Abstract
Metnase, also known as SETMAR, is a SET and transposase fusion protein with an undefined role in mammalian DNA repair. The SET domain is responsible for histone lysine methyltransferase activity at histone 3 K4 and K36, whereas the transposase domain possesses 5′-terminal inverted repeat (TIR)-specific DNA binding, DNA looping, and DNA cleavage activities. Although the transposase domain is essential for Metnase function in DNA repair, it is not clear how a protein with sequence-specific DNA binding activity plays a role in DNA repair. Here, we show that human homolog of the ScPSO4/PRP19 (hPso4) forms a stable complex with Metnase on both TIR and non-TIR DNA. The transposase domain essential for Metnase-TIR interaction is not sufficient for its interaction with non-TIR DNA in the presence of hPso4. In vivo, hPso4 is induced and co-localized with Metnase following ionizing radiation treatment. Cells treated with hPso4-siRNA failed to show Metnase localization at DSB sites and Metnase-mediated stimulation of DNA end joining coupled to genomic integration, suggesting that hPso4 is necessary to bring Metnase to the DSB sites for its function(s) in DNA repair.
Metnase (SETMAR) is a double strand break (DSB)2 repair factor that contains two functional domains: a SET (Su(var)3-9, Enhancer-of-zeste, Trithorax) domain of histone lysine methyltransferase activity at histone 3, lysine 4, and lysine 36 (1) associated with chromatin opening (2-5) and a transposase domain containing the DDE motif (1, 6-8), a conserved acidic motif essential for strand transfer and end joining activities among transposase and retroviral integrase families (9-13). Deletion of either SET or transposase domain abolished Metnase function in vivo (1), suggesting that both domains are likely required for its role in DSB repair and genomic integration. Although Metnase is not an active transposase, it possesses most transposase functions such as sequence-specific DNA binding (6, 11, 14), assembly of paired end complexes (14), and DNA cleavage activity (11, 14). Unlike other transposases, however, Metnase-mediated DNA cleavage was nonprocessive and occurred in the absence of the TIR sequence (11).
Human Pso4 (hPso4) is a human homolog of the protein encoded by the PS04/PRP19 gene in Saccharomyces cerevisiae (15, 16). PSO4 gene is essential for cell survival in yeast (15), and cells harboring a mutant Pso4 showed sensitivity to DNA cross-linking agents, suggesting that PSO4 is an essential DNA repair gene in S. cerevisiae (15). Pso4 is a part of the pre-mRNA splicing complex consisting of Pso4, Cdc5L, Plrg1, and Spf27 (17) and has been previously linked to DNA repair through a direct physical interaction between Cdc5L and WRN, the protein deficient in Werner syndrome (18). Human Pso4 contains six successive WD-40 motifs at the C terminus that is known to form a structural interface for the assembly of multiprotein complexes (19) and has been identified as a component of the nuclear matrix (20). Pso4 is also a U-box protein with associated E3 ubiquitin ligase activity crucial for its function in pre-mRNA splicing in vivo (21-23). Human Pso4 interacts with terminal deoxynucleotidyl transferase (TdT), a protein that adds nucleotides to DNA ends generated during V(D)J recombination that are subsequently processed by proteins involved in general DSB repair pathways (16, 24). Purified hPso4 binds double-stranded DNA in a sequence-independent manner but does not bind single-stranded DNA. Human Pso4 protein is induced in cells by γ-radiation and interstrand cross-linkers, but not by UV treatment (16). A targeted inhibition of hPso4 expression results in accumulation of DNA DSBs and decreased cell survival after DNA damage (16), suggesting that hPso4 plays a unique role in mammalian DNA DSB repair. Nonetheless, the specific role(s) for Pso4 in DSB repair is not known.
In this study, we show that hPso4 is a Metnase (SETMAR)-binding partner that forms a stable complex with Metnase on both TIR and non-TIR DNA. Human Pso4 is induced and co-localized with Metnase following ionizing radiation (IR) treatment. Cells treated with hPso4-siRNA failed to show Metnase localization at DSB sites and Metnase-mediated stimulation of DNA end joining, suggesting that hPso4 forms a stable complex with Metnase and mediates Metnase association at DNA damage sites in vivo.
EXPERIMENTAL PROCEDURES
Cells, Enzymes, and Chemicals—Human embryonic kidney (HEK 293) cells were grown in Dulbecco's modified Eagle's medium/F-12 (Invitrogen) supplemented with 10% fetal calf serum (Invitrogen), penicillin (10 unit/ml; Sigma), and streptomycin (0.1 mg/ml; Sigma). [γ-32P]ATP (3000 Ci/mmol) was from PerkinElmer Life Sciences and Analytical Science (Boston, MA), and Bradford reagents and protein molecular weight markers were purchased from Bio-Rad. An anti-Metnase polyclonal antibody was previously described (1), and an anti-Pso4 polyclonal antibody specific for human Pso4 was obtained from the Calbiochem (Darmstadt, Germany) (16). Anti-FLAG M2 and V5 monoclonal antibodies were from Sigma and Invitrogen, respectively.
Generation of Stable Cell Lines and preparation of Cell Extracts and Immunoblot Analysis—Cells stably overexpressing wt-Metnase or wt-hPso4 were generated by transfecting HEK 293 cells with a vector harboring FLAG-Metnase, V5-Metnase, or FLAG-hPso4 using FuGENE 6 transfection reagent (Roche Applied Science), followed by selection with G418 (Invitrogen) for 14 days, after which a single colony was isolated and amplified. For preparation of cell extracts, the cells were briefly washed with phosphate-buffered saline and lysed in a buffer containing 25 mm HEPES (pH 7.5), 0.3 m NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 0.5% Triton X-100, 20 mm β-glycerolphosphate, 1 mm sodium vanadate, 1 mm dithiothreitol, protease inhibitor cocktails (Sigma). Cell lysates (50 μg) were loaded onto a SDS-PAGE, and following gel electrophoresis, the proteins were transferred to a polyvinylidene difluoride membrane (Millipore, Billerica, MA) and immunoblotted with primary antibody followed by peroxidase-coupled secondary antibody (Amersham Biosciences) and an enhanced chemiluminescence (Amersham Biosciences) reaction prior to visualization on Kodak X-Omat film.
