Abstract
Discoidin domain receptor 1 (DDR1) is a transmembrane receptor tyrosine kinase activated by triple-helical collagen. So far six different isoforms of DDR1 have been described. Aberrant expression and signaling of DDR1 have been implicated in several human diseases linked to accelerated matrix degradation and remodeling, including tumor invasion, atherosclerosis, and lung fibrosis. Here we show that DDR1 exists as a disulfide-linked dimer in transfected as well as endogenously expressing cells. This dimer formation occurred irrespective of its kinase domain, as dimers were also found for the truncated DDR1d isoform. A deletion analysis of the extracellular domain showed that DDR1 mutants lacking the stalk region failed to form dimers, whereas deletion of the discoidin domain did not prevent dimerization. Point mutagenesis within the stalk region suggested that cysteines 303 and 348 are necessary for dimerization, collagen binding, and activation of kinase function. The identification of DDR1 dimers provides new insights into the molecular structure of receptor tyrosine kinases and suggests distinct signaling mechanisms of each receptor subfamily.
Discoidin domain receptors (DDRs)3 are a subfamily of transmembrane collagen-binding receptor tyrosine kinases (RTK). DDRs are distinguished from other RTKs by a discoidin domain in their extracellular region, which functions as a lectin in the slime mold Dictyostelium discoideum (1). There are two genes that code for DDRs in humans, DDR1 and DDR2 (2). Each DDR contains an extracellular region containing the ligand-binding discoidin domain, a stalk region, a single transmembrane region, as well as a cytoplasmic domain consisting of a juxtamembrane region and a tyrosine kinase domain.
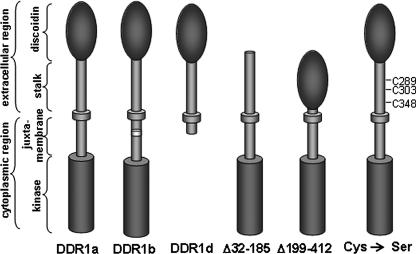
Through alternative splicing in the juxtamembrane or kinase domain of the human gene, at least five isoforms DDR1a– DDR1e are generated (3). The longest DDR1 transcript codes for the c-isoform with 919 amino acids. Compared with the c-isoform, the a-isoform lacks 37 amino acids in the juxtamembrane region, and the b-isoform lacks six amino acids in the kinase domain because of alternative splicing (4, 5). The DDR1d and e-isoforms are truncated variants that either lack the entire kinase region (d-isoform) or parts of the juxtamembrane region and the ATP-binding site (e-isoform (3)). A sixth isoform lacking parts of the extracellular domain has been described from rat testis (6).
DDR1 is expressed in a variety of cell types and tissues, specifically in the mammary gland, brain, kidney, lung, and colon mucosa and has been isolated from several carcinoma cell lines, including MCF7 mammary carcinoma cells, ovarian, lung, and esophageal cancer cells, and primary pediatric brain tumor samples (for review see Ref. 2). Immune cells, including macrophages, vascular smooth muscle cells, as well as oligodendrocytes also express DDR1 (7–10). From experiments with cultured cells, it was found that DDR1 regulates cell migration and branching morphogenesis within collagen-rich matrices (11, 12).
To address the role of DDR1 in an entire organism, gene targeting was used to generate DDR1-null mice. Homozygous mutant animals are smaller in size than control littermates and present with an altered renal basement membrane (13, 14). The majority of DDR1-null females are unable to bear offspring because of defects in blastocyst implantation. Those null females that are able to give birth are unable to lactate, a defect caused by abnormal branching and differentiation of the mammary epithelium (13, 15).
The ligands for DDR1 include various types of native collagen (16, 17). Maximal activation of DDR1 requires up to 18 h of collagen stimulation leading to phosphorylation of several cytoplasmic tyrosine residues (12, 18–20). The presence of the discoidin domain in DDR1 has been shown to be the essential region for collagen binding, and specific residues within the discoidin domain that interact with collagen were identified (21–23). Recently, the structure of the DDR2 discoidin domain has been solved by NMR studies, but such information is as yet not available for DDR1 (24).
In extension of the current model of RTK signaling, it was hypothesized that activation of DDR1 also involves ligand-induced dimerization (25). However, to date, the validity of this ligand-induced RTK dimerization model for DDR1 in particular has not been verified.
In this study, we report the presence of disulfide-linked DDR1 dimers, which potentially contribute to receptor trafficking and ligand-induced receptor activation. We also assess the importance of the stalk region of DDR1, and we found that two cysteines in the stalk region are critical for dimerization.
EXPERIMENTAL PROCEDURES
Cell Culture and Protein Expression—Human embryonic kidney 293 cells and human breast carcinoma MCF-7 and T-47D cells were purchased from the American Tissue Culture Collection and cultivated under the recommended conditions. MCF-7 cells were stably transfected with a mammalian expression vector pIRES-Neo containing the FLAG-tagged human cDNA of DDR1 isoforms a and d. Cells were transfected using Lipofectamine 2000 (Invitrogen) and stably expressing clones isolated. Semi-confluent 293 cells were transfected by calcium phosphate precipitation with 10 μg of plasmid DNA per 10-cm dish. The medium was replaced with serum-free Dulbecco's modified Eagle's medium 18 h after transfection. Twenty four hours later, cells were stimulated with 10 μg/ml rat tail type I collagen (BD Biosciences). Fifteen to 90 min after stimulation, cells were lysed using either Triton X-100 lysis buffer for Western blotting or Tween 20 lysis buffer for ELISA (each containing 1% detergent, 0.1% SDS, 150 mm NaCl, 5 mm EDTA, 50 mm Tris-HCl (pH 7.5), 10 mm NaF, 1 mm phenylmethylsulfonyl fluoride, 1 mm sodium orthovanadate, 10 μg/ml aprotinin). For immunoprecipitation, 293 cells were transfected with plasmids coding for HA-tagged DDR1a and DDR1d. After stimulation, cells were lysed, and immunoprecipitation was performed overnight at 4 °C using an HA-specific monoclonal antibody and protein G-Sepharose (Sigma).
