Abstract
Human HNSC.100 neural stem cells up-regulate expression of GFAP following withdrawal of mitogens. Activation of the ERK signaling pathway prevented the up-regulation of GFAP expression. Incubation of cells with retinoic acid in the absence of mitogens enhanced basal neuronal differentiation that was accompanied by an up-regulation of neuronal gene expression and a down-regulation of GFAP and nestin expression. Retinoic acid treatment changed the histone code of neuronal genes encoding synapsin I, synaptophysin, and synaptotagmins II, IV, and VII from a transcriptionally inactive (methylation of lysine residue 9 of histone 3) to a transcriptionally active state (methylation of lysine residue 4 of histone 3). In contrast, the chromatin structure of the GFAP gene is transformed from a transcriptionally active state in unstimulated neural stem cells to a transcriptionally inactive state in retinoic acid-stimulated cells. Additionally, retinoic acid treatment reduced the binding of histone deacetylase-1 and REST to neuronal genes. The inhibition of histone deacetylase activity induced expression of genes encoding synaptic vesicle proteins in unstimulated neural stem cells. Similarly, neuronal gene transcription was enhanced following expression of a mutant of REST that contained a transcriptional activation domain. These data indicate that in undifferentiated human neural stem cells, neuronal genes encoding synaptic vesicle proteins are accessible for the REST mutant and are sensitive to enhanced histone acetylation.
Neural stem cells are characterized by their self-renewal ability and their capacity to differentiate into neurons, astrocytes, and oligodendrocytes, the major cell types of the central nervous system. Neural stem cells have been isolated from murine brain tissue, in particular from the subventricular zone or the subgranular layer of the hippocampus, i.e. brain regions exhibiting neurogenesis in the adult. Following dissection of the tissue, the dissociated cells are cultured in the presence of the mitogens epidermal growth factor (EGF)2 and bFGF, giving rise to a mixed population of neural progenitor and stem cells (1). The use of neural stem cells as a cellular model to analyze differentiation processes requires the generation of a pure population of neural stem cells in a large enough quantity. This is, in particular for human neural stem cells, no easy task. It is in general difficult to get a reasonable number of human stem cells that maintain a stable phenotype during growth. Therefore, immortalized neural stem cell lines have been established that provide cells that can be cultured for the long term in a proliferative and undifferentiated state (2, 3). Here, we have used HNSC.100 neural stem cells that have been generated by infection of human neural progenitor cells, derived from the diencephalic and telencephalic region of a 10-10.5-week gestational age aborted human fetus, with a v-Myc-encoding retrovirus (4). Like primary neural stem cells, HNSC.100 neural stem cells require mitogens (EGF, bFGF) in the growth medium. Grafting experiments into adult rat brain revealed that the stem cells integrated in a nondisruptive manner into the surrounding tissue (5).
The fact that neural stem cells retain their potential to differentiate into the major cell types of the central nervous system lets them be considered as a useful cellular model system for studying the underlying cellular differentiation process. This includes the identification of transcription factors required for differentiation into a particular neural cell type as well as the analysis of the epigenetic changes that occur during differentiation. Chromatin remodeling, including differentiation-dependent changes in the histone methylation pattern, is known to occur during development and induces cell type-specific gene transcription. Using undifferentiated and differentiated human neural stem cells, we have investigated the regulation of a group of neuronal genes encoding synaptic vesicle proteins. Synaptic vesicles are the key organelle for neurotransmission and neuronal function. Thus, expression of synaptic vesicle proteins mirrors a gain of neuronal character of a particular cellular population. From the genetic point of view, we analyzed the role of the transcription factor REST (6, 7), a dual-specific repressor (8) that induces transcriptional repression via recruitment of histone deacetylases and via gene silencing involving the methyl-CpG-binding protein MeCP2, hetero-chromatin protein-1 (HP-1), G9a histone methyltransferase, and C-terminal-binding proteins CtBP1 and CtBP2 (6, 7, 9). Both the cellular concentration of REST and the cell type-specific structure of the chromatin are key factors in determining whether neuronal genes are transcribed. Most interestingly, REST has recently been shown to regulate the transition from pluripotent to neural stem/progenitor cells and from progenitor cells to mature neurons (10).
Here, we show that HNSC.100 neural stem cells differentiate along the astrocytic lineage when the mitogens EGF and bFGF are withdrawn from the culture medium. The down-regulation of the ERK signaling pathway is crucial for the differentiation into astrocytes. HNSC.100 neural stem cells differentiate along the neuronal lineage when the mitogens are removed from the culture medium and retinoic acid is added. The differentiation is accompanied by a change of the histone code of neuronal genes. While neuronal genes are embedded in a chromatin structure characterized by methylated lysine residue 9 of histone 3 in undifferentiated stem cells, retinoic acid treatment triggers the methylation of lysine residue 4 of histone 3, a mark for open, actively transcribed genes. Accordingly, neuronal genes are expressed. Moreover, retinoic acid treatment leads to a reduced binding of histone deacetylase 1 (HDAC1) and REST to neuronal genes encoding synaptic vesicle proteins. We finally show that a dominant-positive mutant of REST is able to bind to neuronal genes in undifferentiated neural stem cells, indicating that these genes are accessible for transcription factors, despite the nucleosomal microenvironment characterized by the methylated lysine residue 9 of histone 3.
EXPERIMENTAL PROCEDURES
Materials—EGF and bFGF were purchased from Chemicon, Hampshire, UK (cat. no. GF001 and GF003). Trichostatin A (TSA, Sigma T8552) was used at a concentration of 25 ng/ml dissolved in Me2SO. 4-Hydroxytamoxifen (4OHT, Sigma H7904) was dissolved in ethanol and used at a concentration of 1 μm. Retinoic acid was purchased from Calbiochem, Darmstadt, Germany (cat. no. 554720) and was used at a concentration of 1 μm dissolved in ethanol.
Cell Culture—Human HNSC.100 neural stem cells were cultured on poly-l-lysine-coated plates in DMEM-F12 medium (Sigma, cat. no. D-0547) containing 1% N2-Supplements (Invitrogen, cat. no. 17502), 1% bovine serum albumin (Sigma, no. A1470), 20 ng/ml bFGF, 20 ng/ml EGF, 6 g/liter glucose, and 1% non-essential amino acids (Invitrogen, cat. no. 1140-035). Human SH-SY5Y neuroblastoma cells were a kind gift of J. Biedler, Sloan-Kettering Institute for Cancer Research, New York. HaCaT keratinocytes were kindly provided by N. E. Fusenig, Deutsches Krebsforschungszentrum, Heidelberg, Germany. SH-SY5Y, HaCaT, and 293T cells were cultured as described (11-13). For differentiation, one million HNSC.100 neural stem cells were seeded in 100-mm plates for 24 h. The cells were washed with phosphate-buffered saline and incubated in culture medium lacking EGF and bFGF to initiate differentiation. To enhance neuronal differentiation, we added retinoic acid (1 μm) to the culture medium. The medium was changed every 2 days.
Quantification of Neurite Outgrowth—Neurite outgrowth of retinoic acid-stimulated HNSC.100 neural stem cells was quantified using the LUCIA 4.21 program (Laboratory Imaging Ltd). At least 100 cells were analyzed for each condition. The data are depicted using GraphPad Prism 3.02 (Prism). The Student's t test was used to determine the p value.
Transient Transfections and Reporter Gene Assays—The synapsin I promoter/luciferase reporter genes pSyIuc and pSyIlucΔNRSE have been described (14). The transcription units present in these plasmids contain synapsin I promoter sequence from -422 to +47 either with or without the REST-binding site termed neural-restrictive silencing elements (NRSE). The reporter plasmid pSyINRSE2luc contains the luciferase gene under control of a minimal promoter that consists of two copies of the NRSE, derived from the human synapsin I promoter, a TATA box derived from the HIV long terminal repeat, and the initiator element from the adenovirus major late promoter. HNSC.100 cells were transfected using the calcium coprecipitation procedure. Each experiment included four separate transfections for each experimental setting, and the experiments have been repeated at least twice giving consistent results.
