Abstract
Clostridium perfringens is a Gram-positive bacterium responsible for bacteremia, gas gangrene, and occasionally food poisoning. Its genome encodes three sialidases, nanH, nanI, and nanJ, that are involved in the removal of sialic acids from a variety of glycoconjugates and that play a role in bacterial nutrition and pathogenesis. Recent studies on trypanosomal (trans-) sialidases have suggested that catalysis in all sialidases may proceed via a covalent intermediate similar to that of other retaining glycosidases. Here we provide further evidence to support this suggestion by reporting the 0.97Å resolution atomic structure of the catalytic domain of the C. perfringens NanI sialidase, and complexes with its substrate sialic acid (N-acetylneuramic acid) also to 0.97Å resolution, with a transition-state analogue (2-deoxy-2,3-dehydro-N-acetylneuraminic acid) to 1.5Å resolution, and with a covalent intermediate formed using a fluorinated sialic acid analogue to 1.2Å resolution. Together, these structures provide high resolution snapshots along the catalytic pathway. The crystal structures suggested that NanI is able to hydrate 2-deoxy-2,3-dehydro-N-acetylneuraminic acid to N-acetylneuramic acid. This was confirmed by NMR, and a mechanism for this activity is suggested.
Clostridium perfringens is a Gram-positive anaerobic bacterium that causes life-threatening gas gangrene and enterotoxemia in humans. C. perfringens infections are characterized by the release of large amounts of toxins and enzymes that can cause massive destruction of the host tissue, putting the organism into the category of flesh-eating microbes (1). Exo-sialidases are among the virulence factors, two of which (nanH and nanI) had been characterized prior to the sequencing of the complete C. perfringens genome, which revealed a third, nanJ (2). The nanH gene product is a 43-kDa sialidase (3) that is not secreted, whereas the nanI gene product is a 77-kDa sialidase (4) that is secreted. These two sialidases have been extensively characterized and shown to exhibit very different kinetic and biochemical properties (5). The nanJ gene product has yet to be characterized but is predicted to form a 129-kDa sialidase as it contains the conserved catalytic and “bacterial neuraminidase repeat” signatures of a sialidase (6).
Sialidases, or neuraminidases, catalyze the removal of terminal sialic acids from a variety of glycoconjugates and play an important role in pathogenesis, bacterial nutrition, and cellular interactions. Crystal structures of a growing number of exo-sialidases are available from bacteria (7–9), viruses (10–12), trypanosomes (13, 14), leech (15), and man (16). All sialidases share the same six-bladed β-propeller fold for their catalytic domains, with conservation of key catalytic amino acids (17). The nonviral sialidases also have conserved bacterial neuraminidase repeats or Asp boxes ((S/T)XD(X)GXT(W/F)) occurring between one and five times along the sequence. The bacterial neuraminidase repeats occur at topologically identical positions in the β-propeller fold, remote from the active site, but any function beyond dictating a structural fold is unknown. Many sialidases possess domains in addition to the catalytic domain, placed upstream, downstream, or even inserted within the β-propeller domain; the Vibrio cholerae sialidase has two lectin domains flanking the catalytic domain, one of which binds sialic acid (18); Micromonospora viridifaciens sialidase has a galactose-binding domain C-terminal to the catalytic domain and is positioned above the active site (20); the leech sialidase has a lectin-like domain N-terminal to the catalytic domain (15), and the trypanosome (trans-) sialidases have a lectin-like domain C-terminal to the catalytic domain (13, 14). It has been suggested that the presence of these carbohydrate-binding modules increases the catalytic efficiency of the sialidases, particularly in the presence of polysaccharide substrates (21). Many glycoside hydrolases have additional carbohydrate-binding modules, particularly those involved in the degradation of insoluble polysaccharides such as cellulose and starch, and these carbohydrate-binding modules show a great diversity in ligand recognition and folds (22).
Sialidases hydrolyze sialic acids from glycoconjugates with retention of configuration at the anomeric center (23). The early mechanistic and structural studies on the influenza virus sialidase led to the proposal that the mechanism involved distortion of the sialic acid toward an oxocarbenium ion transition state. Structures of the influenza sialidase complexed with the inhibitor 2-deoxy-2,3-dehydro-N-acetylneuraminic acid (Neu5Ac2en),4 a putative transition-state analogue, led to the theory that the positive charge on the transition state could be stabilized by the strictly conserved tyrosine residue sitting directly beneath the C-2 atom of this compound (11). Subsequent complexes of bacterial and trypanosomal sialidases have supported this idea, with the conserved tyrosine sitting close to the C-2 atom of Neu5Ac2en, with a mean distance of ∼2.5 Å (PDB codes 1EUS, 1KIT, and 1MZ6) (13, 24). The proposed mechanism was one in which a conserved aspartic acid acted as a general acid catalyst, facilitating the acid-catalyzed cleavage of the glycosidic bond between the sialic acid and the sub-terminal aglycon. The subsequent oxocarbenium transition state adopts a half-chair configuration, similar to that seen in Neu5Ac2en. In sialidases, a water molecule then acts as the catalytic nucleophile, attacking the positive charge on the C-2 atom, and is facilitated by base catalysis from the conserved aspartic acid, which regains its proton (11, 23).
This mechanism is different from that proposed for other retaining glycosidases, which have been shown to operate via a covalent glycosyl-enzyme intermediate, involving the action of a protein carboxylate nucleophile and an acid/base catalyst (25). The structures of sialidases showed that no carboxylate group was favorably positioned to form a covalent sialyl-enzyme intermediate during catalysis. It was this observation that prompted much of the earlier research to favor an ionic rather than covalent stabilization of the reaction intermediate (11, 23, 27). Recent kinetic work carried out on the Trypanosomal cruzi trans-sialidase using a novel α2,3-difluoro-N-acetylneuraminic acid led to the trapping of a catalytically competent sialyl-enzyme covalent intermediate on the conserved tyrosine residue (28). This led to the proposal that the conserved catalytic tyrosine was in fact the nucleophile for this family of glycosidases. This kinetic study was further supported by an extensive structural investigation of the T. cruzi trans-sialidase revealing snapshots of the Michaelis complex, the covalent sialyl-enzyme intermediate (29), and the complex with Neu5Ac2en (14). It is argued that the choice of a tyrosine as the nucleophile, rather than a carboxylate-containing amino acid, is a consequence of the unfavorable electrostatic interaction that would occur with the carboxylate group of sialic acids (28). An alternative view is that tyrosine is a less reactive leaving group required to compensate for the higher reactivity of sialosides compared with other glycosides (30). The recent structural and kinetic characterization of covalent intermediates with the Trypanosoma rangeli sialidase adds further weight to the idea that all exo-sialidases operate through a similar mechanism involving the transient formation of a covalently sialylated enzyme (31).
