Abstract
Characterization of the basal transcription machinery of mitochondrial DNA (mtDNA) is critical to understand mitochondrial pathophysiology. In mammalian in vitro systems, mtDNA transcription requires mtRNA polymerase, transcription factor A (TFAM), and either transcription factor B1 (TFB1M) or B2 (TFB2M). We have silenced the expression of TFB2M by RNA interference in Drosophila melanogaster. RNA interference knockdown of TF2BM causes lethality by arrest of larval development. Molecular analysis demonstrates that TF2BM is essential for mtDNA transcription during Drosophila development and is not redundant with TFB1M. The impairment of mtDNA transcription causes a dramatic decrease in oxidative phosphorylation and mitochondrial ATP synthesis in the long-lived larvae, and a metabolic shift to glycolysis, which partially restores ATP levels and elicits a compensatory response at the nuclear level that increases mitochondrial mass. At the cellular level, the mitochondrial dysfunction induced by TFB2M knockdown causes a severe reduction in cell proliferation without affecting cell growth, and increases the level of apoptosis. In contrast, cell differentiation and morphogenesis are largely unaffected. Our data demonstrate the essential role of TFB2M in mtDNA transcription in a multicellular organism, and reveal the complex cellular, biochemical, and molecular responses induced by impairment of oxidative phosphorylation during Drosophila development.
The majority of cellular ATP is generated by oxidative phosphorylation (OXPHOS)4 in the inner mitochondrial membrane. The OXPHOS system is composed of five multiprotein complexes that contain subunits encoded in two genomes, nuclear and mitochondrial. As a consequence, the biogenesis of the OXPHOS system is subject to a highly coordinated, dual genetic control (1). Mitochondrial DNA (mtDNA) encodes 13 essential proteins in animals that participate in four of the five OXPHOS complexes (I, III, IV, and V), as well as the genes for the two ribosomal RNAs and 22 tRNAs of the mitochondrial translational machinery.
Both mtDNA and mtRNA metabolism in eukaryotic cells require nuclear-encoded, organelle-specific factors. A dedicated RNA polymerase that is evolutionarily related to the bacteriophage T3, T7, and Sp6 RNA polymerases, and a limited number of auxiliary factors direct transcription of mitochondrial genes (2–4). In the budding yeast, Saccharomyces cerevisiae, the basal transcriptional machinery comprises the yeast mtRNA polymerase, Rpo41p, and a specificity factor, the transcription factor B (Mtf1p), which facilitates the binding of the RNA polymerase to numerous promoters scattered in the yeast mitochondrial genome (5–7). Although Mtf1p was initially considered to be a σ factor, its structure is homologous to bacterial rRNA dimethyltransferases (8).
The mammalian mitochondrial genome differs substantially from yeast mtDNA in several aspects including its small size, lack of introns, and presence of only two promoters located in the non-coding region, also known as the displacement loop region (9). Each promoter directs the synthesis of a polycistronic transcript encompassing all of the genes contained in each strand, the light strand (L strand) and the heavy strand (H strand). The light strand promoter contains a single transcriptional start site, whereas the heavy strand promoter initiates RNA synthesis from two sites, one used specifically for the synthesis of the rRNAs (10). Likewise, the mammalian transcriptional machinery differs from that of yeast. It comprises the mtRNA polymerase that contributes critically to promoter specificity (11), and two transcription factors, TFAM (also known as mtTFA) and TFBM (also known as mtTFB). TFAM is a member of the high mobility group family of proteins and is related structurally to bacterial HU protein (12, 13). TFAM plays a dual role in mtDNA maintenance and transcription (14). There are two isoforms of the TFBM factor in mammals that are encoded by different genes, TFB1M and TFB2M (15). The establishment of an in vitro transcription system employing recombinant proteins showed that both factors can stimulate specific transcription from light and heavy strand promoters, in a reaction that is strictly dependent on TFAM, and arguing that the basal transcription machinery in mammals is composed of mtRNA polymerase, TFAM, and either TFB1M or TFB2M, with the latter being at least 1 order of magnitude more active in the in vitro system (15, 16). Similar to yeast mtTFB, the mammalian TFBM factors are related structurally to bacterial rRNA dimethyltransferases. Recent evidence demonstrates that both TFB1M and TFB2M can bind S-adenosylmethionine in vitro, and can methylate the Escherichia coli small rRNA in vivo (17, 18), suggesting dual roles for both TFBM factors in transcription and translation.
Information about the mechanism of transcription in other animal systems remains more limited. The structure and gene content of the Drosophila melanogaster mitochondrial genome is similar to that of mammals (19). Fly mtDNA is also transcribed polycistronically, most likely from several promoters (20). Furthermore, Drosophila contains genes homologous to mtRNA polymerase, TFAM, TFB1M, and TFB2M, suggesting that the basal transcriptional machinery is conserved from fly to man. We have recently demonstrated by RNAi knockdown that d-TFB2M is essential for mtDNA transcription and replication in Schneider cells (21). In striking contrast, the knockdown of d-TFB1M does not affect either process, but reduces significantly the efficiency of mitochondrial translation (22), suggesting that d-TFB1M is important for mitochondrial protein biosynthesis in vivo.
To date, the data we have reported in Drosophila Schneider cells represent the only in vivo evidence regarding the cellular role of the two TFBM factors, and data evaluating their roles in the whole animal are lacking. To address this question we have silenced by RNAi the expression of d-TFB2M in transgenic flies. Our data show unequivocally that d-TFB2M is essential for mtDNA transcription during Drosophila development. The mitochondrial impairment caused by the decrease in TFB2M level elicits a complex retrograde response including a shift to glycolysis, and an increase in the mitochondrial mass.
EXPERIMENTAL PROCEDURES
Fly Stocks and Culture—Flies were raised on standard yeast-glucose-agar medium at 25 °C and 65% relative humidity in 12-h light/dark cycles unless otherwise indicated. The y1w1118 embryos were microinjected with the appropriated transformation plasmids. To determine the chromosomal location of the transgenes and manipulation of transgenic lines the stock w; CyO/If; TM6B/MKRS was used. Other stocks and GAL4 lines used in this study were: w; UAS-LacZ/UAS-LacZ; w; mioLacZ/mioLacZ; w; da-GAL4/da-GAL4. The driver da-GAL4 was obtained from the Bloomington Drosophila Stock Center at Indiana University (genotype: w*;P{GAL4-da.G32}UH1, #5460). The UAS-LacZ reporter line (23) was a gift from Dr. G. Morata. The mioLacZ/mioLacZ line was a gift of Dr. M. Cervera.
Generation of Transgenic UAS-IRB2 Lines—The constructs needed for RNAi consisted of inverted repeat (IR) fragments from d-TFB2M cDNA cloned into the pUAST Drosophila transformation vector. The IRB2-insert was 365 bp long and obtained as previously described (21). Recombinant pUAST-IR constructs were transformed in Sure competent bacteria (Stratagene) to minimize DNA recombination. Transgenic lines were generated by microinjection of the pUAST-IR plasmids in y1w1118 embryos according to standard procedures (24). Two independent transgenic lines (UAS-IRB2 A-B) were obtained for silencing d-TFB2M. Each of them carried a single independent insertion in chromosome III, which was brought to homozygosis. Chromosome insertion was determined using balancer stocks.