DNA Substrates—DNA substrates were obtained from Integrated DNA Technologies (Coralville, IA). The 32-mer dsDNA containing either the 19-mer core sequence necessary for Metnase binding (TIR32) or a 5-nucleotide mutation at the core (MAR3M) was previously described. For Metnase-induced DNA looping experiment, 118-mer dsDNA containing either two TIR32 sequences in the forward direction separated by 54 bp of random nucleotides (Fwd-Fwd template: 5′-AGG TTG GTG CAA AAG TAA TTG CGG AGC TGG CTA TCG GAA CTC TCG GAA GTT GGG TCA GTT ACA ACG CGC CAC CCG CGC CCC GTA CTG ATA GCA GGG TGC AAA AGT AAT TGC GGA GGT T-3′) or a forward TIR32 sequence and a reverse TIR32 sequence separated by the same 54-bp nucleotides (Fwd-Rev template: 5′-AGG TTG GTG CAA AAG TAA TTG CGG AGC TGG CTA TCG GAA CTC TCG GAA GTT GGG TCA GTT ACA ACG CGC CAC CCG CGC CCC GTA CTG ATA GCA GGG CGT TAA TGA AAA CGT GGA GGT T-3′) were annealed.
Identification of Metnase-associated Proteins—Proteins associated with Metnase were identified by immunoprecipitation with an anti-FLAG M2 antibody (Sigma) and protein G-agarose beads (Upstate, Temecula, CA). Whole cell extracts of 293 cells overexpressing FLAG-Metnase were incubated with 3 μg of antibody at 4 °C for 2 h, followed by the addition of 100 μl of protein G-agarose beads overnight. The complex was washed four times with 25 mm Tris-HCl (pH 7.5) containing 250 mm NaCl prior to running on a 10% SDS-PAGE. After staining the gel with Coomassie Blue, individual bands were excised and immersed in 100 μl of 0.1 m ammonium bicarbonate and 50% acetonitrile (by volume) to completely cover the gel pieces (25). After incubation at 37 °C for 30 min, the gel pieces were dehydrated with 100 μl of acetonitrile for 5 min and rehydrated with 100 μl of 10 mm dithiothreitol at 55 °C for 45 min for reduction. Following alkylation with 100 μl of 55 mm iodoacetamide for 30 min at 37 °C, the gel pieces were briefly dehydrated with 100 μl of acetonitrile and treated with a trypsin solution (Promega V5280, 0.1-0.5 μg) overnight at 37 °C. Following trypsin digestion, the peptides were extracted with 0.1% trifluoroacetic acid and injected onto a 75-μm × 5-cm C-18 reverse-phase column. The peptides were eluted with a gradient from 5 to 45% acetonitrile over 30 min using an Agilent 1100 series nanopump. The column was interfaced with a LTQ ion trap mass spectrometer (Thermo), and the data were collected in the data-dependent neutral loss MS3 mode. Tandem mass spectrometry spectra were searched against the IPI human protein data base with SEQUEST algorithm.
Purification of FLAG-Metnase and FLAG-hPso4—Metnase (or hPso4)-expressing cells (1.6 × 108) were suspended in 20 ml of extraction buffer (TEGDN; 50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 10% glycerol, 5 mm dithiothreitol, 1.0% Nonidet-P40, and mammalian protease inhibitor cocktails containing 0.2 m NaCl) and centrifuged (100,000 × g) for 30 min. The supernatant (S100 fraction) was filtered through a Whatman paper and used for immunoaffinity purification. The S100 fraction was incubated at 4 °C for 60 min with anti-FLAG M2 affinity gel (Sigma) that had been pre-equilibrated with TEGDN buffer containing 0.2 m NaCl. The beads were washed three times with TEGDN, 2.0 m NaCl buffer prior to elution of the protein with TEGDN, 0.2 m NaCl containing FLAG peptide (500 μg/ml). The eluant was diluted with 10 volumes of TEGDN buffer and loaded onto a heparin-Sepharose 6 Fast Flow column (Amersham Biosciences) pre-equilibrated with TEGDN buffer. After washing the column, Metnase (or hPso4) was fractionated using a linear gradient (0-2.0 m NaCl) of TEGDN buffer. The eluted protein was dialyzed against TEGDN buffer containing 50 mm NaCl and stored at -80 °C.
Electrophoretic Mobility Shift Assay of Protein-DNA Interaction—Duplex DNA was labeled with [γ-32P]ATP (3,000 Ci/mmol) and T4 polynucleotide kinase (Roche Applied Science) according to the manufacturer's instructions. The indicated amount of purified Metnase and/or hPso4 was incubated with 200 fmol of 5′-32P-labeled DNA at room temperature for 15 min in a reaction mixture containing 50 mm HEPES-KOH (pH 7.8), bovine serum albumin (0.2 μg/μl), and 50 mm NaCl. The Metnase-DNA complex was analyzed on either 5% polyacrylamide gel (acrylamide:bis-acrylamide 43.2:0.8) in 0.5× TBE or 1% vertical agarose gel in 1× TBE. The gels were dried and exposed to x-ray films (Kodak). For quantification, the bands of interest were excised from the gels and measured for radioactivity using a Beckman Scintillation Counter LS 6500.
Transfection of Cells with siRNA—A control scrambled siRNA and Metnase-specific siRNA were obtained from Dharmacon Research Co. (Lafayette, CO). Human 293 cells (2.0 × 104) were plated on 6-well plates and incubated for 24 h prior to transfection. The cells were washed once with fresh medium, and siRNA (0.2-0.4 μm) was diluted in Dulbecco's modified Eagle's medium/F-12 to a final volume of 200 μl. The diluted siRNA samples were combined with transfection agent (Oligofectamine) and incubated for 20 min at room temperature before adding to cells. After incubation for 4 h, 0.5 ml of culture medium containing 30% serum was added without removing the transfection mixture, and the cells were further incubated for 48-72 h. The cell lysates were prepared and examined for efficacy of siRNA by Western blot analysis.
Laser Scanning Confocal Microscopy—The cells were grown up to 70% confluence on a sterilized Labtek II coverslip, irradiated with Gammacell-40 exactor (Nordion) as a 137Cs source, and further incubated at 37 °C until harvest. Immunofluorescence microscopy was carried out as described previously (26, 27). The images were collected using a Zeiss LSM-510 confocal microscope.
