Abstract
Chemical modification to improve biopharmaceutical properties, especially oral absorption and bioavailability, is a common strategy employed by pharmaceutical chemists. The approach often employs a simple structural modification and utilizes ubiquitous endogenous esterases as activation enzymes, although such enzymes are often unidentified. This report describes the crystal structure and specificity of a novel activating enzyme for valacyclovir and valganciclovir. Our structural insights show that human valacyclovirase has a unique binding mode and specificity for amino acid esters. Biochemical data demonstrate that the enzyme hydrolyzes esters of α-amino acids exclusively and displays a broad specificity spectrum for the aminoacyl moiety similar to tricorn-interacting aminopeptidase F1. Crystal structures of the enzyme, two mechanistic mutants, and a complex with a product analogue, when combined with biochemical analysis, reveal the key determinants for substrate recognition; that is, a flexible and mostly hydrophobic acyl pocket, a localized negative electrostatic potential, a large open leaving group-accommodating groove, and a pivotal acidic residue, Asp-123, after the nucleophile Ser-122. This is the first time that a residue immediately after the nucleophile has been found to have its side chain directed into the substrate binding pocket and play an essential role in substrate discrimination in serine hydrolases. These results as well as a phylogenetic analysis establish that the enzyme functions as a specific α-amino acid ester hydrolase. Valacyclovirase is a valuable target for amino acid ester prodrug-based oral drug delivery enhancement strategies.
Chemical modification through reversible prodrug, modification of a candidate drug, is a frequently employed strategy to improve biopharmaceutical properties of a candidate drug. Notable successes include oseltamivir, enalapril, and capecitabine (1). Membrane transport and absorption are usually thought to be improved by the increased lipophilicity and result in improved passive membrane transport (2). More recently we have shown that prodrugs may be transported by carrier-mediated transport mechanisms (3). A second essential step in effective prodrug therapy is the activation (hydrolysis) of the prodrug to the active therapeutic agent. Carboxylesterase is a common target for lipophilic approaches to improved membrane permeability (4-6). However, often the activation enzymes are unidentified. This manuscript reports the results of structural and biochemical studies on a novel prodrug activating enzyme, human valacyclovirase (VACVase)4 (7).
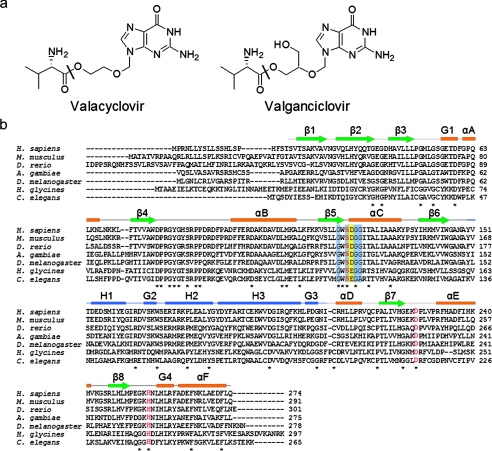
VACVase catalyzes the hydrolytic activation of two clinically important antiviral nucleoside prodrugs, valacyclovir and valganciclovir (Fig. 1a), resulting in l-valine and their corresponding active drugs acyclovir and ganciclovir. Both acyclovir and ganciclovir are polar molecules and are poorly absorbed with low oral bioavailability of 10-20% (8) and 6-9% (9), respectively. The valine ester prodrugs valacyclovir and valganciclovir on the other hand show very significant increases in oral absorption of 3-5-fold (10, 11) and 10-fold (12, 13), respectively. After absorption via intestinal peptide transporters (3, 14, 15), both prodrugs are found to be rapidly activated (10-12), with about 50% of the absorbed dose converted to acyclovir before reaching the portal circulation (16). In addition to activating valacyclovir and valganciclovir, VACVase has been shown to hydrolyze the prodrugs of a broad range of antiviral and anticancer nucleoside analogues such as zidovudine, floxuridine, and gemcitabine (7, 17).
FIGURE 1.
