Abstract
Glycine residues may play functional and structural roles in membrane proteins. In this work we studied the role of glycine residues in EmrE, a small multidrug transporter from Escherichia coli. EmrE extrudes various drugs across the plasma membrane in exchange with protons and, as a result, confers resistance against their toxic effects. Each of 12 glycine residues was replaced by site-directed mutagenesis. Four of the 12 glycine residues in EmrE are evolutionary conserved within the small multidrug resistance family of multidrug transporters. Our analysis reveals that only two (Gly-67 and Gly-97) of these four highly conserved residues are essential for transporter activity. Moreover, two glycine positions that are less conserved, Gly-17 and Gly-90, demonstrate also a nil phenotype when substituted. Our present results identifying Gly-17 and Gly-67 as irreplaceable reinforce the importance of previously defined functional clusters. Two essential glycine residues, Gly-90 and Gly-97, form a protein motif in which glycine residues are separated by six other residues (GG7). Upon substitution of glycine in these positions, the protein ability to form dimers is impaired as evaluated by cross-linking and pull-down experiments.
EmrE is a small (110 residues) multidrug transporter from Escherichia coli that extrudes positively charged aromatic drugs in exchange for two protons, thus rendering bacteria resistant to a variety of toxic compounds (1–3). The protein has been characterized, purified, and reconstituted in a functional form (1, 4). Extensive studies have revealed residues in the protein that are essential for its activity. Glu-14 is the only membrane-embedded acidic residue in EmrE and is involved in both substrate recognition and proton coupling (5). Several highly conserved amino acids in TM14 clustered on the same helical face as Glu-14 have been shown to be functionally important (6). Trp-63 has been shown to be essential and even a conservative replacement by an aromatic residue yielded an inactive transporter (7). Tyr-40 and Tyr-60 can be only replaced with hydroxyamino acids (8).
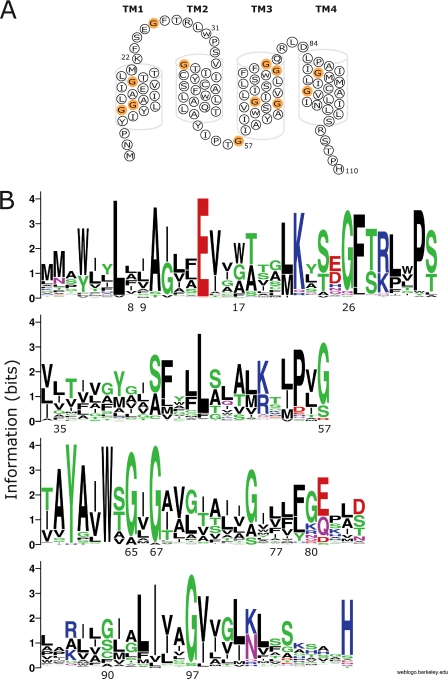
The primary sequence of EmrE is rich in glycine residues: of 110 amino acids in its sequence, 12 are glycines (Fig. 1A). Indeed, widescale genomic analysis of membrane protein families reveals that glycine occurs frequently in the transmembrane helices of membrane proteins (9), and is also considerably more prevalent in conserved positions within transmembrane helices (10). Glycine is a unique amino acid due to its lack of side chain and the conformational flexibility it introduces to the polypeptide backbone of a protein. In various studies glycine residues have been shown to fulfill several roles, both functional and structural, when incorporated in membrane transport proteins. When introduced into lactose permease, glycine residues can confer conformational flexibility to this proton-coupled transporter (11), a glycine residue has been shown to act as a gating hinge in potassium channels (12) and recently, a conserved glycine residue in the neurotransmitter transporter GAT-1 has been demonstrated to be involved in a conformational transition during its transport cycle (13). In the photoreceptor rhodopsin Gly-121 has been shown to form part of the retinal binding pocket and to interact directly with the retinal ligand (14, 15).
FIGURE 1.
Sequence homology analysis of the SMR family. A, secondary structure model of EmrE. Glycine residues that were mutated throughout this work are shown in orange. B, sequence alignment of 113 members of the SMR family, all genes that encode for putative homo-oligomers are represented as sequence logos (24, 29). The scale indicates the certainty of finding a particular amino acid at a given position, and is determined by multiplying the frequency of that amino acid by the total information at that position. The residues at each position are arranged in order of predominance from top to bottom, with the highest frequency residue on top. The height of the symbol within each stack indicates the relative frequency of each amino acid at that position. Sequence logos were generated using the “WebLogo” web based application of Steven E. Brenner. Colors: green, polar; blue, basic; red, acidic, black, hydrophobic.
Sequence motifs that involve glycine residues, mainly the GXXXG (GG4) sequence motif, and the more complex glycine-zipper motif, have been shown to serve as packing and oligomerization motifs in numerous studies (16–19). Statistical analysis of amino acid patterns in transmembrane helices reveal that both sequence motifs, GXXXG (GG4) and GXXXXXXG (GG7), occur more frequently than expected from random probability (9). Taking into account helical geometry of 3.6 residues per turn, these motifs align both glycines on the same face of the helix.
