Abstract
Brd4 is a bromodomain protein that binds to acetylated chromatin. It regulates cell growth, although the underlying mechanism has remained elusive. Brd4 has also been shown to control transcription of viral genes, whereas its role in transcription of cellular genes has not been fully elucidated. Here we addressed the role of Brd4 in cell growth and transcription using a small hairpin (sh) RNA approach. The Brd4 shRNA vector stably knocked down Brd4 protein expression by ∼90% in NIH3T3 cells and mouse embryonic fibroblasts. Brd4 knockdown cells were growth impaired and grew more slowly than control cells. When synchronized by serum starvation and released, Brd4 knockdown cells were arrested at G1, whereas control cells progressed to S phase. In microarray analysis, although numerous genes were up-regulated during G1 in control cells, many of these G1 genes were not up-regulated in Brd4 knockdown cells. Reintroduction of Brd4 rescued expression of these G1 genes in Brd4 knockdown cells, allowing cells to progress toward S phase. Chromatin immunoprecipitation analysis showed that Brd4 was recruited to the promoters of these G1 genes during G0-G1 progression. Furthermore, Brd4 recruitment coincided with increased binding of Cdk9, a component of P-TEFb and RNA polymerase II to these genes. Brd4 recruitment was low to absent at genes not affected by Brd4 shRNA. The results indicate that Brd4 stimulates G1 gene expression by binding to multiple G1 gene promoters in a cell cycle-dependent manner.
Brd4 is a ubiquitously expressed 200-kDa nuclear protein that belongs to the BET family (1, 2). Proteins of this family carry two tandem bromodomains through which they interact with acetylated histones (3-6). Bromodomains are also present in other chromatin-binding proteins such as histone acetylases and chromatin remodeling factors. They also bind to acetylated histones and are involved in transcriptional regulation of many genes. Recent structural analysis indicates that the bromodomain of Brd2, a factor closely related to Brd4, forms a dimmer to bind to acetyl residues of the histone H4 tails (7). Binding of Brd4 and Brd2 to acetylated chromatin persists even during mitosis as well as meiosis when chromatin is highly condensed and transcription is interrupted (2, 5, 6, 8).
Evidence indicates that BET family proteins are multifunctional and regulate cell growth and transcription (3, 4, 9, 10). In line with this evidence, there are reports indicating that Brd4 is involved in cell growth regulation; Brd4-/- embryos fail to grow and die early at around the time of implantation (11). Similarly, Brd4-/- embryonic stem cells do not grow in culture (12). Moreover, in some malignant cells, Brd4 is fused to the NUT gene, and the fusion protein exhibits a growth regulatory activity (13, 14). In addition, overexpression of Brd4 in cultured cells is shown to alter their growth properties, in part due to the interaction of Brd4 with growth regulatory proteins such as RFC140 or Sipa1 (15, 16). The reports that Brd4 facilitates partition of Papillomavirus genomes during mitosis also support the idea that Brd4 plays a role in cell division (17, 18). So far, however, the mechanism(s) by which Brd4 participates in cell growth regulation has remained elusive.
With respect to transcriptional regulation, the yeast BET family protein Bdf1 is shown to be broadly involved in gene transcription (3, 4). Bdf1 is incorporated into the general transcription factor complex and regulates many yeast genes (4). It also inhibits Sir2-mediated gene silencing (3). The mammalian Brd2 is also reported to interact with general transcription factors and takes part in transcription (10). We have previously found that Brd4 complexes with the kinase active form of P-TEFb and enhances human immunodeficiency virus-1 long terminal repeat transcription (19, 20). P-TEFb, composed of cyclin T and Cdk9, phosphorylates the C-terminal domain of RNA polymerase II (pol II)9 and contributes to transcriptional elongation (21, 22). Additionally, Brd4 interacts with components of the Mediator complexes that associates with pol II preinitiation complexes, further suggesting a role in transcription (19, 20, 23). Recently, Brd4 is shown to repress human Papillomavirus gene transcription by interacting with the viral transactivator E2 and blocking the recruitment of TFIID and pol II to the viral promoters (24). Despite the evidence for regulating viral gene transcription, little is known about the role of Brd4 in the transcriptional regulation of cellular genes. Moreover, it has been unclear as to whether cell growth regulation by Brd4 is in any way coupled with its potential role in transcription.
To address the role of Brd4 in cell growth and transcription, we employed a shRNA approach and knocked down Brd4 expression in NIH3T3 cells and primary mouse embryonic fibroblasts (MEFs), non-transformed cells with a normal growth property. Our data indicate that Brd4 has a vital role in promoting cell cycle progression from G0 to G1 and entry into S in these cells. In microarray analysis, a large number of genes showed a marked increase in transcript expression during G0-G1 progression in control cells. However, this extensive G1 gene up-regulation was largely absent in Brd4 knockdown (KD) cells. Chromatin immunoprecipitation (ChIP) analysis showed that Brd4 was recruited to a number of G1 gene promoters, coinciding with up-regulation of G1 gene expression in control cells, but not in Brd4 KD cells. Furthermore, RNA pol II and Cdk9 were recruited to these G1 genes in a Brd4-dependent manner, suggesting that Brd4 facilitates productive transcription of multiple G1 genes. These results provide a mechanistic basis by which Brd4 regulates cell growth.