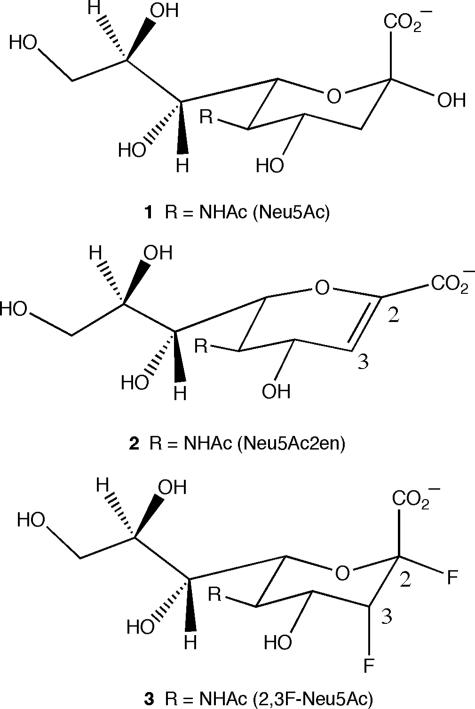
In this study, we report the structure of the catalytic domain (residues 243–694) of the C. perfringens NanI sialidase in its ligand free form and in complex with α-sialic acid (Neu5Ac, Fig. 1, structure 1) at 0.97 Å resolution. Unusually, the Neu5Ac complex was the result of soaking native crystals with Neu5Ac2en (Fig. 1, structure 2) at pH 7, suggesting hydration of the inhibitor by the enzyme, an observation supported by NMR. A complex with the transition-state analogue Neu5Ac2en at 1.5Å resolution was obtained at pH 5. Finally, a covalent intermediate complex at 1.2 Å resolution was obtained by co-crystallizing NanI with 2,3-difluoro-N-acetylneuraminic acid (2,3F-Neu5Ac, Fig. 1, structure 3). These structures confirm the nucleophilic role played by the active site tyrosine and confirm the suggestion that catalysis by sialidases occurs via a very similar mechanism to that of other retaining glycosidases.
FIGURE 1.
Ligands used in this study. Structure 1, Neu5Ac; structure 2, Neu5Ac2en; and structure 3, 2,3-difluoro-N-acetylneuramic acid (2,3F-Neu5Ac).
EXPERIMENTAL PROCEDURES
Data Collection—The 50-kDa catalytic domain (residues 243–694) of the C. perfringens NanI sialidase was subcloned, expressed, crystallized, and cryoprotected as described previously (32). All data were collected on station ID14-EH1 at the ESRF, Grenoble, France, at 100 K and at a wavelength of 0.934 Å. Diffraction data for the ligand free and Neu5Ac complex extended beyond 0.92 Å, but for considerations of completeness, data were limited to 0.97 Å (Table 1). The complex of NanI with Neu5Ac was obtained from a crystal soaked for 30 min in 10 mm Neu5Ac2en in 20% PEG 3350, 0.2 m KNO3, pH 7 (buffer A). A Neu5Ac2en complex was obtained by transferring crystals grown at pH 7 into a solution containing 10 mm Neu5Ac2en in 22% PEG 3350, 0.22 m KNO3, 10 mm sodium acetate, pH 5. The ligand free and Neu5Ac2en-soaked crystals belong to the orthorhombic space group P212121 with one monomer in the asymmetric unit. The covalent complex was obtained by soaking a crystal in 5 mm 2,3-difluoro-N-acetylneuramic acid in buffer A for 2 min, prior to cryoprotection, flash-cryocooling, and data collection in-house. This complex resulted in a breakdown of the orthorhombic symmetry to the subgroup P21 with similar unit cell dimensions, but with two monomers in the asymmetric unit. All crystals were cryoprotected by transfer for a few minutes first to a 12.5% PEG 400 solution and then a 25% PEG 400 solution in appropriate mother liquor. All data were processed with MOSFLM, scaled using SCALA and converted to structure factors with TRUNCATE, all of which are part of the CCP4 software suite (33).
TABLE 1.
Data collection and refinement statistics. Numbers in parentheses refer to the highest resolution shell. Rmerge = Σhkl Σi|Ihkl,i – 〈Ihkl〉|/Σhkl〈Ihkl〉; Rcryst and Rfree = (Σ||Fo| – |Fc||)/(Σ|Fo|); Rfactor and Rfree = (Σ||Fo| – |Fc||)/(Σ|Fo|).