Prodrug substrates and sequences of VACVase. a, chemical structures of valacyclovir and valganciclovir. The sites of hydrolysis are labeled with a black bar. b, sequence alignment of human VACVase and homologues from metazoa. Completely conserved residues are marked with an asterisk. The catalytic triad residues (Ser-122-Asp-227-His-255) are highlighted in red. Conserved residues in the α/β hydrolase-fold characteristic motif (Sm-X-Nu-X-Sm-Sm (Nu = nucleophile, Sm = small residue)) are shaded in light blue. The acidic residue involved in binding the α-amino group of the substrate is shaded in yellow. The elements of the secondary structure from the crystal structure are shown above the amino acid sequence, with helices represented as rectangles, strands as arrows, and coils as lines. The α/β hydrolase domain is in orange and green; the cap domain is in blue.
VACVase contains the consensus sequence motif Sm-X-Nu-X-Sm-Sm (Nu = nucleophile, Sm = small residue) of the α/β hydrolase-fold superfamily (18) as well as a putative catalytic triad, Ser-122—His-255—Asp-227 (Fig. 1b). Further studies have shown that the enzyme exhibits neither peptidase nor protease activity (17). Given the potential importance of VACVase as a target for oral prodrug design, especially for improved oral absorption of polar candidate drugs, the molecular characteristics of this esterase, including its specificity, classification, and mechanisms of catalysis and recognition, need to be elucidated. In the present report our specificity analysis indicated that VACVase is a highly specific α-amino acid ester hydrolase. Crystal structures of the enzyme, two mutants, and a complex with a product analogue combined with biochemical data revealed the key elements in catalysis and substrate recognition.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of VACVase—The gene encoding the mature form of human VACVase (NCB accession number NM_004332, first 60 coding nucleotides excluded) was amplified by standard PCR methodology using the plasmid pET-BPHL (7) as the template and the forward (GATTCTAGAATTCATATGTCGGTAACCTCTGCCAAAGTGG) and reverse (GATTCTGAATTCGGTACCAAGCTTATTGTAGGAAGTCTTCTGC) primers. The PCR product was digested with restriction enzymes NdeI and KpnI and inserted into the corresponding site of the expression vector pET17b. After ligation, the plasmids were transformed into competent Escherichia coli NovaBlue cells. Sequences were verified by DNA sequencing. The plasmid containing the desired gene insert, pET17b-VACVase, was transformed into competent E. coli BL21(DE3) cells, which were then grown in LB medium containing ampicillin (100 μg/ml). Protein expression was induced by isopropyl 1-thio-β-d-galactopyranoside (0.3 mm).
Purification of VACVase was performed at 4 °C with all buffers containing 1 mm dithiothreitol. Cell pellets were suspended in 20 mm sodium phosphate buffer (pH 7.0) (buffer A), disrupted by sonication, and centrifuged (25,000 × g, 20 min). The supernatant was loaded onto an S strong cation exchange column and washed with buffer A until the absorbance at 280 nm reached the base line. VACVase was eluted with an isocratic flow of 90 mm NaCl in buffer A. (NH4)2SO4 was added to the pooled active fractions to a final concentration of 25%. The resulting protein solution was applied to a phenyl-Sepharose hydrophobic interaction column equilibrated with 25% (NH4)2SO4 in 100 mm sodium phosphate buffer (pH 7.4) and eluted with a reverse linear gradient of 25% to 0% (NH4)2SO4 in the same buffer over 8 column volumes. Fractions containing VACVase were collected, dialyzed against 2 liters of 10 mm HEPES (pH 7.4) for 16 h, and concentrated to ∼10 mg/ml.
The selenomethionine form of the enzyme was expressed in the methionine auxotroph E. coli B834(DE3) cells grown in MOPS minimal medium supplemented with 75 mg/liter of dl-selenomethionine. The protein was purified by the same procedures used for the native enzyme except that all buffers contained 5 mm dithiothreitol.
Generation of VACVase Mutants—Point mutations were introduced by PCR using a pair of complementary mutagenic primers. Oligonucleotide primers containing the desired mutation were synthesized by Invitrogen. The methylated wild-type plasmid template pET-VACVase was digested using the DpnI endonuclease, and the nicked plasmid DNA containing the desired mutation was transformed into NovaBlue competent cells. Sequences were verified by DNA sequencing. The mutants were expressed and purified using the same procedures as for the wild-type enzyme.