EXPERIMENTAL PROCEDURES
Cell Culture, Brd4 shRNA, and Rescue Vectors—NIH3T3 cells were cultured in Dulbecco's modified Eagle's medium with 10% donor serum. For synchronization, cells were incubated in 0. 5% donor serum for 60 h and then released in the complete media. MEFs were prepared from embryos of day 13 post-coitus and cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Brd4 shRNA and control shRNA were cloned into the pSUPER retro vector (Oligoengine). The Brd4 shRNA sequence was from +604 to +623 relative to the transcription start site, GATCCCCGCATCAACTTCTCCGCAGATTCAAGAGATCTGCGGAGAAGTTGATGCTTTTTGGAAA. The control siRNA was a shuffled sequence of the above, GATCCCCATGCACGTGCACATATCCCTTCAAGAGAGGGATATGTGCACGTGCATTTTTTGGAAA. BOSC23 packaging cells were transfected with the above vectors, and viral supernatants were harvested 2 days later. NIH3T3 and MEFs were transduced with viral supernatants by spinoculation (2800 rpm, 28 °C, 2 h) in the presence of 4 μg/ml Polybrene (Sigma-Aldrich) and selected by puromycin (5 μg/ml) for 2 days. A retroviral rescue vector expressing a Brd4 mutant resistant to Brd4 shRNA inhibition was constructed from the pMSCVneo vector expressing wild type Brd4 (15) using the oligomer GCTTCTACATCACCGCAGA that changed the Brd4 nucleotide sequence without changing the amino acid sequence (mutations are underlined). NIH3T3 cells were first transduced with the rescue vector (or empty vector) and selected by G418 for 4 days followed by the second transduction with the pSUPERretro vectors containing Brd4 shRNA as above.
Microarray Analysis and Quantitative Real Time PCR (qRT-PCR)—Total RNA was extracted from cells using TRIzol (Invitrogen) and purified by the RNeasy kit (Qiagen), and cDNA was prepared using SuperScript II RNase H-Reverse (Invitrogen). The NIA 15K mouse cDNA microarray system was used (25) (http://lgsun.grc.nia.nih.gov/cDNA/15k.html). cDNA from cells expressing control or Brd4 shRNA and those from reference RNA were labeled with Cy3 and Cy5, respectively, by reverse transcription using the MICROMAX direct cDNA microarray system (PerkinElmer Life Sciences). Twenty-five μg of labeled probes were added to the slides and incubated overnight at 65 °C. Slides were washed in 0.5× SSC (1× SSC 0.15 m NaCl and 0.015 m sodium citrate), 0.01% SDS and 0.06× SSC, 0.01% SDS and 0.06× SSC. Data were analyzed using the ScanArray Express (PerkinElmer Life Sciences). Spots showing signals lower than the average background of the entire Chip were eliminated. Normalization of Cy3 to Cy5 signals was carried out by the global median normalization. To minimize sample to sample variability, hybridization was carried out with duplicate slides, and spots with signals that lie outside the best-fit line were eliminated (26). To reduce biological variability, three independently prepared RNA samples were analyzed, and the spots showing high variability were removed. Microarray signals were represented by a log2 ratio relative to signals by reference RNA. The principal component analysis and hierarchical clustering analysis were performed by the “Cluster” program (27, 28). For qRT-PCR, amplification of sample cDNA was monitored with the SYBR green in a kit along with the ABI Prism 7000 Sequence Detection System (Applied Biosystems) according to the manufacturer's instructions. Transcript levels were normalized by 18 S rRNA levels. Primers used for qRT-PCR are in shown in supplemental Table S1.
Flow Cytometry and Immunoblot—To monitor cell cycle profiles, cells were stained with propidium iodide and analyzed on FACSCalibur interfaced with the Cell Quest software (BD Biosciences) as described in Nishiyama et al. (12). To detect apoptosis, cells were stained with annexin V-fluorescein isothiocyanate (BD Biosciences Pharmingen) according to the manufacturer's instructions. Total cell proteins were extracted by radioimmune precipitation assay buffer, and 40 μg of proteins were resolved on a 4-20% gradient SDS-polyacrylamide gel electrophoresis and immunoblotted with antibodies for cyclin D1, TATA box-binding protein (TBP) (Santa Cruz), cyclin D2, cyclin E, and phosphorylated Rb (Cell Signaling) by the standard procedures. Antibody for unphosphorylated Rb was from Pharmingen. Antibodies for Brd2 and Brd4 were described (5). Acid extraction of total histones and immunoblot analysis were described (12).
ChIP Assay—Chromatin immunoprecipitation was performed using indicated antibodies essentially as described (29). Briefly, control and Brd4 KD NIH3T3 cells (1 × 106) were cross-linked with 1% paraformaldehyde at 37 °C for 10 min and lysed in buffer containing 50 mm Tris HCl, pH 8.0, 10 mm EDTA, 1% SDS, and a protease inhibitor cocktail (Roche Applied Science). Chromatin was sheared by sonication to generate 200-1000-bp DNA fragments and diluted by 10-fold in radioimmune precipitation assay buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA, 0.5 mm EGTA, 1% Triton X, and 0.1% SDS, 0.1% sodium deoxycholate, 140 mm NaCl). Rabbit antibodies for Brd4 (prepared in this laboratory) (5) or Cdk9 (Santa Cruz) or RNA pol II (Covance) or preimmune or normal rabbit IgG were incubated with protein A-coupled paramagnetic beads (Dynalbeads, Invitrogen). Chromatin preparations were then incubated with antibody-precoated magnetic beads above for 2 h to overnight at 4 °C. Immune complexes were washed in radioimmune precipitation assay buffer three times. Antibody bound chromatin was reverse-cross-linked, and DNA was purified by phenol-chloroform and ethanol. DNA was then subjected to quantitative PCR using the primer set compiled in supplemental Table S1. The percentage input was calculated as (2(CT input - CT IP sample)) × 100/CT input. (CT, cycle threshold, IP, immunoprecipitation.)