UAS-GAL4 Experiments—IRB2 fragments were expressed using the UAS-GAL4 system described by Brand and Perrimon (23). The daughterless GAL4 driver (da-GAL4) yields a high and ubiquitous transgene expression throughout development and was used to induce the silencing of d-TFB2M. The homozygous da-GAL4 driver was crossed to homozygous UAS lines and the progeny was subjected to study. Crosses were set with ∼100–150 individuals in standard food vials and were passed daily.
Weight and Size Analysis—Body weight of third instar larvae was measured with a precision scale (max = 410 g, d = 0.001 g; Boeco). About 50 larvae reared under the same growth conditions were collected 7 or 14 days after egg laying (AEL), washed, and transferred to a plastic tube before weighing.
Oligonucleotides—All oligonucleotides were purchased from Isogen (The Netherlands). All sequences are given in the 5′ to 3′ direction. Probes used for Southern and Northern hybridization were PCR products obtained from the amplification of Drosophila nuclear and mitochondrial DNAs using the following primers: d-Cytb (dir): AGGTGTTACTAAAGGAGTTGCTGG; d-Cytb (rev): TACGTAACATCTTTAGCCTCTAATG; d-ND4 (dir): AGGAGCTGCTATATTAGCTG; d-ND4 (rev): CAGCCAGAACGTTTACAAGC; d-RP49 (dir): GACCATCCGCCCAGCATAC; d-RP49 (rev): AGAACGGAGGCGACCGTTG; d-histone (dir): ACACACGGAACACGAATGCTCG; d-histone (rev): AGCGAAGCCAAAGCCTGTAGTAGC.
Oligonucleotides used for quantitative PCR (qPCR) are as follows: d-mtTFB2 (dir): CCCAGAAAGCGTTTACAGAT; d-mtTFB2 (rev): GAGATGTATGTATATGGGTG; d-mtTFB1 (dir): GCACAACAGGATGGCCCAA; d-mtTFB1 (rev): CGCTCGTCCATGAGGAAG; d-RPOLII (dir): GAGTCCGCGTAACACCTATCAAA; d-RPOLII (rev): ACAAGTGGCTTCATCGGATAGTAAAG.
Quantitative RT-PCR Assays—About 10 frozen larvae from each genotype were used for RNA extraction with TRIzol reagent (Invitrogen). From each sample 2 μg of total RNA were reverse transcribed into cDNA according to the manufacturer's instructions using oligo(dT) as a primer and the SuperScript III First-strand Synthesis System for RT-PCR (Invitrogen). The gene encoding the 140-kDa subunit of RNA polymerase II was used as a normalizing internal control (25). Exon-specific oligonucleotide primers for the two genes tested, d-TFB1M and d-TFB2M, were designed with Primer3 software. Real time PCRs were performed in a Rotor Gene thermocycler (Corbett Research) using SYBR Green I (Molecular Probes) as double-stranded DNA binding dye and the following conditions: 94 °C for 3 min; 40 cycles (94 °C for 20 min; 58 °C for 20 min; 72 °C for 20 min; 82 °C for 15 min); 72 °C for 6 min; 50 °C for 3 min; melting ramp. Each sample contained 1× SYBR Green I, 0.2 mm dNTPs, 1× enzyme buffer, 4 mm MgCl2, 0.5 units of Taq polymerase (Biotools), 0.3 μm from each oligonucleotide, and 4 μl of a 1/20 dilution of the reverse transcribed products in a final volume of 20 μl. Three separate samples were collected from each genotype and triplicate measurements were conducted.
Immunoblotting—Mitochondrial protein extracts obtained by differential centrifugation were fractionated by 10.5% SDS-PAGE (20 μg/lane) and transferred to nitrocellulose filters. Filters were preincubated for 1 h with 5% skim milk in PBS, followed by incubation for 1 h with the corresponding primary antibody. Polyclonal antibodies against Drosophila TFB1M and TFB2M were used as previously described (21, 22). A polyclonal antibody against Drosophila hsp60 was used at 1:400 dilution and was a kind gift of Dr. Juan Fernández-Santarén. To detect the mtDNA-encoded COXIII subunit, we used a monoclonal antibody raised against the COXIII yeast subunit (Molecular Probes) at a 1:200 dilution. As loading control, an antibody against Drosophila β-ATPase was used as previously described (26).
DNA Extraction and Southern Blotting—Aliquots of 10 frozen third instar staged larvae were ground in 400 μl of homogenization solution that contained 10 mm Tris-HCl, pH 7.5, 10 mm EDTA, 60 mm NaCl, 0.5% SDS. The precipitates were removed by centrifugation and the supernatants were extracted once with phenol/chloroform (1:1), treated with 10 μg/ml RNase A at 37 °C for 15 min, re-extracted with phenol/chloroform (1:1), and precipitated with 2 volumes of ethanol. DNA precipitates were washed with 80% ethanol and resuspended in 50 μl of H2O. Southern analysis was performed as previously described (21).
RNA Extraction and Northern Blotting—Aliquots of 10 frozen third instar staged larvae were homogenized in TRIzol reagent (Invitrogen) and total RNA was extracted according to the manufacturer's instructions. Using standard procedures, RNA (5 μg/lane) was fractionated on a 1% agarose/formaldehyde gel, blotted onto a Zeta-Probe nylon membrane (Bio-Rad) and hybridized to randomly 32P-labeled probes for each of these genes: ribosomal protein 49 (RP49), cytochrome b (Cytb), and NADH dehydrogenase subunit 4 (ND4). Hybridization, washing, and autoradiography of the membranes was performed as previously described (21).
Mitochondrial Preparations—For ATP synthesis determination, aliquots of fresh third instar larvae were ground in buffer containing 0.22 m sucrose, 0.12 m mannitol, 40 mm Tricine, pH 7.5, and 1 mm EDTA as described previously (27) and mitochondria were isolated by differential centrifugation. For enzymatic measurements, the same procedure was followed except for grounding buffer, which contained 250 mm sucrose, 2 mm EDTA, 100 IU/liter heparin, 10 mm Tris-HCl, pH 7.4, and purified mitochondria were sonicated for 6 s at 4 °C, frozen and thawed. Protein determination was performed using the DC Protein Assay kit (Bio-Rad).
Mitochondrial ATP Synthesis—Mitochondrial ATP synthesis was measured with an Optocomp I BG-1 luminometer (GEM Biomedical Inc.) using the ATP Bioluminiscent Assay Kit (Sigma). Incubation buffer contained 5 mm MgCl2, 10 mm KH2PO4, 0.2% bovine serum albumin, 0.1 mm ADP, and 54 μm APP. Substrates used were glutamate and malate, 10 mm each. Calibration was performed at the end of every measurement by the addition of 100 pmol of ATP.
OXPHOS Activities—Mitochondrial OXPHOS complexes and citrate synthase activities were measured as described previously (28). Incubation temperatures were 30 °C except for complex IV, which was measured at 38 °C. Enzyme activities are expressed in nanomoles of substrate catalyzed per minute and per milligram of protein.