Western Blotting—Prior to Western blot analysis, protein concentration of cell lysates was quantified using the Dc protein assay (Bio-Rad), and equal amounts of each sample were boiled in either reducing Laemmli buffer (187.5 mm Tris-HCl (pH 6.8), 6% SDS, 30% glycerol, 0.01% bromphenol blue, 15% β-mercaptoethanol) or nonreducing Laemmli (lacking β-mercaptoethanol). Samples were subjected to SDS-PAGE on 7.5% polyacrylamide gels. Proteins were transferred to nitrocellulose membrane (Schleicher & Schuell) and immunoblotted using antibodies diluted 1:1000 (monoclonal anti-phosphotyrosine, 4G10 (Upstate Biotechnology, Inc.), anti-HA, or anti-FLAG (Sigma)) or 1:500 (anti-DDR1 C-terminal antibody, Santa Cruz Biotechnology) overnight in NET-Gel (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 5 mm EDTA, and 0.5% gelatin). Western blots were incubated with mouse and rabbit peroxidase-coupled secondary antibodies, respectively (Bio-Rad), and enhanced chemiluminescence (Amersham Biosciences). For re-probing, the membranes were stripped in 70 mm Tris-HCl (pH 6.8), 2% SDS, 0.1% β-mercaptoethanol at 50 °C for 15 min and blocked for 1 h at room temperature.
Site-directed Mutagenesis—Point mutations were introduced into the DDR1b cDNA by PCR using the QuikChange site-directed mutagenesis kit (Stratagene). Point mutations were introduced to make the following single amino acid changes: C289S, C303S, and C348S (primer sequences are available upon request). The numbering refers to full-length DDR1, including its signal peptide. All constructs were sequenced to verify the presence of the respective mutation. The deletion mutants lacking the discoidin domain or stalk region were described previously (21).
ELISA—Type I collagen was diluted in phosphate-buffered saline (PBS) to a concentration of 50 μg/ml and added to 96-well microtiter plates (50 μl/well). Plates were incubated for 1 h at room temperature and washed twice with PBS. Wells were blocked with 150 μl of 1 mg/ml bovine serum albumin in PBS containing 0.05% Tween 20 for 1 h. Wells were washed once with 150 μl of wash buffer (0.5 mg/ml bovine serum albumin in PBS containing 0.05% Tween 20). DDR1-transfected 293 cells were lysed with Tween 20-based lysis buffer, and lysate was added to each well and incubated at room temperature overnight (lysates were normalized for DDR1 expression by Western blot analysis prior to ELISA). Wells were washed six times with wash buffer. 50-μl aliquots of diluted anti-DDR1 C-terminal antibody (1:500 dilution in wash buffer) were added and incubated for 90 min. Wells were washed six times with wash buffer and incubated with horseradish peroxidase-linked anti-rabbit secondary antibody for 90 min. Bound protein was detected using 75 μl/well of 0.5 mg/ml o-phenylenediamine dihydrochloride (Sigma) in 50 mm citrate-phosphate buffer (50 mm sodium citrate and 0.1 m sodium phosphate, pH 5.0) with 0.012% hydrogen peroxide. After 30 min at room temperature, the reaction was stopped using 50 μl/well of 3 m sulfuric acid. Plates were analyzed in an ELISA reader at 490 nm; buffer only values were subtracted, and normalized values were subjected to statistical analysis using Student's t test as calculated by Microsoft Excel. p values are indicated where applicable. All experiments were performed in triplicate.
Collagen-agarose Affinity Purification—For total cell lysates of transfected 293 cells expressing wild type DDR1b, each Cys-Ser mutant and control-transfected cells were normalized for DDR1 expression by Western blotting and incubated overnight at 4 °C with collagen-linked Sepharose beads (Sigma) in HNTG buffer (20 mm HEPES (pH 7.5), 150 mm NaCl, 0.1% Triton X-100, 10% glycerol). Samples were washed three times with HNTG, boiled in Laemmli buffer, and subjected to SDS-PAGE followed by Western blotting. Bound material was detected using anti-DDR1 C-terminal antibody and secondary anti-rabbit horseradish peroxidase.
Cell Surface Biotinylation—293 cells were transfected with wild type DDR1b or the Cys-Ser mutants as outlined above. Cells were washed with PBS, incubated with 0.5 mg/ml sulfo-NHS-biotinate (Pierce) in PBS at 4 °C for 30 min, and washed three times with PBS containing 100 mm glycine. Cells were pelleted, lysed in RIPA buffer containing protease inhibitors, and centrifuged at 15,000 × g at 4 °C for 15 min. Four hundred μg of total lysates were incubated with streptavidin-Sepharose beads overnight at 4 °C. Beads were subsequently washed four times with RIPA buffer, boiled in Laemmli buffer, and subjected to SDS-PAGE followed by Western blotting.