| Crystal | Ligand free | Neu5Ac | Neu5Ac2en | Covalent intermediate |
|---|---|---|---|---|
| Data collection | ||||
| Space group and cell dimensions | P212121 | P212121 | P212121 | P21 |
| a = 96.9 Å, b = 69.0 Å, c = 72.8 Å | a = 97.0 Å, b = 69.4 Å, c = 72.7 Å | a = 98.9Å, b = 69.4Å, c = 71.0Å | a = 69.6Å, b = 97.4Å, c = 72.6Å, β = 91.0° | |
| X-ray source | ESRF ID14-1 | ESRF ID14-1 | ESRF ID14-2 | ESRF ID14-2 |
| Wavelength | 0.934 Å | 0.934 Å | 0.934 | 0.934 |
| Resolution | 20.0-0.97 Å (1.02-0.97 Å) | 20.0-0.97 Å (1.03-0.97 Å) | 28.4-1.50 (1.54-1.50) | 72.5-1.20 (1.27-1.20) |
| Unique reflections | 281,317 | 264,973 | 73,189 | 278,102 |
| Completeness | 98% (95%) | 92% (87%) | 93 (89) | 92 (85) |
| Redundancy | 4.6 (3.1) | 3.8 (3.1) | 3.4 (2.1) | 2.4 (2.2) |
| Rmerge | 0.072 (0.329) | 0.095 (0.186) | 0.055 (0.285) | 0.042 (0.304) |
| I/σI | 15.6 (3.6) | 11.8 (5.0) | 15.5 (3.6) | 11.7 (3.2) |
| Refinement | SHELX | SHELX | REFMAC | REFMAC |
| Reflections used | 276,976 | 264,777 | 69,514 | 272,435 |
| No. of protein atoms | 3,575 | 3,575 | 3,575 | 7,360 |
| No. of water molecules | 935 | 484 | 396 | 1661 |
| Rfactor | 0.115 | 0.113 | .162 | .150 |
| Rfree | 0.126 | 0.130 | .185 | .172 |
| r.m.s. deviation bond lengths | 0.017 Å | 0.016 Å | .018 Å | 0.007 Å |
| r.m.s. deviation bond angles | 2.048° | 2.086° | 1.714° | 1.244° |
| Average B-factors (Å2) | ||||
| All atoms | 8.9 | 8.3 | 13.1 | 12.4/13.9 |
| Ligand atoms | 10.4 | 10.2 | 9.8 | |
| Ramachandran outliers | 0.5% | 0.5% | 0.5 | 0.5 |
| Ramachandran favored | 96.9% | 96.9% | 96.4 | 96.4 |
| PDB code | 2vk5 | 2bf6 | 2vk6 | 2vk7 |
Structure Solution—A molecular replacement solution was readily obtained using CNS (34) with a pruned model of the sialidase from the leech (Macrobdella decora) (PDB code 1sll) as a search model (32). The leech sialidase shares 35% overall sequence identity with NanI and 37% identity within the catalytic domain. The molecular replacement search model consisted of just the β-propeller domain of the leech sialidase minus the irregular β-stranded domain that is inserted between the second and third strands of the second blade of the propeller. CNS was used to rigid body refine the molecular replacement solution using all data to 2.0 Å. ARP/wARP (35) was then used to extend the phases of the model and to build in the missing amino acids using data to 1.6 Å. This procedure built 445 of a possible 451 amino acids, and side chains were built with a 98% confidence level. The starting R and Rfree were 0.42 and 0.44, respectively, and the final R and Rfree were 0.16 and 0.19, respectively.
Refinement—SHELXL (36) was employed to carry out the refinement at 0.97 Å resolution of the structure obtained from the pH 7 Neu5Ac2en-soaked crystal. In total, 15 cycles of conjugate gradient least squares refinement were carried out between rounds of model building with the O program (37). Starting with data to 1.6 Å, one cycle of refinement and one round of solvent divining with SHELXWAT (36) produced 400 waters, and data to 0.97 Å were then included, followed by two rounds of isotropic temperature factor refinement. Anisotropic temperature factor refinement for two cycles lead to a drop of 2% in Rfree and was followed by a second round of solvent divining, and four further rounds of refinement/model building before hydrogens were added in cycle 13, which gave a further 1.3% drop in Rfree. The final round of refinement was used to calculate the standard uncertainties, or estimated standard deviations, of the atomic positions using blocked matrix least squares refinement. The difference maps clearly showed not Neu5Ac2en but Neu5Ac bound in the active site. The ligand free structure was refined in a similar manner with SHELXL using the Neu5Ac complex structure as the starting point. The 1.5 Å Neu5Ac2en complex and 1.2 Å covalent intermediate complex were refined using REFMAC 5.2.0019 from the CCP4 suite (33). The covalent bond between C-2 and Tyr655 was added as a dictionary restraint of length 1.4 Å (31). Refinement of the covalent complex led to a stretching of the covalent bond to 1.7 Å, and negative difference electron density at the C-2 and F-3 positions. In addition, electron density around the C-1–C-2 bond suggested the possibility of Neu5Ac2en being present. Accordingly, Neu5Ac2en and the covalent intermediate were refined each at 50% occupancy resulting in no unexplained difference electron density and a covalent link of 1.41 Å. Refinement statistics, Ramachandran plot analysis of the final structures from MolProbity (38), and PDB codes for deposited atomic coordinates are given in Table 1.
NMR—The hydration of Neu5Ac2en to form Neu5Ac was monitored using 1H NMR spectroscopy. Two solutions of a total volume of 800 μl were prepared for NMR analysis as follows: 1) the NanI enzyme reaction containing 61 μl of NanI (45 mg/ml), 40 μl of Neu5Ac2en (200 mm) in D2O, and 699 μl of 50 mm imidazole buffer in D2O, pH 7; and 2) a control experiment containing 40 μl of Neu5Ac2en (200 mm) in D2O, and 760 μl of 50 mm imidazole buffer in D2O, pH 7. 1H NMR spectra were acquired on a 600-MHz Bruker Avance spectrometer equipped with a cryoprobe at 285 K. Spectra were acquired with 128 scans and a relaxation delay of 2 s, every 24 h for 10 days after the addition of NanI to the Neu5Ac2en reaction mixture. Attenuation of the residual water signal was achieved by continuous low power irradiation of the water signal during the relaxation delay.
RESULTS
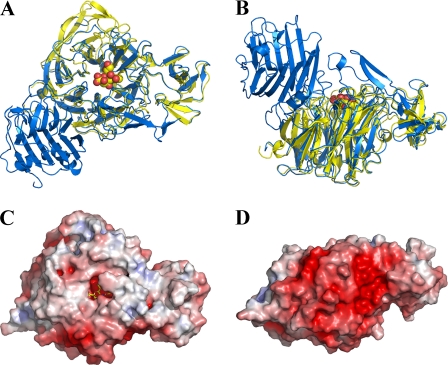
Overall Structure—The structure of the catalytic domain of NanI is described here to a resolution of 0.97 Å in its ligand free form. The NanI structure folds into two distinct domains as follows: a canonical, six-bladed β-propeller catalytic sialidase domain formed by residues 243–359 and 429–691, and a small β-barrel domain formed by residues 360–428 (Fig. 2, A and B). The overall r.m.s. deviation between NanI and the leech sialidase (PDB code 1sll), with which NanI shares 35% sequence identity, is 1.22 Å for 400 C-α positions considered equivalent. The small β-barrel domain in NanI, which was excluded from the molecular replacement model, shows a striking structural similarity to the equivalent domain in the leech sialidase. This domain is considerably shorter in NanI, 68 amino acids compared with 100 in the leech sialidase, with a sequence identity of 33%, and is missing an extended hairpin loop. The r.m.s. deviation for 63 C-α positions considered equivalent is 1.43 Å.
FIGURE 2.
