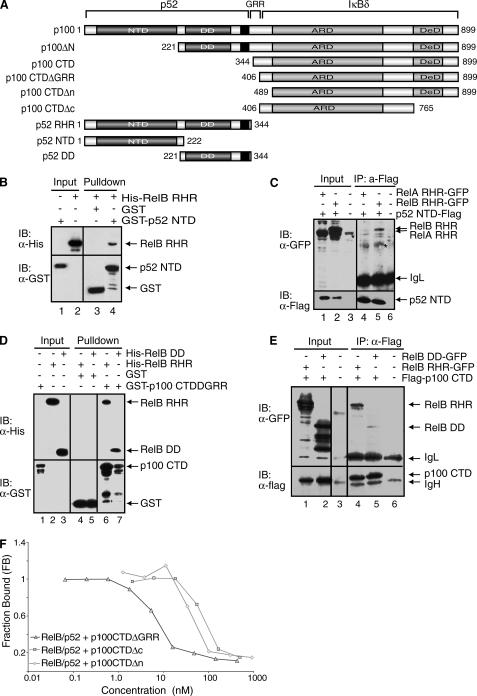
The RelB-p100 and RelB-p52 Complexes Engage Multiple Domains of Both p100 and p52—To understand the mechanism of highly specific complex formation between RelB and p100, we investigated the role of each of the functional and structural domains of p100. p100 can be divided into five regions based on the known domain folds and/or functional importance: the NTD, DD, and the flexible C terminus (CTD, also known as IκBδ), which includes the glycine-rich region (GRR), ankyrin repeat domain (ARD), and the death domain (Fig. 4A). Both in vitro and cell-based co-precipitation experiments reveal that the NTD of p52 interacts with RelB RHR (Fig. 4, B and C). These results are consistent with earlier observations that the p52 NTD plays a role in stabilizing the RelB-p52 heterodimer. However, it should be noted that this interaction is weak, and perhaps the relevance of this weak contact is only appreciated in the context of the entire complex where multiple weak contacts contribute to stabilize the native complex. Furthermore, this interaction appears to be specific to the RelB-p52 complex because no such interaction is observed in the p50-RelA complex (data not shown). To further confirm this, we cotransfected p52 NTD-FLAG and RelA RHR-GFP into HEK 293 cells. As expected, RelA does not interact with the NTD of p52 (Fig. 4C, lane 4).
FIGURE 4.
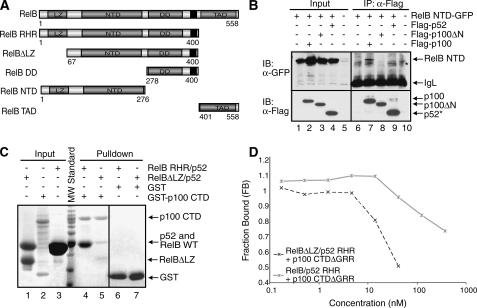
All domains and flexible regions of p100 contact RelB. A, schematic representation of p100 and its deletion mutants. The black regions represent the nuclear localization signals. B, Western blot analysis of in vitro GST pulldown experiments demonstrating the binding interaction between the NTD of p52 and RelB RHR, followed by immunoblot with α-GST or α-His antibodies. C, co-IP experiments showing the interaction between p52 NTD-FLAG and RelB RHR-GFP from the extracts of cotransfected HEK 293 cells. p52 NTD was isolated by IP and analyzed by IB (bottom panel), and the co-precipitated RelB was analyzed by IB (top panel). An asterisk indicates a nonspecific band. D, Western blot analysis of in vitro GST-pulldown experiments showing binding interaction between the GST-tagged CTD of p100 and His-RelB RHR and His-RelB DD. E, co-IP experiments showing the binding interaction between the FLAG tagged CTD of p100 and RelB RHR-GFP and RelB DD-GFP. These experiments were done similarly as described in C. F, fluorescence polarization assay showing the effect of the regions flanking the ARD of p100 in inhibition of DNA binding by the RelB-p52 heterodimer. Constant amounts of RelB-p52 RHR bound to a fluorescently labeled DNA was titrated with increasing amounts of GST-p100 CTD proteins.
