Abstract
The ability of phytochromes (Phy) to act as photointerconvertible light switches in plants and microorganisms depends on key interactions between the bilin chromophore and the apoprotein that promote bilin attachment and photointerconversion between the spectrally distinct red light-absorbing Pr conformer and far red light-absorbing Pfr conformer. Using structurally guided site-directed mutagenesis combined with several spectroscopic methods, we examined the roles of conserved amino acids within the bilin-binding domain of Deinococcus radiodurans bacteriophytochrome with respect to chromophore ligation and Pr/Pfr photoconversion. Incorporation of biliverdin IXα (BV), its structure in the Pr state, and its ability to photoisomerize to the first photocycle intermediate are insensitive to most single mutations, implying that these properties are robust with respect to small structural/electrostatic alterations in the binding pocket. In contrast, photoconversion to Pfr is highly sensitive to the chromophore environment. Many of the variants form spectrally bleached Meta-type intermediates in red light that do not relax to Pfr. Particularly important are Asp-207 and His-260, which are invariant within the Phy superfamily and participate in a unique hydrogen bond matrix involving the A, B, and C pyrrole ring nitrogens of BV and their associated pyrrole water. Resonance Raman spectroscopy demonstrates that substitutions of these residues disrupt the Pr to Pfr protonation cycle of BV with the chromophore locked in a deprotonated Meta-Rc-like photoconversion intermediate after red light irradiation. Collectively, the data show that a number of contacts contribute to the unique photochromicity of Phy-type photoreceptors. These include residues that fix the bilin in the pocket, coordinate the pyrrole water, and possibly promote the proton exchange cycle during photoconversion.
The phytochrome (Phy)5 superfamily encompasses a large and diverse set of photoreceptors present in the plant, fungal, and bacterial kingdoms where they play critical roles in various light-regulated processes (1–3). These processes range from the control of phototaxis, pigmentation, and photosynthetic potential in proteobacteria and cyanobacteria to seed germination, chloroplast development, shade avoidance, and flowering time in higher plants. Phys are unique among photoreceptors in being able to assume two stable, photointerconvertible conformers, designated Pr and Pfr based on their respective absorption maxima in the red and far-red spectral regions. By cycling between Pr and Pfr, Phys act as light-regulated switches in various photosensory cascades.
Phys are homodimeric complexes with each polypeptide containing a single bilin (or linear tetrapyrrole) chromophore, which binds autocatalytically via a thioether linkage to a positionally conserved cysteine (1–3). The photosensing portion typically contains Per/Arndt/Sim (PAS) and cGMP phosphodiesterase/adenyl cyclase/FhlA (GAF) domains, which are essential for bilin binding and Pr assembly, and together comprise the chromophore-binding domain (CBD). The CBD is often followed by a Phytochrome (PHY) domain, which is required for the formation and stability of Pfr. C-terminal to the PHY domain can be a variety of domains that promote signal transmission and/or dimerization. Often histidine kinase or histidine kinase-related domains are present that can direct light-modulated phosphorylation in cis within the homodimer and in trans to other proteins, which typically are response regulator proteins in microorganisms (1, 2, 4, 5).
Biliverdin IXα (BV) is the native chromophore for the proteobacterial and fungal Phys; it is synthesized by oxidative cleavage of heme by a heme oxygenase (HO) (6, 7). BV is then attached to a cysteine residue upstream of the PAS domain via the C32 carbon of the pyrrole ring A vinyl side chain (8, 9). In contrast, cyanobacterial and higher plant Phys incorporate phycocyanobilin (PCB) and phytochromobilin (PΦB), respectively, which are generated from BV, in part, by enzymatic reduction of the C3 vinyl side chain on pyrrole ring A to generate an ethylidene group (1, 10). PCB and PΦB then bind via the C31 carbon of this side chain to a cysteine within the GAF domain, which is predicted to extend toward the same space as the N-terminal binding-site cysteine used by proteobacterial/fungal Phys (8, 11).
Despite intensive study, the unique photochromic nature of Phys remains largely unexplained. Thus far, most of our understanding has been derived from absorption and resonance Raman (RR) spectroscopy, which have collectively identified several distinct but ill-defined steps during Pr → Pfr photoconversion (Fig. 1A). As Pr, the bilin is cationic with all four of the pyrrole ring nitrogens protonated (12, 13). Using plant Phys as the example, photoexcitation with red light converts Pr (absorption maximum of the Q band at 666 nm) within picoseconds to the partially bleached Lumi-R photoproduct (absorption maximum at 688 nm) (14, 15). This conversion is predicted to involve a Z to E isomerization of the C15–C16 methine double bond between the C and D pyrrole rings of the chromophore (16–18). On the microsecond time scale, thermal relaxation steps lead to the formation of the Meta-Ra intermediate with a maximum absorption at 663 nm (19). The Meta-Ra intermediate in turn decays in microto milliseconds to a deprotonated Meta-Rc intermediate with an absorption maximum at 725 nm (20, 21). Finally, the Meta-Rc photoproduct decays to Pfr (absorption maximum at 730 nm) on a millisecond time scale in a process that likely requires reprotonation of the chromophore (12, 16, 17, 22–25). Coupled with these chromophore relaxation steps are conformational changes within the polypeptide that alter the structure of Pfr versus Pr, which in turn are presumed to affect the kinase activity of the photoreceptor and/or its photoreversible interaction(s) with downstream signaling partner(s) (1–3).
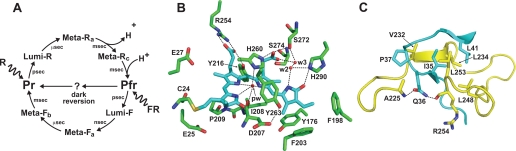
FIGURE 1.
Schematic presentation of the photocycle of Phys (A) and three-dimensional relationships of key amino acids within the CBD structure of DrBphP (B and C). B, region surrounding the BV chromophore. BV is colored cyan. Sulfur, oxygen, and nitrogen atoms are colored yellow, red, and purple, respectively. Pyrrole water (pw), water2 (w2), and water3 (w3) are indicated. C, region surrounding the knot interface. The PAS and GAF polypeptide chains are colored cyan and yellow, respectively. Dashed lines highlight key noncovalent interactions. Three-dimensional structures are from Refs. 8, 11.
Once formed, Pfr will slowly revert nonphotochemically back to Pr or can be photoconverted rapidly back to Pr with far-red light (Fig. 1A). Neither pathway is well understood. The photoinduced Pfr → Pr conversion likely proceeds via a pathway distinct from that for the Pr → Pfr conversion but may use a similar but inverted proton migration cycle (1, 21).
To help understand the photochromicity of Phys at the atomic level, the three-dimensional structures of the Pr and Pfr forms would clearly be helpful. Recently, we accomplished part of this objective by determining the structure of the CBD as Pr, using the sole bacteriophytochrome photoreceptor (BphP) in Deinococcus radiodurans assembled with BV as the source. The original structure solved to 2.5 Å resolution (11), followed by a more refined model at 1.45 Å resolution (8), revealed that the GAF domain forms a deep pocket that cradles the Pr chromophore in a ZZZsyn,syn,anti configuration corresponding to a Z geometry for all methine bridge double bonds and a syn geometry for the A–B and B–C and an anti geometry from the C–D methine bridge single bonds (Fig. 1B). Other than positioning Cys-24 that forms the thioether linkage, the PAS domain has no direct contact with BV in the Pr conformer. Instead, it is connected indirectly to the bilin and the GAF domain through a rare figure-of-eight knot in the polypeptide (Fig. 1C). The structures also identified a heretofore unknown dimerization interface between sister GAF domains that could impact signaling within the Phy dimer and/or photochemical cooperatively between the adjacent bilins (8). A similar crystallographic model was recently described by Yang et al. (26) for BphP3 from the photosynthetic bacterium Rhodopseudomonas palustris, suggesting that the CBDs of all Phys have related tertiary structures.
Inspection of the bilin pocket of the D. radiodurans and R. palustris BphP CBDs revealed a number of amino acids that could be important for covalent binding of the bilin, the unique spectral properties of Pr and Pfr, and the steps required for their interconversion (8, 11, 26) (Fig. 1, B and C). Most of these amino acids are highly conserved throughout the Phy superfamily, further supporting their functional significance (5, 11).
Here we tested the importance of many of these residues through the analysis of engineered protein variants by absorption, fluorescence, and RR spectroscopy. Surprisingly, although nearly all of the DrBphP variants retained their ability to fold correctly, covalently attach BV, and form Pr, a majority failed to properly photoconvert to Pfr. In particular, we demonstrated the importance to Pr → Pfr photoconversion of contacts involving the propionate side chains of BV, the hydrophobic pocket that surrounds the D pyrrole ring, and the highly conserved Asp-207 and His-260 residues, which participate in an extensive hydrogen bond network involving several ordered waters and the bilin. Even subtle substitutions of Asp-207 became trapped in a deprotonated and presumably Meta-Rc-like state after red light irradiation. Collectively, the mutant analysis revealed that although assembly of the Pr form and photoconversion to the Meta-R intermediates are generally insensitive to most changes in the chromophore pocket, complete photoconversion to the Pfr form is strongly compromised, suggesting that the transition from the Meta-R intermediates to Pfr is a dynamic process requiring a number of critical protein/protein and protein/chromophore contacts.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis and Protein Purification—The full-length D. radiodurans BphP (encoding 755 amino acids) gene (27) was PCR-amplified from genomic DNA using primers designed to introduce BamHI and XhoI sites before and after the designated length of coding region, respectively. The BamHI-XhoI-digested PCR products were cloned into pET21b(+) (Novagen, Madison, WI), which was similarly digested, resulting in the addition of codons for an N-terminal T7 tag and codons for a His6 tag just before the stop codon. All site-directed mutations were introduced by the PCR-based QuickChange method (Stratagene, La Jolla, CA). Each coding region was sequenced completely by the dideoxy method to confirm introduction of the appropriate mutation(s) and the absence of secondary mutations.
The DrBphP variants were expressed in Escherichia coli strain Rosetta (DE3) (Novagen, Madison WI) either by themselves or simultaneously with the Synechocystis PCC6803 HO gene cloned into pET 24a(+) (Novagen) (6). After harvesting the cells, all further steps were performed under green safe lights. Cells were homogenized in 30 mm Tris-HCl (pH 8.0), 200 mm NaCl, and 5 mm imidazole, and the resulting extract was clarified at 10,000 × g. To encourage complete chromoprotein assembly, the crude soluble extracts were incubated in darkness for 1 h in at least a 10-fold molar excess of BV or protoporphyrin IXa (PPIXa) prior to affinity purification. The holoproteins were then purified via nickel-nitrilotriacetic acid chromatography (Qiagen, Germantown, MD) using 1 m imidazole and 30 mm Tris-HCl (pH 8.0) for elution. The eluate was brought to 0.3 m ammonium sulfate and subjected to phenyl-Sepharose FPLC (GE Healthcare), using 30 mm Tris-HCl (pH 8.0) for elution. Covalent binding of BV to the apoproteins was monitored by zinc-induced fluorescence of the chromoproteins following SDS-PAGE (6).