RESULTS
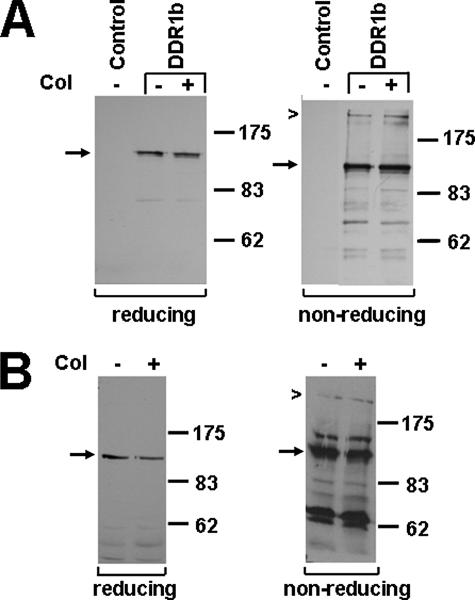
Previous work showed that the full-length DDR1 isoforms a and b are expressed as ∼125-kDa proteins in a number of different cell types, including breast epithelial cells (Fig. 1) (5, 16). To test whether DDR1 forms disulfide-linked dimers, human embryonic kidney 293 cells were transiently transfected with the b-isoform of DDR1 or a vector-only control, stimulated with type I collagen for 90 min, and lysates analyzed by PAGE under reducing or nonreducing conditions. Western blot analysis was performed using an antibody specific to the C terminus of DDR1. Although DDR1 was expressed as single 125-kDa band in the presence of β-mercaptoethanol, an additional band of ∼250 kDa was found in the absence of the reducing agent (Fig. 2A). Quantification of these bands indicated that ∼20% of total DDR1-immunoreactive protein is found as 250-kDa entity. Stimulation with collagen did not notably affect the ratio of 125- versus 250-kDa protein. To test whether the proposed dimer was present in cells expressing DDR1 endogenously, lysates from human breast carcinoma T-47D cells were examined under reducing and nonreducing conditions. In agreement with the data from 293 cells, DDR1 was detected as a 125-kDa monomeric protein under reducing conditions and as both a monomer and 250-kDa dimer under nonreducing conditions (Fig. 2B).
FIGURE 1.
Schematic diagram of various DDR1 receptor isoforms and mutant constructs. The domain organization of the a-, b-, and d-isoform are shown as well as several of the deletion and point mutations within the extracellular domain used for this study.
FIGURE 2.
DDR1b forms covalent dimers. A, 293 cells were transiently transfected with DDR1b or an empty plasmid (control) and stimulated with type I collagen (col) for 90 min. Using an antibody specific for the C terminus of DDR1, Western blot analysis of gels run under reducing conditions (left panel) detected DDR1b as a monomeric protein (arrow), whereas under nonreducing conditions (right panel), DDR1b was present as both a monomer (arrow) and a dimer (chevron). B, T-47D cells that endogenously express DDR1b were stimulated with collagen for 18 h and lysates analyzed by Western blotting under reducing (left panel) and nonreducing conditions (right panel). The experiments were performed three times, and representative data are shown.
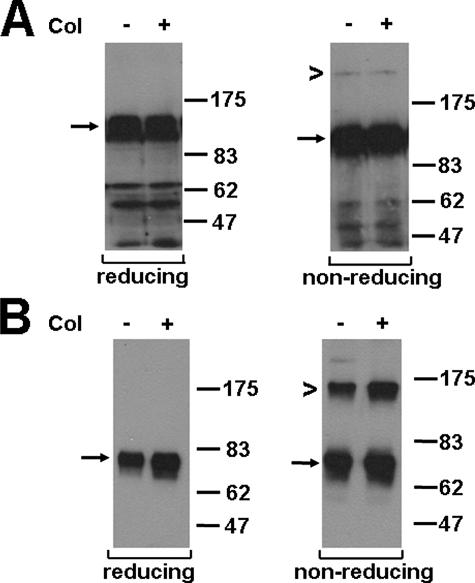
DDR1a and -d were transiently transfected in 293 cells or stably transfected into human breast carcinoma MCF-7 cells (Fig. 1). This was done to determine whether DDR1 covalent dimer formation is also found other than just with the b-isoform. As for DDR1b, dimers of DDR1a and -d were detected by Western blotting of MCF-7 lysates analyzed under nonreducing conditions (Fig. 3). Although the ratio of monomer to dimer for DDR1a was similar to the ratio of DDR1b (Fig. 2A), there was around 40% of total DDR1d found as dimer (Fig. 3B). Similarly to DDR1a or -b, the ratio of DDR1d monomer to dimer was not altered upon collagen exposure. These data suggest that the kinase domain, which is present in both a- and b-isoforms but absent in DDR1d, is not necessary for covalent linkage, and that DDR1 dimerization is likely mediated by the juxtamembrane, transmembrane, and/or extracellular regions. To investigate whether DDR1 could form heterodimers consisting of different isoforms, epitope-tagged DDR1a and d-isoforms were co-transfected using FLAG-tagged DDR1a and HA-tagged DDR1d. We found that these isoforms did not form covalently linked heterodimers, nor did they alter the monomer to homodimer ratio of each individual isoform (data not shown). These findings are in agreement with previous studies that failed to show interaction between full-length and truncated DDR1 isoforms (3).
FIGURE 3.
The monomer to dimer ratio varies between different DDR1 isoforms. MCF7 cells stably transfected with DDR1a-FLAG and DDR1d-FLAG were stimulated with collagen for 18 h and lysed. DDR1 expression was detected by the monoclonal anti-FLAG antibody (1:1000 dilution). A, DDR1a was detected as a monomer (125 kDa, arrow) under reducing conditions and as both a monomer and dimer (250 kDa, chevron) under nonreducing conditions irrespective of collagen (Col) stimulation. B, DDR1d was detected as a monomer (75 kDa, arrow) under reducing conditions and as both a monomer and dimer (150 kDa, chevron) under nonreducing conditions irrespective of collagen stimulation. These experiments were carried out a minimum of three times and very similar results obtained.