In this study, we sought to explore the role of glycine residues in EmrE. Four of the 12 glycine residues in EmrE are highly conserved (above 90% conservation) within the small multi-drug resistance (SMR) family of multidrug transporters (Fig. 1B). Three of the conserved glycines are located in the putative transmembrane region (Gly-65, Gly-67, and Gly-97) and one is located in the first loop (Gly-26). Our analysis reveals that only two (Gly-67 and Gly-97) of these four highly conserved residues are essential for the transporter activity, and once replaced by another amino acid, yield proteins that can no longer confer resistance (nil phenotype). Moreover, two glycine positions that are less conserved, Gly-17 and Gly-90, demonstrate also a nil phenotype when substituted. The two essential glycine residues in TM4 of EmrE, Gly-90 and Gly-97, form a protein motif in which glycine residues are separated by six other residues (GG7). This motif is frequent in transmembrane helices, especially in transporter/channel-like membrane proteins (10). Upon substitution of glycine in these positions, the protein ability to form dimers is impaired as evaluated by cross-linking and pull-down experiments. A different pattern is observed with Gly-17 and Gly-67, suggesting that the essential glycine residues of EmrE fulfill a variety of different roles.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Mutagenesis—E. coli DH5α and TA15 (20) strains were used throughout this work. TA15 strain was previously transformed with plasmid pGP1-2, which codes for the T7 polymerase under the inducible control of the λ PL promoter (21). The plasmids used for EmrE gene expression are pT7-7 (21) derivatives with a His6 tag (EmrE-His, for simplicity will be called EmrE throughout this paper) (4).
The construction of the mutants G8C, G9C, G17C, and G97C was previously described (6, 22, 23, 25). New mutants were obtained by polymerase chain reaction, using the overlap extension procedure as described in Refs. 26 or 27. The template used for all mutants was either CAMY (a His-tagged cysteine-less EmrE that was built with alanine replacements (6)), or CLA (a cysteine-less EmrE without any tags that was built with alanine replacements).
Resistance to Toxic Compounds—For testing resistance to toxic compounds E. coli DH5α cells transformed with pT7-7 EmrE, pT7-7 (vector alone), or with the various mutants were used. 5 μl of serial dilutions of overnight cultures were spotted on a series of LB-ampicillin plates containing 30 mm 1,3-bis-(tris(hydroxymethyl)methylamino)-propane titrated to pH 7, and various EmrE known substrates (200 μg/ml ethidium, 100 μg/ml acriflavine, or 0.2 mm methyl viologen). Growth was analyzed after overnight incubation at 37 °C.
Expression, Purification, and Reconstitution of EmrE into Proteoliposomes—TA15 cells bearing plasmids pGP1-2 and His-tagged EmrE constructs (cloned into pT7-7 expression vector) were used for overexpression. Purification was performed essentially as in Ref. 7. Reconstitution was performed as described (28).
[3H]TPP+ Binding Assay—Tetraphenylphosphonium (TPP+) binding was assayed essentially as described (4). All the experiments were repeated at least twice.
Transport Assay—Uptake of [14C]methyl viologen into proteoliposomes was assayed at 25 °C by dilution of 3 μl of the ammonium chloride-containing proteoliposomes into 200 μl of an ammonium-free solution (1, 28). The latter contained 20 μm [14C]methyl viologen (11.9 mCi/mmol, Sigma), 140 mm KCl, 10 mm Tricine, 5 mm MgCl2, and 10 mm Tris. At given times, the reaction was stopped by dilution with 2 ml of the same ice-cold solution, filtering through Millipore GSWP filters (0.22 μm) and washing with an additional 2 ml of solution. The radioactivity on the filters was estimated by liquid scintillation. In each experiment, the values obtained in a control reaction, with 15 μm nigericin, were subtracted from all experimental points.
Hexamethylene Diisocyanate (HMDC) Cross-linking—Membranes prepared from cells expressing EmrE mutants selectively labeled with [35S]methionine (30) were solubilized in 1% DDM-Na buffer. HMDC (dispersed in 1% DDM-Na buffer) was added to a final concentration of 0.02% (v/v). After 1 h at 25 °C, the reaction was stopped by addition of a mixture containing 600 mm β-mercaptoethanol, 300 mm Tris, pH 6.8, 0.6% bromphenol blue, 12% SDS, and 60% glycerol. The degree of cross-linking was assessed by separation of the samples in 16% acrylamide gels. Radioactive bands were visualized using a Fujifilm FLA-3000 PhosphorImager (Fujifilm, Tokyo).
Pull-down Assays—Membranes from cells expressing tagged-EmrE labeled with [35S]methionine were mixed with increasing amounts of membranes from cells expressing untagged-EmrE labeled with [35S]methionine and solubilized in 1% DDM-Na buffer at 80 °C for 15 min. The extract was centrifuged for 5 min at 20,000 × g at 4 °C to discard precipitates. The supernatant was supplemented with 15 mm imidazole, added to equilibrated Ni-NTA beads (20 μl/assay) and incubated at 4 °C for 1 h. The protein bound to beads was washed three times with 0.08% DDM-Na buffer containing 15 mm imidazole. The bead fraction was then incubated for 10 min at room temperature with 30 μl of sample buffer (450 mm imidazole, 200 mm β-mercaptoethanol, 100 mm Tris, pH 6.8, 0.2% bromphenol blue, 4% SDS, and 20% glycerol) to elute the protein from the Ni-NTA beads. Samples were separated in 16% acrylamide gels and analyzed for radioactive bands using a Fujifilm FLA-3000 PhosphorImager.