Spectrophotometric Analyses—UV-visible absorption spectroscopy was performed with a Lambda 650 spectrophotometer (PerkinElmer Life Sciences). All proteins were diluted with 30 mm Tris-HCl (pH 8.0) so that the Pr absorption maxima at ∼700 nm had an absorbance between 0.25 and 0.6. Spectra of Pr were measured for all proteins following an extended incubation in the darkness. The absorption spectrum of the photoconversion product was obtained immediately following saturating red light (690 nm) irradiation generated using a 10-nm half-bandwidth interference filter (Andover Corp. Salem, NH). Difference spectra were calculated by subtracting the red light-irradiated spectrum from the Pr spectrum. Fluorescence spectra were obtained using a QuantaMaster model C-60/2000 spectrofluorimeter (Photon Technologies International, Birmingham, NJ) with both monochrometers set to a 4-nm band pass. To obtain a more complete excitation spectrum for some of the DrBphP mutants, fluorescence emission was measured at 642 nm in addition to 620 nm.
RR spectra were recorded with 1064-nm excitation (Nd-YAG cw laser, line width <1 cm–1) using Digilab Bio-Rad or an RFS 100/S (Bruker Optics, Ettlinger, Germany) Fourier-transform Raman spectrometers (4 cm–1 spectral resolution). The near-infrared excitation line was sufficiently close to the first electronic transition to generate a strong pre-resonance enhancement of the chromophoric vibrational bands, such that Raman bands of the protein matrix remained very weak in the spectra of the parent states (12, 13). PPIXa also did not contribute to the experimental RR data since it does not gain any resonance enhancement with near-infrared excitation. All spectra were measured at –140 °C using a liquid-nitrogen cooled cryostat (Linkam, Waterfield, Surrey, UK). The laser power at the sample was set at ∼700 milliwatts, which did not damage the chromoproteins as checked by comparing the absorption spectra of the samples obtained before and after RR data acquisition. The total accumulation time was less than 2 h for each spectrum. For all RR spectra shown in this work, the background was subtracted manually.
Photoconversion intermediates were enriched by irradiating the samples with red light for a few minutes at the specified temperatures. Raw RR spectra measured from these irradiated samples included a substantial contribution from residual Pr, which was removed by subtraction, taking the characteristic RR bands of Pr as a reference. Further RR experimental details have been described previously (12, 13).
RESULTS
Site-directed modifications of Phys, either randomly or in the context of amino acid sequence alignments, have been widely employed over the past few decades in attempts to identify domains and residues important for Phy assembly, structure, and function (13, 28–32). However, interpreting the direct effects of these changes has been challenging without three-dimensional templates. Here we exploited our high resolution models of D. radiodurans BphP (Protein Data Bank codes 1ZTU, 2O9B, and 2O9C (8, 11)) as guides to directly test the importance of potentially key and often highly conserved amino acids in the CBD with a special focus on those close to the chromophore or central to the knot (Fig. 1, B and C).
To minimize secondary effects caused by using truncated chromoproteins, all mutations were introduced into the full-length DrBphP polypeptide with its sequential arrangement of PAS, GAF, PHY, and HK domains. RR analysis revealed that the BV geometry of the unmodified full-length chromoprotein as Pr is nearly identical to that bound to the CBD truncation (supplemental Fig. 1A),6 indicating that our CBD models should accurately predict the consequences of the mutations at least for the Pr state. How the mutations might affect Pfr awaits structural resolution of this state. Relative to full-length DrBphP, the CBD chromoprotein becomes trapped in a deprotonated Meta-Rc-like state in red light (supplemental Fig. 1B) (11), indicating the CBD structure by itself is missing key features for full Pr to Pfr photoconversion.
BV assembly was either achieved in vivo following co-expression of the variant BphPs with a HO or in the crude E. coli lysates following the addition of purified BV (6). Although these approaches could generate ample quantities of photoactive holoproteins, they precluded quantitative measurement of BV assembly rates. The complete list of variant proteins (38 substitutions/deletions affecting 16 positions), their solubility, ability to assemble with BV, and some of their photochemical and fluorescence characteristics are presented in Table 1.
TABLE 1.
Spectroscopic and photochemical properties of DrBphP variants
+ or — indicates expression or solubility are good or poor, respectively. NA indicates not applicable and ND indicates not determined.
| Construction | Expression/solubility | BV covalent attachment | Pr λmax | Red-irradiated λmax | Q/Soret ratio | Photoconversion | Porphyrin fluorescence |
|---|---|---|---|---|---|---|---|
| nm | nm | ||||||
| Wild type | +/+ | Yes | 700 | 751 | 2.69 | Pr/Pfr | No |
| ΔN1-20 | +/+ | Yes | 701 | 752 | 2.34 | Intermediate | ND |
| E25A/E27A | +/+ | Yes | 698 | 751 | 2.23 | Pr/Pfr | No |
| 135A | +/+ | Yes | 699 | 751 | 2.34 | Intermediate | ND |
| Q36A | +/- | Yes | NA | NA | NA | NA | NA |
| Q36D | +/+ | Yes | 698 | 752 | 2.47 | Pr/Pfr | ND |
| Q36K | +/- | Yes | NA | NA | NA | NA | NA |
| Q36L | +/+ | Yes | 701 | 750 | 2.62 | Pr/Pfr | ND |
| Q36N | +/- | NA | NA | NA | NA | NA | NA |
| P37A | +/+ | Yes | 700 | 756 | 2.42 | Pr/Pfr | ND |
| Y176H | +/+ | Yes | 696 | 750 | 1.28 | Intermediate | No |
| F203A | +/+ | Yes | 696 | 738 | 2.13 | Intermediate | No |
| F203H | +/+ | Yes | 695 | 742 | 2.16 | Intermediate | No |
| F203W | +/+ | Yes | 701 | 751 | 2.37 | Pr/Pfr | No |
| D207A | +/+ | Yes | 700 | 751 | 1.91 | Intermediate | Yes |
| D207E | +/+ | Yes | 701 | 751 | 2.20 | Intermediate | No |
| D207H | +/+ | Yes | 700 | NA | 0.83 | Locked in Pr | Yes |
| D207K | +/+ | Yes | 700 | NA | 1.95 | Locked in Pr | Yes |
| D207L | +/+ | Yes | 700 | NA | 1.93 | Locked in Pr | Yes |
| D207N | +/+ | Yes | 700 | 751 | 1.74 | Intermediate | Yes |
| D207Q | +/+ | Yes | 701 | 746 | 2.02 | Intermediate | Yes |
| D207S | +/+ | Yes | 701 | NA | 1.59 | Locked in Pr | Yes |
| D207T | +/+ | Yes | 704 | 751 | 1.87 | Intermediate | Yes |
| I208A | +/+ | Yes | 695 | 751 | 2.26 | Pr/Pfr | No |
| P209G | +/+ | Yes | 690 | 745 | 1.97 | Pr/Pfr | ND |
| Y216H | +/+ | Yes | 697 | 748 | 2.42 | Pr/Pfr | No |
| Y216W | +/+ | Yes | 698 | 751 | 0.67 | Intermediate | ND |
| R254A | +/+ | Yes | 700 | 750 | 1.14 | Pr/Pfr | ND |
| R254K | +/+ | Yes | 701 | 750 | 2.45 | Pr/Pfr | ND |
| R254Q | +/+ | No | NA | NA | NA | NA | NA |
| H260A | +/+ | Yes | 698 | 748 | 1.16 | Intermediate | No |
| H260D | +/+ | No | NA | NA | NA | NA | NA |
| H260K | +/+ | No | NA | NA | NA | NA | NA |
| H260N | +/+ | Yes | 698 | 754 | 2.62 | Intermediate | No |
| H260Q | +/+ | Yes | 697 | 748 | 2.35 | Pr/Pfr | No |
| H260S | +/+ | Yes | 694 | 748 | 1.50 | Intermediate | ND |
| Y263H | +/+ | Yes | 698 | 742 | 1.78 | Intermediate | Yes |
| H290N | +/+ | Yes | 699 | 747 | 2.95 | Intermediate | No |
| H290Q | +/+ | Yes | 702 | 746 | 2.58 | Intermediate | Yes |
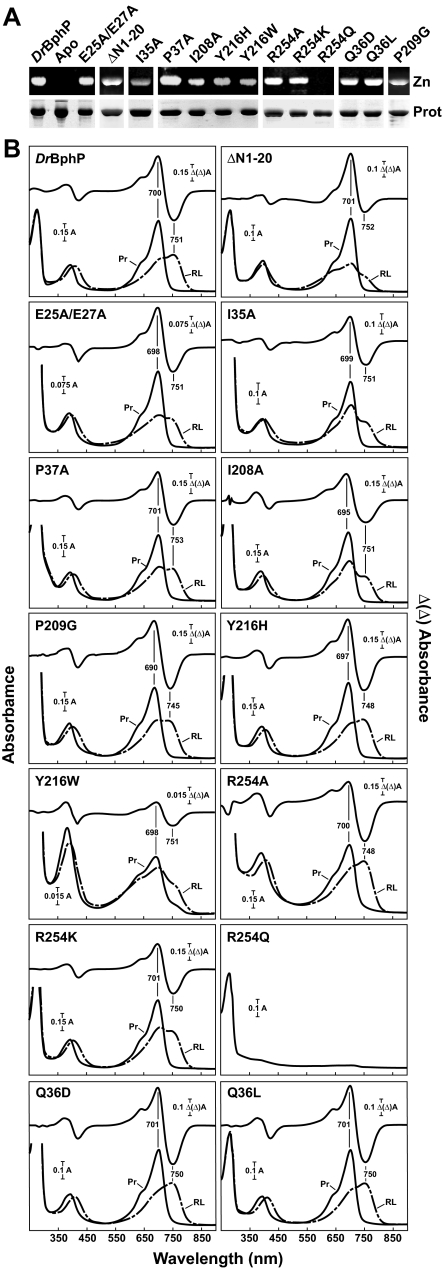
Amino Acids Involved in the Peptide Knot—One unique structural feature of DrBphP, RpBphP3, and presumably all canonical Phys is the figure-of-eight knot that helps connect the PAS and GAF domains (8, 11, 26). To examine the importance of the central knot interface, we substituted several conserved residues in the knot and the upstream PAS domain, including Ile-35 within the hydrophobic core of the knot, Gln-36 that hydrogen bonds with Ala-225 and Arg-254 at the base of the GAF domain lasso, and Pro-37 that kinks the central polypeptide chain following the knot core as it extends toward the PAS domain (Fig. 1C). Ile-35 and Gln-36 are highly conserved throughout the Phy superfamily, whereas Pro-37 is widespread in the proteobacterial, cyanobacterial, and higher plant clades (5). Ile-35 in particular was likely to be important for folding and bilin attachment based on prior mutagenic studies with other Phys (5, 29, 33).