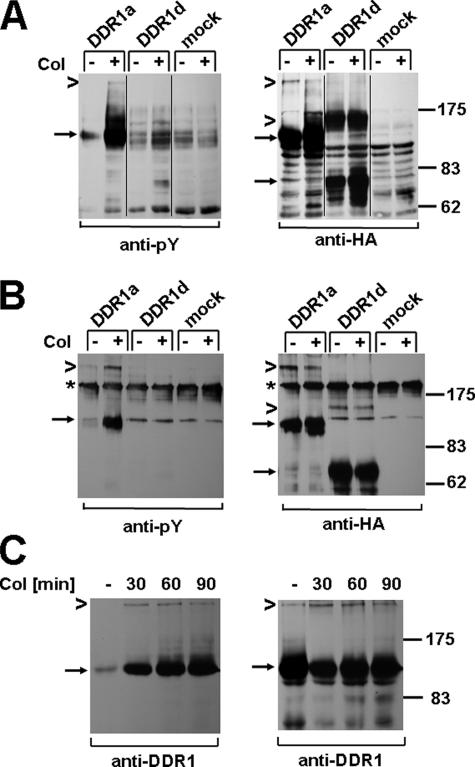
Next, we asked the question whether dimeric DDR1 is phosphorylated upon collagen stimulation. To test this, we transfected 293 cells with plasmids coding for HA-tagged DDR1a and DDR1d and stimulated cells with collagen for 90 min. Anti-phosphotyrosine Western blot analysis of total lysates on nonreducing SDS gels revealed that the 250-kDa dimeric DDR1a entity is indeed phosphorylated upon ligand stimulation (Fig. 4A). To further confirm the identity of the 250-kDa band, we used lysates from 293 cells transfected with HA-tagged DDR1a and DDR1d for immunoprecipitation, followed by nonreducing gel electrophoresis (Fig. 4B). Anti-phosphotyrosine Western blotting revealed phosphorylated 125- and 250-kDa DDR1a species, in a ratio comparable with the one observed with total lysates (Fig. 4A). Equal expression of DDR1a or -d was confirmed by reprobing with an HA-specific antibody (Fig. 4, A and B, right panels) and by additional Western blot analysis of total lysates separated by reducing gel electrophoresis (not shown).
FIGURE 4.
DDR1 dimers are tyrosine-phosphorylated upon collagen stimulation. 293 cells were transfected with DDR1a-HA, DDR1d-HA, or control vector and stimulated with 10 μg/ml type I collagen (Col) for 90 min. A, equal amounts of total cell lysates were resolved by nonreducing gel electrophoresis and Western blots probed with anti-phosphotyrosine (Py) antibody (left panel) and reprobed with anti-HA antibody (right panel). Arrows point to monomeric DDR1a and DDR1d, and chevrons point to dimeric DDR1 forms, respectively. B, cell lysates were subjected to anti-HA immunoprecipitation, and the presence of tyrosine-phosphorylated, dimeric DDR1a was confirmed by anti-phosphotyrosine Western blotting (left panel). The asterisks indicate a nonspecific band, likely IgG oligomers, that showed cross-reactivity under nonreducing conditions. The reprobe of the same blot with anti-HA antibody is shown on the right panel. A set of data representative for two repeats of these experiments is shown.
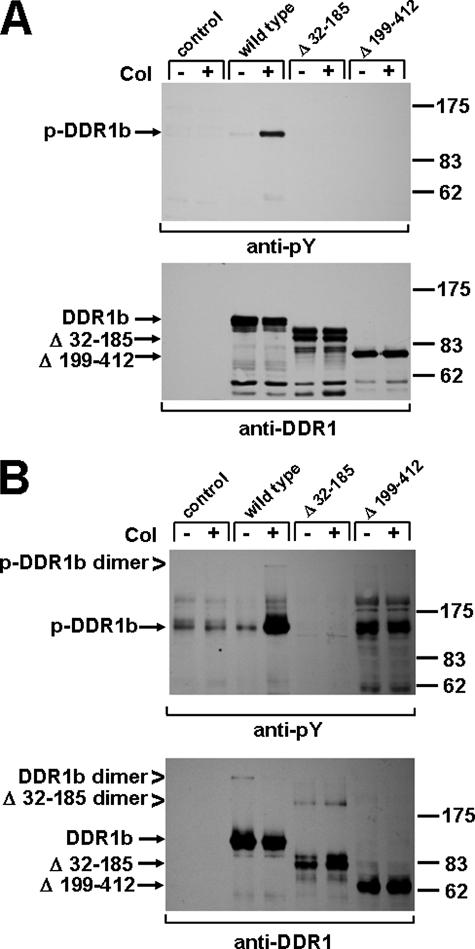
To further define a particular region of DDR1 that may be responsible for dimer formation, we employed deletion mutants, which either lack the discoidin domain (Fig. 1, Δ32–185) or the stalk region (Δ199–412) (21). Under reducing conditions, monomeric receptor species were detected at 125 kDa for full-length DDR1b, at 100 kDa for the Δ32–185 mutant, and at 80 kDa for Δ199–412 (Fig. 5A). As detailed previously, both deletion mutants have lost the ability to undergo collagen-mediated kinase activation (21). Importantly, under nonreducing conditions, additional bands corresponding to dimers were detected for full-length DDR1 (250 kDa) and Δ32–185 (200 kDa) but not for the Δ199–412 (expected at 160 kDa) mutant (Fig. 5B). This finding suggests that a sequence within the stalk region is necessary for the dimerization of DDR1.
FIGURE 5.
The stalk region of DDR1 is essential for dimerization. Human embryonic 293 cells were transiently transfected with full-length DDR1b, a DDR1b mutant lacking the discoidin domain (Δ32–185), or DDR1b lacking the stalk region (Δ199–412). DDR1 phosphorylation was detected under reducing (A) or nonreducing conditions (B) and blots reproved with a DDR1 C-terminal antibody. Dimers were detected for wild type DDR1b (250 kDa) and the Δ32–185 mutant (200 kDa), but not for the Δ199–412 construct (expected size is 160 kDa). Collagen-induced DDR1 dimer phosphorylation was only found for the wild type, but not for either of the deletion mutants. Col, collagen; pY, anti-phosphotyrosine.