DNA End Joining Coupled to Genomic Integration Analysis— Chromosomal integration was analyzed by the ability of cells to pass foreign nonhomologous DNA containing a selectable marker to progeny. pRNA/U6.Hygro plasmid was transfected into 293 cells stably expressing mock (pFLAG2 vector), pFLAG2-wt-Metnase, or pFLAG2-D483A using calcium phosphate transfection method as previously described (1). Forty-eight hours after transfection, varying numbers of cells were plated into 100-mm dishes, and 24 h later selective marker (hygromycin, 0.15 mg/ml) was added. The cells were incubated for 14 days in the presence of hygromycin, washed twice with phosphate-buffered saline, and stained with 0.17% methylene blue in methanol, and colonies defined as greater than 50 cells were counted. All of the experiments were performed in triplicate.
RESULTS
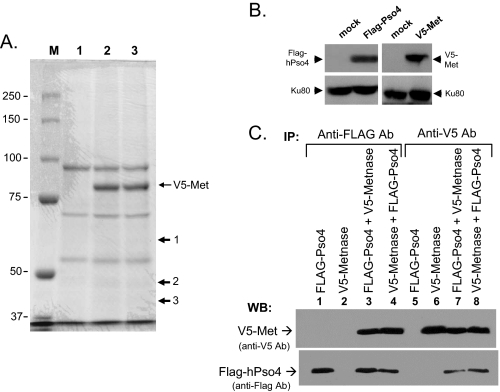
HPso4 Is a Metnase-binding Partner—Although Metnase (SETMAR) preserves most transposase activities including TIR-specific DNA binding, assembly of a paired end complex, and DNA cleavage (6, 11, 14), it unlikely catalyzes transposition in the human genome (14), suggesting that these activities may have a role in other DNA metabolism such as DNA repair (1, 14). Overexpression of Metnase increases both NHEJ and cell survival following IR treatment (1), suggesting that it plays a positive role in DSB repair, although its function in repair is not known. In an effort to find Metnase-binding partner(s), cell extracts from human 293 cells stably expressing FLAG-Metnase were used to pull down Metnase and its associated protein(s) using a FLAG monoclonal antibody. A SDS-PAGE analysis identified three proteins co-immunoprecipitated with Metnase (Fig. 1A). Based on a proteomic analysis, the 55-kDa protein was identified as the human homolog of the PRP19 gene product of S. cerevisiae (PRP19/Pso4, NCBI accession number NP_055317) involved in pre-mRNA splicing and DNA repair (16, 18, 28), and the 50-kDa protein was identified as NP_006506, a cleavage product of Metnase (SETMAR). Interestingly, the 37-kDa was identified as the human homolog of Spf27 (NP_060544), a member of the Prp19 core complex involved in pre-mRNA splicing (18, 28). Because both Metnase and hPso4 are involved in DSB repair (1, 16), we examined whether Metnase directly interacts with hPso4. For this, hPso4 cDNA was put into pFLAG-CMV4 vector to generate a stable cell overexpressing FLAG-hPso4. Human 293 cells stably expressing FLAG-Pso4 or V5-Metnase were transfected with pcDNA3-V5-Metnase or pFLAG-Pso4, respectively (Fig. 1B). Reciprocal protein pulldown experiments confirmed that Metnase directly interacted with hPso4 (Fig. 1C).
FIGURE 1.
hPso4 is a novel Metnase-binding partner. A, SDS-PAGE analysis of Metnase and associated proteins. Whole cell extracts prepared from control 293 cells (lane 1) or cells stably expressing V5-Metnase (lanes 2 and 3) were incubated with 3 μg of V5-monoclonal antibody at 4 °C for 2 h and further incubated overnight following the addition of 100 μl of protein G-agarose beads. The complex was washed four times with a buffer containing 250 mm NaCl prior to running on a 10% SDS-PAGE and staining with Coomassie Blue. Lane M represents molecular weight markers. Proteins 1, 2, and 3 were identified as PRP19 gene product of S. cerevisiae (PRP19/Pso4), a cleavage product of Metnase (SETMAR), and the human homolog of Spf27, respectively. B, Western blot (WB) analysis of cells transfected with vector alone or vector expressing either V5-Metnase- or FLAG-hPso4. Ku80 was used as an internal loading control. C, interaction of Metnase with hPso4. Whole cell extracts (100 μg) from 293 cells overexpressing FLAG-Pso4 (lanes 1 and 5), V5-Metnase (lanes 2 and 6), or both (lanes 3, 4, 7, and 8) were incubated for 2 h with either anti-FLAG (lanes 1-4) or anti-V5 (lanes 5-8) antibody and then for an additional 2 h with protein G-agarose. In lanes 3 and 7, human 293 cells overexpressing FLAG-Pso4 were transfected with a plasmid expressing V5-Metnase, whereas in lanes 4 and 8, cells overexpressing V5-Metnase were transfected with a plasmid expressing FLAG-Pso4. Following immunoprecipitation (IP), the proteins were run on a 10% SDS-PAGE, transferred to membrane, and probed with either anti-V5 monoclonal (top panel) or anti-Pso4 polyclonal (bottom panel) antibody.