Analysis of alanine substitutions of Ile-35 and Pro-37 revealed that neither residue is strictly required for DrBphP assembly. The variant proteins readily assembled with BV, using zinc-induced fluorescence of the chromoprotein following SDS-PAGE as the assay, and generated red/far-red light photoreversible Phys (Fig. 2). Whereas the P37A mutant had Pr and Pfr absorption spectra close to those for the wild-type holoprotein, the I35A mutant as Pfr had reduced absorption of the chromophore Q band at 751 nm relative to the Q band for Pr at 699 nm, indicating that less Pfr was generated at photoequilibrium in red light. In contrast, several substitutions of Gln-36 had dramatic consequences, suggesting a key role for this residue in Phy folding. Engineered proteins harboring Gln-36 to aspartic acid (Q36D) or leucine substitutions (Q36L) expressed well and bound BV covalently, and the resulting holoprotein had relatively normal Pr and Pfr absorption spectra (Fig. 2). However, the conservative substitution of Gln-36 for asparagine (Q36N) was completely insoluble despite high expression in E. coli. The alanine (Q36A) and lysine (Q36K) substitutions were mostly insoluble, implying folding problems, but the residual soluble proteins still bound BV (data not shown).
FIGURE 2.
Assembly and absorption spectra of DrBphP mutants potentially affecting bilin binding and the knot interface. The recombinant full-length and ΔN1–20 truncated apoproteins were incubated with BV and purified by nickel chelate chromatography. A, samples were subjected to SDS-PAGE and either assayed for the bound bilin by zinc-induced fluorescence (Zn) or stained for protein with Coomassie Blue (Prot). Apo, apoprotein prior to BV incubation. B, UV-visible absorption spectra as Pr (Pr, black lines) or following saturating red light irradiation (RL, dashed lines) and red-minus-far red light difference spectra (upper graph) of the apoproteins incubated with BV. Difference spectra maxima and minima are indicated.
Alterations Near the Chromophore-binding Site—BV becomes covalently linked to the DrBphP apoprotein via a thioether bond between Cys-24 and the C32 carbon of the A-ring vinyl side chain (8, 9, 11). Based on the covalent binding of heme to c-type cytochromes, assembly could involve protonation of the C32 carbon via a carbocation mechanism, followed by deprotonated thiol addition (34). Two possible proton donors/acceptors were the carboxylate groups of Glu-25 and Glu-27, which are ∼5.8 and 6.6 Å and ∼6.8 and 8.9 Å away (Fig. 1C), respectively, from the sulfur moiety of Cys-24 and could be closer in the structure of DrBphP apoprotein, which appears to be slightly different from the holoprotein (35). However, analysis of a double alanine substitution affecting Glu-25 and Glu-27 revealed that neither is essential for bilin binding. The E25A/E27A mutant conjugated BV and could reversibly phototransform between Pr and Pfr like the wild-type photoreceptor (Fig. 2). We also examined the importance of the region upstream of Cys-24 via analysis of the ΔN1–20 deletion missing the first 20 amino acids of DrBphP. The ΔN1–20 protein retained its ability to bind BV and generate Pr but was compromised in phototransformation to Pfr. Instead the ΔN1–20 chromoprotein generated a partially bleached species in saturating red light (Fig. 2).
Mutations at the Contacts with the BV Propionate Side Chains—Hanzawa et al. (36) demonstrated using various PΦB derivatives assembled with plant PhyB that the propionic acid side chains help promote efficient association of the bilin with Phy apoproteins. The three-dimensional structures of the DrBphP CBD revealed that these side chains contact a collection of conserved residues deep within the binding pocket that likely lock the chromophore in place prior to covalent attachment (8, 11, 26). Included are Tyr-216 and Arg-254 that interact electrostatically with the B-ring propionate oxygens, and His-260, Ser-272, and Ser-274 that hydrogen bond with the C-ring propionate oxygens and/or the adjacent waters, water2 and water3 (Fig. 1B).
To test whether Tyr-216 helps secure BV in the binding pocket, it was substituted for either a histidine (Y216H), which is smaller but may still electrostatically interact with BV or the more bulky tryptophan (Y216W). Both proteins covalently bound BV (Fig. 2A). The Y216H holoprotein had relatively normal Pr and Pfr absorption spectra except for a slight blue shift of the two spectral forms, suggesting that the hydrogen-bonding pattern was maintained by the replaced histidine (Fig. 2B). In contrast, the absorption spectra of Y216W were substantially altered. The Pr form had a dramatically reduced extinction coefficient for the Q band (698 nm) relative to the Soret band (∼380 nm) and photoconverted poorly to Pfr following saturating red light irradiation. This spectral defect was potentially caused by the bulky side chain of tryptophan restricting penetration of the B-ring propionate into the GAF domain pocket.
Arg-254 interacts with the B-ring propionate via a double salt bridge between the guanidium group of arginine and the carboxylate of the propionate moiety, in addition to its hydrogen bond connection to Gln-36 of the knot through its main chain oxygen (Fig. 1B) (11). The R254K substitution assembled with BV and generated normal Pr and Pfr absorption spectra with maxima at 701 and 750 nm, respectively, indicating that correct bilin/protein interactions can be maintained with just one primary amine interacting with the B-ring propionate group (Fig. 2). Similar results were recently reported for a corresponding arginine to lysine mutation in Synechocystis Cph1 (31).
Replacement of Arg-254 with alanine or glutamine should either abolish the pair of salt bridges to the B-ring propionate (R254A) or potentially generate repulsion between the side chain carbonyl of glutamine and the propionate carboxyl moiety (R254Q). The R254Q polypeptide failed to bind BV either covalently or noncovalently, suggesting that this repulsion effectively blocks entry of BV into the binding pocket (Fig. 2A). In contrast, the R254A polypeptide covalently assembled with BV but its Pr absorbance for Q band (700 nm) relative to the Soret band was markedly lower than that of wild-type DrBphP (Fig. 2), pointing to alterations of the BV conformation in the absence of the B-ring propionate tether. Surprisingly, a relatively normal Pfr-like species was generated after red light excitation of R254A but this species displayed little thermal reversion back to Pr. Whereas DrBphP reverts nonphotochemically from Pfr back to Pr (∼30% in 2 h for wild-type DrBphP), little or no reversion was evident for the R254A variant even after 8 h of darkness (supplemental Fig. 2).
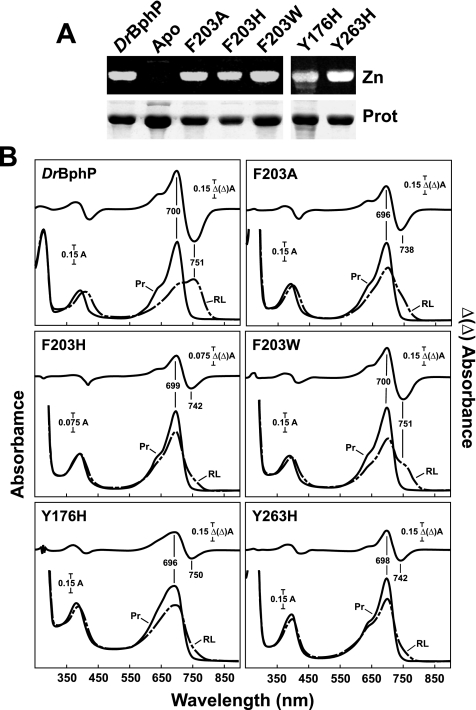
Mutations in the Hydrophobic Pocket for the D Ring—One striking feature of the bilin-binding pocket of DrBphP and presumably other Phys is the hydrophobic cavity that surrounds the D pyrrole ring, created in part by Tyr-176, Tyr-263, Phe-198, and Phe-203 (Fig. 1B). Fischer et al. (30, 37) previously demonstrated that Tyr-176 is photochemically important in Synechocystis Cph1, possibly by maintaining the bilin in an extended conformation, whereas our high resolution model of D. radiodurans CBD (8) suggests that Tyr-263 has the potential to hydrogen bond with the carboxylate of Asp-207, a residue near the epicenter of the A, B, and C pyrrole rings of BV (Fig. 1B). To examine the functions of these two tyrosines, histidine substitutions were analyzed (Y176H and Y263H) that added a positive repulsive charge near the D ring or, in the case of Y263H, could strengthen the interaction of this residue with Asp-207 by replacing the hydrogen bond with a salt bridge. Both variants retained their ability to bind BV covalently and generate Pr (Fig. 3). Whereas the Pr absorption spectrum of Y263H was relatively normal, that for Y176H was substantially broadened in the Q band region, showing that Tyr-176 in particular helps maintain the correct conformation of the bilin as Pr. The Tyr-176 and Tyr-263 variants also failed to properly photoconvert to Pfr upon red light excitation. Instead, a small proportion of Pr was phototransformed to bleached species, indicating that both tyrosines are necessary for the Pr → Pfr photoconversion (Fig. 3B).
FIGURE 3.
Assembly and absorption spectra of amino acid substitutions in DrBphP affecting bulky hydrophobic residues near the D ring of BV. The recombinant full-length apoproteins were incubated with BV and purified by nickel chelate chromatography. A, samples were subjected to SDS-PAGE and either assayed for the bound bilin by zinc-induced fluorescence (Zn) or stained for protein with Coomassie Blue (Prot). Apo, apoprotein prior to BV incubation. B, UV-visible absorption spectra as Pr (Pr, black lines) or following saturating red light irradiation (RL, dashed lines) and red-minus-far red light difference spectra (upper graph) of the apoproteins incubated with BV. Difference spectra maxima and minima are indicated.
In the Pr form, Phe-203 interacts hydrophobically with the D ring vinyl and methyl side chains, an association that is likely dissolved in Pfr after the proposed C15–C16 Z to E isomerization of BV (Fig. 1B) (8, 11). To examine its role, we generated substitutions of Phe-203 for alanine (F203A), which would abolish its aromatic packing with ring D, histidine (F203H), which would change the electrostatic environment around the D ring, or tryptophan (F203W), which would test if another bulky hydrophobic residue can suffice. All three mutant proteins retained the ability to bind BV and generate a nearly normal Pr absorption spectrum (Fig. 3). The Pr maxima of the F203A and F203W holoproteins were slightly blue-shifted, suggesting that the π -conjugation system of BV was altered, but the Q/Soret band absorption ratios were relatively normal. All three Phe-203 variants generated a bleached species after red light irradiation, indicating that Pr → Pfr photoconversion was incomplete (Fig. 3B).