Generation of Heterodimers—Membranes from cells expressing E14C-His or G67C-His and membranes from cells expressing untagged Cys-less Ala EmrE were solubilized in 20 volumes of 0.8% DDM-Na buffer and mixed at 80 °C for 15 min as described (30). The extract was centrifuged for 5 min at 20,000 × g to discard precipitates. [3H]TPP+ binding activity was measured. Where indicated the heterodimer was incubated for 10 min with 1 mm 2-(trimethylammonium)ethylmethanethiosulfonate bromide (MTSET) prior to the binding assay.
RESULTS
Phenotype of Glycine Substitution Mutants—In the amino acid sequence of EmrE there are 12 glycine residues. Ten of those 12 glycine residues are located in putative transmembrane helices (Fig. 1A). Glycine in positions 26, 65, 67, and 97 are highly conserved among the SMR family (Fig. 1B).
In this study, we aimed at characterizing the roles of the different glycine residues in EmrE. For this purpose, we constructed a set of mutants using site-directed mutagenesis. Each of the 12 glycine residues was replaced with cysteine, except for Gly-17, Gly-67, Gly-90, and Gly-97 that were also replaced with alanine and proline.
The ability of the mutated proteins to confer resistance against various toxicants was assessed by testing the ability of cells expressing them to grow under otherwise nonpermissive conditions. Cells from overnight cultures were plated in 5-μl droplets of logarithmic dilutions on solid media containing ethidium (200 μg/ml), acriflavine (100 μg/ml), or methyl viologen (0.2 mm). Cells carrying the vector plasmid without any insert cannot grow on these media, whereas cells expressing either EmrE or Cys-less EmrE (which served as the template for all mutations used in this study) were able to grow at all dilutions. This assay provides us with a qualitative estimate of the in vivo activity of the generated mutants.
Cysteine mutations at most positions were well tolerated (Table 1A). The phenotype of the mutants at positions 9, 35, and 77 did not differ from that of the wild type protein. Mutants at positions 8 and 80 showed a slightly impaired resistance against acriflavine and mutants at positions 26 and 57 showed also a decrease in resistance to ethidium, although overall, all of these mutants demonstrated the ability to confer resistance well above background level. The mutant G65C retained the ability to confer resistance against methyl viologen but could no longer protect the cells against the effect of acriflavine and could only partially confer resistance against ethidium.
TABLE 1.
Growth phenotype of cells expressing Gly mutants For testing resistance to toxic compounds, E. coli DH5α cells transformed with the various mutants were grown overnight at 37 °C on solid media containing toxic compounds as described under “Experimental Procedures.” Growth level of the cells was estimated compared to cell growth of cells transformed with wild type EmrE as positive control and pT7-7 (empty vector) as negative control. The plus symbol is assigned to cells able to grow as well as wild type EmrE under the same conditions. The lack of growth under these conditions is defined as the minus sign. Growth levels between that of wild type EmrE and the empty vector-transformed cells is marked with a +/– sign. Very low, yet detectable resistance is marked as a +/- -.
|
Mutation
|
Growth in presence of substrate
|
||
|---|---|---|---|
| Methyl viologen | Ethidium bromide | Acriflavine | |
| A | |||
| G8C | + | + | +/– |
| G9C | + | + | + |
| G17C | – | – | – |
| G26C | + | +/– | +/– |
| G35C | + | + | + |
| G57C | + | +/– | +/– |
| G65C | + | +/– | – |
| G67C | – | – | – |
| G77C | + | + | + |
| G80C | + | + | +/– |
| G90C | – | – | – |
| G97C | – | – | – |
| B | |||
| G17A/C/P | – | – | – |
| G67A/C/P | – | – | – |
| G90A | + | +/– | – |
| G90C/P | – | – | – |
| G97A | +/- - | – | – |
| G97C/P | – | – | – |
Mutants at four positions, 17, 67, 90, and 97, lost all ability to confer in vivo resistance against all three tested EmrE substrates. To further explore the role of glycine in these positions where a cysteine replacement yielded nonactive proteins, we also replaced the glycines at positions 17, 67, 90, and 97 with either proline or alanine. Strikingly, in two positions, 17 and 67, even the addition of a methyl group as side chain abolished all in vivo activity of the protein. An alanine replacement at position 90 allowed the mutated protein to retain the ability to confer resistance against methyl viologen and to a lower degree also against ethidium, but not to acriflavine. An alanine replacement at position 97 abolished almost all activity, but the mutant could still confer an extremely low, yet above background, resistance against methyl viologen only.
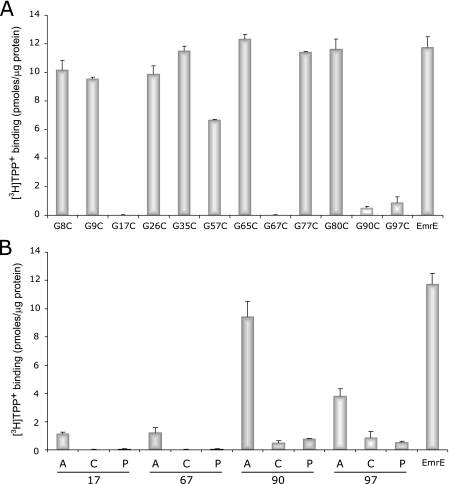
TPP+ Binding and Methyl Viologen Uptake Activity of Glycine Substitution Mutants—To evaluate expression levels and further study their function in more detail, all mutant proteins were purified by Ni-NTA affinity chromatography and assayed for their ability to bind TPP+, a high affinity substrate of EmrE (4). All the mutants tested, including those who failed to confer any resistance, expressed to similar levels (not shown). As expected, all the mutants that supported cell growth in the presence of toxicants bound [3H]TPP+ to considerable levels compared with that of the EmrE control (Fig. 2A). The mutant G65C, displayed a partial resistance profile as compared with the wild type protein, bound TPP+ at the same levels as EmrE. The mutant G57C, which conferred at least partial resistance against all tested toxic compounds in vivo, displayed a reduced TPP+ binding activity (about 50% compared with EmrE).