Overall structure of the NanI sialidase. A and B represent orthogonal views of a comparison of the fold of NanI (yellow) with the leech sialidase (blue). Sialic acid is drawn as spheres to locate the active site. C and D show a surface representation of NanI, in the same orientations as above, colored according to electrostatic potential from -7 kT/e to +7 kT/e, calculated using APBS (26).
The electrostatic potential on the surface of NanI shows a very distinct asymmetry, typical of secreted bacterial sialidases, with the surface remote from the active site carrying a negative charge that may serve to orient the protein toward its negatively charged glycoconjugate substrates (Fig. 2, C and D). The protein contains two potential calcium ions coordinated within the β-propeller fold by six and seven oxygen atoms, respectively. One of the calcium sites is in a very similar location to a key calcium-binding site in the sialidase from Vibriocholerae, a calcium-dependent sialidase (8). The calcium ions in NanI do not appear to function in the same way, in that they play no obvious role in stabilizing active site residues.
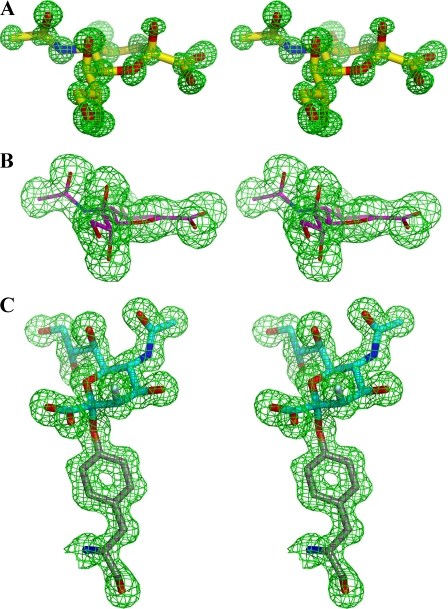
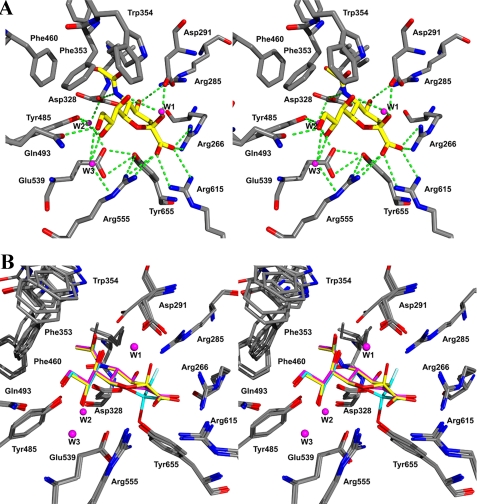
Active Site—Fig. 3 shows the electron density resulting from the individual complexes of NanI individually with Neu5Ac, Neu5Ac2en, and 2,3-difluoro-N-acetylneuramic acid. Fig. 4A shows the details of the interactions of Neu5Ac with the active site, and Fig. 4B shows an overlay of the active sites of the ligand free structure and all three ligand complexes. Although there are a number of significant differences between the active sites of viral, bacterial, and eukaryotic sialidases, a number of key features have been retained throughout their evolution (17) and are shown in Fig. 4A. These include a tri-arginal cluster (Arg266, Arg555, and Arg615) that interacts with the carboxylate group of Neu5Ac. The position of the first arginine (Arg266) is stabilized by a conserved glutamic acid (Glu671). A tyrosine (Tyr655) and a glutamic acid (Glu539) hydrogen bond with each other and sit beneath and close to the C-1–C-2 bond of the substrate. A conserved feature of sialidase active sites is the acid/base catalyst, Asp291. All sialidase active sites have a hydrophobic pocket to accommodate the N-acetyl group of the substrate, but the exact residues that form this pocket are generally not conserved. In the NanI structure, this pocket is made up of mostly aromatic side chains: Phe347, Phe353, Phe460, Thr345, Ile327, and finally Trp354 that forms a cap on the hydrophobic pocket, in an equivalent position to a tyrosine in the T. cruzi trans-sialidase active site.
FIGURE 3.
Stereo images of unbiased Fo - Fc electron density maps contoured at 3σ with the final refined coordinates superimposed. A, Neu5Ac. B, Neu5Ac2en. C, 3-fluoro-2-deoxy-N-acetylneuraminic acid covalently linked to Tyr655.
FIGURE 4.
Stereo views of the NanI active site. A, complex with Neu5Ac showing the hydrogen-bonding interactions as green dotted lines. B, superposition of the three ligand complexes and ligand free structure: Neu5Ac with yellow carbons, Neu5Ac2en with magenta carbons, and the covalent fluorinated ligand with cyan carbons.
All of the interactions between residues in the active site and atoms common to all three ligands are conserved in the different complexes (Table 2), as are three water molecules (W1, W2, and W3 in Fig. 4). W1 shifts position between the complexes as described below, while remaining hydrogen-bonded to the ligand O-7 hydroxyl. W2 does not interact directly with the ligands but bridges the side chains of three critical residues as follows: Asp328 that interacts with the acetamido NH group; Tyr485 that interacts with the O-8 hydroxyl; and Glu539 that plays a key role in the nucleophilic attack. W3 bridges the ligand O-8 and O-9 hydroxyls to the side chains of Thr538 and Arg555.
TABLE 2.