Substitutions of the Highly Conserved Asp-207—Our models of DrBphP CBD place Asp-207 at the focal point of BV geometry and chemistry (8, 11). Via its main chain oxygen, Asp-207 participates an extended hydrogen bond network that also includes the pyrrole N-H groups of the rings A, B, and C, the π -imidazole nitrogen of His-260, and the pyrrole water (Fig. 1B). Its carboxylate moiety can also hydrogen bond with the hydroxyl group of Tyr-263 and may provide a repulsive force that tilts the adjacent A ring 10° relative to the plane of the B and C rings. Such a central role is further supported by the extreme conservation of Asp-207 as part of the Asp-Ile-Pro or DIP motif, which is invariant within the Phy superfamily (5, 11).
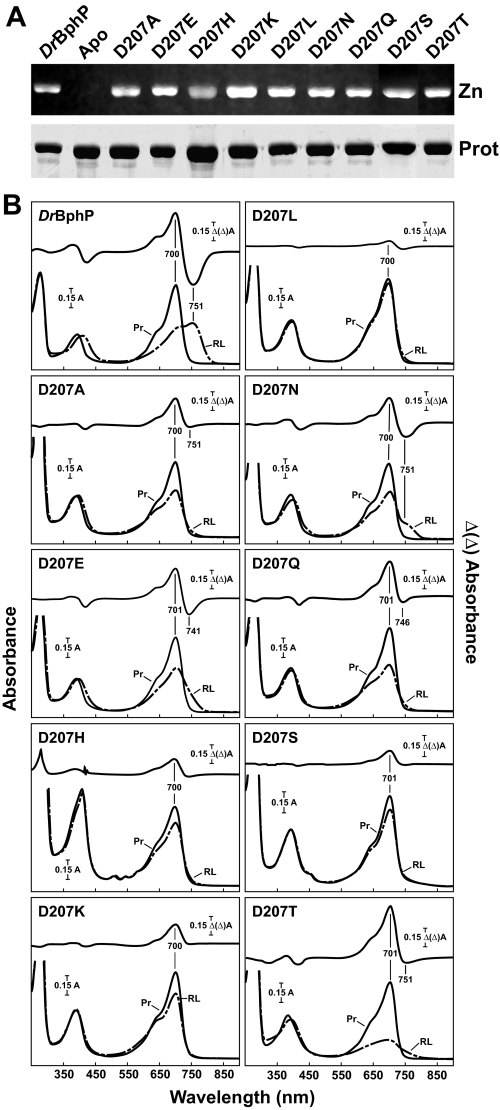
To examine the importance of Asp-207, we analyzed a diverse set of substitutions that included the following: (i) D207A that would abolish the effect of the carboxylate group at this position; (ii) D207E, D207Q, and D207N that tested the importance of size and charge at residue 207; (iii) D207S and D207T that should alter the hydrogen bonding pattern in the pocket by replacing the carboxylate group with alcohol side chains; (iv) D207H and D207K that examined the effects of positive charge and side chain size on bilin ligation and photochemistry; and (v) D207L that introduced an isosteric but hydrophobic residue into the binding pocket. Collectively, these substitutions should not abolish the main chain carbonyl bonding patterns in the Pr state, although subtle changes were possible, but should significantly affect the interaction of Asp-207 with Tyr-263 or other residues outside the CBD in the full-length photoreceptor.
All of the Asp-207 variant proteins expressed robustly in E. coli, covalently bound BV, and generated Pr with relatively normal absorption spectra (Fig. 4). For some (D207H and D207S), a substantially reduced absorption for the Q band relative to the Soret band was observed. In contrast, none of the Asp-207 substitutions properly photoconverted to Pfr upon photoexcitation with red light (Fig. 4B). The most striking behavior was seen for D207L, which displayed little photochromicity even after extended irradiations, followed by D207S, D207H, D207K and D207A, which had reduced absorption at the ∼700 nm Q band after saturating red light irradiation but failed to gain absorption in the far-red region. Even the conservative D207E chromoprotein was spectrally aberrant; it photoconverted to a bleached state in red light. Extended dark incubations or far-red irradiation of the bleached forms regenerated much of the Pr absorbance at ∼700 nm, indicating that the Asp-207 variants retained their ability to convert between these bleached states and Pr (data not shown).
FIGURE 4.
Assembly and absorption spectra of amino acid substitutions in DrBphP affecting Asp-207. The recombinant full-length apoproteins were incubated with BV and purified by nickel chelate chromatography. A, samples were subjected to SDS-PAGE and either assayed for the bound bilin by zinc-induced fluorescence (Zn) or stained for protein with Coomassie Blue (Prot). Apo, apoprotein prior to BV incubation. B, UV-visible absorption spectra as Pr (Pr, black lines) or following saturating red light irradiation (RL, dashed lines) and redminus-far red light difference spectra (upper graph) of the apoproteins incubated with BV. Difference spectra maxima and minima are indicated.
In contrast to the strong effects of Asp-207 substitutions, alanine replacement of the next residue of the DIP motif (Ile-208), which provides hydrophobic packing contacts for the B and C rings, had only a minor impact on DrBphP photochemistry. The I208A protein, which retained the hydrophobic character of the residue, covalently bound BV, generated a normal Pr absorption spectrum, and produced a nearly normal Pfr spectrum after far-red light irradiation (Fig. 2). The only apparent difference was a reduced Pr/Pfr photoequilibrium under saturating red light, suggesting that Pr → Pfr photoconversion was slightly attenuated. We also tested the importance of Pro-209 with a glycine substitution. This P209G substitution bound BV and retained near normal photochromic properties, except for a blue shift of the Pr and Pfr absorption maxima (Fig. 2), indicating that the kink provided by Pro-209 within the DIP motif can be maintained by a glycine or is not essential for folding full-length Phys nor the action of Asp-207.
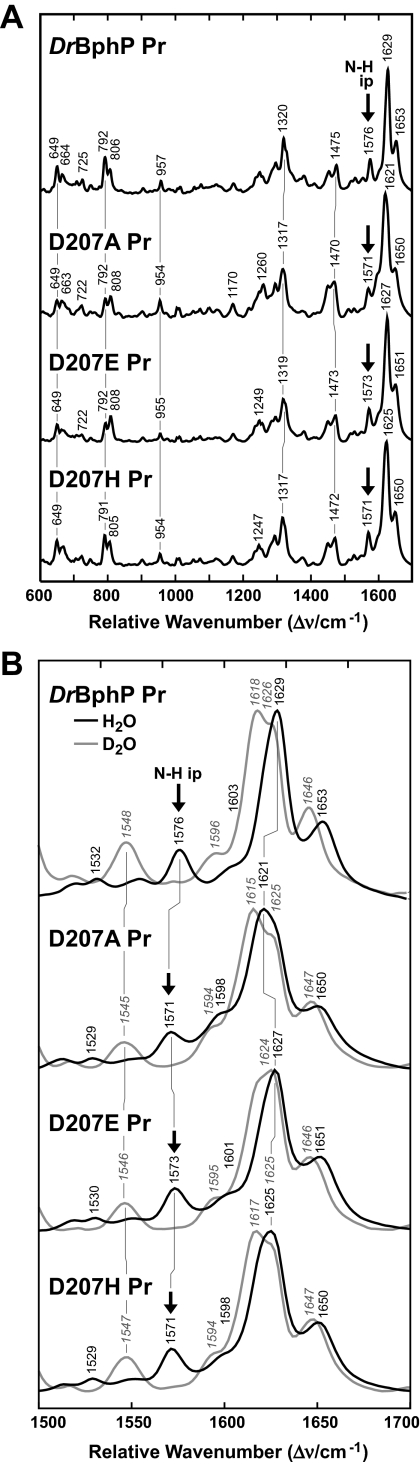
Resonance Raman Spectroscopy of Asp-207 Mutants—To probe the effects of the Asp-207 substitutions on the conformation of BV, we analyzed the D207A, D207E, and D207H variants by RR spectroscopy in H2O and D2O. In particular, RR bands in the region between 1500 and 1700 cm–1 allowed us to directly assess the influence of each amino acid substitution on the methine bridge configurations and conformations (Z/E and syn/anti) and the protonation state of the pyrrole nitrogens (12, 13).
For all three Asp-207 variants, the RR spectra of Pr had the same vibrational band patterns as the wild-type chromoprotein, implying that the methine bridge geometry was not affected by these substitutions (Fig. 5A). Minor spectral differences in the region assigned to the C-H out-of-plane modes (792 and 806 cm–1) indicated only slight deviations of the methine bridge torsional angles. Importantly, each RR spectrum of Pr had a prominent band at ∼1575 cm–1 when measured in aqueous buffer that shifted down to 1080 cm–1 when measured in D2O buffer (Fig. 5B and data not shown). This band represents the N-H in-plane bending (ip) of the rings B and C N-H groups (12), thus demonstrating that each variant, like wild-type DrBphP, is protonated (cationic) as Pr at pH 7.8. The frequency of the N-H ip mode of rings B and C was slightly lowered from 1576 cm–1 in wild-type (Asp-207) to 1573 cm–1 in D207E and to 1571 cm–1 in D207A and D207H (Fig. 5B). In view of theoretical calculations (38), this frequency decrease could reflect a weakening of the hydrogen bonds between the pyrrole water with the N-H groups of rings B and C. The downshift of the N-H ip mode was accompanied by a slight lowering of the frequency of the adjacent peaks at ∼1630 and 1650 cm–1, which originate from modes dominated by the C=C stretching coordinates of the methine bridges (Fig. 5B). Both effects may be related given that alterations in the hydrogen bond network of rings B and C should also affect the electron density of the bilin and thus change the frequency of the C=C stretching modes.
FIGURE 5.
RR spectra of the Pr state of DrBphP and the D207A, D207E, and D207H variants. A, overview RR spectra (600–1700 cm–1) measured from the samples dissolved in H2O. B, expanded view of the spectra in A for the marker band region (1500–1700 cm–1) from samples dissolved in H2O(black lines) and D2O(gray lines). The positions of N-H ip band (arrows) and other diagnostic RR maxima are indicated.
The only subtle difference in the RR spectra between the Asp-207 substitution proteins and wild-type DrBphP as Pr was that for D207A. The intensity of the N-H ip band was decreased relative to those of the C=C stretchings at 1621 and 1650 cm–1 concomitant with a slight increase of the shoulder at 1598 cm–1 (Fig. 5B), which was previously assigned as a marker band for the deprotonated Pr chromophore (13). This finding suggests that the protonated form of D207A was slightly diminished but still the dominant fraction at pH 7.8. von Stetten et al. (13) previously found that an analogous D207A substitution in A. tumefaciens BphP1 (Agp1) (Asp-197 to alanine) also induced deprotonation of Pr chromophore at pH 7.8 (i.e. absence of the N-H ip marker band at ∼1572 cm–1), but in this case almost all of the chromoprotein population was deprotonated.