Retroviral Gene Transfer—The retroviral vector encoding a positive-dominant mutant of REST, fused to the ligand-binding domain of the estrogen receptor, has been described (15). Plasmid pBabepuro3ΔRaf-1:ER, encoding an activated form of the protein kinase Raf-1 as a fusion protein with the hormone-binding domain of the murine estrogen receptor (ER™), was kindly provided by Martin McMahon, Cancer Research Institute and Department of Cellular and Molecular Pharmacology, UCSF, San Francisco (16). The packaging cell line Phoenix-Ampho was obtained from Gary Nolan, Stanford University. Cells were transfected with retroviral vectors using the calcium coprecipitation procedure. Retroviral infection was performed as described (17). Cells were selected with 0.3 mg/ml puromycin. Mass pools of stable transfectants were selected and used for all experiments to eliminate the possibility of specific clonal effects. HNSC.100ΔRaf-1:ER and HNSC.100-DP-REST:ER cells were stimulated with 4OHT (1 mm) for 48 h.
RT-PCR—RT-PCR was performed as previously described (18). The primers are listed in Table 1. The experiments were performed at least three times with consistent results. Quantification and statistical analysis was performed using the program XnView from Pierre e.Gougelet to scan at least three independent photographs for each experiment. The data were quantified using the program LabWorks 4.6 from UVP BioImaging Systems. The data presentation was done with the program GraphPad Prism 3.02 (Prism), showing the S.E.
TABLE 1.
Primers for RT-PCR
| Genes | Forward primer | Reverse primer | Product size | Gene accession number |
|---|---|---|---|---|
| GAPDH | TTCCAGGAGCGAGAT CCC | CACCCATGACGA ACATGGG | 175 | BC83511.1 |
| GFAP | CTGTTGGCCAGAGAT GGAGGTT | TCATCGCTCAGGAGG TCCTT | 382 | BC013596 |
| Synapsin I | GCACGTCCTGGCTGGGTTTCTGGG | AGGCTACCCGTCAGACATCCGTCTC | 248 | NM_006950 |
| Synaptophysin | TGCAGAACAAGTACC GAGAG | CTGTCTCCTTAAACA CGAACC | 297 | NM_003179 |
| Synaptoporin | GGGCCCACTCATTGA CTTCAT | GAGTGCTTCTCCATTGGATCTG | 321 | NM_144642 |
| Synaptotagmin I | AGTCTTTGTGGGCTA CAACAGC | CAGGATTCCAAGTAC CACTGA | 250 | NM_005639 |
| Synaptotagmin II | TGATGGCTGTGTGAGGAGAG | GCTGCAAGTTTGTGCCAGTA | 169 | NM_177402 |
| Synaptotagmin IV | CCAGCACTTCCCTTA CTTCAG | TCCCGAGAGAGGAAT TAGAAC | 400 | NM_020783 |
| Synaptotagmin VII | GATCGTCCATCACCTGACCT | GGCAGAGAAACCACAGCTTC | 179 | NM_004200 |
| Synaptobrevin1 | CCCCTGGCCCTCCTC CTAACA | CACGATGATGGCACA GATGGTTCC | 265 | NM_199245 |
| Synaptobrevin2 | CATCTTGGGAGTGAT TTGCGC | GGAGAGGGACTATTG CATAGC | 406 | NM_014232 |
| REST | TTTGAAGTTGCTTCT ATCTGCTGT | GAATCTGAAGAACAG TTTGTGCAT | 626 | NM_005612 |
Chromatin Immunoprecipitation—Chromatin immunoprecipitation experiments were performed as described (19). The primers are listed in Table 2. The antibodies used for immunoprecipitation were anti-dimethyl H3K9 (Abcam, Cambridge, UK, ab7312), anti-dimethyl H3K4 (Upstate Biotechnology, Lake Placid, NY, 07-030), anti-trimethyl H3K4 (Abcam, ab8580), anti-NRSF (Santa Cruz Biotechnology, Heidelberg, Germany, sc-25398), anti-HDAC1 (Upstate Biotechnology, Lake Placid, NY, 0S-614), and anti-LSD1 (Abcam, ab17721). To detect in vivo binding of DP-REST:ER to DNA, we used M2-agarose (Sigma, A2220) that interacts with the FLAG epitope of the REST mutant as described (15).
TABLE 2.
Primers for chromatin immunoprecipitation
| Genes | Forward primer | Reverse primer | Product size | Gene accession number |
|---|---|---|---|---|
| GFAP | GAGAGGGTCCTCTTGCTTCG | TGAAGGAGTGGGCTAGACTGG | 238 | M 67446 |
| Synapsin I | GGGGAAACAGGATGCGGAG | AAGGTGGCCGGGAAGGGGAGT | 163 | M 55301 |
| Synaptophysin | ATGCTGCTGCTGGCGGACGG | TGGCCACCACCTCCCAGAGTC | 128 | AC009334.8 |
| Synaptotagmin II | ATCCTTCCGTAGCACCCTTGA | TCCATCACCCGAGCACTCAGC | 155 | AC104463.3 |
| Synaptotagmin IV | GAGACGAGCTTCTCAGAAGC | CAGTCCACCCCAGAGTATTCA | 316 | AC087507.7 |
| Synaptotagmin VII | TGTTTCAGCACCAAGGACAA | GCTGGAATGCAGGGTCTAGA | 174 | AP003559.3 |
Antibodies and Immunoblot Analysis—Nuclear extracts were prepared as described (20). 10 μg of protein from nuclear extracts were separated on 10% SDS-PAGE and electrophoretically transferred to nitrocellulose membrane (0.2-μm pore size, Schleicher & Schuell, Dassel, Germany). Membranes were blocked in Tris-buffered saline (TBS) containing 5% nonfat dry milk for 1 h at 37 °C and incubated for 2 h at room temperature with the M2 monoclonal antibody directed against the FLAG epitope (Sigma, F3165), at 1:3000 dilution in TBS. Secondary antibodies (goat anti-mouse peroxidase-conjugated antibody, Jackson ImmunoResearch Laboratories) were incubated for 1 h at room temperature and were used at a dilution of 1:10,000. Immunoreactive bands were detected using the ECL plus system (Amersham Biosciences).
RESULTS
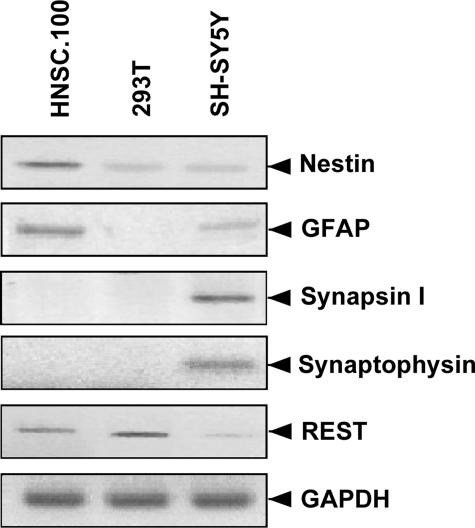
Expression of Marker Genes in Human HNSC.100 Neural Stem Cells—We compared the expression of tissue-specific marker genes in undifferentiated HNSC.100 neural stem cells, human 293T embryonic kidney cells, and dopaminergic SH-SY5Y neuroblastoma cells. HNSC.100 stem cells express nestin, as previously described (4), a marker of neuroepithelial progenitors and stem cells (Fig. 1). Low levels of nestin were also detected in 293T and SH-SY5Y cells. Expression of GFAP, a marker of astrocytes, was observed in HNSC.100 cells. A trace amount of GFAP mRNA was also seen in SH-SY5Y cells, and none was found in 293T cells. The neuronal genes encoding synapsin I and synaptophysin were only expressed in neuroblastoma cells. Similar results were obtained for the genes encoding synaptotagmins II, IV, and VII (data not shown). We also measured the mRNA levels of REST, a transcriptional repressor of neuronal genes. As expected, highest levels of REST were observed in non-neuronal 293T cells. Considerable amounts of REST were also detected in undifferentiated HNSC.100 neural stem cells, and may therefore explain why synapsin I, synaptophysin, and other neuronal genes are not expressed. Trace levels of REST were seen in SH-SY5Y neuroblastoma cells, which already express neuronal genes constitutively. These data show that undifferentiated HNSC.100 stem cells express stem/progenitor cell and glia markers and are not yet differentiated into neurons.