Metabolite Measurements—Determinations of glycolytic intermediates, ATP, ADP, and AMP, was performed as described previously (29) with minor modifications on sample preparation. About 0.5 g of frozen third instar larvae were homogenized in 1 ml of ice-cold 8% perchloric acid, 40% ethanol and further treated as indicated to obtain cytosolic extracts. Metabolites were measured in a UV-visible spectrophotometer UV-1603 (Shimadzu Corp.).
Histochemistry—Standard procedures were used to assay β-galactosidase enzyme activity in w;UAS-LacZ/+; UAS-IRB2/daGAL4 larvae with minor modifications (30). The complete expression pattern was visualized after 10 min of staining at room temperature. The preparations were viewed in a Nikon Eclipse E400 microscope. Images were captured using an Olympus DP50 digital camera and ViewFinder and Studio Lite software.
Neuromuscular Joint Staining—Dissected larvae were fixed in 4% paraformaldehyde for 30 min and then washed in PBS containing 0.4% Triton X-100 every 15 min for 1 h. To visualize neurons, the fixed preparations were blocked in PBS, 5% goat serum, 2% bovine serum albumin, 0.4% Triton X-100 for 30 min, incubated overnight at 4 °C with rabbit anti-horseradish peroxidase (1:200; Jackson ImmunoResearch), washed four times in PBS, 0.4% Triton X-100 for 1 h, and incubated with the secondary antibody goat anti-rabbit IgG coupled to Alexa Fluor 568 (1:400; Molecular Probes) for 2 h at room temperature in the dark. Then they were washed in PBS, 0.4% Triton X-100 every 15 min for 1 h and mounted in Vectashield (Vector Laboratories). Preparations were imaged using a Leica TCS SP2 laser-scanning microscope (Heidelberg, Germany) with ×63 or ×100 oil immersion lenses.
Bromodeoxyuridine (BrdUrd) Labeling in the Larval Central Nervous System—BrdUrd incorporation was performed as previously described (31) with some modifications. Dissected larvae were cultured in Schneider's medium (Invitrogen) containing 100 μg/ml 5′-bromo-2′-deoxyuridine (Sigma) for 30 min at room temperature and fixation was performed in PBS, 5% formaldehyde for 15 min. After a 2.2 n HCl denaturing treatment, larval tissue was neutralized with 100 mm borax for 5 min. Incorporation was monitored with fluorescein isothiocyanate-conjugated mouse anti-BrdUrd (1:20; BD Biosciences) following the suppliers procedures. Larval brains were dissected free from the remainder of the tissue and mounted in Vectashield (Vector Laboratories) containing 1.5 μg/ml 4′,6-diamidino-2-phenylindole for viewing on a Leica TCS SP2 laser-scanning microscope (Heidelberg, Germany) with ×10 or ×63 oil immersion lens. Larval brain areas were measured using LCS Simulator software (Leica Microsystems).
Immunocytochemistry in Imaginal Wing Discs—Imaginal discs from third instar larvae were dissected in PBS and fixed with 4% paraformaldehyde in PBS for 20 min at room temperature. They were blocked in PBS, 1% bovine serum albumin, 0.3% Triton X-100 for 1 h, incubated with the primary antibody overnight at 4 °C (dilution 1:50), washed four times in blocking buffer and incubated with the appropriate secondary antibody for 2 h at room temperature in the dark (dilution 1:200). Finally, they were washed and mounted in Vectashield (Vector Laboratories).
Primary antibodies used were: mouse anti-wingless (Hybridoma Center) and rabbit anti-caspase 3 (Cell Signaling Technology). Secondary antibodies were coupled to the fluorochromes Alexa Fluor 467 (Molecular Probes), fluorescein isothiocyanate, and Rhodamine Red X (Jackson ImmunoResearch).
Preparations were imaged with a Leica TCS SP2 laser-scanning microscope (Heidelberg, Germany). A number of apoptotic cells were measured using analySIS FIVE software (Olympus Digital Imaging Solutions).
Flow Cytometry—Larvae at the third instar stage were dissected in 1× PBS. Cell dissociation of wing discs and larval brains was done as described before (32). An average of 20 wing discs and 10 brains were dissociated. The cell suspension was analyzed using a BD Biosciences FACS Vantage SE and data were processed with BD FACS Diva and Cell Quest Pro software.
Statistical Analysis—Statistical analysis of data were performed with Prism 4.03 software (GraphPad Software). The one-way ANOVA test and the Bonferroni post-test were used to determine statistical significance of the results.
RESULTS
d-TFB2M Is Essential during Drosophila Development—To characterize the functional role of d-TFB2M in Drosophila we adopted a strategy to induce gene silencing by RNAi in the whole animal. An interfering RNA containing IR that form double-stranded RNAs homologous to d-TFB2M (21) was cloned into the pUAST vector and microinjected into Drosophila embryos. We recovered several transgenic lines, and two independent lines with the UAS-IRB2 construct (UAS-IRB2A and -B) were characterized in detail. RNAi-dependent knockdown was induced using the da-GAL4 strain as a driver, which expresses the yeast transcriptional activator GAL4 under control of the daughterless enhancer. da-GAL4 expresses GAL4 throughout development and in most if not all tissues, and has been used extensively to overexpress constitutively transgenes containing UAS binding sites in Drosophila (33).
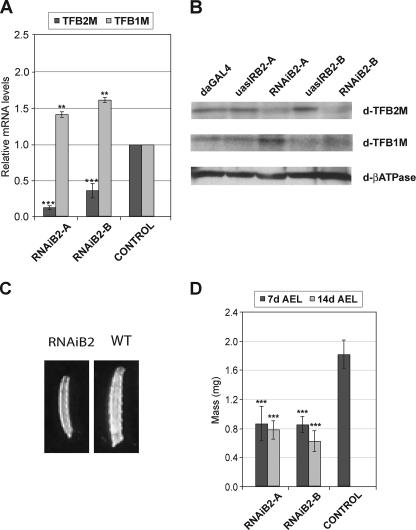
We found that widespread knockdown of d-TFB2M induces 100% lethality. Less than 3% of the larvae entered pupariation, and the remainder died during the third larval stage in both crosses da-GAL4/UAS-IRB2A and da-GAL4/UAS-IRB2B. Quantitative PCR (Fig. 1A) and immunoblot (Fig. 1B) analyses revealed severe reductions (60–80%) in d-TFB2M mRNA and protein. The level of expression of d-TFB1M was evaluated as a control. Interestingly, d-TFB1M expression is increased 1.5-fold at both the mRNA and protein levels (Fig. 1, A and B), suggesting a retrograde compensatory response in nuclear gene expression. These data show that TFB2M is essential in D. melanogaster development, and that it cannot be compensated by TFB1M, even at moderately increased levels of the latter.
FIGURE 1.