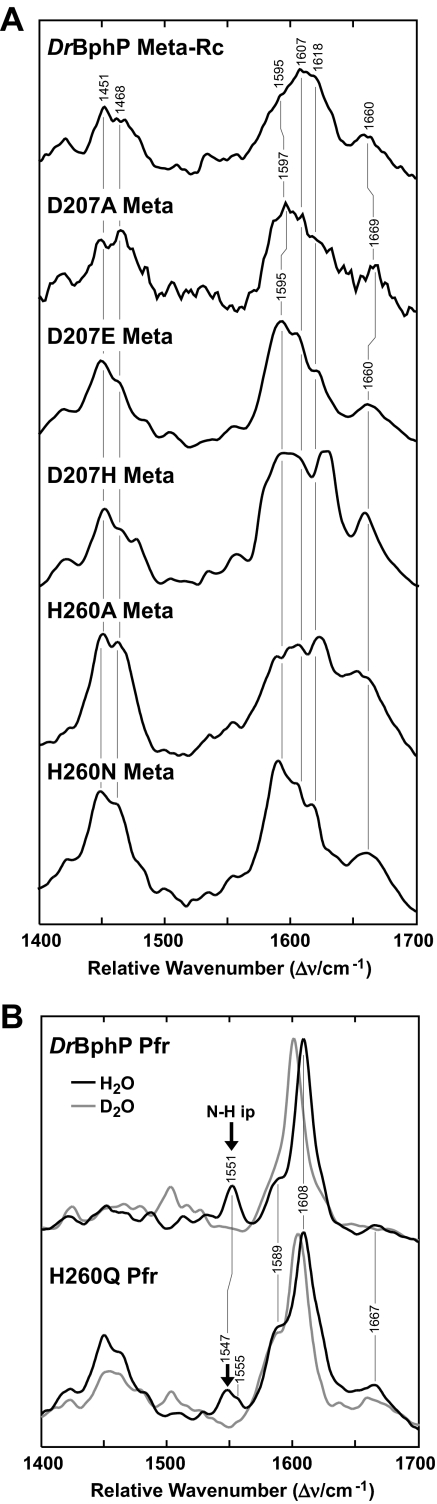
Instead of generating Pfr after red light excitation, the three DrBphP Asp-207 substitutions formed a bleached but still photointerconvertible state (see Fig. 4B). The RR spectra of these species were very similar to that for the Meta-Rc precursor of Pfr that can be cryogenically trapped when wild-type Phys are frozen to –30 °C and then irradiated with red light (Fig. 6A). Together with the observations that most of the BV appeared to be deprotonated in this state (based on the absence of the N-H ip mode band at ∼1575 cm–1), we concluded that the D207E, D207A, and D207H variants stop after red light irradiation at intermediate(s) structurally analogous to Meta-Rc. This scenario is in line with the RR analysis of the Meta-Rc intermediate of A. tumefaciens BphP1 and the photoconversion product of its D197A mutant (22). For the DrBphP mutants, the poorly resolved and broad features in the methine bridge stretching region between 1590 and 1620 cm–1 may reflect the presence of multiple Meta-Rc-type species (Fig. 6A).
FIGURE 6.
RR spectra of the photoconversion products of DrBphP and variants affecting Asp-207 and His-260. A, RR spectra of wild-type DrBphP in the Meta-Rc state trapped at –30 °C during red light irradiation as compared with the meta states of D207A, D207E, D207H, H260A, and H260N variants formed at room temperature during red light irradiation. B, RR spectra of Pfr state of wild-type DrBphP and the H260Q mutant measured from samples either dissolved in H2O(black lines) and D2O(gray lines) and then irradiated at room temperature with red light. Contributions from the residual Pr state were subtracted from each spectrum. The positions of N-H ip band (arrows) and other diagnostic RR maxima are indicated.
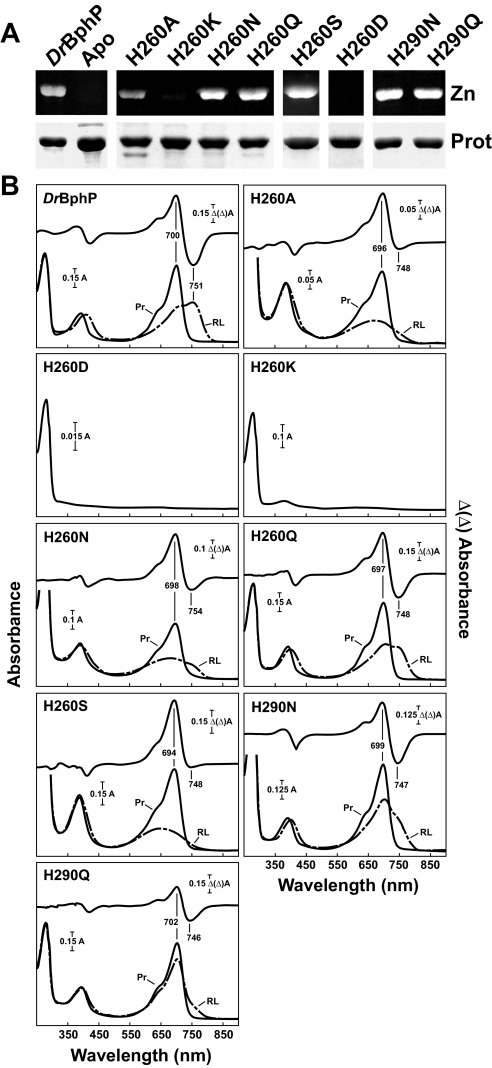
Substitutions of the Highly Conserved His-260—Consistent with the observed interaction of His-260 with the chromophore via the C-ring propionate side chain and the A, B, and C pyrrole rings through the pyrrole water (Fig. 1B), mutational analysis indicated that this residue also has an important role in bilin binding and photochemistry. The H260D and H260K substitutions were among the few soluble proteins tested that completely failed to bind BV either covalently or noncovalently (Fig. 7), indicating that the introduced side chains may: (i) block BV docking, (ii) potentially exclude the pyrrole water, and/or (iii) for the H260K substitution may generate repulsion between this residue and the positive charges of the pyrrole nitrogens. The H260A, H260N, and H260S apoproteins covalently bound BV and had relatively normal Pr absorption spectra (except for increased absorbance of the Soret band relative to the Q band), but photoconverted to a bleached product in red light (Fig. 7). In contrast, DrBphP easily tolerated a glutamine at this position with the H260Q holoprotein generating nearly normal Pr and Pfr absorption spectra.
FIGURE 7.
Assembly and absorption spectra of amino acid substitutions in DrBphP affecting His-260 and His-290. The recombinant full-length apoproteins were incubated with BV and purified by nickel chelate chromatography. A, samples were subjected to SDS-PAGE and either assayed for the bound bilin by zinc-induced fluorescence (Zn) or stained for protein with Coomassie Blue (Prot). Apo, apoprotein prior to BV incubation. B, UV-visible absorption spectra as Pr (black lines) or following saturating red light irradiation (dashed lines) and red-minus-far red light difference spectra (upper graph) of the apoproteins incubated with BV. Difference spectra maxima and minima are indicated.
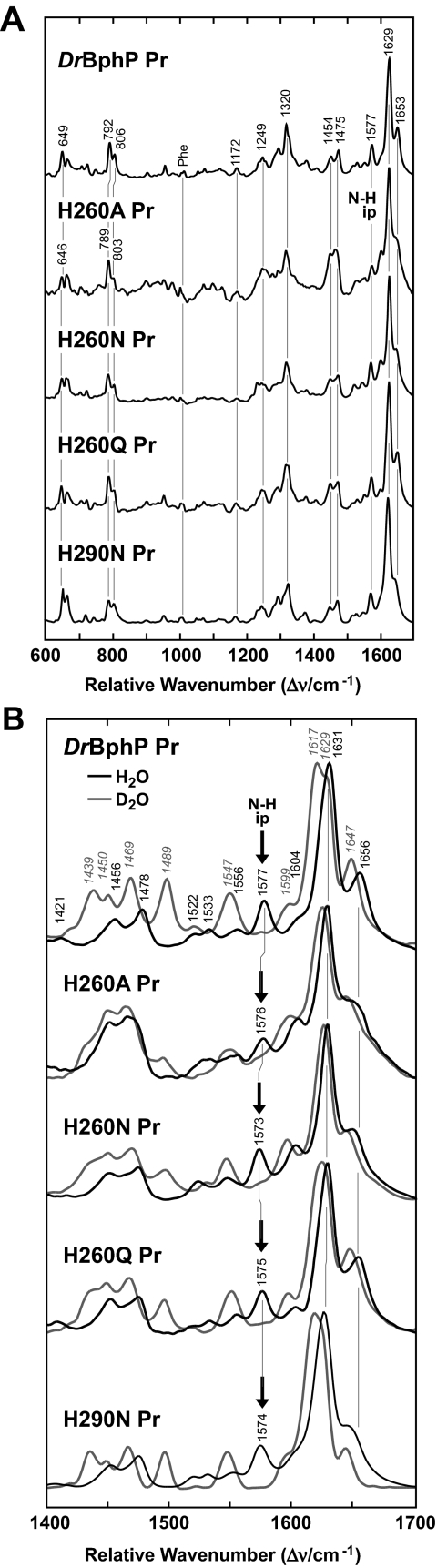
Like substitutions affecting Asp-207, the H260A, H260N, and H260Q chromoproteins had RR spectra similar to wild-type DrBphP as Pr (Fig. 8). For H260Q, the RR spectrum was nearly indistinguishable, implying that the overall chromophore structure and protonation state was unaffected by the histidine to glutamine substitution. More specifically, the hydrogen bond network involving the B and C pyrrole rings was unchanged by the introduced amide moiety based on the identical positions of the N-H ip mode band. Despite aberrant UV-visible absorption spectra, the H260A and H260N proteins also had RR spectra very similar to that of wild type, including obvious N-H ip bands, indicating that these variants remained protonated as Pr (Fig. 8B). The same conclusion was reached for an analogous substitution in A. tumefaciens BphP1 (His-250 to alanine) (13). However, we observed here a substantial broadening of the 1648-cm–1 band for the DrBphP H260A and H260N variants that may reflect an increased flexibility of the chromophore in the vicinity of the A–B methine bridge.
FIGURE 8.