FIGURE 1.
Marker gene expression in undifferentiated HNSC.100 stem cells, 293T cells, and SH-SY5Y neuroblastoma cells. RNA was isolated and analyzed by RT-PCR. The expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as a control.
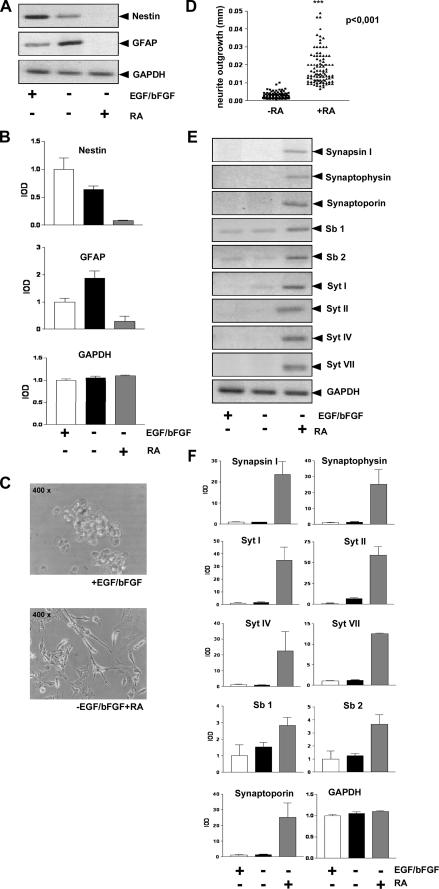
Differentiation Potential of HNSC.100 Neural Stem Cells—The simplest method for induction of neural stem cell differentiation is mitogen withdrawal (1). HNSC.100 neural stem cells require EGF and bFGF in the growth medium. Removal of the mitogens stops the cell cycle and induces differentiation (4). It has been reported that markers of astrocytic and neuronal linages such as GFAP, MAP2, and β-tubulin III are up-regulated 12 days after removal of EGF and bFGF. In addition, the cells dramatically change their morphology (4). We cultured HNSC.100 stem cells for 12 days in medium lacking EGF and bFGF, isolated the RNA and analyzed marker gene expression via RT-PCR. Fig. 2, A and B reveal that GFAP expression is up-regulated in differentiated HNSC.100 neural stem cells. Nestin expression is reduced accordingly. Previous studies have shown that mitogen withdrawal is sufficient enough to generate functional (possibly immature) neurons from immortalized hNSCs, as evaluated by means of protein markers, gene expression data, neurotransmitter production, and functional studies, like neurotransmitter release upon depolarization (21, 22). However, the differentiation induction by mitogen withdrawal did not activate expression of mature neuronal marker genes encoding synaptic vesicle proteins (Fig. 2, E and F), even though the expression of the transcription factor REST was reduced (data not shown).
FIGURE 2.
Differentiation of HNSC.100 neural stem cells. A, withdrawal of EGF and bFGF from the culture medium induces differentiation along the astrocytic lineage. HNSC.100 human neural stem cells were cultured for 12 days without EGF and bFGF in the culture medium. Retinoic acid (1 μm) was added as indicated. RNA was isolated and analyzed by RT-PCR. B, quantification and statistical analysis of the data shown in A. C, retinoic acid stimulation triggers neurite outgrowth of HNSC.100 human neural stem cells. Cells were stimulated with retinoic acid (1 μm) for 12 days. D, quantification for neurite outgrowth (length and number). E, expression of neuronal genes encoding synaptic vesicle proteins in retinoic acid-stimulated neural stem cells. HNSC.100 cells were incubated with retinoic acid in the absence of mitogens for 12 days. RNA was isolated and analyzed by RT-PCR (Sb, synaptobrevin; Syt, synaptotagmin). F, quantification and statistical analysis of the data shown in D.
Retinoic Acid Triggers Transcription of Genes Encoding Synaptic Vesicle Proteins in Human Neural Stem Cells—Treatment of neural stem cells with retinoic acid enhances neuronal differentiation (23, 24). We therefore removed the mitogens EGF and bFGF from the growth medium and added retinoic acid for 12 days. The cells changed their morphology and neurite outgrowth occurred as depicted in Fig. 2C. The quantification of neurite length is shown in Fig. 2D. Furthermore, we detected expression of neuronal genes encoding the synaptic vesicle proteins synapsin I, synaptophysin, synaptoporin, synaptobrevin 2, and synaptotagmins I, II, IV, and VII (Fig. 2, E and F). In contrast, expression of nestin and GFAP was shut-off in retinoic acid-treated human stem cells (Fig. 2, A and B). Together, these data indicate that retinoic acid enhances neuronal differentiation of human HNSC.100 neural stem cells leading to the expression of synaptic vesicle proteins.
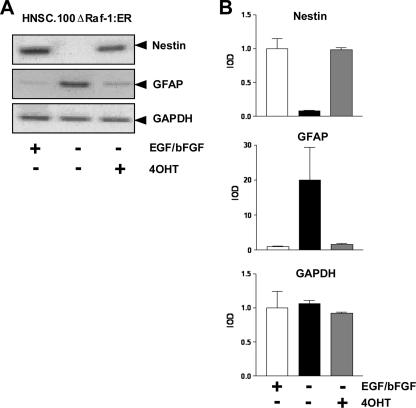
Activation of the ERK Signaling Pathway Blocks the Differentiation of Neural Stem Cells into the Astrocytic Lineage—The experiment depicted in Fig. 2, A and B revealed that withdrawal of EGF and bFGF induced the expression of the astrocytic marker GFAP in HNSC.100 neural stem cells. EGF and bFGF are ligands of receptor tyrosine kinases. Stimulation of these receptors induces the ERK, phosphatidylinositol 3-kinase, and phospholipase C signaling pathways. We tested the impact of the ERK signaling pathway in gliogenesis using HNSC.100 cells expressing an inducible mutant of Raf-1, termed ΔRaf-1:ER. The characterization of the HNSC.100ΔRaf-1:ER cell line will be described elsewhere.3 The cells were cultured in medium with or without EGF and bFGF. In addition, the ΔRaf-1/estrogen receptor fusion protein was activated by 4OHT. Fig. 3, A and B show that activation of the ERK signaling pathway, via 4OHT-triggered activation of ΔRaf-1:ER, blocked the differentiation into glia cells as a result of mitogen withdrawal. In 4OHT-treated cells, the withdrawal of mitogens did not enhance GFAP expression. Similarly, expression of nestin was not reduced. We conclude that a down-regulation of the ERK signaling pathway is essential for neural stem cells to differentiate along the astrocytic/GFAP+ lineage.
FIGURE 3.
Activation of the ERK signaling cascade blocks differentiation along the astrocytic lineage. A, HNSC.100 neural stem cells expressing ΔRaf-1:ER were incubated with or without EGF, bFGF, and 4OHT as indicated. RNA was isolated and analyzed by RT-PCR. B, quantification and statistical analysis of the data shown in A.