Expression analysis and body mass of RNAiB2 larvae. A, d-TFB2M and d-TFB1M transcript levels were determined by qRT-PCR in 7-day-old third instar larvae in the RNAiB2 lines, and normalized to control lines using RNA pol II as an internal control. Error bars indicate average S.D. from a minimum of five independent RT-PCR assays. RNAiB2-A, w; UAS-IRB2-A/+; da-GAL4/+ and RNAiB2-B, w; UAS-IRB2-B/+; da-GAL4/+. The two parental lines bearing the UAS-IRB2-inverted repeats (UASIRB2-A and UASIRB2-B) and the driver da-GAL4 were evaluated as a control. Similar results were obtained with the three control lines. Statistical significance of the results was calculated using the one-way ANOVA test (**, p < 0.01; ***, p < 0.001). B, immunoblot analysis of RNAiB2 and control third instar larvae. Mitochondrial protein extracts (20 μg) were probed with rabbit antiserum against d-TFB2M, d-TFB1M, and d-β-ATPase (as a loading control), as indicated under “Experimental Procedures.” C, photographs of control (WT) and RNAiB2 third instar larvae (7 days AEL). D, total body mass is reduced in RNAiB2 larvae. Larvae of each genotype were weighed and body mass per larva calculated. Error bars indicate S.D. of a minimum of three independent experiments (***, p < 0.001).
d-TFB2M Silencing Produces Long-lived Larvae with a Severe Impairment in mtDNA Transcription—To evaluate further the phenotype produced by a reduction in the level of d-TFB2M, we characterized several morphological and molecular parameters in the knockdown larvae. In control lines (parental and yw), 100% of the larvae entered pupariation 6–9 days AEL. Interestingly the larval stage in both RNAiB2 lines was extended substantially, with most larvae living more than 30 days AEL (data not shown). The RNAiB2 larvae remained close to the food without entering the wandering stage, and never embarked upon pupariation. We confirmed that the larvae reached the third larval instar stage by detecting the presence of two specific morphological markers, mouth hooks and spiracles (data not shown). Interestingly, the duration of the first and second larval instar stages was not extended significantly as compared with those of control larvae, demonstrating a specific arrest at the third larval instar stage induced by d-TFB2M silencing. Finally, a significant reduction in the length and diameter of the RNAiB2 larvae was detected in both lines. Consequently, the total mass of the RNAiB2 larvae was about 50% lower than in the control larvae (Fig. 1, C and D).
To confirm that silencing of d-TFB2M expression was maintained throughout larval development, we measured the abundance of d-TFB2M mRNA by quantitative RT-PCR in RNAiB2 larvae of different ages, and found that it remained below 25–30% of the level detected in control larvae (Fig. 2A). The induction of the expression of d-TFB1M was also maintained between 1.5- and 1.75-fold in larvae of different ages (Fig. 2A). We have shown previously that d-TFB2M regulates mtDNA copy number and transcription in Drosophila Schneider cells (21). To analyze if silencing d-TFB2M expression in the whole animal affected mtDNA transcription, we measured the steady-state levels in RNAiB2 larvae of different ages of the Cytb and ND4 transcripts that were encoded in opposite mtDNA strands. We found a clear reduction in the level of both transcripts, which were present at less than 50% of their levels as compared with the parental lines (Fig. 2B). These data suggest strongly that d-TFB2M is critical for transcription of both mtDNA strands in the animal. We also evaluated mtDNA copy number in the RNAiB2 larvae (Fig. 2C). We found only a moderate reduction in the level of mtDNA, even in the older RNAiB2 larvae (Fig. 2C). Thus, the decrease in d-TFB2M level has a minimal effect on mtDNA replication during larval development.
FIGURE 2.
Effects of TFB2M RNAi knock-down on the levels of TFB2M and TFB1M mRNAs, mitochondrial transcripts, and mtDNA during larval development. A, TFB2M transcript levels during larval development. Total RNA was extracted from larvae of the indicated genotypes at different times AEL. Control lines initiated pupariation at 7–9 days AEL and thereafter, only RNAiB2 larvae were collected. TFB2M transcripts were analyzed by qRT-PCR and normalized to control lines using RNA pol II as an internal control. Data were obtained from at least five independent experiments. Statistical significance of the results was calculated using the one-way ANOVA test (**, p < 0.01; ***, p < 0.001). B, effect of TFB2M silencing on mitochondrial transcription during larval development. Total RNA was extracted from RNAiB2 and control larvae on the days indicated. RNA (5 μg) was fractionated in a 1% agarose/formaldehyde gel, blotted to a nylon membrane, and hybridized with radiolabeled probes for mitochondrial transcripts ND4 and CytB and the nuclear transcript Rp49. Relative transcription efficiency was quantitated by normalizing mitochondrial transcript abundance to that of Rp49. Data were collected from at least three independent experiments and the statistical significance of the results was calculated using the one-way ANOVA test (*, p < 0.05; **, p < 0.01). C, effect of TFB2M silencing on mtDNA copy number during larval development. Total DNA from RNAiB2 and control (CTRL) larvae was hybridized with radiolabeled probes for CytB (mtDNA) and the histone gene cluster (nDNA). mtDNA copy number was quantitated by normalizing mtDNA to nuclear DNA levels (*, p < 0.05).
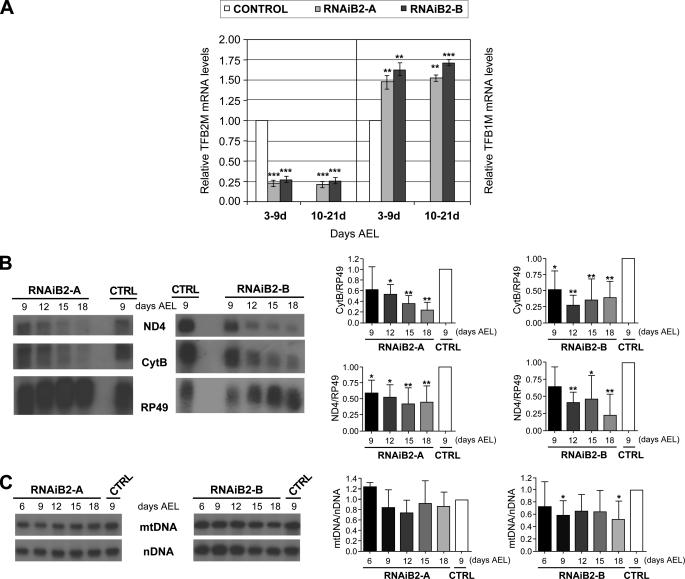
d-TFB2M Silencing Induces OXPHOS Impairment and a Metabolic Shift to Glycolysis—To analyze the biochemical consequences of decreased mtDNA transcription, we measured the enzymatic activities of the respiratory chain complexes in 7- and 14-day-old RNAiB2 larvae. We observed significant decreases in the activities of complexes I, III, and IV as compared with the control lines (Fig. 3A). ATP synthesis capacity measured in isolated mitochondria was also affected severely, with >80% reduction in 14-day-old RNAi larvae (Fig. 3A). The reduction was larger in the 14-day-old larvae, indicating that the OXPHOS impairment phenotype is progressive. Interestingly, the nuclear-encoded complex II and citrate synthase activities are increased substantially (2–3-fold), suggesting strongly a compensatory response of mitochondrial proliferation in the RNAiB2 larvae. Consequently, we found an equivalent increase in the amount of the chaperone hsp60, a reliable marker of mitochondrial mass, and a significant decrease in the ratio of COXIII/hsp60 (Fig. 3B).