RESULTS
Brd4 shRNA Inhibits G1 Progression in NIH3T3 Cells and MEFs— To study the role of Brd4 in cell growth and transcription, we took a shRNA approach to inhibit the expression of Brd4 in fibroblasts. This approach was a practical alternative to studying Brd4-/- cells, which do not grow in culture (11, 12). NIH3T3 cells were transduced with a retroviral vector for Brd4 shRNA or control shRNA. Immunoblot analysis presented in Fig. 1A showed that introduction of the Brd4 shRNA stably knocked down Brd4 protein expression by ∼ 90% as compared with the introduction of control shRNA, which did not affect Brd4 levels. Brd4 expression in Brd4 KD cells remained low at least for 14 days after vector transduction without altering expression of a related protein, Brd2, as well as TBP and other general transcription factors (see below). Several additional control shRNAs including Brd2 shRNA did not change Brd4 expression, supporting specificity of Brd4 shRNA (data not shown). It should be noted here that our shRNA targeted both the long and short forms of Brd4, the latter suggested to exist in some cells (11). So far, however, we have detected only a single Brd4 mRNA (6.5 kilobases) and protein (∼200 kDa) in all cells examined, including NIH3T3 and MEFs (2, 12), suggesting that the short Brd4 isoform occurs rarely, if at all. Brd4 KD cells grew more slowly than cells with control shRNA as tested over a 12-day period after shRNA vector introduction. As shown in Fig. 1B, total cell yields were more than 10-fold lower in Brd4 KD cells compared with control shRNA (hereafter control) cells on both days 8 and 12. To assess the basis of this growth inhibition, cells were synchronized to G0 by 60 h of serum starvation and released into complete media to allow progression to G1, and cell cycle profiles were monitored by flow cytometry. As seen in Fig. 1C, control cells, upon exiting G0, proceeded through G1 and reached S at 16-20 h. By 24 h, control cells moved further to G2, and some cells entered G1 again. In contrast, Brd4 KD cells remained at G1 during this time and did not reach S even at 24 h, indicating solid G1 arrest (see supplemental Fig. S1 for the percentage of cells in G1, S, and G2/M). Confirming the failed S phase entry, Brd4 KD cells did not incorporate [H3]thymidine during this period as presented in Fig. 1D.G1 arrest observed in Brd4 KD cells did not appear to be a simple delay in S phase entry in that a certain fraction of cells underwent apoptosis, as evidenced by increased annexin V uptake by Brd4 KD cells at 24 h (Fig. 1E). Staining with 7-amino-actinomycin D, another apoptosis detecting agent, also supported increased cell death in Brd4 KD cells (data not shown).
FIGURE 1.
Cell growth inhibition and G1 arrest in Brd4 KD cells. A, NIH3T3 cells were transduced with control (Ctrl) or Brd4 shRNA vector (Brd4). Total cell extracts prepared on indicated days after transduction were analyzed for the expression of indicated proteins by immunoblot (I.B.) assay. B, 8 × 104 control and Brd4 KD cells were seeded in a 10-ml plate and successively passed, and cell numbers were counted to estimate total cell yields on indicated days. Values are the average of three determinations. C, control and Brd4 KD cells were synchronized to G0 by serum starvation, released, and incubated in complete media for indicated times (h). Cells were stained with propidium iodide (25 μg/ml), and DNA contents were analyzed by flow cytometry. See supplemental Fig. S1, A and B for the percentage of G1, S, and G2/M cells at each time point. D, cells were pulsed with 1 μCi/ml [3H]thymidine for 1 h at the indicated times after release. Values are the average of three assays ± S.D. E, cells harvested at the indicated times after release were incubated with annexin V-fluorescein isothiocyanate, and stained cells were counted. More than 250 cells were counted for each sample. The values represent the percentage of annexin V-positive cells.
Although NIH3T3 cells are non-transformed cells that exhibit a normal growth property, they are tissue culture-adapted cells. To ascertain if Brd4 knockdown exerts similar growth inhibitory effect on fresh fibroblasts, we tested primary MEFs transduced with Brd4 shRNA vector. Immunoblot data in Fig. 2A confirmed that Brd4 shRNA, but not control shRNA, knocked down Brd4 expression in MEFs without affecting TFIIB and α-tubulin expression. Furthermore, we found that upon G0 synchronization and release, Brd4 KD MEFs were arrested at G1, whereas the majority of control MEFs proceeded to S phase by 16 h (Fig. 2B, supplemental Fig. S1C). These results show that a reduction in Brd4 expression causes G1 arrest in fibroblasts.
FIGURE 2.
G1 arrest in Brd4 KD MEFs and inhibition of G1 protein expression. A, MEFs were transduced with control or Brd4 shRNA vector and synchronized to G0 by serum starvation and released as in Fig. 1C, and expression of the indicated proteins was analyzed by immunoblot. B, control and Brd4 KD MEFs were synchronized as above and monitored for cycle profiles as in Fig. 1C. See the quantification in supplemental Fig S.1C. C, control and Brd4 KD NIH3T3 cells were synchronized and released as above, and total cell extracts were analyzed by immunoblot analysis for indicated proteins. Rb was tested for phosphorylation (pRB) at the indicated residues (arrowheads, under pRb).
Immunoblot analysis was next performed for NIH3T3 cells with Brd4 shRNA to examine the expression of Cyclins involved in G1 progression, namely cyclin D1, D2, E1, and E2, known to play a role in G1/S progression (30-32). As seen in Fig. 2C, their expression was markedly increased during G1 in control cells. However, in Brd4 KD cells they showed a meager increase during the same period. Levels of Rb, cyclin C, and cyclin D3 were similar in control and Brd4 KD cells throughout the period. However, progressive Rb phosphorylation, evident in control cells (see pRb with arrowheads in Fig. 2C), was not observed in Brd4 KD cells, in line with G1 arrest and apoptosis (33). Similarly, in Brd4 KD MEFs, cyclin D1 expression was reduced relative to control cells (Fig. 2A). These results indicate that Brd4 is required for G1 cyclin expression, and Rb phosphorylation is critical for G1 progression.