FIGURE 2.
TPP+ binding activity of the glycine substitution mutants. The results of [3H]TPP+ binding of the various glycine substitution mutants are presented in histograms. The mutants are divided into two groups: cysteine replacements of all 12 native glycine of EmrE (A) and various replacements of glycine at positions 17, 67, and 97 (B). The [3H]TPP+ binding assay was performed as described under “Experimental Procedures.”
Correspondingly to the in vivo phenotype results, the cysteine substitution mutants at positions 17, 67, 90, and 97 displayed significantly impaired binding activity. Whereas mutants G17C and G67C failed to show any measurable binding activity, G90C and G97C displayed a very low, yet detectable, level of TPP+ binding. When comparing the ability of the different substitutions at positions 17, 67, 90, and 97 to bind TPP+, a general tendency is observed where the alanine replacements allow for a better binding activity than the other replacements (Fig. 2B). In the case of G90A, binding is quite robust accounting for ∼80% of the wild type, G97A binds ∼30%, and G17A and G67A bind a small but significant ∼10% compared with wild type. In all the positions the cysteine and proline replacements yield proteins that have either no measurable binding activity (Gly-17 and Gly-67) or a very weak one, slightly above background (Gly-90 and Gly-97) (Fig. 2B).
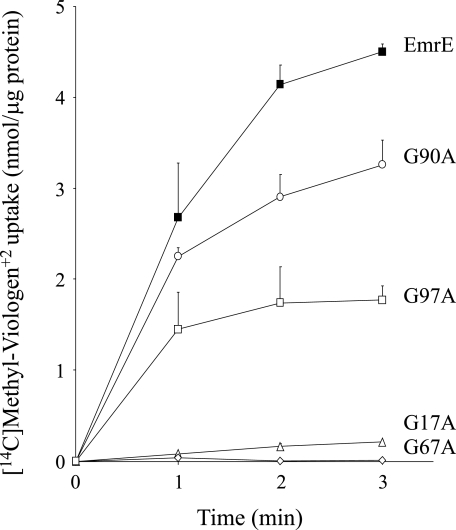
To further analyze the effect of the replacements on the activity of the proteins, all alanine substitution mutants, as well as EmrE, were solubilized, purified, and reconstituted into proteoliposomes and their transport activity was measured. The ΔpH-driven [14C]methyl viologen uptake into the proteoliposomes is presented in Fig. 3. The same tendency observed in the whole cell assay still applies, namely G90A displays a substantial uptake activity, whereas G97A displays a lower, yet significant uptake activity. In contrast, a methyl side chain addition at positions 17 and 67 abolishes uptake activity.
FIGURE 3.
Transport activity of alanine replacement mutants. ΔpH-driven [14C]methyl viologen uptake was assayed in proteoliposomes reconstituted with EmrE (▪), G17A (▵), G67A (⋄), G90A (○), and G97A (□), as described under “Experimental Procedures.”
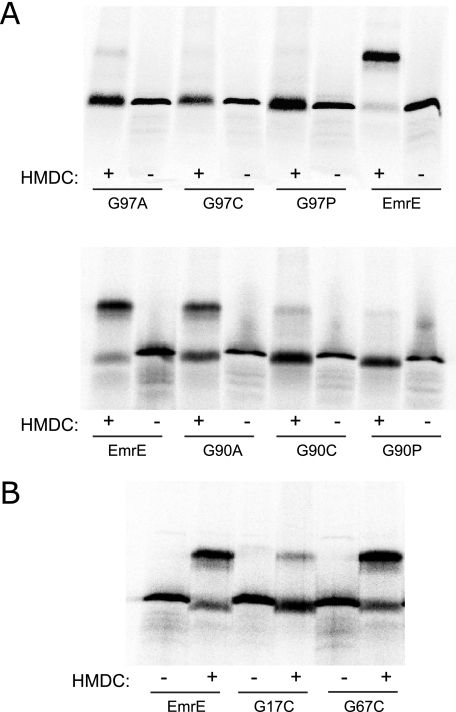
HMDC Cross-linking and Pull-down Experiments—One of the roles of glycine residues in conserved positions in membrane proteins has to do with facilitating helix packing and enabling oligomerization. We, therefore, assessed the dimerization status of the inactive glycine mutants. For this purpose we used HMDC, an irreversible amine-reacting cross-linker. HMDC was shown to cross-link EmrE through a lysine residue at position 22, which is the only native lysine in EmrE and is located on the first loop of the protein (23). The rationale behind this cross-linking strategy was to always use the same residue rather than the mutated one to have a more normalized view of the effect of each mutation on the dimerization.