FIGURE 3.
OXPHOS enzymatic activities and mitochondrial ATP synthesis in RNAiB2 larvae. A, OXPHOS enzymatic activities (complexes I, II, III, and IV), citrate synthase activity, and ATP synthesis capacity using glutamate plus malate as substrate were measured in mitochondrial protein extracts obtained from larvae of each genotype on the days AEL as indicated. Data represent the mean ± S.D. of at least three independent determinations (*, p < 0.05; **, p < 0.01; ***, p < 0.001). B, immunoblot analysis of RNAiB2 and control (UAS-IRB2) third instar larvae. Mitochondrial protein extracts (40 μg) were probed with a monoclonal antibody against COXIII and a polyclonal antibody against d-hsp60 as described under “Experimental Procedures.” The ratio of COXIII/d-hsp60 was calculated from at least three independent experiments. Data represent the mean ± S.D. (*, p < 0.05; ***, p < 0.001). In the control, the data from UASIRB2-A and UASIRB2B were included.
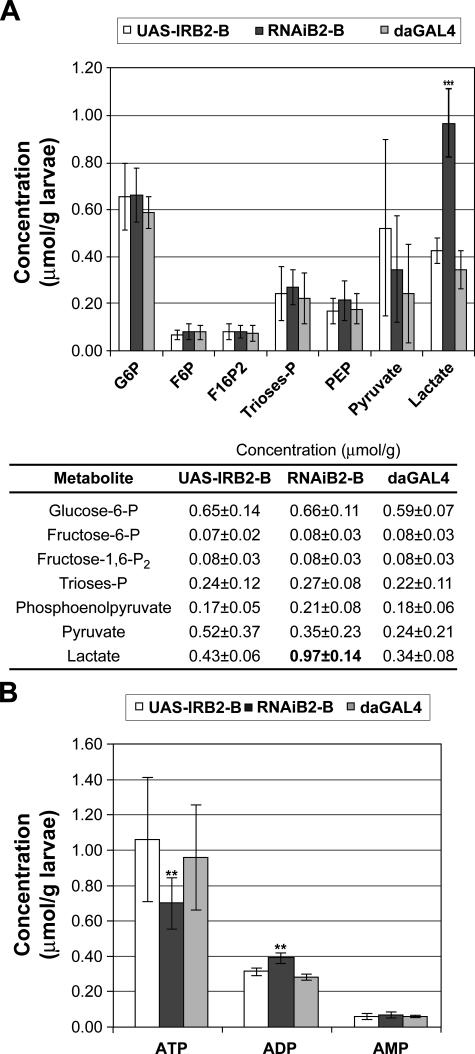
To evaluate further the metabolic state of the larvae with a reduced amount of d-TFB2M, we measured the content of key glycolytic intermediates including glucose 6-phosphate, fructose 6-phosphate, fructose 1,6-biphosphate, triose phosphates, phosphoenolpyruvate, pyruvate, and lactate. No change was observed in the steady-state levels of the metabolites involved in the reactions of the glycolytic enzymes hexokinase, phosphofructokinase, or pyruvate kinase. However, lactate accumulated over 2-fold as compared with the control (Fig. 4A), indicating an increase in the glycolytic flux as a compensatory mechanism for the severe defect in mitochondrial ATP synthesis capacity. Consequently, the RNAiB2 larvae showed only a partial deficit in total ATP levels of 30%, and a modest but significant increase in total ADP concentration (Fig. 4B).
FIGURE 4.
Effect of TFB2M silencing on glycolysis and energy balance. A, glycolysis in RNAiB2 larvae was assessed by determining the content of glycolytic intermediates in cytosolic extracts. Both parental UAS-IRB2 lines were used as controls. Data represent the mean ± S.D. of triplicate measurements from two independent experiments (***, p < 0.001). B, the energy status of RNAiB2 larvae was evaluated by measuring the content of ATP, ADP, and AMP in cytosolic extracts. Both parental UAS-IRB2 lines were used as controls. Data represent the mean ± S.D. of triplicate measurements from two independent experiments (**, p < 0.01).
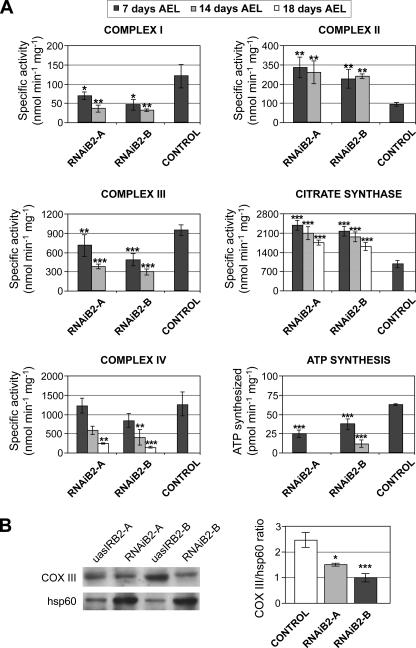
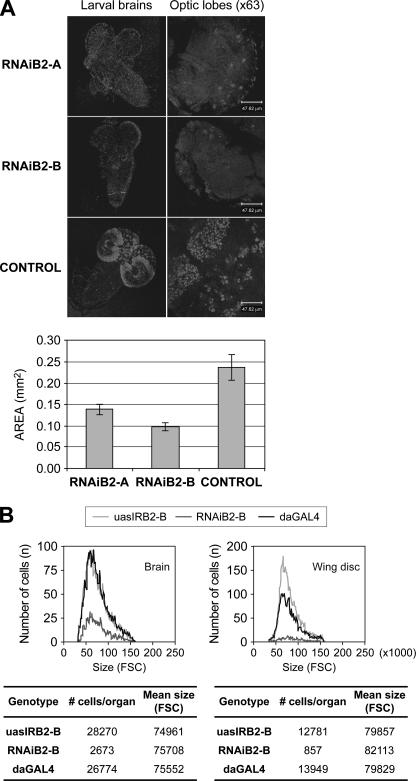
d-TFB2M Silencing Decreases Cell Proliferation without Affecting Cell Growth, and Increases Apoptosis—The RNAiB2 larvae were smaller than control larvae, suggesting a defect in cell proliferation, cell growth, or both. To address this issue we analyzed the incorporation of BrdUrd in isolated larval brains. Compared with the brains of control larvae, the brains isolated from RNAiB2 larvae were significantly smaller. In addition, the incorporation of BrdUrd in the optic lobes was decreased substantially in the brains of the d-TFB2M knockdown larvae (Fig. 5A), revealing a significant defect in cell proliferation. To evaluate the number and size of cells constituting different organs in the RNAiB2 larvae, we isolated wing discs and brains, generated individual cells by treatment with trypsin, stained the cells with Hoechst 33342, and analyzed them by flow cytometry. Fig. 5B shows a drastic reduction in the number of cells in both brain and wing imaginal discs of RNAi larvae. At the same time, the average cell size remained unchanged. Our data provide evidence that d-TFB2M reduction and subsequent OXPHOS impairment cause a defect in cell proliferation without affecting cell growth.