Previous studies showed strong interaction between the CTD of p100 with the RelB-p52 heterodimer (5). Our in vitro GST pulldown experiments show that the CTDΔGRR of p100 interacts with RelB RHR and RelB DD alone (Fig. 4D, lanes 6 and 7). The association has been further confirmed by co-IP using extracts of cells co-expressing FLAG-p100 CTD and RelB RHR-GFP or RelB DD-GFP (Fig. 4E, lanes 4 and 5). A decrease in interaction between RelB DD and the CTD of p100 as compared with RelB RHR implies that the NTD of RelB may be involved in complex formation with p100. We next tested in vitro how the regions flanking the ARD of the CTD of p100 are involved in complex formation with RelB-p52. Because the ARD portion of p100 apparently contributes to most of the binding affinity, we needed a more sensitive DNA binding inhibition assay to test this. We used pure recombinant p100 CTDΔGRR, p100 CTDΔn, and p100 CTDΔc to test their ability to inhibit DNA binding of the RelB-p52 heterodimer in a solution based competition assay. Our results show that the removal of the N- and C-terminal regions reduced the inhibitory activity of the CTD of p100, suggesting that in the context of p100 the entire CTD is involved in contacting the RelB-p52 subcomplex (Fig. 4F).
The LZ Domain Interacts with p100—We next wished to examine how each of the RelB structural domains are involved in the RelB-p100 complex formation. RelB can be divided into three regions: NTD, DD, and the TAD (Fig. 5A). The NTD of RelB contains a nonhomologous 100 residue long LZ segment at the N terminus. As described above for p100, we have tested the binding interactions by two ways: GST pulldown experiments using pure recombinant proteins in vitro and co-IP of proteins expressed in transiently transfected HEK 293 cells. Co-IP confirmed that the RelB NTD associates with p100 but not with p52 or p100ΔN (Fig. 5B). The presence of a cryptic thrombin cleavage site at residue 64 of RelB allowed us to remove the first 64 residues by thrombin treatment and test the role of this region in p100 CTD binding. GST pulldown experiments using pure recombinant proteins revealed a drastic reduction in the binding interaction of the truncated RelB-p52 heterodimer for p100 CTD (Fig. 5C, compare lanes 4 and 5). We further confirmed the role of the LZ domain using a fluorescence-based DNA binding inhibition assay. This assay revealed that the presence of the N-terminal 64 residues was essential for the CTD to efficiently inhibit the DNA binding of the heterodimer (Fig. 5D).
FIGURE 5.
The LZ domain of RelB is involved in p100 binding. A, schematic representations of RelB domains and the deletion mutants of RelB used in the binding experiments. The black regions represent the nuclear localization signals. B, co-IP experiments showing binding interaction between RelB NTD-GFP and wt or deletion mutants of p100 with the FLAG peptide. The cells were cotransfected with RelB NTD-GFP and p100 mutants, extracts were immunoprecipitated by α-FLAG antibody and immunoblotted with FLAG (bottom panel) and GFP (top panel). An asterisk indicates a nonspecific band. C, in vitro GST pulldown experiments were done using equal amounts of pure recombinant proteins showing that the N-terminal LZ domain of RelB is involved in the binding interaction with the CTD of p100. Input and pulldown samples were separated by SDS-PAGE followed by Coomassie staining. Please note that His-RelB RHR and p52 RHR co-migrate in the SDS-PAGE (lanes 3 and 4). At position 64 of RelB, a cryptic thrombin cleavage site is located; therefore the first 64 residues are removed (lanes 1 and 5). D, fluorescence polarization experiments showing the functional role of the LZ domain of RelB in binding the p100 CTDΔGRR. Fluorescence polarization assay was done the same as in Fig. 3F.