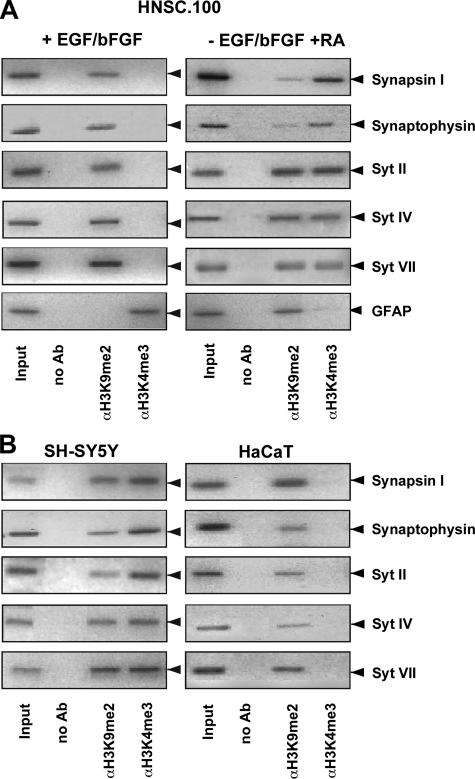
Epigenetic Configuration of Genes Encoding Synaptic Vesicle Proteins in Undifferentiated and Retinoic Acid-treated Neural Stem Cells—The fact that treatment of HNSC.100 neural stem cells with retinoic acid induced the expression of synaptic proteins suggests that genetic and/or epigenetic changes occurred in the environment of these genes leading to neuronal gene transcription. We first analyzed the epigenetic configuration of a group of neuronal genes in HNSC.100 human neural stem cells that encode synaptic vesicle proteins and have been described to be regulated by the transcription factor REST (10, 25-29). We analyzed epigenetic markers that differentiate between actively transcribed or silenced genes. An epigenetic marker for actively transcribed genes is methylation of lysine residue 4 of histone 3 (H3K4) while methylation of lysine residue 9 of histone 3 (H3K9) functions as an epigenetic marker for silenced genes (30). The histone 3 methylation status of neuronal genes encoding synapsin I, synaptophysin, and synaptotagmin II, IV, and VII was analyzed by chromatin immunoprecipitation experiments using antibodies directed either against the trimethylated form of histone H3K4 or against the dimethylated form of histone H3K9. The precipitated DNA was analyzed with primers that amplified the region of synaptic vesicle-encoding genes, encompassing the REST-binding site (NRSE). The results show that in undifferentiated neural stem cells the REST-binding sites of the synapsin I, synaptophysin, and synaptotagmin II, IV, and VII genes are embedded into a nucleosomal context having histone H3 molecules carrying methylated lysine residue 9, a marker that is linked to a condensed form of chromatin and gene silencing. Treatment of the cells with retinoic acid in the absence of mitogens in the culture medium triggered the trimethylation of histone H3 on lysine residue 4 (Fig. 4A), a methylation mark linked to open chromatin and actively transcribed genes. We also observed enhanced dimethylation of H3K4 (data not shown). Additionally, methylation on lysine residue 9 of histone 3 (H3K9) was still observed. As a control, we analyzed the epigenetic profile of the GFAP gene. GFAP is expressed in undifferentiated human neural stem cells and retinoic acid treatment blocked GFAP gene transcription (Fig. 2, A and B). Accordingly, we observed that the proximal region of the GFAP 5′-upstream region is embedded into a chromosomal context with histone H3 molecules methylated on lysine residue 4. Retinoic acid treatment shifted the methylation pattern to methylation of lysine residue 9 of histone H3. We conclude that treatment of neural stem cells with retinoic acid induced a chromatin environment for genes encoding synaptic vesicle proteins that is connected with an open configuration and active gene transcription. In contrast, in retinoic acid-treated human neural stem cells the chromatin environment of the GFAP gene is indicative of gene silencing. These data indicate that gliogenesis and neurogenesis are mutually opposing pathways.
FIGURE 4.
Epigenetic modifications of REST target genes in human neural stem cells, neuroblastoma cells, and keratinocytes. A, HNSC.100 neural stem cells were either cultured in the presence of EGF and bFGF or differentiated for 12 days in a medium that contained 1 μm retinoic acid but lacked EGF and bFGF. Chromatin immunoprecipitations were performed with anti-dimethylated H3K9 and anti-trimethylated H3K4 antibodies. Immunoprecipitated chromatin fragments were amplified with primers encompassing the REST binding sites within the regulatory regions of the genes encoding synapsin I, synaptophysin, and synaptotagmins II, IV, and VII. As a negative control, no primary antibody was added. An aliquot of the total input was also examined by PCR. B, cross-linked and sheared chromatin prepared from human SH-SY5Y neuroblastoma cells and HaCaT keratinocytes was immunoprecipitated with antibodies directed against dimethylated H3K9 or trimethylated H3K4. Immunoprecipitated chromatin fragments were amplified with primers encompassing the REST-binding sites within the regulatory regions of the genes encoding synapsin I, synaptophysin, and synaptotagmins II, IV, and VII. As a negative control, no antibody was added (no Ab). As positive control, an aliquot of the total chromatin in the absence of immunoprecipitation was analyzed by PCR (input).
We compared the epigenetic profiles of genes encoding synaptic vesicle proteins obtained in undifferentiated and retinoic acid differentiated neural stem cells with those in keratinocytes and neuroblastoma cells. Keratinocytes do not express synaptic vesicle proteins. Accordingly, the genes encoding synapsin I, synaptophysin, and synaptotagmins II, IV, and VII are located in a chromatin environment characterized by histone 3 molecules carrying methylated lysine 9 residues. In contrast, these genes are found in neuroblastoma cells in nucleosomes carrying both methylated H3K4 and H3K9 (Fig. 4B). Thus, the methylation pattern observed in keratinocytes resembles that found in undifferentiated neural stem cells. Similarly, the chromatin architecture seen in neuroblastoma cells is similar to the one found in retinoic acid differentiated neural stem cells.
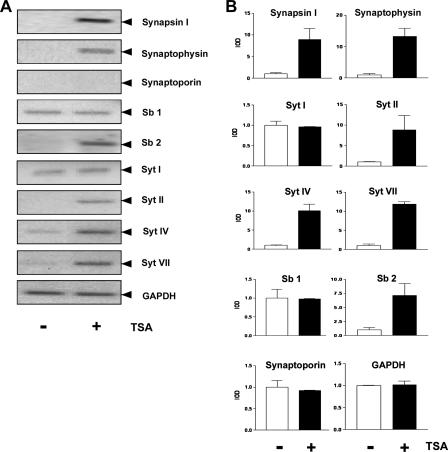
Transcription of Genes Encoding Synaptic Vesicle Proteins as a Result of Histone Deacetylase Inhibition—The acetylation state of histones is regulated by the coordinate activity of histone acetyltransferases and histone deacetylases. Deacetylation of histones by histone deacetylases removes the acetyl group from the ε-amino group of lysine residues of histones allowing ionic interactions between the negatively charged DNA phosphate backbone and the positively charged N termini of the core histones. This results in a more compact chromatin structure that is not easily accessible for the transcriptional machinery. Histone deacetylation is thus connected with repression of transcription. In contrast, histone acetylation is essential for establishing a transcriptionally competent state of chromatin. The level of histone acetylation is increased when histone deacetylase activity is blocked by compounds such as TSA. We tested whether an inhibition of histone deacetylases is sufficient to induce gene transcription of synaptic vesicle encoding genes in human neural stem cells. Cells cultured in the presence of EGF and bFGF were treated for 24 h with the histone deacetylase inhibitor TSA, and gene expression was analyzed by RT-PCR. Fig. 5 shows that expression of synapsin I, synaptophysin, synaptobrevin 2, and synaptotagmins II, IV, and VII is up-regulated as a result of histone deacetylase inhibition. These data indicate that in neural stem cells these genes are regulated via the balance of histone acetylation and deacetylation. TSA-induced inhibition of histone deacetylase activity did not induce the expression of synaptoporin, synaptobrevin 1, and synaptotagmin I indicating that the regulation of these genes is independent of histone acetylation and deacetylation in human neural stem cells.
FIGURE 5.
Inhibition of histone deacetylase activity by TSA up-regulates neuronal gene transcription in HNSC. 100 human neural stem cells. A, HNSC.100 cells were treated for 24 h with the histone deacetylase inhibitor TSA (25 ng/ml) or with the vehicle Me2SO. RNA from Me2SO-treated (denoted -) and TSA-treated (denoted +) cells was isolated and analyzed by RT-PCR. The expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as a control. B, quantification and statistical analysis of the data shown in A.