FIGURE 5.
TFB2M silencing does not affect cell growth but reduces substantially cell proliferation during larval development. A, the effect of TFB2M silencing on cell proliferation was evaluated by BrdUrd labeling in third instar larval brains extracted from RNAiB2 and control larvae. Proliferating cells are organized in proliferation centers that are generally more abundant in the optic lobes. RNAiB2 larval brains are smaller than that of the control line and the number of labeled cells is largely reduced. The graph shows the mean size ± S.D. of five larval brains of each genotype (p < 0.001). B, the effect of TFB2M silencing on cell size was evaluated by flow cytomety. Brains and wing imaginal discs were isolated from third instar larvae of each genotype and disgregated by trypsin treatment in the presence of Hoechst 33342. Samples were analyzed in a cytometer according to their size. The graph and the table show that the mean size of the RNAiB2 cells that formed both organs did not differ substantially with respect to controls but that the number of cells that constituted each RNAiB2 organ was reduced drastically.
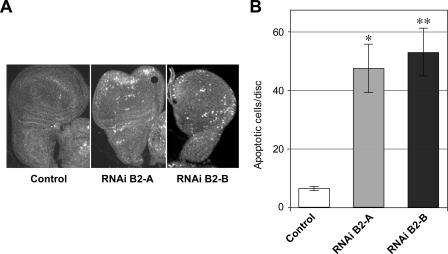
We also investigated the possible effect of d-TFB2M knockdown on apoptosis. We evaluated the level of apoptosis in wing imaginal discs of RNAiB2 larvae by probing them with an antibody that recognizes specifically activated caspase 3. We observed a clear increase in the level of apoptotic cells (Fig. 6), suggesting that a defect in the OXPHOS system induces the apoptotic program, at least in imaginal discs.
FIGURE 6.
TFB2M silencing induces apoptosis in imaginal discs. A, wing maginal discs from third instar larvae from control and RNAiB2 larvae were dissected and fixed. Apoptosis was visualized using an antibody that detects specifically activated caspase 3. B, the number of apoptotic cells were counted in individual wing imaginal discs. The bars shows the number of apoptotic cells per disc ± S.D. of each genotype (n = 10; p < 0.001).
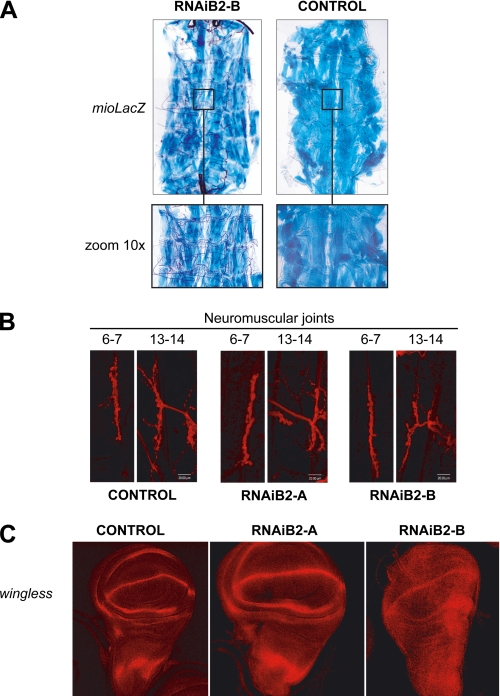
d-TFB2M Silencing Does Not Affect Larval Morphogenesis—We next examined potential effects of d-TFB2M knockdown on the morphogenesis of various tissues during larval development. To examine muscle tissues, we crossed the fly line w;miolacZ/miolacZ; UAS-IRB2-B/UAS-IRB2-B with the da-GAL4 driver, such that the knockdown of d-TFB2M is induced constitutively in the progeny. Here, the larval muscles are identified readily because the β-galactosidase reporter gene is expressed under the control of the myosin enhancer in all of the muscles of the RNAiB2 larvae. We found no significant differences between those of the RNAiB2 larvae as compared with the parental da-GAL4 larvae used as control (Fig. 7A). To visualize the nervous system of the RNAiB2 larvae, we performed immunohistochemistry using an antibody against horseradish peroxidase that stains the peripheral and central nervous systems. Again, we did not detect significant differences in the neuromuscular junction of fibers 6–7 and 13–14 from the third abdominal segment, which showed a pattern that is identical to that observed in the equivalent region of control larvae (Fig. 7B). Finally, we examined the pattern of expression of the wingless (wg) gene in the wing imaginal disc, which is used as a marker of the expression of key genes involved in developmental processes. We observed no differences in the expression pattern of Wg in discs isolated from RNAiB2 larvae as compared with the controls, except for a delay in the establishment of the final pattern (Fig. 7C). In summary, although silencing of d-TFB2M affects cell proliferation and apoptosis, morphogenetic processes are not affected and the various tissues of the RNAiB2 larvae are normal, but smaller as compared with control larvae.
FIGURE 7.
Morphogenesis and patterning in RNAiB2 larvae. A, d-TFB2M silencing does not affect muscular tissue morphogenesis. A transgenic UAS-IRB2 line that expressed the reporter gene LacZ under control of the myosin enhancer was generated to evaluate the morphology of the muscles in RNAiB2 larvae. Reporter expression is revealed by β-galactosidase staining. B, TFB2M silencing does not affect nervous system morphogenesis. Larval central nervous system and neuromuscular joints were visualized by immunostaining with an antibody against horseradish peroxidase (red). Images from neuromuscular joints between muscle fibers 6–7 and 13–14 from the third abdominal segment were obtained by confocal fluorescence microscopy. C, d-TFB2M silencing does not affect the expression pattern of wingless. The expression of the wingless gene was visualized by immunostaining (red) in wing imaginal discs of RNAiB2 larvae and control larvae.
DISCUSSION
In this study, we have evaluated the functional role of d-TFB2M in the context of Drosophila development, and demonstrate that it is not redundant with d-TFB1M. Using the UAS-GAL4 system, we have silenced constitutively the expression of the d-TFB2M gene below 30% of the endogenous level. In the two UAS-IRB2 strains analyzed, most of the animals progress to the third larval stage and fail entering pupariation. Molecular analysis has revealed a large reduction in the abundance of specific mitochondrial transcripts encoded in both strands. Our data demonstrate for the first time that TFB2M plays an essential role in mtDNA transcription during animal development. The RNAi-induced knockdown is specific for TFB2M, because the expression of TFB1M was not reduced and instead, its abundance is increased 1.5-fold, likely due to a compensatory response to the impairment of mitochondrial function. Interestingly, overexpression of human TFB2M in HeLa cells induces an increase in the levels of TFB1M mRNA and protein (34), suggesting the existence of a retrograde signaling pathway from mitochondria to the nucleus, which regulates precisely the expression of these related factors.