Key ligand interactions
|
Ligand/protein atom (resolution)
|
Protein/water atom
|
Distance
|
||
|---|---|---|---|---|
| α-Neu5Ac (0.97 Å) | Neu5Ac2en (1.5 Å) | Covalent intermediate (1.2 Å) | ||
| Å | Å | Å | ||
| O-1A | Arg615-N-η2 | 2.91 | 2.86 | 2.75 |
| Arg555-N-η1 | 3.15 | 3.14 | 3.13 | |
| Arg555-N-η2 | 3.11 | 3.09 | 3.18 | |
| H2O | 2.95 | |||
| O-1B | Arg615-N-η1 | 2.92 | 2.89 | 2.89 |
| Arg266-N-η1 | 3.01 | 2.86 | 2.99 | |
| Arg266-N-η2 | 3.02 | 2.99 | 2.91 | |
| H2O | 2.90 | |||
| O-2 | Asp291-O-δ2 | 2.57 | ||
| H2O (W1) | 2.93 | |||
| F-1 | Arg266-N-η1 | 2.96 | ||
| Asp291-O-δ1 | 2.87 | |||
| O-4 | Arg285-N-η2 | 2.97 | 2.92 | 2.95 |
| Asp328-O-δ1 | 2.68 | 2.71 | 2.69 | |
| O-7 | H2O (W1) | 3.15 | 2.73 | |
| H2O | 2.71 | |||
| H2O | 2.83 | |||
| O-8 | Tyr485-O | 2.82 | 2.77 | 2.74 |
| H2O (W3) | 2.79 | 2.74 | 2.78 | |
| O-9 | Gln493-O-ε1 | 2.73 | 2.76 | 2.97 |
| H2O (W3) | 3.06 | 3.13 | 2.99 | |
| H2O | 2.65 | 3.17 | ||
| O-10 | H2O | 2.84 | 2.74 | 2.80 |
| N-5 | Asp328-O-δ2 | 2.86 | 2.88 | 2.83 |
| C-1 | Tyr655-O | 2.82 | 2.87 | 2.36 |
| C-2 | Tyr655-O | 2.96 | 2.64 | 1.41 |
| Glu539-O-ε1 | Tyr655-O | 3.13 | 3.08 | 3.25 |
| Glu539-O-ε2 | Tyr655-O | 2.62 | 2.67 | 2.78 |
| Arg555-N-η1 | Tyr655-O | 2.91 | 2.90 | 2.97 |
Neu5Ac Complex—The 0.97 Å resolution structure of the Neu5Ac complex shows that it is bound in its α-anomeric form, with the ring adopting a distorted B2,5 boat conformation (Fig. 4A). The O-2 hydroxyl of Neu5Ac forms a close hydrogen bonding interaction (2.57 Å) with the acid/base catalyst, Asp291, and with a water molecule (W1) also coordinated by O-7. The O-4 hydroxyl of Neu5Ac interacts with Arg285 and Asp328, residues that are conserved among bacterial and mammalian sialidases. The N-acetyl group sits in a hydrophobic pocket with the amide of the N-acetyl group also interacting with Asp328, an interaction that has been seen previously in complexes of bacterial sialidases with Neu5Ac2en. The O-10 hydroxyl makes a single hydrogen bond to a water molecule sitting above the hydrophobic pocket. The Neu5Ac glycerol side chain makes a total of five hydrogen bonds. The O-7 hydroxyl makes a single hydrogen bond to the water (W1) coordinated also with the O-2 hydroxyl, and O-8 makes two hydrogen bonds to Tyr485 and a water molecule (W3). The terminal hydroxyl, O-9, makes two hydrogen bonds, one to the same water (W3) coordinated by O-8 and Gln493. Finally, beneath the anomeric carbon is the hydroxyl group of the catalytic nucleophile, Tyr655, which is poised for attack on the anomeric carbon at a distance of 2.96 Å. The O-ε2 of Glu539 forms a close hydrogen bond interaction (2.62 Å) with the hydroxyl of Tyr655.
Neu5Ac2en Complex—The 1.5 Å resolution structure shows that Neu5Ac2en adopts a 4H5 half-chair conformation. The distance between the hydroxyl of Tyr655 and C-2 of the ligand shortens to 2.64 Å, compared with 2.96Å in the Neu5Ac complex. Water W1 shifts position by 0.6 Å compared with the Neu5Ac complex, remains hydrogen bonded to O-7 of the ligand, and forms a hydrogen bond with the acid/base catalyst Asp291.
Covalent Intermediate—The 1.2 Å resolution structure clearly shows 3-fluoro-β-N-acetylneuraminic acid (3F-β-Neu5Ac) covalently linked to Tyr655 in an unstrained 2C5 conformation as observed in the T. cruzi trans-sialidase (29) and T. rangeli sialidase (31) covalent complexes. Careful refinement suggests that there is a 50:50 mixture of 3-F-β-Neu5Ac and Neu5Ac2en to account for the difference electron density. Water W1 shifts position 0.8 Å, compared with the Neu5Acen complex, and 1.3 Å, compared with the Neu5Ac complex. It remains hydrogen bonded to O-7 of the ligand and to the acid/base catalyst Asp291. Of particular note are the appearances of seven water molecules, compared with only two in the Neu5Ac complex. The relaxed conformation of the ring places the C-1 carboxylate group into the equatorial position, and this now hydrogen bonds to two of these waters.
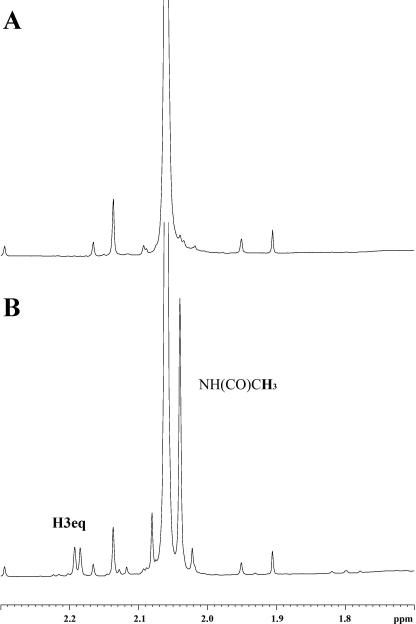
NMR Monitoring of Neu5Ac2en Hydration—The hydration of Neu5Ac2en to form Neu5Ac at pH 7 was monitored as a time course reaction using 1H NMR spectroscopy. Examination of 1H NMR spectra containing NanI and Neu5Ac2en revealed that the integration of the H3 signal of Neu5Ac2en at 5.69 ppm progressively decreased over a period of 10 days by ∼15%. Concomitant with the decrease in intensity of the H3 signal of Neu5Ac2en was the appearance of a doublet (d = 2.19 ppm, J = 5.05 Hz) in the aliphatic region of the spectrum because of the H3eq proton of β-Neu5Ac and the appearance of an additional N-acetamido methyl signal at 2.03 ppm as shown in Fig. 5. The appearance of the doublet at 2.19 ppm is consistent with Neu5Ac2en being attacked in the axial position (deuteration in 100% D2O resulting in a Dax) resulting in the formation of β-(H3eqD3ax)-Neu5Ac. The observed coupling constant of 5.05 Hz is because of coupling of the H3eq with the H4 proton. A control experiment performed under identical conditions but in the absence of NanI showed no decrease in intensity of the H3 proton of Neu5Ac2en nor the appearance of the H3eq doublet at 2.19 ppm or N-acetamido methyl signal of Neu5Ac. Taken together these results suggest that NanI sialidase can catalyze the hydration of Neu5Ac2en to produce Neu5Ac. The formation of Neu5Ac2en from Neu5Ac has previously been demonstrated for the influenza virus neuraminidase (11) and the V. cholerae sialidase (18). It has also previously been demonstrated that Arthrobacter sialophilus sialidase can catalyze the formation of Neu5Ac by hydration of C-2–C-3 double bond of Neu5Ac2en (39).