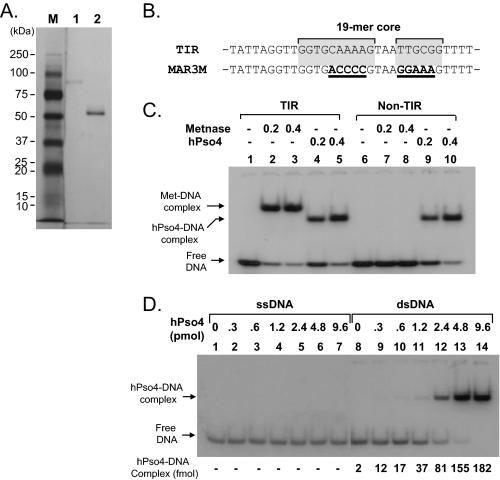
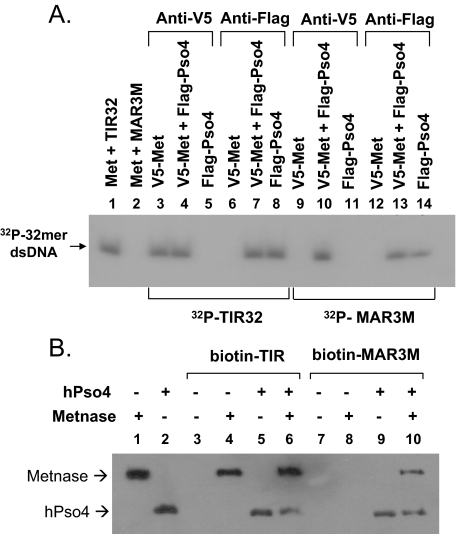
Metnase Associates with Non-TIR DNA in the Presence of hPso4—Human Pso4 is not only a Metnase-binding partner but also a dsDNA-binding protein that plays a major but undefined role in mammalian DNA repair (16). We therefore examined whether the Metnase-DNA interaction is influenced by hPso4. Using affinity-purified proteins (Fig. 2A), we first examined DNA binding activity of Metnase and hPso4. Unlike the TIR-specific DNA binding activity of Metnase (6, 14, 29) (Fig. 2C), hPso4 showed dsDNA binding activity with both TIR and non-TIR DNA (Fig. 2, C and D) (16). On the basis of a titration experiment, we calculated an approximate KD value of 9.0 × 10-8 m for the hPso4-dsDNA interaction (Fig. 2D). In a reciprocal co-immunoprecipitation experiment, Metnase was able to associate with non-TIR DNA (MAR3M) in the presence of hPso4, but it alone did not (Fig. 3A). To see whether Metnase association with non-TIR DNA represents the Metnase-hPso4-DNA complex, we used biotin-labeled duplex DNA bound to streptavidin beads for a pulldown of the protein(s)-DNA complex. In keeping with the co-immunoprecipitation experiment (Fig. 3A), Metnase interacted with non-TIR DNA in the presence of hPso4, whereas Metnase alone failed to associate with non-TIR DNA (Fig. 3B), suggesting that Metnase associates with non-TIR DNA via interaction with hPso4.
FIGURE 2.
Interaction of Metnase or hPso4 with DNA. A, SDS-PAGE of immunoaffinity purified FLAG-Metnase (lane 1) and FLAG-hPso4 (lane 2) used in this study. B, DNA sequence of the TIR and the mutant (MAR3M). The bipartite Metnase binding core site (19-mer) is shaded gray. In MAR3M, mutation sites are indicated as bold type with underlines. C, interaction of Metnase or hPso4 with TIR32 or non-TIR (MAR3M) DNA. The reaction mixtures (20 μl) containing indicated amount of Metnase (or hPso4) and 5′-32P-labeled DNA (200 fmol; ∼3,000cpm/fmol) were incubated for 15 min at 25 °C. After incubation, the samples were analyzed by 5% native PAGE. D, interaction of hPso4 with dsDNA and not single-stranded DNA. Reaction mixtures (20 μl) containing increasing amounts of hPso4 and 200 fmol of 5′-32P-labeled single-stranded DNA (top strand of MAR3M) or dsDNA (MAR3M duplex DNA) were incubated for 15 min at 25 °C and analyzed by 5% PAGE.
FIGURE 3.
Metnase associates with non-TIR DNA in the presence of hPso4. A, V5-wt-Metnase and/or FLAG-wt-hPso4 were immunoprecipitated with cell extracts (100 μg) overexpressing V5-Metnase (lanes 3, 6, 9, and 12), FLAG-Pso4 (lanes 5, 8, 11, and 14), or both (lanes 4, 7, 10, 13) using either anti-V5 (lanes 3-5 and 9-11) or -FLAG (lanes 6-8 and 12-14) monoclonal antibody. Following immunoprecipitation, either 5′-32P-labeled TIR32 (lanes 3-8) or 32P-MAR3M (lanes 9-14) was added to the mixtures for interaction of Metnase/hPso4 with 32P-labeled DNA. Following 10% SDS-PAGE, gel was dried and exposed to x-ray film. In control lanes (lanes 1 and 2), purified FLAG-Metnase (100 ng) was incubated with either TIR32 or MAR3M prior to immunoprecipitation using an anti-FLAG antibody. B, purified FLAG-wt-Metnase and/or FLAG-wt-hPso4 were incubated for 15 min prior to addition of either biotin-labeled TIR (lanes 3-6) or MAR3M (lanes 7-10). Following the addition of Streptavidin-agarose beads, the protein-DNA complexes were precipitated, washed, and analyzed by 10% SDS-PAGE. Western blotting was done using an anti-FLAG antibody.