We found previously in Schneider cells that d-TFB2M levels correlate with mtDNA abundance (21), and similar results have more recently been reported for human HeLa cells (34). Those data are compatible with a role for TFB2M in RNA primer synthesis during mtDNA replication (35). In larvae expressing only 20% of endogenous d-TFB2M, mtDNA copy number was also affected, although only modestly, even in long-lived larvae (30 days old) that showed a drastic reduction in mtRNA levels. Variability in the reduction of mtDNA levels in cell culture and animals likely results from several factors. RNAiB2 larvae express 20% of TFB2M, which may be not rate-limiting for RNA primer synthesis; it is important to keep in mind that during larval development, cell proliferation is low and mtDNA replication is only needed to offset mtDNA turnover. In mammals, two different mechanisms for mtDNA replication have been described: strand-asymmetric synthesis (36, 37) and coupled leading and lagging strand synthesis (38, 39). If such a situation occurs in invertebrate mitochondria, one might invoke the alternative mechanisms to explain the differences found in Schneider cells and larvae. However, we have found previously that the response to overexpression of the catalytic subunit of DNA polymerase γ, the sole DNA polymerase found in animal mitochondrial, also varies in cell culture versus the whole animal (40). Schneider cells overexpressing the catalytic core of polymerase γ grow well and show normal mtDNA replication. In contrast, a similarly high level of overexpression in transgenic flies induces a severe mtDNA depletion due to defective mtDNA replication, which provokes a wide range of phenotypes including pupal lethality (40).
The most obvious phenotype of the knockdown flies is an arrest in larval development, with extension of the third larval stage of more than 30 days, as compared with 5–7 days for control flies. RNAiB2 larvae are shorter and thinner than control larvae, and show a substantial reduction in body mass. We explored the effect of reduced mitochondrial transcript levels in the energy metabolism of the long-lived larvae, and found a large and progressive decline in the activity of respiratory complexes I, III, and IV, each containing subunits encoded in the mitochondrial genome. As a consequence, the capacity of mitochondria to synthesize ATP is severely impaired. Using a mix of malate and glutamate as substrates, we showed that mitochondria isolated from 14-day-old RNAiB2 larvae synthesize less than 20% of ATP as compared with mitochondria isolated from control larvae. To explain how RNAiB2 larvae can survive for long periods of time with this drastic impairment in OXPHOS function, we determined the levels of several glycolytic intermediate metabolites, and found a 2.5-fold increase in the level of lactate. Our results suggest strongly that RNAiB2 larvae shift their energy metabolism to increase the flux of the glycolytic anaerobic pathway. Accordingly, the total amount of ATP in RNAiB2 larvae was maintained at 70% of control larvae, such that the defect in mitochondrial synthetic capacity is largely compensated by glycolysis. Furthermore, we found that RNAiB2 larvae exhibit a substantial increase in the activity of citrate synthase and complex II activities, and in the steady-state level of hsp60, which are considered to be markers of mitochondrial mass (33). We interpret these findings to suggest that OXPHOS impairment due to a reduced amount of TFB2M induces a compensatory response that increases mitochondrial mass. Most interestingly, the level of d-TFB1M in RNAiB2 larvae is also increased 1.5-fold as compared with controls, although it did not allow maintenance of normal mitochondrial transcript levels. Altogether, our data suggests a compensatory retrograde response that affects the expression of a wide number of nuclear-encoded mitochondrial genes, and that among them, d-TFB1M too, serves an important function in mitochondrial physiology. As mentioned before, the increase in mitochondrial biogenesis is not accompanied by a similar increase in mtDNA copy number, which in fact is slightly decreased, a situation that is frequently found in human mitochondrial diseases (41).
Development in multicellular animals is a process that involves strict regulation of cell number and cell size. This regulation is critical to produce an animal in which each organ and the whole body are of appropriate size. We found that the RNAiB2 third instar larvae with impaired mtDNA transcription show a substantial reduction in organ and body size, suggesting that they are defective in cell number (cell proliferation), cell size (cell growth), or both and as a result, they remain in the third larval instar without entering pupariation. Brains and imaginal discs in RNAiB2 larvae show a severe reduction in total cell number, yet cell size is unaffected. This suggests strongly that OXPHOS impairment results in a specific arrest of the cell cycle in the extensive growth period during larval development. Such a failure in cell proliferation has also been observed in lack of function mutants of genes encoding other Drosophila proteins involved in OXPHOS biogenesis such as the two subunits of DNA polymerase γ (42, 43), the single-stranded DNA-binding protein (44) and the cytochrome oxidase subunit Va (45).
We found that wing imaginal discs of RNAiB2 larvae showed a significant increase in apoptosis, strongly suggesting that the severe growth defects in the RNAiB2 larvae are most likely due to a combination of cell cycle arrest and induction of apoptosis. Morphogenesis and cell differentiation processes remained normal throughout larval development in the RNAiB2 larvae despite their lower ATP content as compared with control larvae. The larval muscles showed a normal morphology and complex morphological processes, such as the establishment of neuromuscular junctions, were not affected. During the development of imaginal discs a variety of developmental genes are expressed in a defined and well coordinated, spatiotemporal pattern to establish different cellular territories. We found that the expression pattern of wingless was not altered in the RNAiB2 larvae, but shows a temporal delay. In summary, we report that the OXPHOS impairment induced by a defect in mtDNA transcription reduces the level of ATP, causes a specific defect in cell proliferation, and induces apoptosis but under these conditions, cell growth, cell differentiation, and morphogenesis are largely unaffected.
Acknowledgments
We thank Jesús Romero for help in neuromuscular junction staining and critical reading of the manuscript. We acknowledge the valuable help of V. Sánchez in some experiments and E. Sánchez-Herrero for critical reading of the manuscript.