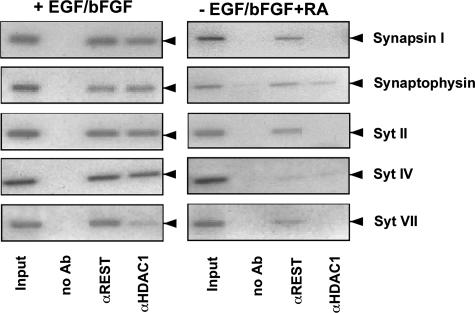
Binding of REST and HDAC1 to Neuronal Genes Is Reduced in Retinoic Acid-treated Neural Stem Cells—The transcription factor REST functions as a repressor of a group of neuronal genes. The fact that neuronal gene expression is induced in neural stem cells treated with retinoic acid suggests that the transcriptional repression activity of REST is reduced as a result of the treatment. We analyzed under physiological conditions the binding of REST to synaptic vesicle protein-encoding genes in neural stem cells in the presence or absence of retinoic acid. REST recruits HDAC1 to its target genes; thus, catalyzing the removal of acetyl groups from histone molecules, leading to transcriptional silencing. Therefore, we also analyzed the binding of HDAC1 to neuronal genes in undifferentiated and retinoic acid-differentiated neural stem cells. Fig. 6 reveals that in undifferentiated neural stem cells REST and HDAC1 interacted with the regulatory region of neuronal genes encoding synapsin I, synaptophysin, and synaptotagmins II, IV, and VII. Treatment of the cells with retinoic acid reduced the binding of REST to these genes and reduced or abolished the recruitment of HDAC1. We conclude that retinoic acid treatment triggered a reduction of REST and HDAC1 binding to the synapsin I, synaptophysin, and synaptotagmin II, IV, and VII genes and thus allowed neuronal/synaptic gene expression.
FIGURE 6.
Binding of REST and HDAC1 to neuronal genes encoding synaptic vesicleproteinsinhumanneuralstemcells.Chromatinimmunoprecipitations were performed with anti-REST and anti-HDAC1 antibodies. Immunoprecipitated chromatin fragments were amplified with primers encompassing the REST binding site within the synapsin I, synaptophysin, and synaptotagmins II, IV, and VII genes. As a negative control, chromatin immunoprecipitation was performed with protein A-Sepharose, without addition of antibodies (no Ab). As a positive control an aliquot of the total chromatin in the absence of immunoprecipitation was analyzed by PCR (input).
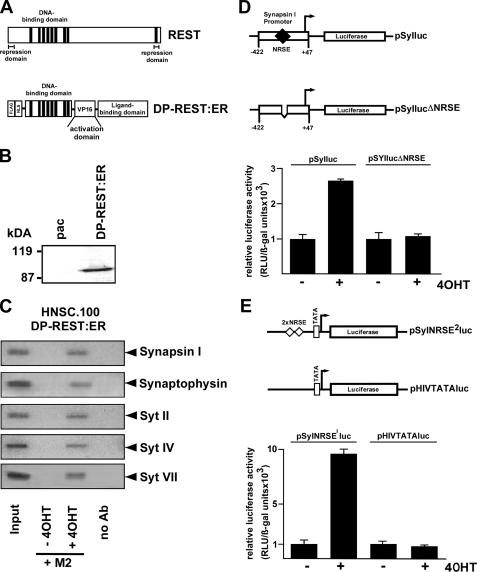
Expression of a Dominant-positive Mutant of REST in Human Neural Stem Cells—The previous experiments revealed that down-regulation of REST in retinoic acid-treated neural stem cells may be essential for induction of neuronal gene transcription. We then went a step further and asked whether we could directly activate neuronal gene expression in undifferentiated neural stem cells via expression of a mutant form of REST. This inducible dominant-positive mutant of REST termed DP-REST:ER activated transcription of REST-responsive genes following stimulation with 4OHT (15). The modular structure of REST and the REST mutant DP-REST:ER is depicted in Fig. 7A. DP-REST:ER contains the DNA-binding domain of REST, and therefore binds to the identical site on DNA. The N- and C-terminal repression domains of REST are deleted. The mutant contains instead a transcriptional activation domain, derived from the herpes simplex virus protein VP16. In addition, the dominant-positive mutant of REST contains the hormone binding domain of the estrogen receptor to confer regulation by 4OHT as described for the ΔRaf1:ER fusion protein. DP-REST:ER is expressed in an inactive state in the absence of 4OHT, but can be activated by the addition of 4OHT. We infected human HNSC.100 neural stem cells with a recombinant retrovirus encoding DP-REST:ER. As a control, cells were infected with recombinant retroviruses encoding the selection marker puromycin acetyltransferase (HNSC.100pac cells). The expression of the transgene that occurred under control of the murine stem cell virus long terminal repeat was verified in Western blot experiments using antibodies specific for the FLAG epitope (Fig. 7B). The DP-REST:ER fusion protein could be immunologically detected in infected human HNSC.100 neural stem cells, but not in the cells only expressing puromycin acetyltransferase (pac).
FIGURE 7.
Generation of HNSC.100 human neural stem cells expressing DP-REST:ER, a dominant positive mutant of REST. A, schematic representation of the modular structure of REST and the REST mutant DP-REST:ER. The functional domains for DNA binding, transcriptional repression and activation are indicated. The activation domain present in the REST mutant DP-REST:ER is derived from the herpes simplex virus protein VP16. DP-REST:ER additionally contains the ligand binding domain of the murine estrogen receptor (ER). B, expression of DP-REST:ER in HNSC.100 cells. Nuclear extracts of DP-REST:ER-expressing cells were prepared and analyzed by immunoblotting for FLAG immunoreactivity. As a control, we analyzed cells expressing the selection marker puromycin acetyltransferase (pac). Molecular mass markers in kDa are shown on the left. C, chromatin immunoprecipitation reveals binding of DP-REST:ER to the synapsin I, synaptophysin, and synaptotagmin II, IV, and VII genes in the presence of 4OHT.HNSC.100 cells were cultured in the presence or absence of 4OHT (1 μm) for 24 h. The chromatin was isolated, cross-linked, sheared, and immunoprecipitated with M2-agarose to selectively precipitate the FLAG-tagged DP-REST:ER mutant. Immunoprecipitated chromatin fragments were amplified with primers encompassing the REST-binding site of the synapsin I, synaptophysin, and synaptotagmin II, IV, and VII genes. As a negative control, chromatin immunoprecipitation was performed without adding M2-agarose (no Ab). As positive control an aliquot of the total chromatin in the absence of immunoprecipitation was analyzed by PCR (Input). D, one of the reporter plasmids pSyIluc or pSyIlucΔNRSE was transfected into HNSC.100 neural stem cells expressing DP-REST:ER together with the reference plasmid pRSVβ (1 mg/plate) that encodes β-galactosidase under the control of the Rous sarcoma virus long terminal repeat. Cells were stimulated with 4OHT or treated with vehicle for 24 h. Cell extracts were prepared, and the β-galactosidase and luciferase activities determined. Luciferase activities were normalized for transfection efficiency by dividing luciferase light units by β-galactosidase activities. Two separate transfection experiments were performed in quadruplicate, and the mean ± S.D. is depicted. E, reporter plasmids pSyNRSE2luc or pHIVTATA-luc were transfected into HNSC.100/DP-REST:ER neural stem cells. Cells were stimulated with 4OHT or treated with vehicle for 24 h. The cells were harvested and the relative luciferase activity was determined.