HMDC cross-linking was applied on the inactive mutants G17C, G67C, G90C, and G97C and the results are presented in Fig. 4, A and B. Interestingly, the mutants exhibit a diverse cross-linking pattern, which implies that although substitutions of those four glycine positions yield a shared phenotype, their role in EmrE is probably different. Whereas EmrE yielded high levels of cross-linking when subjected to HMDC, G97C showed no measurable cross-linking and G90C yielded only borderline levels of cross-linking. The cross-linking level of G67C was as high as that of EmrE. G17C showed a lower, yet reproducible, level of HMDC cross-linking. All the substitutions at positions 90 and 97 generated mutants that exhibit extremely low (G97A, G90C, and G90P) or no cross-linking at all (G97C and G97P), except for the alanine replacement in position 90, the only mutant that exhibited a very significant cross-linking level (Fig. 4A). This is also a mutant that displays almost wild type levels of transport activity.
FIGURE 4.
HMDC cross-linking of inactive glycine mutants. Membranes prepared from cells expressing EmrE glycine mutants selectively labeled with [35S]methionine were solubilized in 0.8% DDM-Na buffer, treated with HMDC as described under “Experimental Procedures,” and separated by SDS-PAGE. The lower bands (∼ 13 kDa) correspond to the monomer and the higher bands (∼26 kDa) to the cross-linked dimer. Radioactive protein was visualized using a FLA-3000 PhosphorImager.
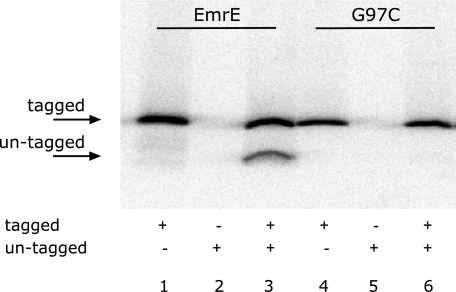
To further support the above results we tested oligomerization using a pull-down assay where one of the monomers (bait) was tagged with His6 residues and the other (prey) was not (30). [35S]Met-labeled tagged and un-tagged proteins (either EmrE-His and EmrE, or G97C-His and G97C) were mixed and treated with a short heat treatment that reversibly dissociates the oligomer. The protein mixture was immobilized on Ni-NTA beads, washed, eluted, separated on SDS-PAGE, and analyzed for radioactivity. Binding to Ni-NTA beads is completely dependent on the presence of a His-tagged protein (Fig. 5, lanes 1 and 2, 4 and 5). Because only His-tagged monomers can bind to the Ni-NTA beads, only the untagged monomers that interact with tagged ones were pulled down and detected after separation by SDS-PAGE. As previously shown, tagged EmrE monomers interact with untagged EmrE monomers and pull them down (Fig. 5, lane 3). However, no pull-down was observed with the G97C mutants. Tagged G97C monomers did not pull down untagged G97C monomers although the latter were in excess in the reaction (Fig. 5, lane 6).
FIGURE 5.
Pull-down of untagged EmrE by tagged EmrE-His. Membranes prepared from cells expressing tagged or untagged EmrE glycine mutants selectively labeled with [35S]methionine were solubilized in 0.8% DDM-Na buffer, mixed, and heat treated as described under “Experimental Procedures.” The tagged and untagged mixed proteins were then immobilized on Ni-NTA beads, washed, eluted, and separated by SDS-PAGE. Radioactive protein was visualized using a FLA-3000 PhosphorImager.
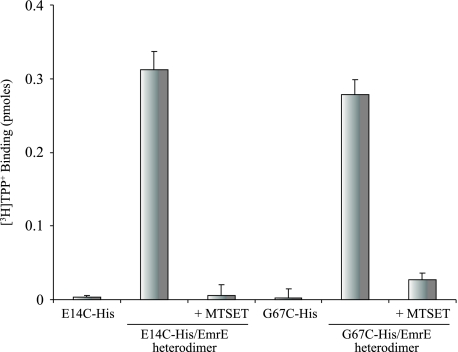
Inhibition of Heterodimer Activity by a Thiosulfonate Reagent—The study of essential residues is by nature limited because replacement mutants do not display any activity. In the case of an oligomeric protein such as EmrE we have developed an approach to circumvent this problem by generating heterodimers between the inactive His-tagged mutant and untagged wild type EmrE. This results in functional complementation yielding a protein with partial but robust activity. Glu-14 is one of the essential residues in the binding domain. When the tagged inactive E14C mutant is mixed with an untagged wild type protein, the heterodimer binds ligand with a lower affinity (Fig. 6 and Ref. 30). The thiosulfonate derivative MTSET that inserts a positive charge inhibits the activity implying that a positive charge in one monomer at position 14 has a deleterious effect on the activity of the dimer even though in the other monomer at the equivalent position there is a carboxyl. Also the inactive G67C interacts productively with a wild type protein yielding a functional heterodimer (Fig. 6). The other Cys substitutions (G17C, G90C, and G97C) failed to show functional complementation, most likely because of their inability to form heterodimers (data not shown). Similar to what was observed with the E14C/EmrE dimer, the activity of the G67C/EmrE heterodimer is inhibited by MTSET (Fig. 6) supporting the suggestion that Gly-67 is in the binding pocket or in its vicinity.
FIGURE 6.
Inhibition of heterodimer activity by a thiosulfonate reagent. Membranes from cells expressing CLA (untagged Cys-less EmrE) were solubilized with 0.8% DDM/Na buffer and mixed at 80 °C with solubilized membranes of E14C or G67C (∼100 ng of tagged protein per assay), inactive His-tagged mutants. The extract was centrifuged for 5 min at 14,000 × g to discard precipitates and allowed to bind to Ni-NTA beads. MTSET at a final concentration of 1 mm was added for 10 min and then substrate binding activity was tested as described above (10 nm [3H]TPP+ in a 200-μl reaction).