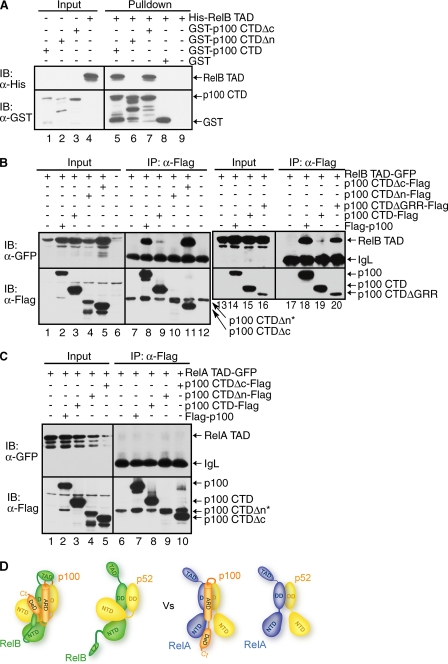
TAD of RelB Binds to p100 at or near the Site of Processing—We next tested the role of the RelB TAD in p100 binding. GST pulldown experiments show that p100 CTD interacts specifically with the TAD of RelB. Interestingly, the C-terminal 134 residues, which contain the death domain plus the sites of induced phosphorylation, are not important for stable interaction with RelB TAD (Fig. 6A, lane 7). In contrast, deletion of the segment N terminus to the ARD of p100 abolishes binding (Fig. 6A, lane 6). Co-IP experiments further confirmed in vitro binding interactions. The TAD of RelB is involved in complex formation with both wt p100 and the CTD (Fig. 6B, lanes 8 and 9). However, the RelB TAD binds more weakly to p100 CTD than p100 (Fig. 6B, compare lanes 18–20). Therefore, it is possible that the GRR in the isolated CTD negatively affects binding. This is supported by strong binding interactions observed between the TAD of RelB and p100 CTDΔGRR. When the CTD of p100 was further deleted to the beginning of the ARD, no binding to RelB TAD was observed (Fig. 6B, compare lanes 8 and 10). We conclude that the TAD of RelB directly interacts with the processing region of p100 (the segment bracketing the GRR and ARD). RelA TAD does not show any binding interaction with p100, suggesting that RelB interacts with p100 in a unique manner (Fig. 6C). In all, these results clearly demonstrate that all known functional segments/domains of RelB participate in the complex formation with multiple segments/domains of p100 (Fig. 6D).
FIGURE 6.
The TAD of RelB contacts the p100 processing site. A, Western blot analysis of in vitro GST pulldown experiments showing the interaction between the TAD of RelB with the GST-tagged CTD of p100. Input and pulldown samples were separated by SDS-PAGE followed by IB. B, co-IP showing the same interaction as in A but in cotransfected HEK 293 cells. FLAG-p100 or mutants were co-expressed with RelB TAD. IPed extracts were separated by SDS-PAGE, and the presence of co-precipitated RelB was analyzed by IB (top panel). The level of p100 and p100 CTD were analyzed by IB (bottom panel). An asterisk indicates that p100 CTDΔn migrates as the same size as the IgH band. C, the TAD of RelA does not bind p100. D, model of the interactions between RelB and p52/p100 as compared with the interactions between p52/p100 and p65.
DISCUSSION
NF-κB dimers are highly stable with the exception of RelB. Intriguingly, it is not the nonhomologous LZ and TAD domains that render RelB unstable, instead the RHR of RelB appears to be the primary reason for its lack of stability. Unstable proteins are degraded by cellular degradation machinery of which one of the most prominent protein degradation enzymes is the proteasome. We show that the proteasome is at least partly responsible for RelB degradation. However, it is possible that other cellular proteases are involved in RelB degradation. RelB is protected from degradation by p100, its processed product p52 and p50. There might also be other specific RelB-interacting proteins, such as aryl hydrocarbon, which has recently been shown to specifically interact with RelB through its RHR (28). The protection of flexible regions in the complex suggests a simple mechanism by which RelB avoids degradation by cellular proteases.
Thermodynamic stabilization of proteins through association with their partners is a common regulatory mechanism adopted by eukaryotic cells. Cell cycle inhibitor p21, ornithine decarboxylase, and p53 have been shown to undergo degradation unless they are self-associated or bound to partner proteins (29–31). An example of partner-mediated protein stabilization can also be drawn from the NF-κB-IκB system. In wt cells, IκBα is primarily bound by RelA and c-Rel dimers, and in their absence IκBα protein levels are significantly reduced (32).