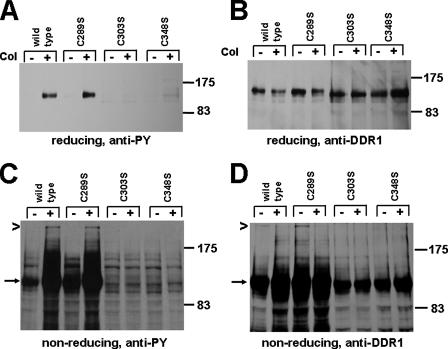
To explore this observation further, we identified a total of three cysteines at position 289, 303, and 348 in the stalk region (Fig. 1). These are a total of seven cysteines in the extracellular domain of DDR1. Four cysteines are located in the discoidin domain and are suggested to be disulfide-linked based on our studies using molecular modeling (22). Site-directed mutagenesis was performed to introduce single amino acid exchanges switching each cysteine in the stalk region to serine. The resulting mutant constructs were expressed in 293 cells and total lysates subjected to SDS-PAGE under reducing or nonreducing conditions. Anti-phosphotyrosine Western blotting of reducing gels showed strong ligand-triggered phosphorylation of wild type DDR1 and C289S mutant, but no signal was detected for the C303S and C348S mutants (Fig. 6, A and B). In the absence of β-mercaptoethanol, DDR1 dimers (250 kDa) were detected for wild type DDR1a and for the C289S mutant but not for C303S and C348S mutants suggesting that switching these residues to serines affected the dimerization of the receptor (Fig. 6C). Furthermore, we noted on Western blots obtained from nonreducing gels that C303S and C348S were expressed somewhat lower than wild type DDR1 or C289S (Fig. 6D). These data suggest that the cysteines at position 303 and 348 are involved in covalent dimerization and/or ligand-induced receptor activation.
FIGURE 6.
Cys-303 and Cys-348 residues in the stalk region are critical for the activation and dimerization of DDR1. DDR1b wild type and point mutants on cysteines 289, 303, or 348 were expressed in 293 cells and stimulated with collagen (Col) for 90 min. Cell lysates were analyzed under reducing and nonreducing conditions. A, probing a Western blot from reducing gel with anti-phosphotyrosine (PY) antibodies was first used to assess DDR1 activity, followed by anti-DDR1 (C-terminal antibody) blotting (B). DDR1b wild type and C289S were activated upon collagen stimulation, whereas C303S and C348S were not. C and D, dimeric DDR1 was detected for wild type and the C289S mutant but not C303S and C348S mutants.
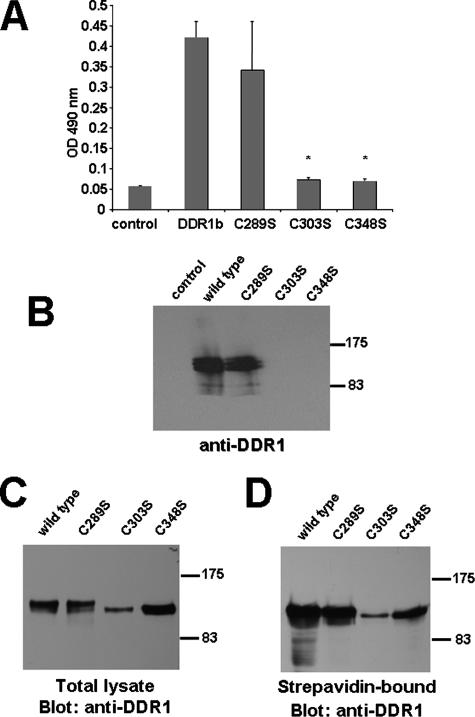
To verify these differences in the DDR1-collagen interaction, we assessed the ability of the stalk region cysteine mutants to bind collagen in vitro. First, a collagen-binding assay was performed where a collagen I-coated 96-well microtiter plate was incubated with cell lysates normalized for equal amounts of wild type DDR1, C289S, C303S, or C348S (Fig. 7A). We found that wild type and C289S bound to immobilized collagen at comparable levels, whereas C303S and C348S mutants had a significantly lower affinity, relative to wild type DDR1 (p < 0.001). Second, lysates from transfected cells were incubated with agarose beads that are coupled with type I collagen. DDR1b and C289S constructs bound to collagen-agarose beads equally well, whereas C303S and C348S mutants had no detectable affinity (Fig. 7B). Therefore, mutations at position Cys-303 or Cys-348 in the stalk region of DDR1 appear to change the topology of the receptor rendering it incapable of binding to collagen. As we noticed that C303S and C348S mutants ran at a slightly lower molecular weight on SDS-PAGE than the wild type (Fig. 6B), we wanted to confirm that both mutants undergo similar post-translational modification. Treating transfected cells with tunicamycin, an inhibitor of protein glycosylation (26), showed that all three cysteine mutants had a glyco moiety comparable with the wild type (data not shown). At this time, however, we cannot rule out that these mutants could be differentially glycosylated compared with wild type DDR1. Furthermore, the inability of the C303S and C348S mutants to be activated by collagen could be attributed to incorrect cellular trafficking. Therefore, we performed surface biotinylation and found that cell surface localization of neither C303S nor C348S is impaired compared with the wild type or C289S mutant (Fig. 7, C and D).
FIGURE 7.
DDR1 mutants C303S and C348S fail to bind to type I collagen. A, lysates from transfected 293 cells were normalized for wild type DDR1, C289S, C303S, or C348S expression, incubated with immobilized collagen I, and detected by ELISA (C-terminal antibody). DDR1 and C289S bound to collagen at comparable levels, whereas C303S and C348S mutants bound to collagen significantly less, relative to wild type DDR1 (n = 3, p < 0.001). B, normalized amounts of 293 cell lysates were mixed with collagen immobilized on agarose beads. Bound material was detected by anti-DDR1 (C-terminal antibody) Western blotting. C and D, similar amounts of C303S and C348S mutants are localized on the cell surface. Transfected 293 cells were surface-biotinylated and lysed. Total protein (C) or streptavidin-Sepharose-bound protein (D) was analyzed by anti-DDR1 (C-terminal) Western blotting. A set of data representative for two repeats of these experiments is shown.
In conclusion, our data provide evidence for the existence of a disulfide-linked, dimeric form of DDR1 in human cells, which is activated upon collagen stimulation. We further show that two cysteines in the stalk region of the receptor are most likely necessary not only for the formation of covalent dimers but also for proper receptor post-translational maturation.