FIGURE 5.
The 1H NMR spectrum (600 MHz, 285 K) of Neu5Ac2en in the presence of NanI 0 h after preparation of the sample (A), and the same sample after 10 days (B).
DISCUSSION
The structures presented here provide high resolution snapshots of the catalytic cycle of the NanI sialidase. The Neu5Ac complex reveals the sugar ring in a strained and distorted B2,5 conformation that is induced upon formation of the Michaelis complex. A similar sialic acid ring conformation was seen in the Michaelis complex between an aspartic acid mutant of T. cruzi trans-sialidase and α(2,3)-sialyl-lactose (29). This conformation of the sugar ring results in a pseudo-axial orientation of the glycosidic bond being cleaved, bringing it within 2.6 Å of the acid catalyst Asp291 for proton transfer. The transition state, mimicked by the complex with Neu5Ac2en, changes the sugar ring to a half-chair 4H5 conformation, reducing the distance between C-2 and the hydroxyl of the nucleophile Tyr655 from 2.96 to 2.64 Å. Formation of the covalent intermediate and the concomitant release of the aglycon moiety promotes further changes in the conformation of the sugar ring. The sugar ring is seen to relax into the 2C5 chair conformation, with a β-linkage to the catalytic nucleophile Tyr655. This conformational change in the sugar ring accommodates the change in relative position of Tyr655 and C-2 of the substrate that is required to form the covalent bond, with the anomeric carbon moving 1.4 Å from its position in the Neu5Ac complex, as well as the hydroxyl of Tyr655 moving by 0.3 Å.
Concomitant with the covalent bond formation is a small shift in the position of one of the ε oxygens of the general base Glu539 by 0.5 Å, extending its distance from the Tyr655 side chain oxygen to 2.8 Å from 2.62 Å in the Neu5Ac complex. The atomic resolution of the Neu5Ac complex allows very accurate determination of bond lengths from which protonation states can be inferred. This technique has been used previously to assign protonation states to key catalytic residues in xylanase Xyn10A (40). In the Neu5Ac complex the bond lengths of both C-δ–C-ε bonds of Glu539 are 1.26 Å and strongly suggest that the charge on this carboxylate group is delocalized and that the residue is not protonated. In all sialidases studied to date, the carboxylate group of this conserved glutamic acid forms a short hydrogen bond with the hydroxyl group of the catalytic tyrosine. The T. cruzi trans-sialidase study suggested that a deprotonated glutamic acid would act as a base catalyst for the attack of the tyrosine on the anomeric center, therefore acting in a charge relay system (29). This idea has also received support from a recent study on tyrosine mutants of the bacterial sialidase from M. viridifaciens (30), which showed that the rate of glycosidic bond cleavage in the wild type enzyme is dependent on the protonation state of the glutamic acid.
Of particular note is the appearance of seven water molecules in the covalent complex, compared with only two in the Neu5Ac complex. In the covalent complex, the relaxed conformation of the ring places the C-1 carboxylate group into the equatorial position, and this now hydrogen bonds to two of these waters. Several studies have indicated that water rearrangement plays an important role in carbohydrate-protein interactions (41, 42). The ordering of five extra water molecules in the covalent complex would be expected to decrease the entropy, and subsequently increase the free energy of the system. It has been suggested that each water molecule that is ordered at an interface can contribute -0.5 to -3.0 kcal mol-1 to ΔH (43). This increase in free energy would serve to destabilize the covalent intermediate and may act to drive the reaction forward to product formation. Unique to NanI, compared with other sialidases, is Trp354 acting as a lid on the hydrophobic pocket but extending over the glycerol group of sialic acid. It may shield this part of the active site from bulk solvent and help contribute to the strength of the hydrogen bonds made by Gln493 and Tyr485 to the glycerol group through desolvation effects.
Neu5Ac2en is an inhibitor of most sialidases, with Ki values ∼10-6 m. All previously studied bacterial and viral sialidases have been crystallized in complex with this compound (7–9, 11, 27), as have the trypanosomal sialidases from T. rangeli (13) and the trans-sialidase from T. cruzi (14). These complexes have helped characterize the active sites of these enzymes and aided in the understanding of the hydrogen bonding interactions made by the transition-state intermediate. It was known previously that sialidases could dehydrate Neu5Ac to form Neu5Ac2en as a by-product of their catalytic mechanism (11, 18). This study shows that the reverse reaction is possible, namely that NanI is able to hydrate the C-2–C-3 double bond of Neu5Ac2en to form Neu5Ac. Neu5Ac acts as a poor inhibitor of the influenza viral neuraminidases (Ki ∼ 10-3 m), and as such allowed a complex to be observed crystallographically (44). However, this is the first report of Neu5Ac being observed in the active site of a bacterial sialidase, which tends to have high turnover rates in comparison with the influenza virus enzyme.
Glycals are well known to be hydrated by retaining glycosidases to generate a deoxysugar product (normally a 2-deoxysugar) via a glycosyl enzyme intermediate (45). Early stereochemical studies involving NMR analyses in D2O demonstrated that protonation occurred from the same face as the incoming catalytic carboxylic acid nucleophile (46, 47). Indeed it seems likely that initial addition of the carboxylic acid across the double bond occurs in a syn fashion. The only exception observed to date has been the hydration of N-acetyl-d-glucal by a retaining hexosaminidase (19). In that case protonation occurred on the opposite face and was likely effected by the normal acid/base catalyst. The observed stereochemical outcome was a necessary consequence of the mechanism for such enzymes, in which the 2-acetamido group acts as an intramolecular nucleophile and cleaves the glycosidic bond with formation of an oxazolinium ion intermediate. This species then undergoes hydrolysis via attack at the anomeric center. It was therefore of considerable interest to determine the stereochemical outcome of hydration by the NanI sialidases because it also does not use a carboxylic acid nucleophile but is not stereochemically constrained around C-3, which bears no substituent.