This work was supported in part by Ministerio de Educación y Ciencia, Spain, Grants BFU2004-04591 (to R. G.) and BFU2005-09071 (to J. J. A.), Instituto de Salud Carlos III, Redes de centros RCMN Grant C03/08 (to R. G. and J. J. A.), Temáticas Grant G03/011 (to R. G.), the Centro de Investigación Biomédica en Red de Enfermedades Raras, and National Institutes of Health Grant GM45295 (to L. S. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: OXPHOS, oxidative phosphorylation; AEL, after egg laying; BrdUrd, 5′-bromo-2′-deoxyuridine; da, daughterless; IR, inverted repeat; TFB1/2M, mitochondrial transcription factor B1/2; TFAM, mitochondrial transcription factor A; UAS, upstream activating sequences; RNAi, RNA interference; qPCR, quantitative PCR; d, Drosophila; PBS, phosphate-buffered saline; RT, reverse transcriptase; ANOVA, analysis of variance; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
References
- 1.Garesse, R., and Vallejo, C. G. (2001) Gene (Amst.) 263 1-16 [DOI] [PubMed] [Google Scholar]
- 2.Gaspari, M., Larsson, N. G., and Gustafsson, C. M. (2004) Biochim. Biophys. Acta 1659 148-152 [DOI] [PubMed] [Google Scholar]
- 3.Shadel, G. S. (2004) Trends Genet. 20 513-519 [DOI] [PubMed] [Google Scholar]
- 4.Bonawitz, N. D., Clayton, D. A., and Shadel, G. S. (2006) Mol. Cell 24 813-825 [DOI] [PubMed] [Google Scholar]
- 5.Schinkel, A. H., Groot Koerkamp, M. J., and Tabak, H. F. (1988) EMBO J. 7 3255-3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jang, S. H., and Jaehning, J. A. (1991) J. Biol. Chem. 266 22671-22677 [PubMed] [Google Scholar]
- 7.Shadel, G. S., and Clayton, D. A. (1995) Mol. Cell. Biol. 15 2101-2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schubot, F. D., Chen, C. J., Rose, J. P., Dailey, T. A., Dailey, H. A., and Wang, B. C. (2001) Protein Sci. 10 1980-1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clayton, D. A. (1992) J. Inherit. Metab. Dis. 15 439-447 [DOI] [PubMed] [Google Scholar]
- 10.Martin, M., Cho, J., Cesare, A. J., Griffith, J. D., and Attardi, G. (2005) Cell 123 1227-1240 [DOI] [PubMed] [Google Scholar]
- 11.Gaspari, M., Falkenberg, M., Larsson, N. G., and Gustafsson, C. M. (2004) EMBO J. 23 4606-4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parisi, M. A., and Clayton, D. A. (1991) Science 252 965-969 [DOI] [PubMed] [Google Scholar]
- 13.Parisi, M. A., Xu, B., and Clayton, D. A. (1993) Mol. Cell. Biol. 13 1951-1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsson, N. G., Wang, J., Wilhelmsson, H., Oldfors, A., Rustin, P., Lewandoski, M., Barsh, G. S., and Clayton, D. A. (1998) Nat. Genet. 18 231-236 [DOI] [PubMed] [Google Scholar]
- 15.Falkenberg, M., Gaspari, M., Rantanen, A., Trifunovic, A., Larsson, N. G., and Gustafsson, C. M. (2002) Nat. Genet. 31 289-294 [DOI] [PubMed] [Google Scholar]
- 16.McCulloch, V., Seidel-Rogol, B. L., and Shadel, G. S. (2002) Mol. Cell. Biol. 22 1116-1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seidel-Rogol, B. L., McCulloch, V., and Shadel, G. S. (2003) Nat. Genet. 33 23-24 [DOI] [PubMed] [Google Scholar]
- 18.Cotney, J., and Shadel, G. S. (2006) J. Mol. Evol. 63 707-717 [DOI] [PubMed] [Google Scholar]
- 19.Garesse, R., and Kaguni, L. S. (2005) IUBMB Life 57 555-561 [DOI] [PubMed] [Google Scholar]
- 20.Berthier, F., Renaud, M., Alziari, S., and Durand, R. (1986) Nucleic Acids Res. 14 4519-4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsushima, Y., Garesse, R., and Kaguni, L. S. (2004) J. Biol. Chem. 279 26900-26905 [DOI] [PubMed] [Google Scholar]
- 22.Matsushima, Y., Adán, C., Garesse, R., and Kaguni, L. S. (2005) J. Biol. Chem. 280 16815-16820 [DOI] [PubMed] [Google Scholar]
- 23.Brand, A. H., and Perrimon, N. (1993) Development 118 401-415 [DOI] [PubMed] [Google Scholar]
- 24.Rubin, G. M., and Spradling, A. C. (1982) Science 218 348-353 [DOI] [PubMed] [Google Scholar]
- 25.Marin, M. C., Rodriguez, J. R., and Ferrus, A. (2004) Mol. Biol. Cell 15 1185-1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peña, P., and Garesse, R. (1993) Biochem. Biophys. Res. Commun. 195 785-791 [DOI] [PubMed] [Google Scholar]
- 27.Toivonen, J. M., O'Dell, K. M., Petit, N., Irvine, S. C., Knight, G. K., Lehtonen, M., Longmuir, M., Luoto, K., Touraille, S., Wang, Z., Alziari, S., Shah, Z. H., and Jacobs, H. T. (2001) Genetics 159 241-254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez-Carreras, M., Del Hoyo, P., Martin, M. A., Rubio, J. C., Martin, A., Castellano, G., Colina, F., Arenas, J., and Solis-Herruzo, J. A. (2003) Hepatology 38 999-1007 [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez-Mateos, F., Gomez, M., Garcia-Salguero, L., Sanchez, V., and Aragon, J. (1993) J. Biol. Chem. 268 7809-7817 [PubMed] [Google Scholar]
- 30.Marco-Ferreres, R., Vivar, J., Arredondo, J. J., Portillo, F., and Cervera, M. (2005) Mech. Dev. 122 681-694 [DOI] [PubMed] [Google Scholar]
- 31.Truman, J. W., and Bate, M. (1988) Dev. Biol. 125 145-157 [DOI] [PubMed] [Google Scholar]
- 32.Neufeld, T. P., de la Cruz, A. F., Johnston, L. A., and Edgar, B. A. (1998) Cell 93 1183-1193 [DOI] [PubMed] [Google Scholar]
- 33.Zordan, M. A., Cisotto, P., Benna, C., Agostino, A., Rizzo, G., Piccin, A., Pegoraro, M., Sandrelli, F., Perini, G., Tognon, G., De Caro, R., Peron, S., Kronnie, T. T., Megighian, A., Reggiani, C., Zeviani, M., and Costa, R. (2006) Genetics 172 229-241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cotney, J., Wang, Z., and Shadel, G. S. (2007) Nucleic Acids Res. 35 4042-4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pham, X. H., Farge, G., Shi, Y., Gaspari, M., Gustafsson, C. M., and Falkenberg, M. (2006) J. Biol. Chem. 281 24647-24652 [DOI] [PubMed] [Google Scholar]
- 36.Clayton, D. A. (1982) Cell 28 693-705 [DOI] [PubMed] [Google Scholar]
- 37.Brown, T. A., Cecconi, C., Tkachuk, A. N., Bustamante, C., and Clayton, D. A. (2005) Genes Dev. 19 2466-2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, M. Y., Bowmaker, M., Reyes, A., Vergani, L., Angeli, P., Gringeri, E., Jacobs, H. T., and Holt, I. J. (2002) Cell 111 495-505 [DOI] [PubMed] [Google Scholar]
- 39.Reyes, A., Yang, M. Y., Bowmaker, M., and Holt, I. J. (2005) J. Biol. Chem. 280 3242-3250 [DOI] [PubMed] [Google Scholar]
- 40.Lefai, E., Calleja, M., Ruiz de Mena, I., Lagina, A. T., 3rd, Kaguni, L. S., and Garesse, R. (2000) Mol. Gen. Genet. 264 37-46 [DOI] [PubMed] [Google Scholar]
- 41.Spinazzola, A., and Zeviani, M. (2007) Biosci. Rep. 27 39-51 [DOI] [PubMed] [Google Scholar]
- 42.Iyengar, B., Luo, N., Farr, C. L., Kaguni, L. S., and Campos, A. R. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 4483-4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iyengar, B., Roote, J., and Campos, A. R. (1999) Genetics 153 1809-1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maier, D., Farr, C. L., Poeck, B., Alahari, A., Vogel, M., Fischer, S., Kaguni, L. S., and Schneuwly, S. (2001) Mol. Biol. Cell 12 821-830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mandal, S., Guptan, P., Owusu-Ansah, E., and Banerjee, U. (2005) Dev. Cell 9 843-854 [DOI] [PubMed] [Google Scholar]