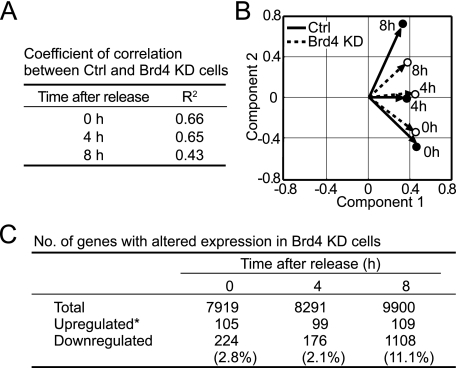
Microarray Analysis Reveals Deficiency in G1 Gene Expression in Brd4 KD Cells—To study the role of Brd4 in genome wide gene expression, microarray analysis was performed with synchronized control and Brd4 KD cells. We analyzed cells at G0 and early G1 rather than those at later stages. We felt that gene expression at early stages would reflect the effect of Brd4 knockdown on gene expression more directly than at later stages or in unsynchronized cells. Given that cell cycle status becomes different in control and Brd4 KD cells toward S phase (Fig. 1C), gene expression at later stages may be influenced by secondary effects such as differences in cell cycle stages more extensively than at an earlier stage. Furthermore, a preliminary microarray analysis with unsynchronized cells did not show a consistent, large scale difference in control and Brd4 KD cells, presumably due to mixed cell populations (data not shown). RNA samples from control and Brd4 KD cells at 0, 4, and 8 h after release were analyzed using the NIA 15 K mouse cDNA array (25). After normalization by the reference RNA and removal of spots giving high variability in three independent sets of synchronized samples, 8,000-10,000 genes could be compared between control and Brd4 KD cells at all three time points. Signals higher or lower than >0.8-fold by log2 were regarded as significant and analyzed further. Control and Brd4 KD cells exhibited a large difference in gene expression profiles at 8 h, whereas differences were less at 0 and 4 h. This was verified by the lower coefficients of correlation at 8 h relative to 0 and 4 h in scatter plot analysis (Fig. 3A). Similarly, expression profiles within control or KD cells differed most at 8 h in the principal component analysis that allows classifying expressed genes into groups of similar biological status and phenotype (Fig. 3B) (28). Thus, many genes were up-regulated at 8 h in control cells, in line with the induction of many genes during G1 (30, 32). In Brd4 KD cells, however, this large scale up-regulation was largely absent (Fig. 3C, supplemental Fig. S2); as many as 1108 genes (11%) were underexpressed in Brd4 KD cells at 8 h relative to control cells. At 0 and 4 h, about 200 genes showed underexpression in Brd4 KD cells relative to control cells. Contrary to the large number of genes underexpressed in Brd4 KD cells, only ∼100 genes were scored overexpressed in KD cells relative to control cells at all three time points. This score is likely to be an overestimate, because in the subsequent qRT-PCR analysis many of these genes did not reveal reduced expression in Brd4 KD cells (see the legend to Fig. 3C). This may be due to the conservative normalization we adopted for our microarray analysis. Collectively, these results indicate that Brd4 KD cells fail to up-regulate expression of many G1 genes, consistent with the idea that Brd4 has a role in the expression of multiple G1 genes. Our data also suggest that Brd4 represses relatively few genes during G0/G1 progression.
FIGURE 3.
Global gene expression patterns in control and Brd4 KD NIH3T3 cells. A, coefficient of correlation (R2) was calculated based on scatter plot analysis of microarray results. B, biplot scores for control and Brd4 KD cells at 0, 4, and 8 h were obtained according to the first two components of the principal component analysis. C, the number of genes whose expression was altered by Brd4 shRNA. *, the number of up-regulated genes listed was most likely an overestimate, since in qRT-PCR only 2 of 20 genes in this category showed higher expression in Brd4 KD cells compared with control cells. This discrepancy may be due to a normalization procedure used in this work.
Genes underexpressed in Brd4 KD cells during G1 were of diverse functions and included those important for G1/S progression, such as Ccnd1 and Ccnd2 (cyclin D1 and cyclin D2), as illustrated in hierarchical clustering analysis in supplemental Fig. S2. In addition, Orcl2 (Orc2) and Mcm2, components of the origin recognition complex, Dnttip, Dhfr, Top2a, Pcna, critical for DNA replication, were underexpressed in Brd4 KD cells (34). Furthermore, genes involved in chromatin regulation during G1/S transition, such as Chaf1a (known as CAF-1), Hmgb1, and And-1 (35) as well as other G1/S genes, Ranbp1, Pop1, Nid1, Msh2, Pms2, and Xab1 (36), were down-regulated in Brd4 KD cells.
To confirm microarray data, qRT-PCR was performed with separate RNA preparations for 35 selected genes (marked with asterisks in supplemental Fig. S2). All genes scored underexpressed in Brd4 KD cells by microarray analysis also showed reduced expression in qRT-PCR analysis (Fig. 4B; data not shown). We noted that the underexpressed gene group in our microarray analysis did not include some well characterized G1 genes. This included E2f genes, known to be critical for S phase entry and up-regulated during G1 (30, 31). qRT-PCR analysis showed that transcript levels of E2f1, and E2f3 through E2f6 as well as Tfdp1/Tfdp2 (Dp1/Dp2) were similar in control and Brd4 KD cells, suggesting that these genes did not require Brd4 or required it to a lesser degree (supplemental Fig. S3). However, transcripts for E2f2 and E2f7 were consistently underexpressed in Brd4 KD cells. Similarly, Myc and Jun, also important for G1 progression, were not scored lower in Brd4 KD cells in this microarray analysis but were nevertheless underexpressed in Brd4 KD cells, as revealed by qRT-PCR analysis (supplemental Fig. S3). Exclusion of these genes from the present microarray analysis may be due to the relatively high cut-off line and strict normalization procedure we used. These results further support the role of Brd4 in the expression of multiple G1 genes.
FIGURE 4.
Restoration of G1 gene expression and S phase entry upon Brd4 re-expression. A, NIH3T3 cells were transduced with control or Brd4 rescue vector plus Brd4 shRNA vector (Ctrl, Brd4 KD, and Rescue). Cells were synchronized by serum starvation, released, and incubated for 12 h. Expression of indicated proteins was analyzed by immunoblot. pRb, phosphorylated Rb. B, cells transduced as above (Ctrl, Brd4 KD, Rescue) were synchronized and allowed to proceed through G1 for the indicated times (h) and analyzed for expression of indicated genes by qRT-PCR. Values were normalized to 18 S rRNA. C, cells (Ctrl, Brd4 KD, Rescue) were allowed to exponentially grow for 14 days, and total cell yields were estimated as in Fig. 1B. D, Ctrl, Brd4 KD, and Rescue cells were synchronized as above, and cell cycle profiles were examined by flow cytometry as in Fig. 1C. See the quantification in supplemental Fig. S1D.