Chromatin Immunoprecipitation Experiments Reveal Binding of DP-REST:ER to REST Target Genes in 4OHT-treated Human Neural Stem Cells—We tested the binding of DP-REST:ER to REST target genes in HNSC.100 cells by chromatin immunoprecipitation. Cross-linked and sheared chromatin prepared from unstimulated HNSC.100-DP-REST:ER cells and HNSC.100-DP-REST:ER cells stimulated with 4OHT was immunoprecipitated with M2-agarose. Fig. 7C shows that DP-REST:ER was able to bind in vivo to the regulatory regions of the synapsin I, synaptophysin, and synaptotagmin II, IV, and VII genes when the cells had been stimulated with 4OHT. In contrast, we did not detect binding of DP-REST:ER to neuronal genes in non-stimulated neural stem cells. These data indicate that the ER domain not only masks the VP16 activation domain within the DP-REST:ER molecule, but furthermore blocked the DNA binding of the fusion protein. Interestingly, DP-REST:ER was able to bind to the synapsin I, synaptophysin, and synaptotagmin II, IV, and VII genes in cells that had been cultured in the presence of EGF and bFGF. Under these conditions, these genes have been shown to be embedded in a nucleosomal context with histone H3 methylated on lysine residue 9. Thus, despite a chromosomal environment that is linked to “closed” chromatin structure, DP-REST:ER had access to these genes in 4OHT-treated neural stem cells.
The REST Mutant DP-REST:ER Activates NRSE-containing Reporter Genes in Human Neural Stem Cells—To test the biological activity of the endogenously expressed DP-REST:ER fusion protein we transfected the DP-REST:ER-expressing cells with the reporter plasmids pSyIluc and pSyIlucΔNRSE (Fig. 7D). Plasmid pSyIluc contains the human synapsin I promoter sequence from -422 to -22. As a control for REST function, a deletion from -234 to -200 had been introduced into the synapsin I promoter forming plasmid pSylucΔNRSE. The deletion includes the REST-binding site. Transient transfections were performed with HNSC.100 cells expressing DP-REST:ER. Co-transfection of a β-galactosidase-encoding plasmid under the control of the Rous sarcoma virus long-terminal repeat was used to correct for variations in transfection efficiencies. The results show that treatment with 4OHT-activated reporter gene transcription in HNSC.100 cells when a functional NRSE was present in the promotor (plasmid pSyIluc). DP-REST:ER had no effect on a transcription unit lacking an NRSE (plasmid pSyIlucΔNRSE) (Fig. 7D). Similar results were obtained in the analysis of a transcription unit having two NRSE motifs upstream of a minimal promoter (Fig. 7E).
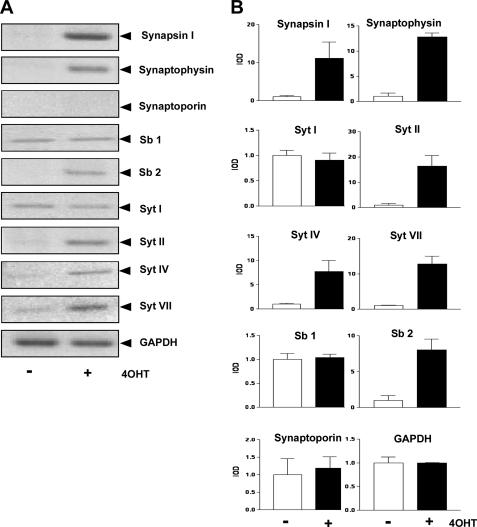
Activation of a Dominant-positive Mutant of REST Enhances Transcription of Genes Encoding Synaptic Vesicle Proteins in Human Neural Stem Cells—Next, we tested whether DP-REST:ER is able to enhance transcription of genes encoding synaptic vesicle proteins in neural stem cells. HNSC.100 cells expressing DP-REST:ER were treated with vehicle (-) or 4OHT (+) for 24 h. RNA was prepared, and analyzed by RT-PCR. Fig. 8 shows that expression of synapsin I, synaptophysin, synaptobrevin 2, and synaptotagmins II, IV, and VII was stimulated in 4OHT-treated HNSC.100 cells expressing DP-REST:ER. In contrast, no stimulation was observed for the genes encoding synaptotagmin I, synaptobrevin 1, and synaptoporin, indicating that these genes are not regulated by REST in human neural stem cells. This experiment underlines the importance of REST as a key transcription factor controlling expression of neuronal genes in neural stem cells.
FIGURE 8.
Up-regulation of neuronal/synaptic gene expression in HNSC.100 human neural stem cells following activation of DP-REST:ER, a dominant-positive mutant of REST. A, HNSC.100 cells expressing DP-REST:ER were incubated for 24 h with 4OHT (1 μm) or ethanol (vehicle). Total RNA from ethanol-treated (denoted -) and 4OHT-treated (denoted +) cells was isolated and analyzed by RT-PCR. The expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as a negative control. B, quantification and statistical analysis of the data shown in A.
DISCUSSION
Neural stem cells offer a cell culture model to study differentiation into glial cells or neurons. The objective of this study was to investigate the genetic and epigenetic changes of neural stem cells forced to differentiate into neurons. We focused our analysis on the regulation of a group of neuronal genes encoding synaptic vesicle proteins that are key markers of the neuronal phenotype. On the genetic level, we concentrated on the transcription factor REST, a transcriptional repressor that regulates many neuronal genes and is thought to be important for the establishment of the neuronal phenotype (6, 27). On the epigenetic level, we analyzed the methylation pattern of histone H3 to distinguish between actively transcribed and condensed chromatin structures. Moreover, we analyzed the role of histone deacetylation in neuronal gene expression.
As a cellular model, we chose the human HNSC.100 neural stem cell line described to retain the basic features of neural stem cells. The cells express markers of neurons, astrocytes, and oligodendrocytes following mitogen withdrawal-induced differentiation (4). Cells maintained in the undifferentiated state, i.e. cultured in medium containing EGF and bFGF, expressed the astrocytic marker GFAP, indicating the glia-like character of the HNSC.100 neural stem cells. Neural stem/progenitor cells with characteristics of glia cells have already been described (31, 32). Upon mitogen withdrawal, the expression of GFAP was up-regulated in HNSC.100 stem cells. Neuronal genes encoding synaptic vesicle proteins were not or only marginally expressed in undifferentiated HNSC.100 neural stem cells. Surprisingly, synapsin I and synaptotagmin VII are already expressed in MHP36 neural stem cells (28). Using the expression of synaptic vesicle proteins as mature neuronal markers we did not detect an up-regulation of neuronal gene expression in HNSC.100 neural stem cells as a result of mitogen withdrawal. Rather, the up-regulation of GFAP expression indicates that the cells mostly differentiated along the astrocytic lineage in the absence of EGF and bFGF in the culture medium. Interestingly, we have shown here that the impairment of the ERK signaling pathway is responsible for this differentiation. The up-regulation of GFAP expression in the absence of EGF and bFGF was prevented when the ERK signaling cascade was activated via activation of a ΔRaf-1/estrogen receptor fusion protein. Similarly, nestin expression was not down-regulated under these circumstances. To enhance neuronal differentiation we stimulated the cells with retinoic acid in the absence of EGF and bFGF in the culture medium. We detected an up-regulation of neuronal gene expression, including elevated mRNA levels of synapsin I, synaptophysin, synaptoporin, synaptobrevin 1 and 2, and synaptotagmins I, II, IV, and VII. In contrast to immortalized human bipotent progenitor cells (3) retinoic acid treatment did not enhance astrocytic differentiation. Rather, expression of GFAP and nestin was shut-off in retinoic acid-treated HNSC.100 cells, indicating that neuronal differentiation was accompanied by a down-regulation of markers specific for astrocytes and stem/progenitor cells.
The expression of neuronal genes in retinoic acid-stimulated human neural stem cells suggests that genetic and epigenetic changes occur during differentiation. The analysis of the chromatin structure of the genes encoding synapsin I, synaptophysin, and synaptotagmin II, IV, and VII revealed that treatment with retinoic acid induced the methylation of H3K4, a marker of transcriptionally active genes. In addition, methylation of H3K9 that marks a transcriptional inactive state was detected. Thus, the pattern observed following retinoic acid treatment is an intermediate in development and we conclude that with both histone methylations observed, expression wins over silencing. In MHP36 neural stem cells, methylation of both H3K4 and H3K9 has been reported (28), suggesting that these cells are already differentiated along the neuronal lineage. This assumption is in accordance with the expression of synapsin I and synaptotagmin VII in these cells (28). A comparison of the methylation pattern of neuronal genes in neuroblastoma cells, i.e. cells that started to differentiate along the neuronal lineage, showed a similar methylation pattern as seen in retinoic acid-differentiated human neural stem cells. In contrast, in undifferentiated neural stem cells as well as in keratinocytes, analyzed as a control for a differentiated nonneuronal cell type, only methylation of H3K9 was detected. In MHP36 neural stem cells, methylation of both H3K4 and H3K9 has been reported (28), indicating that these cells are already differentiated along the neuronal lineage. This assumption is in accordance with the expression of synapsin I and synaptotagmin VII in these cells (28).