What causes the instability of RelB? The structure of the RelB DD homodimer revealed a lack of surface hydrogen bonds in these domains, which at least in part explains why the molecule might not be able to fold properly and consequently forms a domain swapped homodimer in the crystal (9). We also observe similar sparse surface hydrogen bonds in the RelB NTD x-ray crystal structure (26). These observations are consistent with previous reports showing that the NTD of RelB induces its degradation in cells (33). Altogether these structural and theoretical predictions suggest that the entire RHR of RelB is susceptible to degradation because of its low folding stability. It is likely that the lack of folding stability of RelB enable it to interact with its partners more efficiently, which would not be possible if RelB was a more stable protein. Alternatively, the presence of RelB in the absence of its partners could result in unwanted interactions between RelB and other cellular proteins that could be detrimental to the cell.
Of all the partner proteins of RelB, p100 appears to be the most significant in its ability to protect RelB from degradation. It does so by forming a highly stable complex through extensive binding interactions between RelB and p100. Results from our binding experiments suggest a complex mode of interaction between RelB and p100, where different structural domains and flexible regions of both proteins participate in the complex formation. The unique complex between RelB and p100 (Fig. 5D) also suggests that RelB may play a role in the processing of p100 into p52. The interaction between the TAD of RelB and the site of processing of p100 suggest that RelB may inhibit p100 processing, causing the RelB-p100 complex to be an inactive complex. This interaction is unique to the RelB-p100 complex because the similar interaction is not observed in the RelA-p100 complex. The compact complex of RelB-p100 also suggests an inhibitory role for RelB in p100 processing. Consistent with this notion, our ongoing experiments clearly suggest that RelB influences p100 processing. Moreover, the specific domain arrangement in the RelB-p100 complex might serve as a platform to bind other NF-κB and IκB proteins. This large inhibitory complex in uninduced cells would liberate specific NF-κB dimers through degradation of p100. An example that has recently been shown is the RelA-p50 and RelB-p50 heterodimers through the degradation of p100 (25).
RelB also interacts in a stable manner with p52. However, the RelB-p52 heterodimer is not expected to be as stable as the RelB-p100 complex because the TAD and LZ of RelB are exposed in these complexes. How then are these RelB domains stabilized in the RelB-p52 heterodimer? How do they protect RelB from degradation? It is likely that these RelB heterodimers exist only transiently in vivo as free dimers. They localize to the nucleus and associate with DNA and are involved in transcriptional regulation. Therefore, these transcriptionally active dimers remain mostly in DNA-bound states, where the NTD interacts with target DNA and the TAD presumably interacts with other proteins. The unique interactions between the p52 NTD and RelB RHR create a puzzling situation. This interaction would keep the DNA-binding domains of both subunits in a closed conformation, a state that is incompatible with DNA binding. However, in the cases of non-RelB NF-κB dimers, the two NTDs remain in dynamic states, which stabilize upon DNA binding. There are two likely possibilities by which the NTDs of RelB-p52 heterodimer can dissociate to bind target DNAs. It is possible that the p52 NTD only binds weakly and dynamically to the RelB RHR primarily through electrostatic contacts and upon encountering DNA, stronger interactions between the dimers and DNA alters the equilibrium to the open conformation. It is also possible that an active process such as phosphorylation of any of these proteins may force the release of the p52 NTD from the RelB RHR. To this end it is important to mention that p52 NTD has been shown to be phosphorylated by IKKα (16). Although we do not know whether these sites in p52 might play a direct or indirect role in the binding interaction with RelB, such a possibility exists.
p105 and p50 also stabilize RelB through stable interactions similar to p52. We observe that the general binding mode is similar in both the RelB-p105 and RelB-p50 complexes. That is, the primary contacts are through the RHR of p50 and RelB. Stable complex formation between RelB and p50 explains how RelB can carry out functions that are independent of p100. For example, hyperinflammation is seen only in mice deficient in relb but not in mice deficient in nfkb2. The RelB-p50 heterodimer may function as a modulator of key inflammatory genes and such activity of RelB does not require p100.