The NMR data show that Neu5Ac2en is indeed hydrated to Neu5Ac by NanI in solution. Furthermore, the data clearly show that protonation (deuteration in this case) occurs from the bottom face. Clearly in this case neither the nucleophile, Tyr655 nor its partner Glu539, is acting as proton donor. As was the case with the hexosaminidase it is likely that the normal acid catalyst, Asp291, plays this role. This seems quite reasonable given the fact that the Tyr655 cannot provide concerted acid catalysis and nucleophilic attack in the way that a carboxylic acid can.
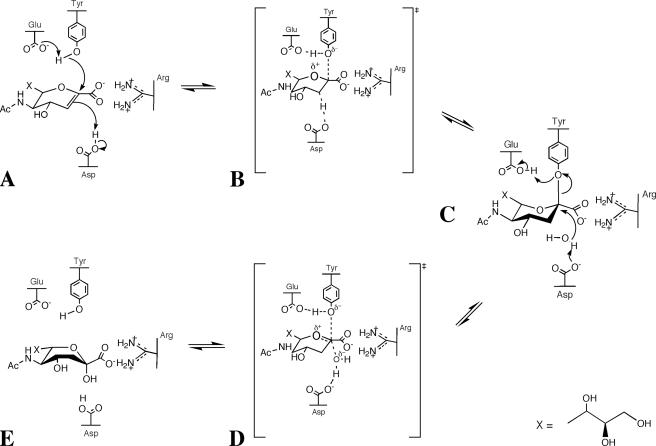
Measurement of the affinity of Neu5Ac2en for NanI gave a Ki of 40 and 5 μm at pH 5 and 7, respectively (data not shown). The observation of Neu5Ac in the crystal at pH 7 could be related to a slow off rate for Neu5Ac at pH 7. The mechanism for the hydration of Neu5Ac2en could proceed as shown in Fig. 6. The stronger interactions of the protein with the glycerol group may contribute to ring strain in the Neu5Ac2en. Once destabilized, the electrons in the nucleophilic π bond could attack the suitably positioned proton on Asp291 in an electrophilic addition reaction (Fig. 6A). In the complex of Neu5Ac with NanI, the distance from the anomeric carbon to carboxylate of Asp291 is 3.12 Å. This would result in a carbocation (oxocarbenium ion) intermediate, with the positive charge delocalized between the anomeric carbon and the endocyclic oxygen (Fig. 6B). The structure of the covalent complex with NanI shows that such an intermediate could be stabilized through a covalent bond with the nucleophilic Tyr655 sitting directly beneath (Fig. 6C). Asp291 could then activate an incoming water molecule to attack the positive charge on the anomeric carbon, creating a protonated alcohol intermediate (Fig. 6D). Loss of a H+ from this protonated alcohol back to Asp291 gives the product Neu5Ac (Fig. 6E).
FIGURE 6.
Proposed mechanism for the hydration of Neu5Ac2en to Neu5Ac.
These studies have provided a detailed insight into the catalytic cycle of a sialidase at an unprecedented level of detail that adds to our understanding of this enzyme superfamily that plays a key role in the pathogenesis of many important diseases.
Acknowledgments
We thank the staff at the European Synchrotron Radiation Facility and the European Union for funds to access the facility. The facilities of the Scottish Structural Proteomics Facility were also used to complete this work. We thank Jim Naismith for useful discussions.
The atomic coordinates and structure factors (codes 2vk5, 2vk6, 2vk7) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by the Biotechnology and Biological Sciences Research Council and the Royal Society of Edinburgh. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Neu5Ac, N-acetylneuramic acid; Neu5Ac2en, 2-deoxy-2,3-dehydro-N-acetylneuraminic acid; PDB, Protein Data Bank; PEG, polyethylene glycol; r.m.s., root mean square.
References
- 1.Rood, J. I. (1998) Annu. Rev. Microbiol. 52 333-360 [DOI] [PubMed] [Google Scholar]
- 2.Shimizu, T., Ohtani, K., Hirakawa, H., Ohshima, K., Yamashita, A., Shiba, T., Ogasawara, N., Hattori, M., Kuhara, S., and Hayashi, H. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 996-1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kruse, S., Kleineidam, R. G., Roggentin, P., and Schauer, R. (1996) Protein Expression Purif. 7 415-422 [DOI] [PubMed] [Google Scholar]
- 4.Traving, C., Schauer, R., and Roggentin, P. (1994) Glycoconj. J. 11 141-151 [DOI] [PubMed] [Google Scholar]
- 5.Roggentin, P., Kleineidam, R. G., and Schauer, R. (1995) Biol. Chem. Hoppe-Seyler 376 569-575 [DOI] [PubMed] [Google Scholar]
- 6.Roggentin, P., Kleineidam, R. G., and Schauer, R. (1989) Glycoconj. J. 6 349-353 [DOI] [PubMed] [Google Scholar]
- 7.Crennell, S. J., Garman, E. F., Laver, W. G., Vimr, E. R., and Taylor, G. L. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 9852-9856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crennell, S. J., Garman, E., Laver, G., Vimr, E. R., and Taylor, G. (1994) Structure (Camb.) 2 535-544 [DOI] [PubMed] [Google Scholar]
- 9.Gaskell, A., Crennell, S. J., and Taylor, G. (1995) Structure (Camb.) 3 1197-1205 [DOI] [PubMed] [Google Scholar]
- 10.Varghese, J. N., Laver, W. G., and Colman, P. M. (1983) Nature 303 35-40 [DOI] [PubMed] [Google Scholar]
- 11.Burmeister, W. P., Henrissat, B., Bosso, C., Cusack, C., and Ruigrok, R. W. H. (1993) Structure (Camb.) 1 19-26 [DOI] [PubMed] [Google Scholar]
- 12.Crennell, S. J., Takimoto, T., Portner, A., and Taylor, G. (2000) Nat. Struct. Biol. 7 1068-1074 [DOI] [PubMed] [Google Scholar]
- 13.Buschiazzo, A., Tavares, G. A., Campetella, O., Spinelli, S., Cremona, M. L., Paris, G., Amaya, M. F., Frasch, A. C., and Alzari, P. M. (2000) EMBO J. 19 16-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buschiazzo, A., Amaya, M. F., Cremona, M. L., Frasch, A. C., and Alzari, P. M. (2002) Mol. Cell 10 757-768 [DOI] [PubMed] [Google Scholar]