DISCUSSION
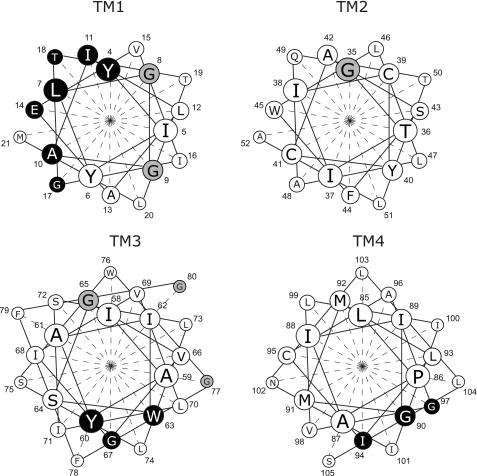
Previous studies have identified residues in EmrE essential for activity grouped in two distinct clusters in TM1 and TM3 (1, 6–8) (Fig. 7). The major cluster in TM1 includes Glu-14, the only membrane-embedded charged residue in EmrE, which has been shown to play a pivotal role in both substrate binding and proton coupling (1, 6). Mutations of residues in the same face of the helix of Glu-14 have effects on the catalytic activity of the protein, whereas activity is not affected when residues on the other face of the helix are mutated (6, 31). Gly-17, one of the four essential glycine residues, is located one helix turn from and on the same face of the helix as Glu-14. A second cluster of functionally important residues is located in helix 3 and is comprised of the highly conserved aromatics Tyr-60 and Trp-63 (7, 8). Kinetic studies of the substrate-induced quenching of the Trp-63 fluorescence strongly support the contention that this residue is directly involved in substrate binding (7, 32). Gly-67 is located one helix turn from Trp-63, in the same face of the helix as the aromatic cluster (Fig. 7). Our present results identifying Gly-17 and Gly-67 as irreplaceable reinforce the functional importance of the previously defined clusters. Gly-65 is located on the opposite face from the aromatic cluster and can be replaced even though it is highly conserved. The functionally important residues in TM1 and TM3 are also evolutionary conserved (Fig. 1B) and the pattern of conservation already implies a helical pattern in which one face of the helix is highly conserved.
FIGURE 7.
Clusters of functionally important residues in EmrE. Helical wheel projection of the four transmembrane helices of EmrE. Residues that once mutated yield transporters with severely impaired, or no activity at all, are in black. Glycine residues that once mutated still yield an active transporter are in gray.
Replacement of essential residues such as Gly-17 and Gly-67 with Cys generates inactive proteins and limits their further characterization. We have developed an approach to circumvent this problem by generating heterodimers between inactive mutants and wild type. In many cases, this results in functional complementation yielding a protein with partial but robust activity. Our previous work with E14C supports the notion that the functional properties of the dimer are a result of the interaction between individual monomers. In this work the functional heterodimer was generated so that it contains Cys residues only in the inactive monomer. When this species was challenged with a sulfhydryl reagent, which can react only with the inactive subunit, a dose-dependent inhibition was observed that was due to a decrease in the affinity to TPP+ (30). Here we used the thiosulfonate MTSET and we showed that it completely inhibits the activity of a G67C/EmrE dimer. We have previously shown that G17C has a very low affinity (Kd at least 2 orders of magnitude higher than wild type) and indeed the substrate protects very poorly (25). In addition Gly-17 did not show functional complementation when mixed with wild type protein (25). This may be due to a decreased dimerization as a result of the mutation or incorrect folding of the heterodimer but may also hint that the role of Gly-17 is related to the flexibility of the protein rather than substrate binding.
A third cluster identified here is located in TM4 and is comprised of two positions identified in the present study, namely Gly-90 and Gly-97 (Fig. 7). Gly-97 is highly conserved, whereas Gly-90 is less conserved (about ∼41%) (Fig. 1B). At positions 93 and 94 large hydrophobic amino acids are found in most of the SMRs. Interestingly, replacement of Ile-94 with cysteine is tolerated and the mutant still retains part of its activity. However, a replacement with a small side chain such as alanine is not tolerated and yields an inactive mutant that can no longer confer resistance to any of the toxicants tested and do not bind [3H]TPP+ (data not shown). It has been previously shown that the functional unit of EmrE is a dimer and here we identify one motif involved in this dimerization and demonstrate a clear correlation between dimerization status and functionality. The structural information available for this protein suggests that the TM4s are directly involved in the interdimeric contacts. However, the two earlier x-ray crystallography papers for the protein have been retracted (33). Their publication sparked a controversy regarding the relative topology of the protomers in the functional dimer and this controversy is still ongoing (34–36). The claim for an antiparallel topology was supported by a reinterpretation of the electron density maps of two-dimensional crystals of EmrE that showed that parts of the structure are related by quasisymmetry (37). A Cα model of the transmembrane region was constructed by considering the evolutionary conservation pattern of each helix (34) and was supported by a re-evaluation of the data published in the retracted papers (38). Our own work demonstrates that dimers with parallel topology are functional and the functionality of antiparallel dimers remains to be established (36, 39, 40). Therefore, we cannot use the available structural information to learn whether TM4s are indeed directly involved in the interdimeric interface. Peptides that correspond to TM4 of the Halobacterium salinarum protein Hsmr, an homologue of EmrE, have been shown to interact strongly in vitro (41). Our early cross-linking analysis suggests proximity of TM4s from the two monomers (23). We conclude that additional experimentation is needed to resolve the subject of whether the residues in this motif directly participate in the dimerization interface or whether they affect packing needed for dimerization.