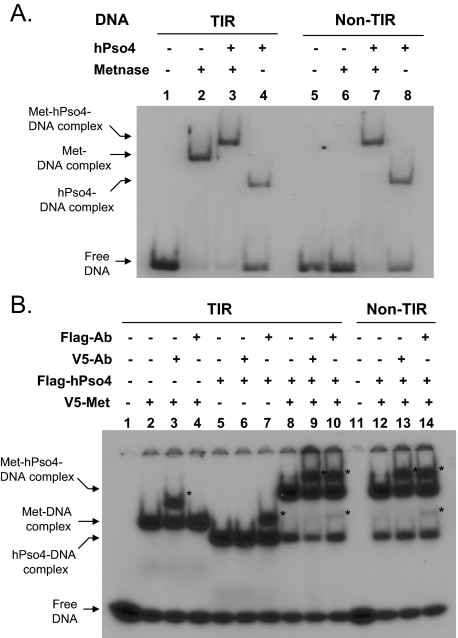
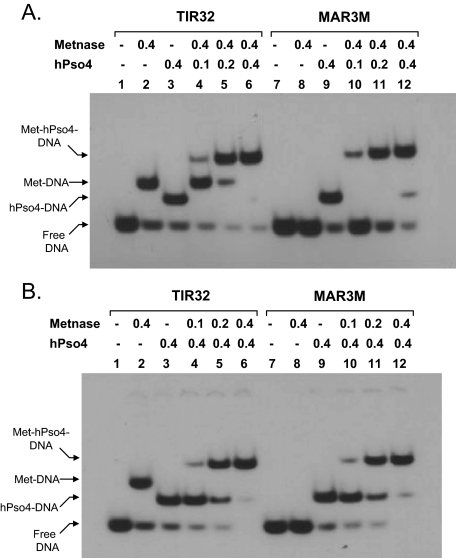
Metnase Forms a Stable Complex with hPso4 on Both TIR and Non-TIR DNA—To further understand how hPso4-Metnase interaction influences Metnase DNA binding activity, we examined the interaction of Metnase with hPso4 on DNA using a gel mobility shift assay. Metnase showed TIR-specific DNA binding activity, whereas hPso4 formed a stable complex with both TIR and non-TIR DNA (Fig. 4A). A slower migrating band was observed when Metnase and hPso4 was incubated together in the presence of TIR or non-TIR DNA (Fig. 4A, lanes 3 and 7), suggesting that Metnase forms a stable complex with hPso4 on DNA. To confirm that the slower migrating band truly represents the Metnase-hPso4 complex on DNA, we carried out a supershift assay where the complex of V5-Metnase and/or FLAG-hPso4 with 32P-labled DNA was identified by a supershift in the presence of either V5 or FLAG antibody. A more slowly migrating band observed with V5-Metnase, FLAG-hPso4, and DNA (TIR32 or MAR3M) was supershifted in the presence of either V5 or FLAG antibody (Fig. 4B, lanes 9, 10, 13, and 14), which verifies a stable complex of Metnase and hPso4 on both TIR and non-TIR DNA. We next examined how Metnase and hPso4 influence their binding to TIR and non-TIR DNA. In a gel mobility shift assay, the Metnase-hPso4-DNA complex was proportionally increased with increasing amounts of hPso4 (Fig. 5A) or Metnase (Fig. 5B) in the presence of either TIR or non-TIR (MAR3M) DNA, confirming that hPso4 mediates the Metnase-non-TIR DNA interaction via physical interaction. The result also suggests that Metnase and hPso4 form a single complex that appears to have one hPso4 tetramer (23) and one Metnase dimer (Ref. 14 and data not shown) on dsDNA.
FIGURE 4.
Metnase forms a stable complex with non-TIR DNA via interaction with hPso4. A, interaction of Metnase and/or hPso4 with TIR32 (lanes 1-4) or non-TIR (MAR3M) DNA (lanes 5-8). Reaction mixtures (20 μl) containing Metnase (0.2 μg) and/or hPso4 (0.2 μg), and 200 fmol of 5′-32P-labeled DNA were incubated for 15 min at 25 °C and analyzed by 5% native PAGE. B, interaction of V5-Metnase and/or FLAG-hPso4 with DNA in the presence of V5 or FLAG antibody. Reaction mixtures (20 μl) containing Metnase (0.2 μg) and/or hPso4 (0.2 μg) were incubated with 1 μg of V5 or FLAG antibody for 30 min at 25 °C prior to the addition of 5′-32P-labeled DNA (200 fmol). After 15 min of incubation, samples were analyzed by 5% native PAGE. The antibody-protein-DNA complexes (supershift) are indicated as asterisks.
FIGURE 5.
Formation of the Metnase-hPso4 complex on TIR and non-TIR DNA. Reaction mixtures (20 μl) were the same as those indicated in Fig. 2C, except that varying amounts of hPso4 (A) or wt-Metnase (B) were included. Individual protein-DNA complexes are marked on the left side of the figure.
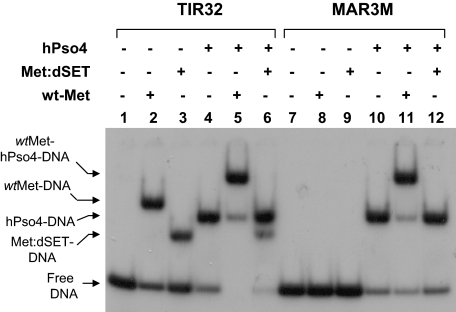
The Transposase Domain Is Not Sufficient for Formation of the Metnase-hPso4 Complex on DNA—To further understand the Metnase-hPso4 interaction on DNA, we compared wt-Metnase with a mutant lacking the SET domain (Met:dSET) for interaction with hPso4 on DNA. Although Met:dSET retains its TIR binding activity, it failed to form a complex with hPso4 on TIR or non-TIR DNA (Fig. 6), suggesting that the transposase domain essential for Metnase-TIR interaction is not sufficient for the Metnase-hPso4 interaction with either TIR or non-TIR DNA. The SET domain may be directly responsible for interaction with hPso4, although we were not able to test a mutant lacking transposase domain because it was unstable and quickly degraded in cell extracts (data not shown).
FIGURE 6.
The transposase domain is not sufficient for formation of the Metnase-hPso4 complex on DNA. A, wt-Metnase or a mutant lacking the SET domain (Met:dSET) was incubated with 32P-TIR (lanes 1-6) or non-TIR (32P-MAR3M, lanes 7-12) in the absence or the presence of hPso4. Following 15 min of incubation, protein-DNA complexes were analyzed by 5% nondenaturing PAGE. Individual protein-DNA complexes are marked on the left side of the figure.
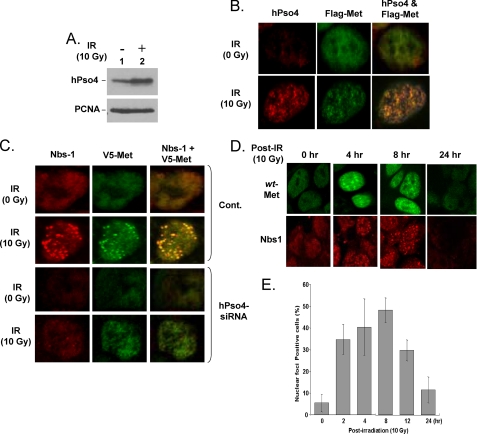
Human Pso4 Co-localizes with Metnase and Is Necessary for Metnase Localization at DSB Sites—Because Metnase and hPso4 form a stable complex on nonTIR DNA, we examined whether Metnase co-localizes with hPso4 at DSB sites. For this, human 293 cells stably expressing FLAG-Metnase were treated with IR (0 and 10 Gy), fixed, and examined for co-localization of the two proteins using anti-FLAG monoclonal and -hPso4 polyclonal antibodies. HPso4 was induced (Fig. 7A) and formed distinct nuclear foci following IR treatment (Fig. 7B, first column), and Metnase also formed distinct nuclear foci following IR treatment (Fig. 7B, second column). A merged image of Metnase and hPso4 showed that Metnase co-localized with hPso4 following IR (Fig. 7B, third column). We next examined whether hPso4 is necessary to form Metnase foci at DSB sites. Human 293 cells stably expressing FLAG-Metnase were transfected with a control or hPso4-siRNA, and 48 h later, the cells were treated with IR (10 Gy) and examined for formation of nuclear foci of Metnase and Nbs-1 at DSB sites. Cells treated with a control siRNA showed formation of distinct Metnase foci co-localized with Nbs-1 following IR treatment (Fig. 7C, second row). Cells treated with hPso4-siRNA also showed somewhat less formation of both Nbs-1 and Metnase foci following IR treatment; however, Metnase failed to co-localize with Nbs-1 (Fig. 7C, fourth row), suggesting that hPso4 is necessary for formation of discrete Metnase foci at DSB sites. A kinetic study showed that distinct Metnase foci were formed within 2 h and peaked at 8 h following IR exposure (Fig. 7, D and E), which was very similar to the kinetics of Nbs-1 foci formation (Fig. 7D) (30, 31).