In conclusion, our results clearly show that RelB is an unstable protein in vivo and requires highly specific partners for its stabilization. However, the apparent lack of RelB stability is not unusual, because many cellular proteins are stabilized in a partner-dependent manner. The unique complex between RelB and p100/p52 and p50 explain RelB specificity toward p100, p52, and p50 in cells. Future studies are needed to determine the residues involved and the physical-chemical mechanism underlying RelB destabilization and the role stable interactions between RelB and p100 have on the processing of p100.
Acknowledgments
We thank Erika Mathes and Sutapa Chakrabarti for critically reading the manuscript and the Keck computer facility for supporting in the structural and graphical work. We thank Dustyn Miller for running the electrophoretic mobility shift assay.
This work is supported by funding from the National Institutes of Health and the Universitywide AIDS Research Program (to G. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: RHR, Rel homology region; NTD, N-terminal domain; TAD, transcriptional activation domain; IKK, IκB kinase; LZ, leucine zipper; DD, dimerization domain; GST, glutathione S-transferase; IB, immunoblotting; GFP, green fluorescent protein; HEK, human embryonic kidney; wt, wild type; MEF, mouse embryonic fibroblast; IP, immunoprecipitation; GRR, glycine-rich region; ARD, ankyrin repeat domain.
References
- 1.Baldwin, A. S., Jr. (1996) Annu. Rev. Immunol. 14 649-683 [DOI] [PubMed] [Google Scholar]
- 2.Ghosh, S., May, M. J., and Kopp, E. B. (1998) Annu. Rev. Immunol. 16 225-260 [DOI] [PubMed] [Google Scholar]
- 3.Ghosh, S., and Karin, M. (2002) Cell 109 (Suppl.) S81-S96 [DOI] [PubMed] [Google Scholar]
- 4.Karin, M., and Ben-Neriah, Y. (2000) Annu. Rev. Immunol. 18 621-663 [DOI] [PubMed] [Google Scholar]
- 5.Solan, N. J., Miyoshi, H., Carmona, E. M., Bren, G. D., and Paya, C. V. (2002) J. Biol. Chem. 277 1405-1418 [DOI] [PubMed] [Google Scholar]
- 6.Lernbecher, T., Kistler, B., and Wirth, T. (1994) EMBO J. 13 4060-4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruben, S. M., Klement, J. F., Coleman, T. A., Maher, M., Chen, C. H., and Rosen, C. A. (1992) Genes Dev. 6 745-760 [DOI] [PubMed] [Google Scholar]
- 8.Ryseck, R. P., Bull, P., Takamiya, M., Bours, V., Siebenlist, U., Dobrzanski, P., and Bravo, R. (1992) Mol. Cell Biol. 12 674-684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang, D. B., Vu, D., and Ghosh, G. (2005) Structure (Camb.) 13 1365-1373 [DOI] [PubMed] [Google Scholar]
- 10.Xiao, G., Harhaj, E. W., and Sun, S. C. (2001) Mol. Cell 7 401-409 [DOI] [PubMed] [Google Scholar]
- 11.Senftleben, U., Cao, Y., Xiao, G., Greten, F. R., Krahn, G., Bonizzi, G., Chen, Y., Hu, Y., Fong, A., Sun, S. C., and Karin, M. (2001) Science 293 1495-1499 [DOI] [PubMed] [Google Scholar]
- 12.Dejardin, E., Droin, N. M., Delhase, M., Haas, E., Cao, Y., Makris, C., Li, Z. W., Karin, M., Ware, C. F., and Green, D. R. (2002) Immunity 17 525-535 [DOI] [PubMed] [Google Scholar]
- 13.Claudio, E., Brown, K., Park, S., Wang, H., and Siebenlist, U. (2002) Nat. Immunol. 3 958-965 [DOI] [PubMed] [Google Scholar]