A recent report showed that in murine embryonic stem cells the synaptotagmin IV gene is embedded into a chromosomal context characterized by methylation of H3K4. In this study, methylation of H3K9 for the synaptotagmin IV locus was only detected in fibroblasts (10). Similarly, nucleosomes of active and inactive protein-coding genes are trimethylated on H3K4 in human embryonic stem cells (33), showing a lack of correlation between methylation of H3K4 and actively transcribed genes in embryonic stem cells. These observations fit with the hypothesis that in the pluripotent state epigenetic repression of genetic information is removed (34). In contrast, in HNSC.100 human stem cells the analysis of genes encoding either synaptic vesicle proteins or GFAP revealed a clear-cut correlation between this epigenetic mark and active gene transcription. Moreover, the fact that human SH-SY5Y neuroblastoma cells that already express genes encoding synaptic vesicle proteins carry both lysine residue 4 and 9 methylation marks indicates that methylation of H3K9 is not always connected with silenced genes. Similar observations have been reported by others (35). Interestingly, the occurence of an embryonic stem cell-specific bivalent state has been proposed that is characterized by the coexistence of both active and suppressive chromatin modifications. During differentiation, this bivalent state resolves to either an active or a suppressive state (36). According to this hypothesis, the bivalent state seen in differentiated human neural stem cells as well as in neuroblastoma cells indicates that the cells are caught in the process of differentiation that has yet to be resolved in one direction.
For comparison, we analyzed the chromatin structure of the GFAP promoter in undifferentiated and retinoic acid-differentiated neural stem cells. The analysis revealed that the chromatin structure of the GFAP gene resembled its expression pattern. GFAP is expressed in undifferentiated neural stem cells, and the gene is embedded into chromatin characterized by methylation of H3K4. Following retinoic acid treatment, the GFAP expression is prevented. Accordingly, the gene is found in nucleosomes carrying histone H3 molecules with methylated lysine residue 9. Future work will identify the histone methyltransferases and demethylases responsible for this dynamic methylation pattern. The binding of the histone demethylase LSD1 to the genes encoding synapsin I, synaptophysin, and synaptotagmin II, IV, and VII was shown by chromatin immunoprecipitation experiments, but no changes have been observed following retinoic acid-induced differentiation.4 LSD1 is known to require cofactors such as CoREST, HDAC1/2, and BHC80 for its function (37). Thus, the analysis of these cofactors as well as other H3K4 demethylases such as JARID/SMCX will be included in future studies.
Chromatin immunoprecipitation experiments revealed that binding of REST to its neuronal target genes decreased in retinoic acid-treated neural stem cells. Moreover, in vivo binding of the histone deacetylase HDAC1 that is recruited by REST to the regulatory region of neuronal genes decreased or even disappeared following differentiation of neural stem cells with retinoic acid. REST recruits histone deacetylases to neuronal genes resulting in removal of acetyl groups from the core histones. Consequently, the neuronal genes are embedded into more tightly packed chromatin that is inaccessible for transcriptional activators. Neuronal genes normally repressed by REST are expressed following inhibition of histone deacetylases (26, 38). However, the de-repression of neuronal genes by histone deacetylase inhibitors depends on the cell type and the chromatin structure of the gene under investigation (15). The analysis of REST target gene transcription in human neural stem cells in the presence of TSA revealed that many synaptic vesicle protein-encoding genes are sensitive to decompaction of the chromatin in neural stem cells. This includes the genes encoding synapsin I, synaptophysin, synaptobrevin 2, and synaptotagmin II, IV, and VII. Synaptoporin, synaptobrevin 1, and synaptotagmin 1 expression was not changed by increased histone acetylation. Together, these experiments reveal that a group of neuronal genes is regulated by the balance between histone acetylation and deacetylation in neural stem cells. In support of this conclusion, enhanced synapsin I expression has been reported in neural progenitor cells or H-2Kb-tsA58 transgenic mouse hippocampal neuroepithelial cells stimulated with valproic acid or TSA (28, 39).
The fact that binding of REST and HDAC1 to neuronal genes decreased in the process of neuronal differentiation indicates that a de-repression of neuronal genes occurred in retinoic acid-treated neural stem cells that finally allowed expression of these genes. Similarly, binding of REST to its target genes was observed in cortical progenitor cells while REST binding was no longer observed in postmitotic neurons (10). To directly investigate the impact of REST on REST target gene transcription, we have expressed a mutant of REST, DP-REST:ER, in neural stem cells and induced transcription of REST target genes. Therefore, we were able to directly test whether neuronal genes are accessible. Furthermore, we analyzed whether DP-REST:ER transactivated these genes in its natural chromosomal context. In fact, synapsin I, synaptophysin, synaptobrevin 2, and synaptotagmin II, IV, and VII gene transcription was up-regulated in neural stem cells following activation of DP-REST:ER by 4OHT. These data reveal that these genes are bona fide targets for REST in human neural stem cells. However, activation of DP-REST:ER failed to stimulate synaptoporin and synaptobrevin 1 transcription. Furthermore, DP-REST:ER did not enhance synaptotagmin I expression, indicating that the synaptotagmin I gene is not (in contrast to a previous report, Ref. 40) regulated by REST in neural stem cells.
In summary, we have shown that HNSC.100 human neural stem cells are a cellular model for studying differentiation along the astrocytic or neuronal lineage. Using the GFAP gene as a cellular marker for astrocytes we have shown that undifferentiated neural stem cells already express GFAP and this expression is up-regulated following the withdrawal of EGF and bFGF. This expression profile perfectly correlated with nucleosomal modifications that distinguish active versus inactive genes. The neuronal lineage was analyzed using the expression of synaptic vesicle proteins as a marker for a gain in neuronal character. We decided to analyze the expression of this group of neuronal genes to identify similarities and differences in gene regulation. We show that the genes encoding synapsin I, synaptophysin, synaptobrevin 2, and synaptotagmins II, IV, and VII are regulated in a similar fashion in human neural stem cells. The genes are not or only marginally expressed in undifferentiated stem cells. Accordingly, the genes carry the epigenetic marker for silenced genes. However, the loci are not permanently blocked as has been shown for differentiated non-neuronal cells such as fibroblasts or hepatocytes (15).4 Rather, inhibition of histone deacetylases was sufficient to induce neuronal gene transcription in human neural stem cells. Moreover, a dominant-positive mutant of REST was able to bind to neuronal genes in undifferentiated neural stem cells, indicating that the loci were accessible. We further showed that retinoic acid treatment induced neuronal gene expression in human neural stem cells. The treatment of the cells with retinoic acid induced the methylation of H3K4, an epigenetic marker for actively transcribed genes, within the loci of neuronal genes. Similarly, binding of REST and HDAC1 was reduced. Using the dominant-positive mutant of REST, we were able to correlate down-regulation of REST with increased expression of neuronal genes. In fact, we could directly show that the genes encoding synapsin I, synaptophysin, synaptobrevin 2, and synaptotagmins II, IV, and VII are bona fide targets of REST in human neural stem cells.
Acknowledgments
We thank Karl Bach for excellent technical help, and Libby Guethlein and Oliver Rössler for critical reading of the manuscript. We would also like to thank the excellent technical help of Bárbara Sesé Cobos at the CBMSO, for culture and handling of the cells.