FIGURE 7.
hPso4 is necessary for formation of Metnase foci at DSB sites. A, Western blot analysis of hPso4 expression following IR treatment. Proliferating cell nuclear antigen was used as a loading control. B, co-localization of Metnase with hPso4 following DSB damage in vivo. Human 293 cells overexpressing FLAG-Metnase were treated with 0 Gy (top panels) or 10 Gy (bottom panels) of IR. Following 8 h of incubation, the cells were fixed and examined for cellular localization of Metnase and hPso4 using anti-FLAG monoclonal (Sigma) and anti-hPso4 polyclonal (Calbiochem) antibodies. The images were obtained following double labeling of fixed cells with polyclonal antibody to hPso4 and monoclonal antibody to FLAG epitope with two different fluorochromes, Texas Red-conjugated anti-rabbit antibody, and fluorescein-conjugated anti-mouse antibody. A co-localization of Metnase and hPso4 was examined using a Zeiss LSM-510 confocal microscope. C, effect of hPso4 on formation of discrete nuclear foci of Metnase at DSB sites. Human 293 cells stably expressing FLAG-Metnase were treated with a control (Cont.) siRNA or hPso4-siRNA (5′-ACCACAGGCUGGCCUCAUUTT-3′, 5′-AAUGAGGCCAGCCUGUGGUTT-3′) for 48 h. The cells were then treated with 0 or 10 Gy of IR and examined for formation of Metnase and/or Nbs-1 foci. D, kinetic analysis of Metnase foci formation following IR treatment. Human 293 cells expressing FLAG-wt-Metnase were treated with IR (10 Gy) and harvested samples at various times for nuclear foci formation of Metnase and Nbs-1. The cells were fixed, labeled with an anti-FLAG or anti-Nbs-1 antibody, and images were collected using a Zeiss LSM-510 confocal microscope. E, cells (%) with IR-induced Metnase foci were calculated from the experiment in Fig. 7D. For each time point, three samples were analyzed.
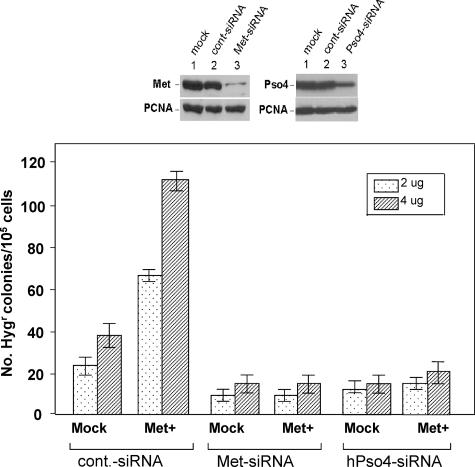
Cells Treated with hPso4-siRNA Fail to Show Metnase-mediated Stimulation of DNA End Joining Coupled to Genomic Integration—Pso4 is a dsDNA-binding protein that plays a major but undefined role in mammalian DNA repair (16). Our confocal microscopic study (Fig. 7) suggested that one of the roles of hPso4 in DSB repair is to bring Metnase to the DSB sites through physical interaction. This prompted us to test whether hPso4 influences the function of Metnase in DSB repair in vivo. Human 293 cells (1.0 × 105/plate) either mock or overexpressing Metnase were transfected with Metnase-siRNA, hPso4-siRNA, or a control siRNA, and examined how a targeted inhibition of hPso4 expression affects Metnase-mediated stimulation of in vivo end joining coupled to genomic integration. For selection to occur, the linearized U6 Hygro plasmid containing the Hygr marker must be end-joined and then integrated within the genome. Following incubation at 37 °C for 14 days, the two-step NHEJ/genomic integration process was measured by the number of Hygr colonies/105 cells/plate (1). Overexpression of wt-Metnase in human 293 cells did not affect plating efficiency (data not shown) but increased integration of a plasmid DNA carrying Hygr marker by 2-3-fold, whereas cells treated with hPso4-siRNA inhibited colony formation in both mock and Metnase-overexpressing cells (Fig. 8), suggesting that hPso4 is necessary to bring Metnase to the DSB sites for its function(s) in DNA repair.
FIGURE 8.
A targeted inhibition of hPso4 expression abolished Metnase-mediated stimulation of DNA end joining coupled to genomic integration. Top panels, effect of Metnase- or hPso4-specific siRNA on protein expression. Human 293 cells were treated with mock (lane 1), scrambled siRNA (lane 2), or Metnase- or hPso4-specific siRNA (lane 3). After 48 h, the cells were harvested and analyzed for Metnase or hPso4 expression by Western blotting. Proliferating cell nuclear antigen was used as a loading control. Bottom panel, effect of Metnase or hPso4 siRNA on DNA end joining coupled to genomic integration. Human 293 cells (1.0 × 105/plate) stably transfected with pFLAG2 (mock) or pFLAG2-Metnase were treated with control-siRNA, Metnase-siRNA, or hPso4-siRNA. Forty-eight hours later, the cells were transfected with indicated amounts (2 and 4 μg) of KpnI-linearized pRNA/U6.Hygro plasmid. Following incubation at 37 °C for 14 days, the number of Hygr colonies were measured for assessment of the in vivo end joining efficiency (1). For statistical analysis, six plates/sample were analyzed using the paired t test.