Genomic scale analysis reveals that glycine is frequent in transmembrane helices of membrane proteins (9). Glycine is also considerably more prevalent in conserved positions relatively to its average occurrence in transmembrane helices (10). Furthermore, its occurrence in conserved positions in transporter families is twice as high (10). The GG7 motif was shown to be prevalent in putative transmembrane helices, especially in transporter/channel-like membrane proteins (10). The high frequency of glycine residues in transmembrane helices suggests a structural role, which is distinct from that in soluble proteins as suggested by studies on model peptides (43). The structural role of glycine residues in a GG4 motif has been well studied in monotopic membrane proteins. In these proteins, the GG4 motif is involved in protein dimerization through specific packing interactions (44). In tetraspanin proteins a GG7 motif very similar to the one found in EmrE, with two large hydrophobic residues in positions 3 and 4 has been shown to participate in helix packing (45). Here we present experimental evidence that supports a similar role in EmrE. Cross-linking results using the irreversible amine cross-linker HMDC show that upon mutation of glycines at either position 90 or 97, cross-linking of EmrE monomers is practically abolished. The only mutant in these positions that allows significant cross-linking is G90A, and this is in good correlation with the results showing that this mutant is the only one that retained partial activity. The cross-linking results are supported by the pull-down experiments that do not involve chemical modification of the protein and also show failure to dimerize in the mutant G97C. According to our results Gly-90 and Gly-97 play a role in facilitating helix-helix interaction and mediating dimer formation of EmrE. In addition, the large hydrophobic residues in position 93 and 94 define a motif (GXX(large hydrophobic)2XXG) that may suggest a knob-notch interaction in helical packing.
In enzymatic reactions, glycine has practically nil catalytic propensity (42). In membrane proteins involved in vectorial metabolism glycine residues have been shown to play several roles, both structural and functional, some of them already described in the Introduction. Our results described in this paper suggest that the four essential glycine residues identified in EmrE fulfill several roles. We speculate that Gly-17 and Gly-67 that are located in close vicinity to catalytically crucial residues may contribute to the binding pocket architecture, either by direct interaction, or simply by forming a cavity needed to prevent steric hindrance with the substrate molecule. In addition, they could be involved in conformational changes necessary for an alternate access of the binding sites to both sides of the membrane. On the other hand our results suggest that Gly-90 and Gly-97 play a role in helix-helix packing and, as a result, changes in these positions prevent interactions between EmrE monomers that are essential for function.
This work was supported in part by National Institute of Health Grant NS16708 and Israel Science Foundation Grant 119/04. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TM, transmembrane; DDM, n-dodecyl-β-maltoside; TPP+, tetraphenylphosphonium; HMDC, hexamethylene diisocyanate; Ni-NTA, nickel-nitrilotriacetic acid; MTSET, 2-(trimethylammonium)ethylmethanethiosulfonate bromide; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine; SMR, small multidrug resistance.
References
- 1.Yerushalmi, H., Lebendiker, M., and Schuldiner, S. (1995) J. Biol. Chem. 270 6856-6863 [DOI] [PubMed] [Google Scholar]
- 2.Schuldiner, S., Granot, D., Mordoch, S. S., Ninio, S., Rotem, D., Soskin, M., Tate, C. G., and Yerushalmi, H. (2001) News Physiol. Sci. 16 130-134 [DOI] [PubMed] [Google Scholar]
- 3.Schuldiner, S., Granot, D., Steiner, S., Ninio, S., Rotem, D., Soskin, M., and Yerushalmi, H. (2001) J. Mol. Microbiol. Biotechnol. 3 155-162 [PubMed] [Google Scholar]
- 4.Muth, T. R., and Schuldiner, S. (2000) EMBO J. 19 234-240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yerushalmi, H., and Schuldiner, S. (2000) J. Biol. Chem. 275 5264-5269 [DOI] [PubMed] [Google Scholar]
- 6.Gutman, N., Steiner-Mordoch, S., and Schuldiner, S. (2003) J. Biol. Chem. 278 16082-16087 [DOI] [PubMed] [Google Scholar]
- 7.Elbaz, Y., Tayer, N., Steinfels, E., Steiner-Mordoch, S., and Schuldiner, S. (2005) Biochemistry 44 7369-7377 [DOI] [PubMed] [Google Scholar]
- 8.Rotem, D., Steiner-Mordoch, S., and Schuldiner, S. (2006) J. Biol. Chem. 281 18715-18722 [DOI] [PubMed] [Google Scholar]
- 9.Senes, A., Gerstein, M., and Engelman, D. M. (2000) J. Mol. Biol. 296 921-936 [DOI] [PubMed] [Google Scholar]