DISCUSSION
Data presented here are the first to report that DDR1 exists as disulfide-linked dimers. These dimers are found in cells transfected with DDR1 as well as in epithelial cells endogenously expressing DDR1. The three major isoforms of DDR1, -a, -b, and DDR1d, all demonstrated covalent dimerization. The ratio of monomers to dimers was similar between DDR1a and DDR1b, whereas DDR1d showed a greater proportion of dimers. Because DDR1d does not contain the tyrosine kinase domain, our data suggest a role for this region in regulating the extent of dimerization.
In exploring the DDR1 dimer formation in more detail, we found that dimers do not form in the absence of the stalk region. Upon further analysis of the stalk region, mutation of cysteines 303 or 348 to serine resulted in a receptor that did not form covalent dimers and was no longer activated upon ligand stimulation. In addition, C303S and C348S mutants were unable to bind collagen in either an ELISA or a solid phase binding assay. On nonreducing gels, the expression level of these two mutants was somewhat reduced compared with C289S or the wild type receptor (Fig. 6D), possibly indicating altered cellular maturation and/or trafficking.
Several previous studies on the mechanism of DDR signaling provided a basis for this study. Curat et al. (21) examined the role of the discoidin domain and the stalk region for DDR1 activation and collagen binding. Although the discoidin domain was essential for the interaction with collagen, omission of the stalk region resulted in DDR1 molecules still being able to bind to collagen. Conversely, deletion of the discoidin domain resulted in a receptor species that could neither bind to nor be activated by collagen. Additional mechanistic evidence for DDR1 signaling was obtained using an Fc affinity-tagged construct of the DDR1 ectodomain (23). It was shown that DDR1 recognized collagen only as a dimeric and not as a monomeric construct, indicating a requirement for noncovalent receptor dimerization in DDR1-collagen signaling. Furthermore, the expression of recombinant discoidin as well as ectodomain in insect cells revealed that the ectodomain had an about 10-fold higher affinity to collagen than the discoidin domain alone (22). Following completion of this study, Noordeen et al. (27) reported on the formation of DDR1 dimers as well. Using chemical cross-linking and immunoprecipitation, these authors found ligand-independent DDR1 dimers and monomers in a ratio very similar to this study. In contrast to this study, they failed to identify any cysteine residues in the stalk region responsible for dimer formation. Based on bacterial expression of the isolated DDR1 transmembrane region, Noordeen et al. (27) proposed that a leucine zipper motif within this region might facilitate DDR1 dimer formation.
Members of the RTK family are believed to adhere to a similar activation model, the ligand-induced RTK dimerization and activation model. In this model, ligand binding to the extracellular domain of the RTK leads to conformational changes that induce and stabilize receptor dimerization, leading to increased kinase activity and trans-phosphorylation of tyrosine residues (reviewed in Ref. 28). However, more recent work suggested that specific subgroups of RTKs possess unique modes of activation. Of all RTKs, the EGFR and insulin receptor subfamily have been most intensely investigated; however, the actual contribution of individual receptor domains to dimerization and activation is still very much under debate (29, 30). It is believed that EGFR and the three other members of the ErbB family are monomeric in resting cells. Binding of growth factors induces homo- or heterodimerization leading to trans-autophosphorylation and activation of numerous downstream signaling pathways (31). Dimerization is a transient event, and several cellular mechanisms are in place to auto-inhibit or rapidly down-regulate ErbB signaling in nontransformed cells (32). Although the aggregation state of the EGFR in the absence of ligand is still under debate, it is well accepted that addition of ligand favors the formation of dimers/oligomers by facilitating either monomeric unfolding or realignment of pre-existing dimers (33). Furthermore, recent crystallographic evidence suggested asymmetric dimer formation of EGFR, with one kinase domain, drives activation of the other domain through an allosteric interaction (34).
For the EGFR, it was found that preformed dimers included ∼10% of the total receptor detected in the absence of ligand; however, no covalent linkage has been shown for these dimers as yet (35). Although these dimers comprise a relatively small fraction of the cell's total EGFR amount, they closely reflected our results for DDR1 where about one-tenth of receptors formed ligand-independent dimers in endogenously expressing cells. It has been reported previously that EGFRs appear in two different affinity classes on the cell surface as follows: 2–5% of the receptors bind epidermal growth factor with high affinity, whereas 92–95% bind with lower affinity (36). The affinity classes are thought to represent different receptor conformations and/or states of oligomerization. Although unconfirmed, it is tempting to speculate that the high affinity receptors may be identical to these EGFR dimers. The presence of pre-formed dimers may be of particular importance in cells that express EGFR at low levels on the cell surface. Although it is possible that the presence of ligand-independent DDR1 dimers may mediate more efficient signaling in terms of the time necessary for individual receptors to find partners, the activation time line of DDR1 differs significantly from the EGFR. DDR1 achieves maximal phosphorylation between 1.5 and 18 h after collagen stimulation, whereas the EGFR can demonstrate significant phosphorylation less than 1 min after ligand stimulation. Therefore the physiological function of DDR1 dimers may differ from that of EGFR dimers.
In contrast to the EGFR, the insulin receptor forms a covalently linked tetramer. An insulin receptor precursor is cleaved intracellularly into α and β subunits, of which the α subunit localizes to the extracellular environment and binds ligand, whereas the β subunit contains a short extracellular region, a single transmembrane domain, and a cytoplasmic region responsible for signal transduction (37). A disulfide bond links the two subunits through amino acid Cys-647 in the α subunit and Cys-872 in the β subunit. Dimer formation between two αβ monomers requires the presence of at least one of the following disulfide bonds: Cys-524, Cys-682, Cys-683, or Cys-685 (38–40). Dimerization of the insulin receptor takes place in the endoplasmic reticulum as the endoplasmic reticulum has an oxidizing environment, and the disulfide isomerases and chaperones calnexin and calreticulin are present (41, 42). The current model for insulin receptor activation proposes that insulin induces a conformational change in the “inverted V”-shaped extracellular domain of the receptor that causes the two intracellular domains to move into closer proximity thereby increasing receptor kinase activity. Potentially, analogous changes in the orientation of dimeric DDR1 extracellular domains are critical for ligand-induced kinase activation.