DISCUSSION
Metnase is a SET and transposase fusion protein with histone lysine methyltransferase (1), a sequence (TIR)-specific DNA binding (6, 11, 14), assembly of paired end complex (14), and DNA cleavage activities (14, 32). Although Metnase is involved in DSB repair (1), it is not clear how a protein with sequence-specific DNA binding activity functions in DNA repair. In this study, we show that a damage-induced DNA repair factor, hPso4 is the Metnase-binding partner that mediates Metnase interaction with non-TIR DNA such as DNA damage sites necessary for the function of Metnase in DSB repair.
HPso4 is a human homolog of the PS04/PRP19 gene in S. cerevisiae that has pleiotropic functions in DNA recombination and error-prone repair (15, 16, 18, 28, 33-38). Specific role(s) for Pso4 in DSB repair is not known; however, it has been identified as a component of the nuclear matrix (20). Although hPso4 is a direct binding partner of Metnase, our immunoprecipitation of Metnase experiment also pulled down the human homolog of Spf27, a member of the Prp19 core complex involved in pre-mRNA splicing. Because the Pso4 is a part of the pre-mRNA splicing complex consisting of Pso4, Cdc5L, Plrg1, and Spf27 (17), Metnase may interact with a portion of the pre-mRNA splicing complex. In fact, the presplicing complex was shown to link to DNA repair through a direct physical interaction of Cdc5L with WRN, a DSB repair factor deficient in Werner syndrome (18, 28). It is possible that the Metnase-hPso4 interaction is only a part of the much bigger complex that participates in DSB repair in vivo.
Metnase recognizes the 19-mer core of the TIR of the Hsmar1 element (6, 11, 14). Given that about half of the human genome derives from transposable elements (39), and there are over 7,000 potential Metnase-binding sites in the human genome sequence (11, 14),3 the recruitment of the Metnase to the TIR sites may form a gene regulatory network that affects global gene expression (8). Upon DNA damage, however, hPso4, another DSB repair factor with dsDNA binding activity was induced (16) (Fig. 7A) and formed a stable complex with Metnase on both TIR and non-TIR DNA (Figs. 4 and 5). Following IR treatment, Metnase formed discrete nuclear foci and co-localized with the MRN complex (Mre11-Rad50-Nbs-1). Given that the MRN complex is a damage sensor that recognizes the DSB sites and recruits other key factors such as ATM (40), IR-induced Metnase foci likely formed at DSB sites. From our finding that the treatment of cells with hPso4-siRNA abolished co-localization of Metnase and Nbs-1 at DSB sites (Fig. 7C), the interaction of hPso4-Metnase likely plays a crucial role in the recruitment of Metnase to the DSB sites. The transposase domain crucial for Metnase-TIR interaction was not sufficient for interaction with hPso4 on DNA, indicating that the SET domain is likely responsible for interaction with hPso4. Given that the SET domain of Metnase is essential for its function in DNA repair (1), the SET domain may have a unique role in its localization at DSB sites. It remains to be seen whether the SET domain contributes to the function of Metnase in DSB repair via histone lysine methyltransferase, interaction with hPso4, or both.
Metnase and hPso4 share common properties. First, both are DNA-binding proteins involved in DSB repair (1, 6, 11, 14, 16, 18), and their chromatin associations are enhanced following DNA damage (18),4 suggesting that Metnase and hPso4, once forming a complex, likely function together at DSB sites for DNA repair. Our study described here showed that the Metnase-hPso4 interaction modulates the DNA binding activity of Metnase to allow the localization of Metnase at non-TIR region such as DNA damage sites. It would be interesting to see whether hPso4 also affects the other biochemical activities of Metnase such as DNA cleavage and histone lysine methyltransferase. Additionally, Pso4/Prp19 is a U-box protein with associated E3 ubiquitin ligase activity that is critical for its function in pre-mRNA splicing in vivo (21-23). Given that ubiquitylation is rapidly induced at repair foci in response to DNA damage and can be detected using an antibody that specifically detects conjugated ubiquitin (41, 42), hPso4 E3 ligase activity may also play a role in DSB repair. It remains to be seen whether Metnase is a physiological substrate of hPso4.
This work was supported by National Institutes of Health Grant CA92111 (to S.-H. L.), National Institutes of Health Predoctoral Training Grants T32 DK0075-20 (to B. D. B.) and T32 CA111198-01 (to Y. R.), United StatesArmy Grant DAMD17-00-1-0295 (to S.-H. L.), and funds from the Walther Cancer Institute and the IU Simon Cancer Center. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: DSB, double strand break; TIR, 5′-terminal inverted repeat sequence; hPso4, human Pso4; E3, ubiquitin-protein isopeptide ligase; wt, wild type; ds, double-stranded; siRNA, small interfering RNA; Gy, gray; IR, ionizing radiation.