This work was supported in part by a grant from the Zentrale Forschungskommison der Universität des Saarlandes (2006) and HOMFOR (B/2004/38). The work at the Center of Molecular Biology Severo Ochoa was supported by grants from the following Spanish Agencies: Ministerio de Educacíon y Ciencia Project SAF 2004-03405, Foundation La Caixa BM05-20, Instituto de Salud Carlos III, RD06/0010/0009, and the institutional grant to the Centro de Biología Molecular Severo Ochoa from the Foundation Ramon Areces. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: EGF, epidermal growth factor; ERK, extracellular signal-regulated kinase; ER, estrogen receptor; GFAP, glial fibrillary acidic protein; NRSE, neural-restrictive silencer element; 4OHT, 4-hydroxytamoxifen; REST, RE-1 silencing transcription factor; TSA, trichostatin A; bFGF, basic fibroblast growth factor; RT, reverse transcriptase.
M. Ekici, A. Martínez-Serrano, and G. Thiel, manuscript in preparation.
M. Ekici, and G. Thiel, unpublished observations.
References
- 1.Gage, F. H. (2000) Science 287 1433-1438 [DOI] [PubMed] [Google Scholar]
- 2.Kitchens, D. L., Snyder, E. Y., and Gottlieb, D. I. (1994) J. Neurobiol. 25 797-807 [DOI] [PubMed] [Google Scholar]
- 3.Sah, D. W. Y., Ray, J., and Gage, F. H. (1997) Nat. Biotech. 15 574-580 [DOI] [PubMed] [Google Scholar]
- 4.Villa, A., Snyder, E. Y., Vescovi, A., and Martínez-Serrano, A. (2000) Exp. Neurol. 161 67-84 [DOI] [PubMed] [Google Scholar]
- 5.Rubio, F. J., Bueno, C., Villa, A., Navarro, B., and Martínez-Serrano, A. (2000) Mol. Cell. Neurosci. 16 1-13 [DOI] [PubMed] [Google Scholar]
- 6.Thiel, G., and Hohl, M. (2006) in Transcription Factors in the Nervous System - Development, Brain Function and Disease (Thiel, G., ed), pp. 113-128, Wiley-VCH, Weinheim, Germany
- 7.Ooi, L., and Wood, I. C. (2007) Nat. Rev. Genet. 8 544-554 [DOI] [PubMed] [Google Scholar]
- 8.Thiel, G., Lietz, M., and Hohl, M. (2004) Eur. J. Biochem. 271 2855-2862 [DOI] [PubMed] [Google Scholar]
- 9.Ballas, N., and Mandel, G. (2006) Curr. Opin. Neurobiol. 15 1-7 [DOI] [PubMed] [Google Scholar]
- 10.Ballas, N., Grunseich, C., Lu, D. D., Speh, J. C., and Mandel, G. (2005) Cell 121 645-657 [DOI] [PubMed] [Google Scholar]
- 11.Cibelli, G., Policastro, V., Rössler, O., and Thiel, G. (2002) J. Neurosci. Res. 67 450-460 [DOI] [PubMed] [Google Scholar]
- 12.Rössler, O. G., and Thiel, G. (2004) Am. J. Physiol. Cell Physiol. 286 C1118-C1129 [DOI] [PubMed] [Google Scholar]
- 13.Al-Sarraj, A., Day, R. M., and Thiel, G. (2005) J. Cell. Biochem. 94 153-167 [DOI] [PubMed] [Google Scholar]
- 14.Lietz, M., Bach, K., and Thiel, G. (2001) Eur. J. Neurosci. 14 1303-1312 [DOI] [PubMed] [Google Scholar]
- 15.Hohl, M., and Thiel, G. (2005) Eur. J. Neurosci. 22 2216-2230 [DOI] [PubMed] [Google Scholar]
- 16.McMahon, M. (2001) Methods Enzymol. 332 401-417 [DOI] [PubMed] [Google Scholar]
- 17.Rössler, O. G., Giehl, K. M., and Thiel, G. (2004) J. Neurochem. 88 1240-1252 [DOI] [PubMed] [Google Scholar]
- 18.Bauer, I., Al Sarraj, J., Vinson, C., Larsen, R., and Thiel, G. (2007) J. Cell. Biochem. 100 242-255 [DOI] [PubMed] [Google Scholar]
- 19.Rössler, O. G., Henβ, I., and Thiel, G. (2008) Arch. Biochem. Biophys. 470 93-102 [DOI] [PubMed] [Google Scholar]
- 20.Kaufmann, K., and Thiel, G. (2002) J. Cell. Biochem. 85 381-391 [DOI] [PubMed] [Google Scholar]
- 21.Liste, I., García-García, E., and Martínez-Serrano, A. (2004) J. Neurosci. 24 10786-10795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liste, I., García-García, E., Bueno, C., and Martínez-Serrano, A. (2007) Cell Death Diff. 14 1880-1892 [DOI] [PubMed] [Google Scholar]
- 23.Hsieh, J., Aimone, J. B., Kaspar, B. K., Kuwabara, T., Nakashima, K., and Gage, F. G. (2004) J. Cell Biol. 164 111-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi, Y., Lie D. C., Taupin, P., Nakashima, K., Ray, J., Yu, R. T., Gage, F. H., and Evans, R. M. (2004) Nature 427 78-83 [DOI] [PubMed] [Google Scholar]
- 25.Thiel, G., Lietz, M., and Cramer, M. (1998) J. Biol. Chem. 273 26891-26899 [DOI] [PubMed] [Google Scholar]
- 26.Lietz, M., Hohl, M., and Thiel, G. (2003) Eur. J. Biochem. 270 2-9 [DOI] [PubMed] [Google Scholar]
- 27.Bruce, A. W., Krejci, A., Ooi, L., Deuchars, J., Wood, I. C., Dolezal, V., and Buckley, N. J. (2006) J. Neurochem. 98 1828-1840 [DOI] [PubMed] [Google Scholar]
- 28.Greenway, D. J., Street, M., Jeffries, A., and Buckley, N. J. (2007) Stem Cells 25 354-363 [DOI] [PubMed] [Google Scholar]
- 29.Otto, S. J., McCorkle, S. R., Hover, J., Conaco, C., Han, J.-J, Impey, S., Yochum, G. S., Dunn, J. J., Goodman, R. H., and Mandel, G. (2007) J. Neurosci. 27 6729-6739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin, C., and Zhang, Y. (2005) Nat. Rev. Mol. Cell Biol. 6 838-849 [DOI] [PubMed] [Google Scholar]
- 31.Doetsch, F. (2003) Nat. Neurosci. 6 1127-1134 [DOI] [PubMed] [Google Scholar]
- 32.Garcia, A. D. R., Doan, N. B., Imura, T., Bush, T. G., and Sofroniew, M. V. (2004) Nat. Neurosci. 7 1233-1241 [DOI] [PubMed] [Google Scholar]
- 33.Guenther, M. G., Levine, S. S., Boyer, L. A., Jaenisch, R., and Young, R. A. (2007) Cell 130 77-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niwa, H. (2007) Genes Dev. 21 2671-2676 [DOI] [PubMed] [Google Scholar]
- 35.Vakoc, C. R., Mandat, S. A., Olenchock, B. A., and Blobel, G. A. (2005) Mol. Cell 19 381-391 [DOI] [PubMed] [Google Scholar]
- 36.Nishikawa, S.-I., Jakt, L. M., and Era, T. (2007) Nat. Rev. Mol. Cell Biol. 8 502-507 [DOI] [PubMed] [Google Scholar]
- 37.Shi, Y.-J., Matson, C., Lan, F., Iwase, S., Baba, T., and Shi, Y. (2005) Mol. Cell 19 857-864 [DOI] [PubMed] [Google Scholar]
- 38.Huang, Y., Myers, S. J., and Dingledine, R. (1999) Nat. Neurosci. 2 867-872 [DOI] [PubMed] [Google Scholar]
- 39.Hsieh J., Nakashima, K., Kuwabara, T., Mejia, E., and Gage, F. H. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 16659-16664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Su, X., Kameoka, S., Lentz, S., and Majumder, S. (2004) Mol. Cell. Biol. 24 8018-8025 [DOI] [PMC free article] [PubMed] [Google Scholar]