RR spectra of the Pr state of amino acid substitutions in DrBphP affecting His-260 and His-290. A, overview RR spectra (600–1700 cm–1) measured from the samples dissolved in H2O. B, expanded view of the spectra in A for the marker band region (1400–1700 cm–1) from samples dissolved in H2O(black lines) and D2O(gray lines). The positions of N-H ip band (arrows) and other diagnostic RR maxima are indicated.
The RR spectra of the bleached species formed upon irradiation of the H260A and H260N variants with red light (Fig. 6A) were similar to those observed for the Asp-207 variants of DrBphP and the H250A variant of A. tumefaciens BphP1 (13). Only minor disagreements were evident in the details in the C=C stretching band profile, which likely reflect different distributions among various Meta-(sub)states. We did not detect the N-H ip band for H260A and H260N chromoproteins upon red light irradiation, indicating that BV was deprotonated (Fig. 6A). Thus, just like Asp-207 substitutions, the alanine and asparagine substitutions of His-260 become trapped in a Meta-Rc-like state after photoconversion.
By contrast, the RR spectrum of the photoconversion product of the H260Q substitution displayed a similar vibrational band pattern as the Pfr state of wild-type DrBphP (Fig. 6B), in agreement with their similar UV-visible absorption spectra. Even though the positions of the main RR peaks were unchanged in the marker band region, more subtle changes in peak intensities were evident. Specifically, the N-H ip mode, which gives rise to a well defined band at 1551 cm–1 in the wild-type chromoprotein, was split into two components at 1547 and 1555 cm–1 for the H260Q mutant. This splitting was reminiscent of the antenna pigment α-C-phycocyanin for which a similar band pair has been attributed to the N-H ip bendings of two bilin species with different hydrogen bond interactions (38). Furthermore, the intensity increase of the 1589-cm–1 band and the loss of intensity of the C-H out-of-plane mode of the C–D methine bridge suggest that the H260Q variant had slightly different torsional angles for the methine bridges and increased chromophore flexibility as compared with wild-type DrBphP (Fig. 6B). In contrast to the wild-type chromoprotein, the cryogenically trapped, bleached meta state of H260Q had a substantially sharper RR band at ∼1600 cm–1, suggesting that the distribution of meta states in this mutant is narrower than in wild type (data not shown).
Substitutions of His-290 in the Vicinity of Pyrrole Ring D—The primary Pr → Pfr photoprocess of Phys is thought to involve a Z to E isomerization of the C–D-ring methine bridge (16–18, 22). His-290 might play a central role in this movement because it hydrogen bonds with the D-ring carbonyl and with water2 as Pr, thereby helping stabilize the 44° out-of-plane rotation of ring D relative to rings B and C (8, 11). This His-290 D-ring contact must then be ruptured to complete the Z to E rotation of the D ring, a motion that may be encouraged by the large torsional angle of the C–D methine bridge double bond.
When we replaced His-290 with asparagine (H290N) or glutamine (H290Q), either of which could alter the hydrogen bonding network and/or add more space around the D-ring, the apoproteins still retained the ability to bind BV and generate normal Pr spectra with a maximum at 699 nm (Fig. 7, A and B). RR spectroscopy demonstrated that the H290N and H290Q chromoproteins also remained protonated as Pr (Fig. 8 and data not shown). Both mutant proteins were phototransformed by red light, but the absorption spectra after saturating red irradiations reveal striking differences. Whereas the spectrum of the H290Q substitution was reminiscent of a bleached meta state, the spectrum of H290N appeared to be a composite of a meta state and Pfr (Fig. 7B). The differences between H290N and H290Q were supported by RR spectroscopy. The RR spectrum of the photoconversion product of H290Q displayed features similar to that of the Pfr state of wild-type DrBphP, whereas the H290Q variant appeared to better fit a mixture of the Meta-Rc and Pfr states (data not shown).
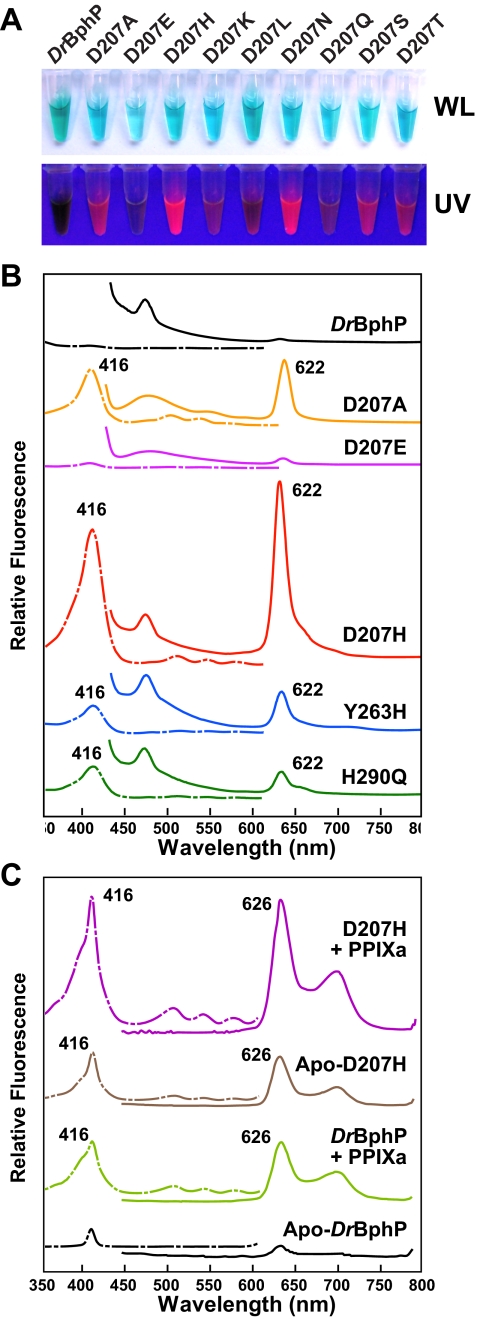
Mutants Affecting Asp-207, Tyr-263, and His-290 Are Highly Red Fluorescent—In addition to their ability to impair photochemistry, one or more DrBphP mutants affecting Asp-207, Tyr-263, and His-290 were highly red fluorescent when exposed to UV or 416-nm light (Fig. 9A and data not shown). Surprisingly, their fluorescence excitation and emission spectra, which were nearly identical to each other, did not match expectations based on the Pr absorption spectrum of DrBphP or fluorescence mutants generated in Synechocystis Cph1 by replacement of Tyr-176 with a histidine (30, 37). The excitation spectra had a maximum at ∼416 nm, which likely represented absorption by the Soret band of a bilin/porphyrin-type chromophore, but the spectra were missing the Q band absorption maximum near 698 nm expected for BV (Fig. 9B). The emission spectra were also substantially blue-shifted from the absorption of BV bound to DrBphP with a sharp peak at ∼622 nm that was not flanked by an obvious excitation maximum around 600 nm.
Figure 9.
Asp-207, His-290, and Tyr-263 mutants bind porphyrin and become red fluorescent. The UV-vis absorption spectra of each variant assembled with BV are shown in Figs. 3, 4, and 7. A, purified solutions of the DrBphP and the various Asp-207 variants in white light (WL) or upon irradiation with UV light. The concentration of each sample was adjusted to have an absorbance at ∼700 nm of 0.5 for the Pr form. B, fluorescence excitation (dashed lines) and emission spectra (solid lines) of the wild-type and mutant DrBphP after in vivo assembly with BV. Sample concetrations were adjusted to have an absorbance at 700 nm of 0.6 for the Pr form. C, fluorescence excitation (dashed lines) and emission spectra (solid lines) of purified wild-type DrBphP and the D207H mutant before (Apo) and after incubation with PPIXa in vitro. Sample concentrations were adjusted to have an absorbance at 280 nm of 2.
Based on the report of Fischer et al. (37) that a Tyr-176 to arginine substitution in Synechocystis Cph1 can bind cyclic porphyrins and become similarly red fluorescent, we tested whether the fluorescent species observed here were porphyrin-bound chromoproteins. One likely candidate was PPIXa, which is the immediate precursor of heme. Using the D207H apoprotein as an example, we found that it covalently bound purified PPIXa in vitro as judged by orange fluorescence of the polypeptide after SDS-PAGE with or without zinc staining of gels (data not shown).
Following re-purification, the PPIXa-D207H chromoprotein displayed excitation and emission maxima nearly identical to those obtained with the Asp-207, Tyr-263, and His-290 mutant collection (Fig. 9B). The only substantive difference was the appearance of a second emission peak at ∼690 nm for the PPIXa-assembled samples. The absence of this fluorescence peak in the BV-assembled samples was likely caused by strong absorption of this fluorescence by Q band of the BV-containing chromoproteins. In agreement, this second fluorescent peak at ∼690 progressively appeared as we diluted the D207H samples assembled with BV (data not shown). Wild-type DrBphP could also bind PPIXa and become red fluorescent but with much reduced binding efficiency as compared to the mutants (Fig. 9B). Limited amounts of recombinant D207H holoprotein bearing a PPIXa-like chromophore were even generated without co-expression of the HO or subsequent addition of PPIXa in vitro, indicating that variant (as possibility others) can spontaneously assemble with porphyrins either produced by E. coli or present in the growth medium.
DISCUSSION
By inspecting the available crystal structures for two bacterial CBDs (8, 11, 26), we identified a number of highly conserved residues that should play prominent roles in Phy folding, chromophore assembly, and Pr → Pfr photoconversion. Prior mutagenic studies on DrBphP confirmed the importance of Cys-24 and Met-259 in BV ligation (5, 8). Here, we extended this analysis to 16 other positions, using UV-visible absorption, fluorescence, and RR spectroscopy to probe the impact of individual substitutions/deletions on chromophore binding and geometry and the mechanics of Pr → Pfr photoconversion.
In general, the data revealed that apoprotein solubility, BV binding, the chromophore structure of the Pr state, and possibly the primary photochemical process in red light that generates Lumi-R are not substantially affected by many substitutions introduced individually. Evidently, these properties are remarkably immune to structural and electrostatic perturbations within the binding pocket. In contrast, photoconversion of Pr to Pfr is very sensitive to most amino acid substitutions, indicating that the final relaxation step(s) from Meta-Ra/Rc to Pfr is more complicated than expected and may critically depend on a number of structural elements working together. The results also highlight the need of a three-dimensional structure for the Phy under analysis to accurately interpret the consequences of various mutations. For example, by comparing our data from mutants affecting Tyr-176 and Asp-207 with those from comparable mutants in Synechocystis Cph1 (30, 37) and Agrobacterium tumefaciens BphP1 (13), respectively, it appears that the roles of some amino acids can be context-dependent, thus complicating extrapolations from one Phy to another.