Brd4 Reintroduction Rescues G1 Gene Expression and S Phase Entry—The above data suggest that under-expression of multiple G1 genes partly accounts for G1 arrest in Brd4 KD cells. To test this possibility, rescue experiments were performed with a retroviral vector harboring a mutated Brd4 rendered resistant to Brd4 shRNA (see “Experimental Procedures”). NIH3T3 cells were sequentially transduced with control or rescue vector plus Brd4 shRNA vectors. Immunoblot analysis in Fig. 4A showed that the Brd4 rescue vector, but not the empty vector, led to re-expression of Brd4 protein in Brd4 KD cells at a level similar to that in control cells (transduced with control shRNA) without affecting the levels of TBP and TFIIB. Furthermore, introduction of the Brd4 rescue vector increased expression of cyclin D1 and D2 and restored Rb phosphorylation during G1. To test whether rescue of Brd4 also rescues G1 gene expression, qRT-PCR was performed for cyclin D1 and D2, Mcm2, Ranbp1, Nid1, Orc2, Pop1 in synchronized cells (Fig. 4B). As expected, expression of these genes was increased in control cells during G1, whereas the expression remained low in Brd4 KD cells without Brd4 rescue. Importantly, with Brd4 rescue, all of these genes increased their expression in Brd4 KD cells when cells progressed from G0 to G1. The levels and kinetics of transcript expression in rescued cells closely paralleled those of control cells, suggesting that Brd4 re-expression directly affected expression of these genes. Furthermore, Brd4 re-expression led to the restoration of cell growth. The growth curve analysis in Fig. 4C showed that cell yields in Brd4-rescued cells were similar to those of control cells tested on days 4, 10, and 14, whereas Brd4 KD cells without rescue grew more slowly through the same period. The restoration of cell growth was at least partly attributed to the rescue of S phase entry. In flow cytometry analysis presented in Fig. 4D and supplemental Fig. S1D, Brd4 KD cells without rescue remained arrested at G1, as expected. In contrast, a significant fraction of Brd4-rescued cells entered S as observed at 20 h. As anticipated, control cells entered S phase at 16 h and progressed further at 20 h. These data support the idea that proper levels of Brd4 expression are critically required for expression of multiple G1 genes and progression toward S phase.
Brd4 Is Recruited to G1 Genes—It was of importance to address the mechanism by which Brd4 regulates transcription of multiple G1 genes. We tested the possibility that Brd4 binds to the promoters of individual G1 genes in a cell cycle-dependent manner. Chromatin was prepared from synchronized control and Brd4 KD cells at 0 and 4 h upon release and precipitated with anti-Brd4 antibody. Precipitated DNA was amplified for the promoter fragments (containing transcription start site) of G1 genes by quantitative PCR. We first examined seven G1 genes whose expression was strongly dependent on Brd4 (Fig. 4B). Results of cyclin D1, D2, Orc2, Mcm2, and Pop1 are in Fig. 5A, and Ranbp1 and Nid1 are in supplemental Fig. S4. Brd4 binding to these promoters (red bar) was low at 0 h but substantially increased at 4 h in control cells. This increase was significant, because binding of control IgG (white bar) was low at both 0 and 4 h. Contrary to increased Brd4 recruitment in control cells, Brd4 binding in Brd4 KD cells was near background at 0 h and showed virtually no increase at 4 h. Thus, Brd4 was recruited to multiple G1 genes as cell cycle progressed from G0 to G1. To ascertain specificity of increased Brd4 recruitment, ChIP analysis was performed for Dp1, E2f1, and E2f4 genes whose expression was not affected by Brd4 shRNA, although their transcript expression was increased during G1. Brd4 binding to Dp1 (Fig. 5A) and E2f1 and E2f4 (supplemental Fig. S4) was low at 0 h and showed only a minor increase at 4 h in control cells. In Brd4 KD cells, Brd4 binding was at background both at 0 and 4 h. As another control, ChIP was performed for the Tlr9 gene that encodes an immune cell-specific Toll-like receptor not expressed in NIH3T3 cells (supplemental Fig. S4). Brd4 binding was again low at 0 h and showed a negligible increase at 4 h in control and Brd4 KD cells. These results indicate that Brd4 recruitment correlated well with Brd4-dependent G1 gene expression, supporting its role in G1 gene transcription.
FIGURE 5.
Cell cycle-dependent recruitment of Brd4 to G1 gene promoters. A, ChIP analysis was performed with control or Brd4 KD cells synchronized to G0 (0) and released for 4 h (4). Chromatin was precipitated with rabbit antibody for Brd4 and Cdk9 or for RNA pol II (N20) along with rabbit IgG as a control (IgG). Precipitated chromatin was analyzed for the indicated genes by quantitative PCR by detecting a region containing/near transcription initiation site. Values represent the average of three determinations ± S.D. Similar results were observed with three independent chromatin preparations. B, immunoblot analysis was performed for total extracts prepared from control and Brd4 KD cells synchronized to G0 and released at 0 h and 4 h.