- 10.Liu, Y., Engelman, D. M., and Gerstein, M. (2002) Genome Biol. 3 research0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinglass, A. B., Smirnova, I. N., and Kaback, H. R. (2001) Biochemistry 40 769-776 [DOI] [PubMed] [Google Scholar]
- 12.Jiang, Y., Lee, A., Chen, J., Cadene, M., Chait, B. T., and MacKinnon, R. (2002) Nature 417 523-526 [DOI] [PubMed] [Google Scholar]
- 13.Zhou, Y., and Kanner, B. I. (2005) J. Biol. Chem. 280 20316-20324 [DOI] [PubMed] [Google Scholar]
- 14.Han, M., Lin, S. W., Smith, S. O., and Sakmar, T. P. (1996) J. Biol. Chem. 271 32330-32336 [DOI] [PubMed] [Google Scholar]
- 15.Han, M., Groesbeek, M., Sakmar, T. P., and Smith, S. O. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 13442-13447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russ, W. P., and Engelman, D. M. (2000) J. Mol. Biol. 296 911-919 [DOI] [PubMed] [Google Scholar]
- 17.Arbely, E., Granot, Z., Kass, I., Orly, J., and Arkin, I. T. (2006) Biochemistry 45 11349-11356 [DOI] [PubMed] [Google Scholar]
- 18.Kim, S., Jeon, T. J., Oberai, A., Yang, D., Schmidt, J. J., and Bowie, J. U. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 14278-14283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, S., Chamberlain, A. K., and Bowie, J. U. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 5988-5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldberg, E. B., Arbel, T., Chen, J., Karpel, R., Mackie, G. A., Schuldiner, S., and Padan, E. (1987) Proc. Natl. Acad. Sci. U. S. A. 84 2615-2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabor, S., and Richardson, C. C. (1985) Proc. Natl. Acad. Sci. U. S. A. 82 1074-1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mordoch, S. S., Granot, D., Lebendiker, M., and Schuldiner, S. (1999) J. Biol. Chem. 274 19480-19486 [DOI] [PubMed] [Google Scholar]
- 23.Soskine, M., Steiner-Mordoch, S., and Schuldiner, S. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 12043-12048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider, T. D., and Stephens, R. M. (1990) Nucleic Acids Res. 18 6097-6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharoni, M., Steiner-Mordoch, S., and Schuldiner, S. (2005) J. Biol. Chem. 280 32849-32855 [DOI] [PubMed] [Google Scholar]
- 26.Ho, S. N., Hunt, H. D., Horton, R. M., Pullen, J. K., and Pease, L. R. (1989) Gene (Amst.) 77 51-59 [DOI] [PubMed] [Google Scholar]
- 27.Li, F., and Mullins, J. I. (2002) Methods Mol. Biol. 182 19-27 [DOI] [PubMed] [Google Scholar]
- 28.Yerushalmi, H., Mordoch, S. S., and Schuldiner, S. (2001) J. Biol. Chem. 276 12744-12748 [DOI] [PubMed] [Google Scholar]
- 29.Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004) Genome Res. 14 1188-1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rotem, D., Salman, N., and Schuldiner, S. (2001) J. Biol. Chem. 276 48243-48249 [DOI] [PubMed] [Google Scholar]
- 31.Ninio, S., and Schuldiner, S. (2003) J. Biol. Chem. 278 12000-12005 [DOI] [PubMed] [Google Scholar]
- 32.Adam, Y., Tayer, N., Rotem, D., Schreiber, G., and Schuldiner, S. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 17989-17994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang, G., Roth, C. B., Reyes, C. L., Pornillos, O., Chen, Y.-J., and Chen, A. P. (2006) Science 314 1875. [DOI] [PubMed] [Google Scholar]
- 34.Fleishman, S. J., Harrington, S. E., Enosh, A., Halperin, D., Tate, C. G., and Ben-Tal, N. (2006) J. Mol. Biol. 364 54-67 [DOI] [PubMed] [Google Scholar]
- 35.Rapp, M., Seppala, S., Granseth, E., and von Heijne, G. (2007) Science 315 1282-1284 [DOI] [PubMed] [Google Scholar]
- 36.Soskine, M., Mark, S., Tayer, N., Mizrachi, R., and Schuldiner, S. (2006) J. Biol. Chem. 281 36205-36212 [DOI] [PubMed] [Google Scholar]
- 37.Ubarretxena-Belandia, I., Baldwin, J. M., Schuldiner, S., and Tate, C. G. (2003) EMBO J. 22 6175-6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen, Y. J., Pornillos, O., Lieu, S., Ma, C., Chen, A. P., and Chang, G. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 18999-19004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schuldiner, S. (2007) Trends Biochem. Sci. 32 252-258 [DOI] [PubMed] [Google Scholar]
- 40.Steiner-Mordoch, S., Soskine, M., Solomon, D., Rotem, D., Gold, A., Yechieli, M., Adam, Y., and Schuldiner, S. (2008) EMBO J. 27 17-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rath, A., Melnyk, R. A., and Deber, C. M. (2006) J. Biol. Chem. 281 15546-15553 [DOI] [PubMed] [Google Scholar]
- 42.Holliday, G. L., Almonacid, D. E., Mitchell, J. B., and Thornton, J. M. (2007) J. Mol. Biol. 372 1261-1277 [DOI] [PubMed] [Google Scholar]
- 43.Li, S. C., and Deber, C. M. (1992) FEBS Lett. 311 217-220 [DOI] [PubMed] [Google Scholar]
- 44.Lemmon, M. A., and Engelman, D. M. (1994) Q. Rev. Biophys. 27 157-218 [DOI] [PubMed] [Google Scholar]
- 45.Kovalenko, O. V., Metcalf, D. G., DeGrado, W. F., and Hemler, M. E. (2005) BMC Struct. Biol. 5 11. [DOI] [PMC free article] [PubMed] [Google Scholar]