DrBphP Assembly with BV—Surprisingly, only 1 of the 38 mutants tested here was completely insoluble (Q36N), and only three others failed to covalently bind BV in vitro (H260D, H260K, and R254Q). Neither removal of the first 20 amino acids upstream of Cys-24 nor alanine substitutions of Glu-25 and Glu-27 blocked formation of the Cys-24-BV thioether adduct, thus ruling out any of these residues as essential to the binding reaction. Instead, critical parameters appear to be amino acids that control the precise positioning/conformation of BV and the proper fold of the chromophore-binding pocket. Among them are Arg-254 and His-260, which form salt bridges with the propionate side chains of rings B and C, respectively (Fig. 1B). Even though hydrogen bond interactions with the ring C propionate are predicted to be qualitatively altered or impaired in the H260A, H260N, H260Q and H260S variants, they still bound BV. Only upon introduction of a stronger and longer cationic side chain provided by the H260K substitution was BV binding severely inhibited. This observation suggests that correct electrostatics in the vicinity of B and C pyrrole rings and/or the pyrrole water are strong determinants for correct chromophore fit. As an example, introduction of a polar amino acid with partial charge reversal (R254Q) near the B-ring propionate carboxyl group was sufficient to block BV binding.
We expected that substantial changes within the figure-of-eight knot would also have dramatic effects on DrBphP folding and BV attachment (11). Two central residues are Ile-35, which provides van der Waals interactions with Val-232, Leu-234, Leu-248, and Leu-253 within the three-stranded anti-parallel β-sheet that forms the knot core, and Glu-36, which hydrogen bonds with the main chain carbonyl of Arg-254 and the amide of Ala-225, thus forming a lasso that condenses the knot around Ile-35 (see Fig. 1C). Our observations that the I35A variant is relatively normal with respect to apoprotein solubility, BV binding, and Pr/Pfr photochemistry indicate that subtle changes in the knot center can be tolerated. By comparison, Gln-36 appears to be important but its role is complex. Some substitutions of Gln-36 were either completely insoluble (Q36N) or had limited solubility (Q36A and Q36K), suggesting that its contacts with Arg-254 and Ala-225 are vital to fold the nascent Phy polypeptide. However, the Q36L mutant that cannot not form similar hydrogen bonds folded well and was photochemically normal, indicating that other parameter(s) at position 36 are also key (e.g. size). Whatever its exact role, sequence alignments of the Phy superfamily indicate that Gln-36 is important. Whereas Ile-35 can sometimes be replaced by valine, all Phys we have analyzed phylogenetically to date have glutamine as the next amino acid (5, 11).
Chromophore Structure in the Pr State—Intuitively, we expected that substitutions of many amino acids in close contact with the bilin would affect the spectral properties of Pr given the importance of the apoprotein to its absorption spectrum (1). However, analyses by us and others indicate that the Pr chromophore is relatively tolerant to individual mutations (13, 30, 31). Remarkable exceptions for DrBphP are the Y176H, Y216W, and R254A variants, which alter hydrophobic interaction with ring D or hydrogen bonding with the B-ring propionate. For Y176H, we note considerable changes in the UV-visible absorption spectrum for Pr, suggesting conformational heterogeneity. For all other CBD substitutions, including Y216H, the only impact on the spectral properties of Pr was an increase in the Soret/Q band absorbance ratio for some mutants (Table 1). Taken together, it appears that the bilin structure in the Pr conformer is either generated by the ensemble of amino acids in the CBD working together such that the effects of most single alterations are insignificant or that the chromophore is flexible within the pocket and thus tolerant to most substitutions.
This conclusion also holds for substitutions of Asp-207, His-260, and His-290, which are involved in an extended hydrogen bond network with BV and several waters (see Fig. 1B) (8, 11). Compared with the wild-type DrBphP, only subtle differences were found in the RR spectra of the various Asp-207, His-260, and His-290 variants, indicating that most features of the Pr chromophore geometry are preserved. Asp-207 has been proposed to be critical for maintaining the protonated (cationic) nature of BV as Pr (11, 13). For DrBphP, RR spectroscopy demonstrated that at pH 7.8 the chromophore remains largely protonated upon substitution of Asp-207 by Glu, His, and Ala. Only in the D207A mutant did we detect a small increase in unprotonated BV. Consequently, we conclude that carboxylate group of Asp-207 is not essential to stabilize the positive charge of the ground state chromophore. This conclusion agrees with the DrBphP CBD structural models where only the main chain oxygen of Asp-207 hydrogen bonds with the pyrrole water (Fig. 1C). It is interesting to note that the same substitution of the analogous amino acid in A. tumefaciens BphP1 (D197A) leads to a larger fraction of deprotonated Pr chromophore at pH 7.8 (13). Such differences imply that the acid-base equilibrium of the bilin depends on subtle structural and electrostatic details inside the binding pocket, which can differ somewhat even among highly related Phys.
The Role of Key Amino Acids in the Meta-R → Pfr Photoconversion—Whereas the Pr state is relatively insensitive toward alterations in the chromophore-binding pocket, single substitutions, even rather conservatives ones, are deleterious for complete photoconversion to Pfr. This is specifically true for most substitutions near the bilin. Previous studies on A. tumefaciens BphP1 identified one key event during the Pr → Pfr transition that involves transient proton release from the bilin to the external medium to form Meta-Rc followed by reprotonation of the bilin upon Pfr formation (13, 22). This protonation cycle requires a well positioned group to accept a proton from the A, B, and C pyrrole rings. Based on its central position in the DrBphP CBD structure (8, 11) and our prior analysis with a comparable aspartic acid to alanine variant in A. tumefaciens BphP1 (13), the acidic side chain of Asp-207 is a likely candidate, using the pyrrole water as a proton conduit. In this study, we provide further support through the analysis of an enlarged the set of Asp-207 substitutions. Strikingly, most cannot proceed through the Meta-Ra/Rc to Pfr relaxation following photoexcitation with several (D207L, D207H, D207K, and D207S) potentially blocked at the earlier steps. The failure to undergo the proton-coupled transition from the Meta-Rc to the Pfr states even holds for D207E variant, implying that the precise positioning of the Asp-207 carboxylate relative to the pyrrole water, BV, and/or PHY domain is a prerequisite for the final relaxation step.
How Asp-207 maintains its position next to BV and the pyrrole water is not yet clear. It could involve the known electrostatic interaction of Asp-207 with Tyr-263 (Fig. 1B) or yet to be defined interactions with other residue(s) in the full-length Phy homodimer given that the carboxylate moiety of Asp-207 is solvent-exposed in the structure of only the CBD (8, 11). Of potential significance is that the histidine substitution of Tyr-263 induces the same photochemical defects as a number of mutants affecting Asp-207, including a failure to generate Pfr in red light and an increased affinity for PPIXa. Thus, the defects of the Y263H chromoprotein could reflect an altered placement of Asp-207. Likely other candidates fixing Asp-207 in place are one or more invariant basic residues in the PHY domain given the importance of this domain for completing the relaxation of Meta-Rc to Pfr and for the thermal stabilization of Pfr (1, 2). Such a strict positioning is further illustrated by the extreme conservation of Asp-207 and its neighboring isoleucine and proline residues as part of the invariant DIP motif (5, 11). However, the roles of Ile-208 and Pro-209 must be subtle given that substitutions of these residues had little impact on the spectral properties of Pr or Pfr. Clearly, the exact role of Asp-207 will require continued structural analysis of several Asp-207 mutants (e.g. D207L and D207H) and three-dimensional models of DrBphP as both Pr and Pfr that include the PHY domain.
It is likely that the transient proton transfer associated with the formation and the decay of Meta-Rc further depends on the extended hydrogen bond network that embeds the chromophore. His-260, which hydrogen bonds with the pyrrole water and the propionate side chain of ring C, is probably also key to this network (13). His-260 can be adequately replaced by glutamine based on the nearly normal UV-visible and RR spectra of the H260Q holoprotein as Pr and Pfr. However, more significant substitutions such as H260D and H260K fail to bind BV, whereas the H260A, H260N, and H260S substitutions induce severe spectral defects. The H260A and H260N variants become locked in a bleached and deprotonated state after red light irradiation that resembles Meta-Rc.
His-290 also participates in the Pr → Pfr photoprocess with the H290Q chromoprotein stalled in a Meta-Rc-like state upon red light irradiation and the H290N chromoprotein forming Pfr with a slightly different chromophore structure (Figs. 7 and 8 and data not shown). Because the amide moiety in both substitutions may still hydrogen bond with the carbonyl of ring D, these mutants could retain the rotational strain on the D ring that may be essential for phototransformation (8, 11). The differential effects of asparagine and glutamine imply that the distance between the amide moiety of His-290 and ring D is critical. The ultimate appreciation of His-290 in the Pr/Pfr photocycle will clearly require further experimental analysis.
The first step in Pr → Pfr photoconversion is the Z to E photoisomerization of the methine bridge between the C and D pyrrole rings (18). This photoisomerization is then predicted to substantially alter the position of the D ring such that the D ring may sterically clash with one or more bulky hydrophobic residues that line the D-ring pocket (e.g. His-290, Tyr-176, Tyr-263, and Phe-203 (Fig. 1B)) (8, 11). Consequently, it is not surprising that substitutions at these residues substantially alter photochromicity of DrBphP. Unfortunately, without a three-dimensional structure of Pfr, the exact roles of these hydrophobic amino acids and how they might move during photoisomerization remain unclear. The defects induced by the Y176H change are strikingly different from those observed by Fisher et al. (30, 37) using an analogous substitutions in Synechocystis Cph1. Whereas the Y176H mutant in DrBphP is only weakly fluorescent (supplemental Fig. 2), the same substitution in SynCph1 is strongly red fluorescent (30), indicating that the Tyr-176 can have distinct roles/properties depending on its local environment or the nature of the chromophore (BV versus PCB). Our mutational analysis of Tyr-263 and Phe-203 demonstrated that these aromatic residues are particularly important for proper photochemistry of DrBphP. As discussed above, Tyr-263 may be especially critical given its dual role in lining the pocket near the C–D-ring methine bridge and helping fix Asp-207 in place (11, 18). One can imagine that Tyr-263 must also move within the binding pocket as BV swings during its red light-induced isomerization.
One surprising observation is that Arg-254, which interacts with the propionate side chain of ring B and the knot, has little appreciable role in Pfr formation after BV is attached. Both the R254A and R254K variants have near normal Pfr absorption spectra, although in the former case the Pr spectrum is slightly compromised. Given the proposal that Pr → Pfr photoconversion involves a (partial) rotation around the A–B methine bridge single bond in addition to the Z to E isomerization of the C–D methine bridge double bond (24, 39), a stronger effect on photoconversion might have been expected. It is intriguing that the R254A chromoprotein has a substantially dampened rate of dark reversion from Pfr back to Pr. We speculate that Arg-254 through its contact with the ring B propionate helps strain the Pfr chromophore in a way that lowers the barrier for thermal E to Z isomerization of the C–D methine bridge needed to regenerate Pr.