Brd4 Increases Binding P-TEFb and RNA Pol II to G1 Genes— We sought to address the significance of Brd4 recruitment in G1 gene expression. It seemed possible that Brd4 binding facilitates binding of P-TEFb and RNA pol II complexes to the promoters, since Brd4 interacts with P-TEFb and components of the pol II Mediator (19, 23). P-TEFb is a Cdk9/cyclin T heterodimer essential for transcriptional elongation (21). ChIP analysis was performed with antibodies for Cdk9 and pol II (N20, recognizing total pol II) for the above genes (gray and black bars, respectively, in Fig. 5A and supplemental Fig. S4). For seven Brd4-dependent G1 genes (cyclin D1, D2, Orc2, Mcm2, Pop1, Ranbp1, and Nid1), binding of Cdk9 and pol II closely paralleled that of Brd4; their binding was noticeably increased at 4 h in control cells but not in Brd4 KD cells. Their binding to the Dp1, E2f1, E2f4, and Tlr9 genes, although appeared slightly increased at 4 h, was distinctly lower than those in the above Brd4-dependent genes. These results indicate that Brd4 promotes binding of P-TEFb and pol II to many G1 genes.
Immunoblot analysis in Fig. 5B confirmed that Cdk9 and pol II were expressed at comparable levels in control and Brd4 KD cells, and Brd4 levels were lower in Brd4 KD cells than control cells, as expected. Given that their levels remained unchanged during 0 and 4 h, it is likely that all three proteins were induced to bind to these G1 genes as cell cycle progressed from G0 to G1.
Brd4 Recruitment and Histone Acetylation—Brd4 binds to acetylated histones H3 and H4 (5, 12). Given that histone acetylation is generally associated with gene activation (37), it was of interest to study whether histone acetylation increases in G1 genes during G0-G1 progression and whether it correlates with Brd4 recruitment. ChIP analysis was performed with antibodies for diacetyl H3 (ac-K9, -K14) and tetra-acetyl H4 (Ac-K5, -K8, -K12, -K16) for Brd4-dependent G1 genes above (supplemental Fig. S5A). There was no clear increase in histone acetylation levels in these genes during G0-G1 progression. Only cyclin D1 showed an increase in H3/H4 acetylation during G1, but other genes gave variable acetylation patterns that showed no apparent correlation with Brd4 recruitment. We also examined genome-wide histone acetylation patterns during G1 by immunoblot analysis with antibodies for acetylated H3 and H4 at specific sites which showed no clear, consistent changes during G1 progression (supplemental Fig. S5B). These results indicate that Brd4 recruitment to G1 genes is not directly caused by increased histone acetylation.
DISCUSSION
This study began with the observations that Brd4 has a critical role in G1 progression and S phase entry. Microarray data showed that many G1 genes, although up-regulated in control cells, were not up-regulated in Brd4 KD cells, indicating its function in G1 gene transcription. Supporting a role in G1 gene transcription, Brd4 was recruited to individual G1 genes during G0-G1 progression. Furthermore, Brd4 recruitment correlated well with enhanced binding of P-TEFb and pol II to G1 genes, pointing to an underlying mechanism for Brd4-regulated G1 gene transcription and cell growth.
Brd4-dependent Growth Regulation and G1 Gene Expression— Brd4 knockdown by specific shRNA led to inhibition of cell growth both in NIH3T3 cells and MEFs. This inhibition was in part accounted for by G1 arrest in Brd4 KD cells. Indicating a G1 block rather than a delay in S phase entry, almost all Brd4 KD cells remained arrested at G1 even after prolonged incubation, with a fraction of cells undergoing apoptosis. The crucial role of Brd4 in G1 progression observed in this work may explain why Brd4-/- embryos die early and Brd4-/- cells fail to grow in culture (11, 12). The subsequent microarray work reinforced the role for Brd4 in G1 progression and provided further evidence that Brd4 KD cells, although capable of exiting G0, were unable to proceed with a normal, ordered course of G1 gene expression; although some well characterized G1 genes, such as E2fs and Dp1/Dp2 were expressed comparably in control and Brd4 KD cells, expression of many other G1 genes was markedly inhibited in Brd4 KD cells (30, 31). As many as 11% of genes (>1000 genes) studied here were underexpressed in Brd4 KD cells during G1 compared with control cells. This figure may be an underestimate, since other G1 genes not picked up by our microarray assays were nevertheless found underexpressed in Brd4 KD cells by qRT-PCR, including Myc and Jun. This omission is likely to be due to the conservative criteria we used to score down-regulation. Nevertheless, some G1 genes were clearly unaffected by Brd4 knockdown; particularly of interest are the E2f family of genes, many of which showed normal induction during G1 in Brd4 KD cells (supplemental Fig. S3). Thus, although Brd4 is critical for expression of many G1 genes, there are some G1 genes that apparently do not depend (or depend less) on Brd4 for their expression. Another interesting aspect of our data is that few genes were up-regulated as a result of Brd4 knockdown during this period, suggesting that Brd4 primarily functions to stimulate rather than to repress gene expression during G1, although Brd4 is shown to be capable of acting as a transcriptional repressor for a human Papillomavirus gene (24). Given that many genes underexpressed in Brd4 KD cells are critical for G1 progression, it is reasonable to assume that the global deficiency in G1 gene transcription is a main cause of G1 arrest in Brd4 KD cells. For example, the defects in cyclin D1 and D2 expression may lead to insufficient Rb phosphorylation, contributing not only to G1 arrest but apoptosis (33). Further supporting this view is that reintroduction of Brd4 rescued G1 gene expression, which led to rescue of S phase entry in Brd4 KD cells. However, in light of the reports that Brd4 overexpression also leads to cell growth inhibition (15, 16), Brd4 may regulate cell growth through other mechanisms as well.
Brd4 Recruitment and Increased Binding of Pol II and P-TEFb to G1 Promoters—ChIP data showed that Brd4 was recruited to all G1 genes whose expression depended on Brd4 as the cell cycle moved from G0 to G1. Illustrating a correlation between Brd4 recruitment and G1 gene expression, Brd4 was not recruited in Brd4 KD cells. Reinforcing this correlation, Brd4 recruitment was low to negligible for those G1 genes whose expression was not affected by Brd4 knockdown or not expressed in the cells. These data indicate that induced Brd4 recruitment is a basis of subsequent transcription of many G1 genes. This idea is strengthened by Brd4-dependent enhancement of P-TEFb and pol II binding to G1 genes. The importance of Brd4 in enhanced P-TEFb and pol II binding was evident, since it was noticeably lower in Brd4 KD cells. It can be surmised that enhanced P-TEFb binding to G1 genes is due to a direct, physical interaction between Brd4 with P-TEFb (19, 20). By recruiting P-TEFb, Brd4 may impact on transcriptional elongation of many G1 genes. However, P-TEFb may be recruited to some promoters in a Brd4-independent manner given that some DNA-specific transcription factors are shown to interact with P-TEFb (38-40). Furthermore, recent studies present evidence that P-TEFb is not required for some pol II-dependent genes (41, 42). Thus, co-recruitment of P-TEFb may not be a universal requirement for Brd4-dependent transcription.