Based on our observation that point mutation of cysteines 303 or 348 abrogate the formation of covalent dimers, we propose that intermolecular disulfide linkage of these residues causes DDR1 dimer formation, in a similar fashion as seen in the insulin receptor. However, because no structural information on the DDR1 stalk region is presently available, it is unknown whether dimerization is achieved by linking identical (303-303 and 348-348) or nonidentical (303-348 and 348-303) residues. Importantly, the reduced ability of the C303S and C348S compared with the C289S mutant to bind to collagen in vitro (Fig. 7B) suggests that these cysteines need to be in an oxidized state to allow proper domain folding and/or trafficking to the plasma membrane. Based on the slightly lower apparent molecular weight of these two cysteine mutants, currently we cannot exclude the possibility that altered intracellular maturation and processing contribute to the effects we observed here. In summary, our current model of DDR1 receptor maturation suggests that Cys-303 and Cys-348 require either interor intramolecular linkage to allow full processing through the endoplasmic reticulum and trafficking to the membrane. Finally, the third cysteine in the stalk region of DDR1, Cys-289 may reside as reduced sulfhydryl group, most likely buried within the stalk folding domain. Compared with DDR1, similar mutational studies of the insulin receptor revealed notable differences. Mutations made to the four cysteines responsible for insulin receptor dimerization (Cys-524, Cys-682, Cys-683, and Cys-685) abolished covalent interaction; however, unlike DDR1, the insulin receptor was still able to noncovalently dimerize and transduce signal in the presence of insulin ligand (41). In agreement with our findings for DDR1, it is of note that about 10 times lower expression level has been reported for a dimerization-deficient mutant of the insulin receptor, where one of the cysteines was mutated not into a serine but a more bulky tryptophan reside (C524W).
What roles could covalently linked DDR1 have? Such covalent dimers could represent a “primed” state that facilitates accelerated ligand recognition and formation of higher order receptor clusters, which may be necessary for receptor activation and phosphorylation of noncovalently linked dimers. However, it may also be possible that DDR1 transduces signals through a mechanism independent of phosphorylation/kinase activation. Of note, DDR1 has recently been shown to trigger cytoskeletal responses leading to enhanced cell migration (8, 12). Conformational changes of the receptor folding induced by ligand binding to covalently linked dimers may result in transduction of a mechanical signal with more immediate effects than phosphorylation. Future studies involving inhibition of DDR1 ligand-independent dimer formation in migrating cells will help to test this as a potential role of dimers.
A detailed understanding of homo- and heterodimerization of the ErbB family or the mechanistics of insulin receptor function were instrumental for the development of specific ligand analogues or blocking reagents, which are now cornerstones in a targeted therapy of many disease entities (43, 44). With a better understanding of the mechanism of dimerization, ligand binding, and activation, similar applications can be envisioned for DDR1.
This work was supported in part by grants from the Association for International Cancer Research, the National Cancer Institute of Canada, and the Canada Research Chair Program (to W. F. V.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: DDR, discoidin domain receptor; ELISA, enzyme-linked immunosorbent assay; RTK, receptor tyrosine kinase; EGFR, epidermal growth factor receptor; PBS, phosphate-buffered saline; HA, hemagglutinin.
References
- 1.Johnson, J. D., Edman, J. C., and Rutter, W. J. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 5677–5681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogel, W. F., Abdulhussein, R., and Ford, C. E. (2006) Cell. Signal. 18 1108–1116 [DOI] [PubMed] [Google Scholar]
- 3.Alves, F., Saupe, S., Ledwon, M., Schaub, F., Hiddemann, W., and Vogel, W. F. (2001) FASEB J. 15 1321–1323 [DOI] [PubMed] [Google Scholar]
- 4.Playford, M. P., Butler, R. J., Wang, X. C., Katso, R. M., Cooke, I. E., and Ganesan, T. S. (1996) Genome Res. 6 620–627 [DOI] [PubMed] [Google Scholar]
- 5.Alves, F., Vogel, W., Mossie, K., Millauer, B., Hofler, H., and Ullrich, A. (1995) Oncogene 10 609–618 [PubMed] [Google Scholar]
- 6.Mullenbach, E., Walter, L., and Dressel, R. (2006) Gene (Amst.) 372 53–61 [DOI] [PubMed] [Google Scholar]
- 7.Kamohara, H., Yamashiro, S., Galligan, C., and Yoshimura, T. (2001) FASEB J. 15 2724–2726 [DOI] [PubMed] [Google Scholar]
- 8.Hou, G., Vogel, W., and Bendeck, M. P. (2001) J. Clin. Investig. 107 727–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avivi-Green, C., Singal, M., and Vogel, W. F. (2006) Am. J. Respir. Crit. Care Med. 174 420–427 [DOI] [PubMed] [Google Scholar]