- 15.Lou, Y., Li, S.-C., Chou, M.-Y., Li, Y.-T., and Lou, M. (1998) Structure (Camb.) 6 521-530 [DOI] [PubMed] [Google Scholar]
- 16.Chavas, L. M., Tringali, C., Fusi, P., Venerando, B., Tettamanti, G., Kato, R., Monti, E., and Wakatsuki, S. (2005) J. Biol. Chem. 280 469-475 [DOI] [PubMed] [Google Scholar]
- 17.Taylor, G. (1996) Curr. Opin. Struct. Biol. 6 830-837 [DOI] [PubMed] [Google Scholar]
- 18.Moustafa, I., Connaris, H., Taylor, M., Zaitsev, V., Wilson, J. C., Kiefel, M. J., von Itzstein, M., and Taylor, G. (2004) J. Biol. Chem. 279 40819-40826 [DOI] [PubMed] [Google Scholar]
- 19.Lai, E. C., and Withers, S. G. (1994) Biochemistry 33 14743-14749 [DOI] [PubMed] [Google Scholar]
- 20.Newstead, S. L., Watson, J. N., Bennet, A. J., and Taylor, G. (2005) Acta Crystallogr. Sect. D Biol. Crystallogr. 61 1483-1491 [DOI] [PubMed] [Google Scholar]
- 21.Thobhani, S., Ember, B., Siriwardena, A., and Boons, G.-J. (2002) J. Am. Chem. Soc. 125 7154-7155 [DOI] [PubMed] [Google Scholar]
- 22.Boraston, A. B., Bolam, D. N., Gilbert, H. J., and Davies, G. J. (2004) Biochem. J. 382 769-781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chong, A. K., Pegg, M. S., Taylor, N. R., and von Itzstein, M. (1992) Eur. J. Biochem. 207 335-343 [DOI] [PubMed] [Google Scholar]
- 24.Taylor, G., Crennell, S. J., Thompson, C., and Chuenkova, M. (1999) in Sialobiology and Other Novel Forms of Glycosylation (Inuoue, Y., Lee, Y. C., and Troy, F. A., eds) pp. 187-195, Gakushin Publishing Co., Osaka, Japan
- 25.Rye, C. S., and Withers, S. G. (2000) Curr. Opin. Chem. Biol. 4 573-580 [DOI] [PubMed] [Google Scholar]
- 26.Baker, N., Sept, D., Joseph, S., Holst, M., and McCammon, J. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 10037-10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lou, Y., Li, S. C., Li, Y. T., and Lou, M. (1998) J. Mol. Biol. 285 323-332 [DOI] [PubMed] [Google Scholar]
- 28.Watts, A. G., Damager, I., Amaya, M. F., Buschiazzo, A., Alzari, P. M., Frasch, A. C., and Withers, S. G. (2003) J. Am. Chem. Soc. 125 7523-7533 [DOI] [PubMed] [Google Scholar]
- 29.Amaya, M. F., Watts, A. G., Damager, I., Wehenkel, A., Nguyen, T., Buschiazzo, A., Paris, G., Frasch, A. C., Withers, S. G., and Alzari, P. M. (2004) Structure (Camb.) 12 775-784 [DOI] [PubMed] [Google Scholar]
- 30.Watson, J. N., Dookhun, V., Borgford, T. J., and Bennet, A. J. (2003) Biochemistry 42 12682-12690 [DOI] [PubMed] [Google Scholar]
- 31.Watts, A. G., Oppezzo, P., Withers, S. G., Alzari, P. M., and Buschiazzo, A. (2006) J. Biol. Chem. 281 4149-4155 [DOI] [PubMed] [Google Scholar]
- 32.Newstead, S., Chien, C. H., Taylor, M., and Taylor, G. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2063-2066 [DOI] [PubMed] [Google Scholar]
- 33.CCP4 (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760-763 [DOI] [PubMed] [Google Scholar]
- 34.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 35.Perrakis, A., Morris, R., and Lamzin, V. S. (1999) Nat. Struct. Biol. 6 458-463 [DOI] [PubMed] [Google Scholar]
- 36.Sheldrick, G. M., and Schneider, T. R. (1997) Methods Enzymol. 277 319-343 [PubMed] [Google Scholar]
- 37.Jones, T. A., Zou, J.-Y., Cowan, S. W., and Kjeldgaard, M. (1991) Acta Cryst. Sect. A 47 110-119 [DOI] [PubMed] [Google Scholar]
- 38.Lovell, S. C., Davis, I. W., Arendall, W. B., III, de Bakker, P. I., Word, J. M., Prisant, M. G., Richardson, J. S., and Richardson, D. C. (2003) Proteins 50 437-450 [DOI] [PubMed] [Google Scholar]
- 39.Flashner, M., Kessler, J., and Tanenbaum, S. W. (1983) Arch. Biochem. Biophys. 221 188-196 [DOI] [PubMed] [Google Scholar]
- 40.Gloster, T. M., Williams, S. J., Roberts, S., Tarling, C. A., Wicki, J., Withers, S. G., and Davies, G. J. (2004) J. Chem. Soc. Chem. Commun. 16 1794-1795 [DOI] [PubMed] [Google Scholar]
- 41.Ladbury, J. E. (1996) Chem. Biol. 3 973-980 [DOI] [PubMed] [Google Scholar]
- 42.Clarke, C., Woods, R. J., Gluska, J., Cooper, A., Nutley, M. A., and Boons, G. J. (2001) J. Am. Chem. Soc. 123 12238-12247 [DOI] [PubMed] [Google Scholar]
- 43.Holdgate, G. A., Tunnicliffe, A., Ward, W. H., Weston, S. A., Rosenbrock, G., Barth, P. T., Taylor, I. W., Pauptit, R. A., and Timms, D. (1997) Biochemistry 36 9663-9673 [DOI] [PubMed] [Google Scholar]
- 44.Varghese, J. N., McKimm-Brechkin, J. L., Caldwell, J. B., Kortt, A. A., and Colman, P. M. (1992) Proteins 14 327-332 [DOI] [PubMed] [Google Scholar]
- 45.Legler, G. (1990) Adv. Carbohydr. Chem. Biochem. 48 319-384 [DOI] [PubMed] [Google Scholar]
- 46.Wentworth, D. F., and Wolfenden, R. (1974) Biochemistry 13 4715-4720 [DOI] [PubMed] [Google Scholar]
- 47.Hehre, E. J., Genghof, D. S., Sternlicht, H., and Brewer, C. F. (1977) Biochemistry 16 1780-1787 [DOI] [PubMed] [Google Scholar]