Novel Fluorescent Derivatives of DrBphP—An unusual feature of the DrBphP mutants replacing Asp-207, His-290, or Tyr-263 is their greater ability to assemble with PPIXa, and possibility related porphryins, and become red fluorescent. Some porphyrin incorporation was evident even when the mutant polypeptides were co-expressed with excess BV. In a similar way, the Tyr-176 to arginine substitution of Synechocystis Cph1 was previously reported to bind porphyrins and become fluorescent (37). Given the tight fit of linear bilins in the CBD (8, 11, 26), it might seem surprising that porphyrins are accommodated in the binding pocket. However, stop-flow assembly studies suggest that Phys first associate with linear bilins arranged in a more cyclic porphryin-like conformation before the chromophores are converted to the more extended ZZZsyn, syn, anti conformation (40). One intriguing possibility is that Asp-207, His-290, and Tyr-263 work in concert to limit fully cyclic porphyrins from the CBD, a barrier that is lessened in the suite of mutants affecting these residues. Asp-207 and Tyr-263 in particular through their hydrogen bonding with each other and their positions above and below the binding pocket (8, 11, 26) may provide a clamp at the opening of the pocket that prohibits this access. Some Asp-207 mutants are more promiscuous in binding PPIXa (e.g. D207H versus D207E), suggesting that they have a greater impact on perturbing this clamp. Since most if not all bacteria synthesize porphyrins like PPIXa, such interference may have been a necessity during Phy evolution to encourage the binding of linear bilins like BV that are a prerequisite for the R/FR photochromicity of Phy-type photoreceptors.
Supplementary Material
Acknowledgments
We thank Drs. Andrew Ulijasz and David Anstrom for helpful advice and Dr. J. Clark Lagarias for his suggestion that the fluorescing DrBphP variants contain a porphyrin.
This work was supported by National Science Foundation Grants MCB 04240 62 and 0719153 (to R. D. V. and K. T. F.) and Deutsche Forschungsgemeinschaft Grant SFB498 (to P. H. and M. A. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: Phy, phytochrome; BphP, bacteriophytochrome; BV, biliverdin IXα; CBD, chromophore-binding domain; GAF, cGMP phosphodiesterase/adenylcyclase/FhlA; HO, heme oxygenase; ip, in-plane bending; PAS, Per/Arndt/Sim; PCB, phycocyanobilin; PΦB, phytochromobilin; PHY, phytochrome domain; PPIXa, protoporphyrin IXa; Pr, red light-absorbing state of phytochrome; Pfr, far red light-absorbing state of phytochrome; RR, resonance Raman.
D. von Stetten, J. R. Wagner, J. Zhang, R. D. Vierstra, and P. H. Hildebrandt, unpublished data.
References
- 1.Rockwell, N. C., Su, Y. S., and Lagarias, J. C. (2006) Annu. Rev. Plant Biol. 57 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vierstra, R. D., and Karniol, B. (2005) in Handbook of Photosensory Receptors (Briggs, W. R., and Spudich, J. L., eds) pp. 171–196, Wiley-VCH Press, Weinheim, Germany
- 3.Quail, P. H. (2002) Nat. Rev. Mol. Cell Biol. 3 85–93 [DOI] [PubMed] [Google Scholar]
- 4.Yeh, K. C., and Lagarias, J. C. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 13976–13981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karniol, B., Wagner, J. R., Walker, J. M., and Vierstra, R. D. (2005) Biochem. J. 392 103–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhoo, S. H., Davis, S. J., Walker, J., Karniol, B., and Vierstra, R. D. (2001) Nature 414 776–779 [DOI] [PubMed] [Google Scholar]
- 7.Froehlich, A. C., Noh, B., Vierstra, R. D., Loros, J., and Dunlap, J. C. (2005) Eukaryot. Cell 4 2140–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner, J. R., Zhang, J., Brunzelle, J. S., Vierstra, R. D., and Forest, K. T. (2007) J. Biol. Chem. 282 12298–12309 [DOI] [PubMed] [Google Scholar]
- 9.Lamparter, T., Carrascal, M., Michael, N., Martinez, E., Rottwinkel, G., and Abian, J. (2004) Biochemistry 43 3659–3669 [DOI] [PubMed] [Google Scholar]
- 10.Lagarias, J. C., and Rapoport, H. (1980) J. Amer. Chem. Soc. 102 4821–4828 [Google Scholar]
- 11.Wagner, J. R., Brunzelle, J. S., Forest, K. T., and Vierstra, R. D. (2005) Nature 438 325–331 [DOI] [PubMed] [Google Scholar]
- 12.Kneip, C., Hildebrandt, P., Schlamann, W., Braslavsky, S. E., Mark, F., and Schaffner, K. (1999) Biochemistry 38 15185–15192 [DOI] [PubMed] [Google Scholar]
- 13.von Stetten, D., Seibeck, S., Michael, N., Scheerer, P., Mroginski, M. A., Murgida, D. H., Krauss, N., Heyn, M. P., Hildebrandt, P., Borucki, B., and Lamparter, T. (2007) J. Biol. Chem. 282 2116–2123 [DOI] [PubMed] [Google Scholar]
- 14.Andel, F., Hasson, K. C., Gai, F., Anfinrud, P. A., and Mathies, R. A. (1997) Biospect. 3 421–433 [Google Scholar]
- 15.Bischoff, M., Hermann, G., Rentsch, S., and Strehlow, D. (2001) Biochemistry 40 181–186 [DOI] [PubMed] [Google Scholar]
- 16.Andel, F., 3rd, Lagarias, J. C., and Mathies, R. A. (1996) Biochemistry 35 15997–16008 [DOI] [PubMed] [Google Scholar]
- 17.Mizutani, Y., Tokutomi, S., and Kitagawa, T. (1994) Biochemistry 33 153–158 [DOI] [PubMed] [Google Scholar]
- 18.Rudiger, W., Thummler, F., Cmiel, E., and Schneider, S. (1983) Proc. Natl. Acad. Sci. U. S. A. 80 6244–6248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tu, S.-L., and Lagarias, J. C. (2005) in Handbook of Photosensory Receptors (Briggs, W. R., and Spudich, J. L., eds) pp. 121–149, Wiley-VCH Press, Weinheim, Germany
- 20.Foerstendorf, H., Benda, C., Gartner, W., Storf, M., Scheer, H., and Siebert, F. (2001) Biochemistry 40 14952–14959 [DOI] [PubMed] [Google Scholar]
- 21.Braslavsky, S. E. (2003) in Photochromisms, Molecules and Systems (BrouasLaurent, D. H., and BrouasLaurent, H., eds) pp. 738–755, Elsevier Science BV, Amsterdam
- 22.Borucki, B., von Stetten, D., Seibeck, S., Lamparter, T., Michael, N., Mroginski, M. A., Otto, H., Murgida, D. H., Heyn, M. P., and Hildebrandt, P. (2005) J. Biol. Chem. 280 34358–34364 [DOI] [PubMed] [Google Scholar]
- 23.Andel, F., 3rd, Murphy, J. T., Haas, J. A., McDowell, M. T., van der Hoef, I., Lugtenburg, J., Lagarias, J. C., and Mathies, R. A. (2000) Biochemistry 39 2667–2676 [DOI] [PubMed] [Google Scholar]
- 24.Mroginski, M. A., Murgida, D. H., von Stetten, D., Kneip, C., Mark, F., and Hildebrandt, P. (2004) J. Am. Chem. Soc. 126 16734–16735 [DOI] [PubMed] [Google Scholar]
- 25.Kneip, C., Schlamann, W., Braslavsky, S. E., Hildebrandt, P., and Schaffner, K. (2000) FEBS Lett. 482 252–256 [DOI] [PubMed] [Google Scholar]
- 26.Yang, X., Stojkovic, E. M., Kuk, J., and Moffat, K. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 12571–12576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis, S. J., Vener, A. V., and Vierstra, R. D. (1999) Science 286 2517–2520 [DOI] [PubMed] [Google Scholar]
- 28.Cherry, J. R., Hondred, D., Walker, J. M., Keller, J. M., Hershey, H. P., and Vierstra, R. D. (1993) Plant Cell 5 565–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhoo, S. H., Hirano, T., Jeong, H.-Y., Lee, J.-G., Furuya, M., and Song, P.-S. (1997) J. Am. Chem. Soc. 48 11717–11718 [Google Scholar]
- 30.Fischer, A. J., and Lagarias, J. C. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 17334–17339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hahn, J., Strauss, H. M., Landgraf, F. T., Gimenez, H. F., Lochnit, G., Schmieder, P., and Hughes, J. (2006) FEBS J. 273 1415–1429 [DOI] [PubMed] [Google Scholar]
- 32.Stockhaus, J., Nagatani, A., Halfter, U., Kay, S., Furuya, M., and Chua, N. H. (1992) Genes Dev. 6 2364–2372 [DOI] [PubMed] [Google Scholar]
- 33.Zhao, K. H., Ran, Y., Li, M., Sun, Y. N., Zhou, M., Storf, M., Kupka, M., Bohm, S., Bubenzer, C., and Scheer, H. (2004) Biochemistry 43 11576–11588 [DOI] [PubMed] [Google Scholar]
- 34.Stevens, J. M., Daltrop, O., Allen, J. W., and Ferguson, S. J. (2004) Acc. Chem. Res. 37 999–1007 [DOI] [PubMed] [Google Scholar]
- 35.Lamparter, T., Esteban, B., and Hughes, J. (2001) Eur. J. Biochem. 268 4720–4730 [DOI] [PubMed] [Google Scholar]
- 36.Hanzawa, H., Inomata, K., Kinoshita, H., Kakiuchi, T., Jayasundera, K. P., Sawamoto, D., Ohta, A., Uchida, K., Wada, K., and Furuya, M. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 3612–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fischer, A. J., Rockwell, N. C., Jang, A. Y., Ernst, L. A., Waggoner, A. S., Duan, Y., Lei, H., and Lagarias, J. C. (2005) Biochemistry 44 15203–15215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mroginski, M. A., Murgida, D. H., and Hildebrandt, P. (2007) Acc. Chem. Res. 40 258–266 [DOI] [PubMed] [Google Scholar]
- 39.Inomata, K., Noack, S., Hammam, M. A., Khawn, H., Kinoshita, H., Murata, Y., Michael, N., Scheerer, P., Krauss, N., and Lamparter, T. (2006) J. Biol. Chem. 281 28162–28173 [DOI] [PubMed] [Google Scholar]
- 40.Borucki, B., Otto, H., Rottwinkel, G., Hughes, J., Heyn, M. P., and Lamparter, T. (2003) Biochemistry 42 13684–13697 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.