As for Brd4-stimulated pol II binding, an interaction of Brd4 with components of the Mediator complex may serve as part of its mechanism (19, 23). At present, it is not clear whether Brd4 increases assembly of the preinitiation complex or stability/continuity of RNA pol II binding to the promoter. Considering that Mediator complexes affect post-assembly events and that an antibody that detected total pol II (N20), the latter possibility may be more likely. It is of note here that although pol II binding to many G1 genes was reduced in Brd4 KD cells, there was above-background binding of pol II at 0 and 4 h both in control and Brd4 KD samples. This trend was found even for the genes not affected by Brd4. This may be due to nonproductive occupancy of pol II that does not result in elongation, found broadly on a large fraction of promoters in human ES and other cells (43). Given that those genes whose expression was not significantly reduced in Brd4 KD showed overall low binding for Brd4, P-TEFb, and pol II, their expression may rely more heavily on transcriptional processing or message stability than de novo transcription.
Our attempt to correlate Brd4 recruitment with histone H3/H4 acetylation at G1 genes did not reveal a positive outcome; (a) histone acetylation did not increase during G0 to G1 progression in most G1 genes tested, (b) acetylation patterns in these genes displayed no discernible correlation with Brd4 recruitment patterns, and (c) genome-wide histone acetylation patterns were similar during G0 to G1 progression. Thus, chromatin acetylation does not appear to directly control Brd4 recruitment to G1 promoters. However, based on the requirement of the bromodomains for increasing human immunodeficiency virus-1 promoter activity, acetyl chromatin binding of Brd4 is likely to be required, although not sufficient for Brd4 recruitment (19). As an additional requirement, Brd4 may undergo signal-dependent post-translational modification or it may change interacting partners, promoting its ability to bind to specific genes.
In summary, Brd4 is recruited to a number of G1 genes during G0-G1 progression and stimulates binding of P-TEFb and pol II to the promoters, ultimately resulting in transcriptional elongation. By regulating a large set of G1 genes, Brd4 plays a critical role in proper G1 progression. This work adds Brd4 to a list of chromatin binding general transcription factors that broadly regulate gene expression relevant to cell growth. During revision of this paper, Yang et al. (44) published a paper reporting that expression of several G1 genes was dependent on Brd4 in HeLa cells.
Supplementary Material
Acknowledgments
We thank Drs. D. Price, Y. Su, and M. Ko for critical discussion of our results and P. Thotakura for microarray analysis.
This work was supported in part by the Intramural Program of NICHD, National Institutes of Health, and the Intramural AIDS targeted antiviral program (IATAP) program of National Institutes of Health. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1-S5.
Footnotes
The abbreviations used are: RNA pol II, RNA polymerase II; shRNA, small hairpin RNA; ChIP, chromatin immunoprecipitation; KD, knockdown; MEF, mouse embryonic fibroblasts; qRT-PCR, quantitative reverse transcription PCR; Rb, retinoblastoma; Ctrl, control.
References
- 1.Jeanmougin, F., Wurtz, J. M., Le Douarin, B., Chambon, P., and Losson, R. (1997) Trends Biochem. Sci. 22 151-153 [DOI] [PubMed] [Google Scholar]
- 2.Dey, A., Ellenberg, J., Farina, A., Coleman, A. E., Maruyama, T., Sciortino, S., Lippincott-Schwartz, J., and Ozato, K. (2000) Mol. Cell. Biol. 20 6537-6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ladurner, A. G., Inouye, C., Jain, R., and Tjian, R. (2003) Mol. Cell 11 365-376 [DOI] [PubMed] [Google Scholar]
- 4.Matangkasombut, O., and Buratowski, S. (2003) Mol. Cell 11 353-363 [DOI] [PubMed] [Google Scholar]
- 5.Dey, A., Chitsaz, F., Abbasi, A., Misteli, T., and Ozato, K. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 8758-8763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanno, T., Kanno, Y., Siegel, R. M., Jang, M. K., Lenardo, M. J., and Ozato, K. (2004) Mol. Cell 13 33-43 [DOI] [PubMed] [Google Scholar]
- 7.Nakamura, Y., Umehara, T., Nakano, K., Jang, M. K., Shirouzu, M., Morita, S., Uda-Tochio, H., Hamana, H., Terada, T., Adachi, N., Matsumoto, T., Tanaka, A., Horikoshi, M., Ozato, K., Padmanabhan, B., and Yokoyama, S. (2007) J. Biol. Chem. 282 4193-4201 [DOI] [PubMed] [Google Scholar]
- 8.Nagashima, T., Maruyama, T., Furuya, M., Kajitani, T., Uchida, H., Masuda, H., Ono, M., Arase, T., Ozato, K., and Yoshimura, Y. (2007) Mol. Hum. Reprod. 13 141-148 [DOI] [PubMed] [Google Scholar]
- 9.Denis, G. V., Vaziri, C., Guo, N., and Faller, D. V. (2000) Cell Growth Differ. 11 417-424 [PMC free article] [PubMed] [Google Scholar]
- 10.Crowley, T. E., Kaine, E. M., Yoshida, M., Nandi, A., and Wolgemuth, D. J. (2002) Mol. Endocrinol. 16 1727-1737 [DOI] [PubMed] [Google Scholar]