X. Li, unpublished data.
Y. J. Lee, unpublished data.
References
- 1.Lee, S. H., Oshige, M., Durant, S. T., Rasila, K. K., Williamson, E. A., Ramsey, H., Kwan, L., Nickoloff, J. A., and Hromas, R. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 18075-18080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamamoto, R., Furukawa, Y., Morita, M., Iimura, Y., Silva, F. P., Li, M., Yagyu, R., and Nakamura, Y. (2004) Nat. Cell Biol. 6 731-740 [DOI] [PubMed] [Google Scholar]
- 3.Krogan, N. J., Kim, M., Tong, A., Golshani, A., Cagney, G., Canadien, V., Richards, D. P., Beattie, B. K., Emili, A., Boone, C., Shilatifard, A., Buratowski, S., and Greenblatt, J. (2003) Mol. Cell Biol. 23 4207-4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao, T., Hall, H., Kizer, K. O., Shibata, Y., Hall, M. C., Borchers, C. H., and Strahl, B. D. (2003) Genes Dev. 17 654-663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang, X., Yang, Z., Khan, S. I., Horton, J. R., Tamaru, H., Selker, E. U., and Cheng, X. (2003) Mol. Cell 12 177-185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cordaux, R., Udit, S., Batzer, M. A., and Feschotte, C. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 8101-8106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robertson, H. M., and Zumpano, K. L. (1997) Gene (Amst.) 205 203-217 [DOI] [PubMed] [Google Scholar]
- 8.Jordan, I. K. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 7941-7942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craigie, R. (2001) J. Biol. Chem. 276 23213-23216 [DOI] [PubMed] [Google Scholar]
- 10.Skalka, A. M., and Katz, R. A. (2005) Cell Death Differ. 12 (Suppl. 1) 971-978 [DOI] [PubMed] [Google Scholar]
- 11.Roman, Y., Oshige, M., Lee, Y. J., Goodwin, K., Georgiadis, M. M., Hromas, R. A., and Lee, S. H. (2007) Biochemistry [DOI] [PMC free article] [PubMed]
- 12.Leschziner, A. E., Griffin, T. J. T., and Grindley, N. D. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 7345-7350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vos, J. C., and Plasterk, R. H. (1994) EMBO J. 13 6125-6132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, D., Bischerour, J., Siddique, A., Buisine, N., Bigot, Y., and Chalmers, R. (2007) Mol. Cell Biol. 27 1125-1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grey, M., Dusterhoft, A., Henriques, J. A., and Brendel, M. (1996) Nucleic Acids Res. 24 4009-4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahajan, K. N., and Mitchell, B. S. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 10746-10751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ajuh, P., Kuster, B., Panov, K., Zomerdijk, J. C., Mann, M., and Lamond, A. I. (2000) EMBO J. 19 6569-6581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, N., Kaur, R., Lu, X., Shen, X., Li, L., and Legerski, R. J. (2005) J. Biol. Chem. 280 40559-40567 [DOI] [PubMed] [Google Scholar]
- 19.Gotzmann, J., Gerner, C., Meissner, M., Holzmann, K., Grimm, R., Mikulits, W., and Sauermann, G. (2000) Exp. Cell Res. 261 166-179 [DOI] [PubMed] [Google Scholar]
- 20.Gerner, C., and Sauermann, G. (1999) J. Cell Biochem. 72 470-482 [PubMed] [Google Scholar]
- 21.Ohi, M. D., Vander Kooi, C. W., Rosenberg, J. A., Chazin, W. J., and Gould, K. L. (2003) Nat. Struct. Biol. 10 250-255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loscher, M., Fortschegger, K., Ritter, G., Wostry, M., Voglauer, R., Schmid, J. A., Watters, S., Rivett, A. J., Ajuh, P., Lamond, A. I., Katinger, H., and Grillari, J. (2005) Biochem. J. 388 593-603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vander Kooi, C. W., Ohi, M. D., Rosenberg, J. A., Oldham, M. L., Newcomer, M. E., Gould, K. L., and Chazin, W. J. (2006) Biochemistry 45 121-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahajan, K. N., Gangi-Peterson, L., Sorscher, D. H., Wang, J., Gathy, K. N., Mahajan, N. P., Reeves, W. H., and Mitchell, B. S. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 13926-13931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen, J. Y., Shen, C., Yan, Z., Brown, D. P., and Wang, M. (2006) Comput. Syst. Bioinformatics Conf, 389-398 [PubMed]
- 26.Park, S. J., Ciccone, S. L., Freie, B., Kurimasa, A., Chen, D. J., Li, G. C., Clapp, D. W., and Lee, S. H. (2004) J. Biol. Chem. 279 6046-6055 [DOI] [PubMed] [Google Scholar]
- 27.Murti, K. G., He, D. C., Brinkley, B. R., Scott, R., and Lee, S. H. (1996) Exp. Cell Res. 223 279-289 [DOI] [PubMed] [Google Scholar]
- 28.Lu, X., and Legerski, R. J. (2007) Biochem. Biophys. Res. Commun. 354 968-974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roman, Y., Oshige, M., Lee, Y. J., Goodwin, K., Georgiadis, M. M., Hromas, R. A., and Lee, S. H. (2007) Biochemistry 46 11369-11376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maser, R. S., Mirzoeva, O. K., Wells, J., Olivares, H., Williams, B. R., Zinkel, R. A., Farnham, P. J., and Petrini, J. H. (2001) Mol. Cell Biol. 21 6006-6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirzoeva, O. K., and Petrini, J. H. (2001) Mol. Cell Biol. 21 281-288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park, J. S., Wang, M., Park, S. J., and Lee, S. H. (1999) J. Biol. Chem. 274 29075-29080 [DOI] [PubMed] [Google Scholar]
- 33.Brendel, M., Bonatto, D., Strauss, M., Revers, L. F., Pungartnik, C., Saffi, J., and Henriques, J. A. (2003) Mutat. Res. 544 179-193 [DOI] [PubMed] [Google Scholar]
- 34.Brendel, M., and Henriques, J. A. (2001) Mutat. Res. 489 79-96 [DOI] [PubMed] [Google Scholar]
- 35.da Silva, K. V., de Morais Junior, M. A., and Henriques, J. A. (1995) Curr. Genet. 27 207-212 [DOI] [PubMed] [Google Scholar]
- 36.de Andrade, H. H., Marques, E. K., Schenberg, A. C., and Henriques, J. A. (1989) Mol. Gen. Genet. 217 419-426 [DOI] [PubMed] [Google Scholar]
- 37.Henriques, J. A., Vicente, E. J., Leandro da Silva, K. V., and Schenberg, A. C. (1989) Mutat. Res. 218 111-124 [DOI] [PubMed] [Google Scholar]
- 38.Meira, L. B., Fonseca, M. B., Averbeck, D., Schenberg, A. C., and Henriques, J. A. (1992) Mol. Gen. Genet. 235 311-316 [DOI] [PubMed] [Google Scholar]
- 39.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., Funke, R., Gage, D., Harris, K., Heaford, A., Howland, J., et al. (2001) Nature 409 860-921 [DOI] [PubMed] [Google Scholar]
- 40.Lee, J. H., and Paull, T. T. (2005) Science 308 551-554 [DOI] [PubMed] [Google Scholar]
- 41.Polanowska, J., Martin, J. S., Garcia-Muse, T., Petalcorin, M. I., and Boulton, S. J. (2006) EMBO J. 25 2178-2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris, J. R., and Solomon, E. (2004) Hum. Mol. Genet. 13 807-817 [DOI] [PubMed] [Google Scholar]