- 10.Franco-Pons, N., Virgos, C., Vogel, W. F., Urena, J. M., Soriano, E., del Rio, J. A., and Vilella, E. (2006) Neuroscience 140 463–475 [DOI] [PubMed] [Google Scholar]
- 11.Ferri, N., Carragher, N. O., and Raines, E. W. (2004) Am. J. Pathol. 164 1575–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang, C. Z., Hsu, Y. M., and Tang, M. J. (2005) J. Cell Physiol. 203 295–304 [DOI] [PubMed] [Google Scholar]
- 13.Vogel, W. F., Aszodi, A., Alves, F., and Pawson, T. (2001) Mol. Cell. Biol. 21 2906–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gross, O., Beirowski, B., Harvey, S. J., McFadden, C., Chen, D., Tam, S., Thorner, P. S., Smyth, N., Addicks, K., Bloch, W., Ninomiya, Y., Sado, Y., Weber, M., and Vogel, W. F. (2004) Kidney Int. 66 102–111 [DOI] [PubMed] [Google Scholar]
- 15.Faraci-Orf, E., McFadden, C., and Vogel, W. F. (2006) J. Cell. Biochem. 97 109–121 [DOI] [PubMed] [Google Scholar]
- 16.Vogel, W., Gish, G. D., Alves, F., and Pawson, T. (1997) Mol. Cell 1 13–23 [DOI] [PubMed] [Google Scholar]
- 17.Shrivastava, A., Radziejewski, C., Campbell, E., Kovac, L., McGlynn, M., Ryan, T. E., Davis, S., Goldfarb, M. P., Glass, D. J., Lemke, G., and Yancopoulos, G. D. (1997) Mol. Cell 1 25–34 [DOI] [PubMed] [Google Scholar]
- 18.Wang, C. Z., Su, H. W., Hsu, Y. C., Shen, M. R., and Tang, M. J. (2006) Mol. Biol. Cell 17 2839–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogel, W., Brakebusch, C., Fassler, R., Alves, F., Ruggiero, F., and Pawson, T. (2000) J. Biol. Chem. 275 5779–5784 [DOI] [PubMed] [Google Scholar]
- 20.Koo, D. H., McFadden, C., Huang, Y., Abdulhussein, R., Friese-Hamim, M., and Vogel, W. F. (2006) FEBS Lett. 580 15–22 [DOI] [PubMed] [Google Scholar]
- 21.Curat, C. A., Eck, M., Dervillez, X., and Vogel, W. F. (2001) J. Biol. Chem. 276 45952–45958 [DOI] [PubMed] [Google Scholar]
- 22.Abdulhussein, R., McFadden, C., Fuentes-Prior, P., and Vogel, W. F. (2004) J. Biol. Chem. 279 31462–31470 [DOI] [PubMed] [Google Scholar]
- 23.Leitinger, B. (2003) J. Biol. Chem. 278 16761–16769 [DOI] [PubMed] [Google Scholar]
- 24.Ichikawa, O., Osawa, M., Nishida, N., Goshima, N., Nomura, N., and Shimada, I. (2007) EMBO J. 26 4168–4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlessinger, J. (1997) Cell 91 869–872 [DOI] [PubMed] [Google Scholar]
- 26.Tkacz, J. S., and Lampen, O. (1975) Biochem. Biophys. Res. Commun. 65 248–257 [DOI] [PubMed] [Google Scholar]
- 27.Noordeen, N. A., Carafoli, F., Hohenester, E., Horton, M. A., and Leitinger, B. (2006) J. Biol. Chem. 281 22744–22751 [DOI] [PubMed] [Google Scholar]
- 28.Schlessinger, J. (2000) Cell 103 211–225 [DOI] [PubMed] [Google Scholar]
- 29.Li, E., and Hristova, K. (2006) Biochemistry 45 6241–6251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward, C. W., Lawrence, M. C., Streltsov, V. A., Adams, T. E., and McKern, N. M. (2007) Trends Biochem. Sci. 32 129–137 [DOI] [PubMed] [Google Scholar]
- 31.Dawson, J. P., Berger, M. B., Lin, C. C., Schlessinger, J., Lemmon, M. A., and Ferguson, K. M. (2005) Mol. Cell. Biol. 25 7734–7742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Normanno, N., De Luca, A., Bianco, C., Strizzi, L., Mancino, M., Maiello, M. R., Carotenuto, A., De Feo, G., Caponigro, F., and Salomon, D. S. (2006) Gene (Amst.) 366 2–16 [DOI] [PubMed] [Google Scholar]
- 33.Gan, H. K., Walker, F., Burgess, A. W., Rigopoulos, A., Scott, A. M., and Johns, T. G. (2007) J. Biol. Chem. 282 2840–2850 [DOI] [PubMed] [Google Scholar]
- 34.Zhang, X., Gureasko, J., Shen, K., Cole, P. A., and Kuriyan, J. (2006) Cell 125 1137–1149 [DOI] [PubMed] [Google Scholar]
- 35.Walker, F., Orchard, S. G., Jorissen, R. N., Hall, N. E., Zhang, H. H., Hoyne, P. A., Adams, T. E., Johns, T. G., Ward, C., Garrett, T. P., Zhu, H. J., Nerrie, M., Scott, A. M., Nice, E. C., and Burgess, A. W. (2004) J. Biol. Chem. 279 22387–22398 [DOI] [PubMed] [Google Scholar]
- 36.Klein, P., Mattoon, D., Lemmon, M. A., and Schlessinger, J. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 929–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boni-Schnetzler, M., Rubin, J. B., and Pilch, P. F. (1986) J. Biol. Chem. 261 15281–15287 [PubMed] [Google Scholar]
- 38.Sparrow, L. G., McKern, N. M., Gorman, J. J., Strike, P. M., Robinson, C. P., Bentley, J. D., and Ward, C. W. (1997) J. Biol. Chem. 272 29460–29467 [DOI] [PubMed] [Google Scholar]
- 39.Wu, J. J., and Guidotti, G. (2002) J. Biol. Chem. 277 27809–27817 [DOI] [PubMed] [Google Scholar]
- 40.Lu, K., and Guidotti, G. (1996) Mol. Biol. Cell 7 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu, J. J., and Guidotti, G. (2004) J. Biol. Chem. 279 25765–25773 [DOI] [PubMed] [Google Scholar]
- 42.Bass, J., Chiu, G., Argon, Y., and Steiner, D. F. (1998) J. Cell Biol. 141 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Meyts, P., and Whittaker, J. (2002) Nat. Rev. Drug Discov. 1 769–783 [DOI] [PubMed] [Google Scholar]
- 44.Fruehauf, J. (2006) J. Exp. Ther. Oncol. 5 231–246 [PubMed] [Google Scholar]