- 11.Houzelstein, D., Bullock, S. L., Lynch, D. E., Grigorieva, E. F., Wilson, V. A., and Beddington, R. S. (2002) Mol. Cell. Biol. 22 3794-3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishiyama, A., Dey, A., Miyazaki, J., and Ozato, K. (2006) Mol. Biol. Cell 17 814-823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.French, C. A., Kutok, J. L., Faquin, W. C., Toretsky, J. A., Antonescu, C. R., Griffin, C. A., Nose, V., Vargas, S. O., Moschovi, M., Tzortzatou-Stathopoulou, F., Miyoshi, I., Perez-Atayde, A. R., Aster, J. C., and Fletcher, J. A. (2004) J. Clin. Oncol. 22 4135-4139 [DOI] [PubMed] [Google Scholar]
- 14.Haruki, N., Kawaguchi, K. S., Eichenberger, S., Massion, P. P., Gonzalez, A., Gazdar, A. F., Minna, J. D., Carbone, D. P., and Dang, T. P. (2005) J. Med. Genet. 42 558-564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maruyama, T., Farina, A., Dey, A., Cheong, J., Bermudez, V. P., Tamura, T., Sciortino, S., Shuman, J., Hurwitz, J., and Ozato, K. (2002) Mol. Cell. Biol. 22 6509-6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farina, A., Hattori, M., Qin, J., Nakatani, Y., Minato, N., and Ozato, K. (2004) Mol. Cell. Biol. 24 9059-9069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.You, J., Croyle, J. L., Nishimura, A., Ozato, K., and Howley, P. M. (2004) Cell 117 349-360 [DOI] [PubMed] [Google Scholar]
- 18.Baxter, M. K., McPhillips, M. G., Ozato, K., and McBride, A. A. (2005) J. Virol. 79 4806-4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jang, M. K., Mochizuki, K., Zhou, M., Jeong, H. S., Brady, J. N., and Ozato, K. (2005) Mol. Cell 19 523-534 [DOI] [PubMed] [Google Scholar]
- 20.Yang, Z., Yik, J. H., Chen, R., He, N., Jang, M. K., Ozato, K., and Zhou, Q. (2005) Mol. Cell 19 535-545 [DOI] [PubMed] [Google Scholar]
- 21.Price, D. H. (2000) Mol. Cell. Biol. 20 2629-2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chao, S. H., and Price, D. H. (2001) J. Biol. Chem. 276 31793-31799 [DOI] [PubMed] [Google Scholar]
- 23.Wu, S. Y., and Chiang, C. M. (2007) J. Biol. Chem. 282 13141-13145 [DOI] [PubMed] [Google Scholar]
- 24.Wu, S. Y., Lee, A. Y., Hou, S. Y., Kemper, J. K., Erdjument-Bromage, H., Tempst, P., and Chiang, C. M. (2006) Genes Dev. 20 2383-2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka, T. S., Jaradat, S. A., Lim, M. K., Kargul, G. J., Wang, X., Grahovac, M. J., Pantano, S., Sano, Y., Piao, Y., Nagaraja, R., Doi, H., Wood, W. H., III, Becker, K. G., and Ko, M. S. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 9127-9132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadota, K., Miki, R., Bono, H., Shimizu, K., Okazaki, Y., and Hayashizaki, Y. (2001) Physiol. Genomics 4 183-188 [DOI] [PubMed] [Google Scholar]
- 27.Eisen, M. B., Spellman, P. T., Brown, P. O., and Botstein, D. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 14863-14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landgrebe, J., Wurst, W., and Welzl, G. (2002) Genome Biology 3 RESEARCH0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dahl, J. A., and Collas, P. (2007) Front. Biosci. 12 4925-4931 [DOI] [PubMed] [Google Scholar]
- 30.Sherr, C. J., and Roberts, J. M. (2004) Genes Dev. 18 2699-2711 [DOI] [PubMed] [Google Scholar]
- 31.Harbour, J. W., and Dean, D. C. (2000) Genes Dev. 14 2393-2409 [DOI] [PubMed] [Google Scholar]
- 32.Massague, J. (2004) Nature 432 298-306 [DOI] [PubMed] [Google Scholar]
- 33.Harbour, J. W., and Dean, D. C. (2000) Nat. Cell Biol. 2 65-67 [DOI] [PubMed] [Google Scholar]
- 34.DePamphilis, M. L. (2005) Cell Cycle 4 70-79 [DOI] [PubMed] [Google Scholar]
- 35.Hoek, M., and Stillman, B. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 12183-12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Meijden, C. M., Lapointe, D. S., Luong, M. X., Peric-Hupkes, D., Cho, B., Stein, J. L., van Wijnen, A. J., and Stein, G. S. (2002) Cancer Res. 62 3233-3243 [PubMed] [Google Scholar]
- 37.Strahl, B. D., and Allis, C. D. (2000) Nature 403 41-45 [DOI] [PubMed] [Google Scholar]
- 38.Barboric, M., Nissen, R. M., Kanazawa, S., Jabrane-Ferrat, N., and Peterlin, B. M. (2001) Mol. Cell 8 327-337 [DOI] [PubMed] [Google Scholar]
- 39.Kanazawa, S., Okamoto, T., and Peterlin, B. M. (2000) Immunity 12 61-70 [DOI] [PubMed] [Google Scholar]
- 40.Kanazawa, S., Soucek, L., Evan, G., Okamoto, T., and Peterlin, B. M. (2003) Oncogene 22 5707-5711 [DOI] [PubMed] [Google Scholar]
- 41.Gomes, N. P., Bjerke, G., Llorente, B., Szostek, S. A., Emerson, B. M., and Espinosa, J. M. (2006) Genes Dev. 20 601-612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luecke, H. F., and Yamamoto, K. R. (2005) Genes Dev. 19 1116-1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guenther, M. G., Levine, S. S., Boyer, L. A., Jaenisch, R., and Young, R. A. (2007) Cell 130 77-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, Z., He, N., and Zhou, Q. (2008) Mol. Cell. Biol. 28 967-976 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.