- 14.Coope, H. J., Atkinson, P. G., Huhse, B., Belich, M., Janzen, J., Holman, M. J., Klaus, G. G., Johnston, L. H., and Ley, S. C. (2002) EMBO J. 21 5375-5385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonizzi, G., Bebien, M., Otero, D. C., Johnson-Vroom, K. E., Cao, Y., Vu, D., Jegga, A. G., Aronow, B. J., Ghosh, G., Rickert, R. C., and Karin, M. (2004) EMBO J. 23 4202-4210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao, G., Fong, A., and Sun, S. C. (2004) J. Biol. Chem. 279 30099-30105 [DOI] [PubMed] [Google Scholar]
- 17.Fong, A., and Sun, S. C. (2002) J. Biol. Chem. 277 22111-22114 [DOI] [PubMed] [Google Scholar]
- 18.Weih, F., Carrasco, D., Durham, S. K., Barton, D. S., Rizzo, C. A., Ryseck, R. P., Lira, S. A., and Bravo, R. (1995) Cell 80 331-340 [DOI] [PubMed] [Google Scholar]
- 19.Miyawaki, S., Nakamura, Y., Suzuka, H., Koba, M., Yasumizu, R., Ikehara, S., and Shibata, Y. (1994) Eur. J. Immunol. 24 429-434 [DOI] [PubMed] [Google Scholar]
- 20.Ishikawa, H., Carrasco, D., Claudio, E., Ryseck, R. P., and Bravo, R. (1997) J. Exp. Med. 186 999-1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franzoso, G., Carlson, L., Poljak, L., Shores, E. W., Epstein, S., Leonardi, A., Grinberg, A., Tran, T., Scharton-Kersten, T., Anver, M., Love, P., Brown, K., and Siebenlist, U. (1998) J. Exp. Med. 187 147-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu, Y. X., and Chaplin, D. D. (1999) Annu. Rev. Immunol. 17 399-433 [DOI] [PubMed] [Google Scholar]
- 23.Shinkura, R., Kitada, K., Matsuda, F., Tashiro, K., Ikuta, K., Suzuki, M., Kogishi, K., Serikawa, T., and Honjo, T. (1999) Nat. Genet. 22 74-77 [DOI] [PubMed] [Google Scholar]
- 24.Malek, S., Huxford, T., and Ghosh, G. (1998) J. Biol. Chem. 273 25427-25435 [DOI] [PubMed] [Google Scholar]
- 25.Basak, S., Kim, H., Kearns, J. D., Terganokar, V., O'Dea, E., Werner, S. L., Benedict, C. A., Ware, C. F., Ghosh, G., Verma, I. M., and Hoffmann, A. (2007) Cell 128 369-381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moorthy, A. K., Huang, D. B., Wang, V. Y., Vu, D., and Ghosh, G. (2007) J. Mol. Biol. [DOI] [PMC free article] [PubMed]
- 27.Toledano, M. B., Ghosh, D., Trinh, F., and Leonard, W. J. (1993) Mol. Cell Biol. 13 852-860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vogel, C. F., Sciullo, E., Li, W., Wong, P., Lazennec, G., and Matsumura, F. (2007) Mol. Endocrinol. 21 2941-2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheaff, R. J., Singer, J. D., Swanger, J., Smitherman, M., Roberts, J. M., and Clurman, B. E. (2000) Mol. Cell 5 403-410 [DOI] [PubMed] [Google Scholar]
- 30.Asher, G., Bercovich, Z., Tsvetkov, P., Shaul, Y., and Kahana, C. (2005) Mol. Cell 17 645-655 [DOI] [PubMed] [Google Scholar]
- 31.Asher, G., Lotem, J., Sachs, L., Kahana, C., and Shaul, Y. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13125-13130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann, A., Levchenko, A., Scott, M. L., and Baltimore, D. (2002) Science 298 1241-1245 [DOI] [PubMed] [Google Scholar]
- 33.Marienfeld, R., Berberich-Siebelt, F., Berberich, I., Denk, A., Serfling, E., and Neumann, M. (2001) Oncogene 20 8142-8147 [DOI] [PubMed] [Google Scholar]