Enzyme Assays—The standard esterase activity against valacyclovir was measured with 4 mm valacyclovir in 50 mm HEPES (pH 7.4) in a 300-μl reaction mixture at 37 °C. After preincubation for 3 min, the reaction was initiated by the addition of enzyme and quenched with an equal volume of 10% (w/v) ice-cold trichloroacetic acid. The initial rate was determined by measuring product appearance by HPLC. One unit of enzyme activity is defined as the release of 1 μmol of acyclovir per min at 37 °C. The effect of nonenzymatic hydrolysis of substrates was corrected through control reactions. Measurements were carried out in triplicates.
Various other esters were tested as potential substrates of VACVase using the standard esterase assay method unless otherwise stated. The initial rate was determined by measuring substrate disappearance by HPLC. The concentration of the ester compounds was 4 mm unless otherwise specified.
The concentrations of substrates or products in the assay mixture were determined using a Waters HPLC system on a reverse phase column with water and acetonitrile (0.1% trifluoroacetic acid) as the mobile phase and a flow rate of 1 ml/min. Samples were eluted with either a linear gradient or an isocratic flow. The UV absorbance of the column effluent was monitored at 254 nm for acyclovir, 257 nm for the substrates containing a benzyl group, 270 nm for l-Asn p-nitrobenzyl ester, 275 nm for l-Tyr benzyl ester, 278 nm for l-Trp methyl ester, and 220 nm for l-Met methyl ester.
Crystallization and Data Collection—Protein crystals were grown by the sitting-drop vapor diffusion method. 2 μl of VACVase solution was mixed with 2 μl of reservoir solution containing 12-16% PEG4000, 0.1 m MOPS (pH 7.0), and 0.1 m MnSO4 and equilibrated against 1 ml of reservoir solution. Selenomethionine-substituted proteins were crystallized similarly with 5 mm dithiothreitol in the reservoir solution. Crystals were cryoprotected by soaking for 1 min in a cryo solution containing 20% (v/v) glycerol in reservoir solution followed by flash-cooling under liquid nitrogen.
To obtain the VACVase-l-tyrosine amide (TYC) complex, crystals of native VACVase were transferred to a stabilizing solution containing 18% PEG4000, 40 mm Tris (pH 7.0), and 60 mm MnSO4 and then soaked with 50 mm TYC for 10 h before cryoprotection.
Synchrotron data collection was performed at 100 K under a nitrogen stream on a Mar165 CCD detector at COM-CAT or a Mar300 CCD detector at GM/CA-CAT of the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). Multiwavelength anomalous diffraction data were collected to 2.05 Å at wavelengths corresponding to the inflection point (λi), high energy remote (λh), low energy remote (λl), and peak point (λp) of the selenium x-ray absorbance spectrum from a single crystal of selenomethionine-labeled VACVase. Data were processed with D*TREK (19) or HKL2000 (20) (Table 1).
TABLE 1.
Data collection, phasing, and refinement statistics
| Se-Met VACVase | Native | S122A | D123N | Native + TYC | |||||
|---|---|---|---|---|---|---|---|---|---|
| Data collection | |||||||||
| Space group | P62 | P62 | P62 | P62 | P62 | ||||
| Cell dimensions | |||||||||
| a, b, c (Å) | 88.62 | 88.97 | 88.79 | 88.99 | 88.60 | ||||
| 88.62 | 88.97 | 88.79 | 88.99 | 88.60 | |||||
| 86.58 | 86.34 | 86.01 | 86.35 | 86.71 | |||||
| α, β, γ (°) | 90 | 90 | 90 | 90 | 90 | ||||
| 90 | 90 | 90 | 90 | 90 | |||||
| 90 | 120 | 120 | 120 | 120 | |||||
|
Peak
|
Inflection
|
Remote 1
|
Remote 2
|
||||||
| Wavelength | 0.9791 | 0.9793 | 0.9565 | 1.0029 | 1.0029 | 1.0000 | 1.0000 | 0.9792 | |
| Resolution (Å) | 2.05 | 2.05 | 2.05 | 2.05 | 1.75 | 1.90 | 1.85 | 1.90 | |
| Rmerge | 0.077 (0.165) | 0.067 (0.156) | 0.068 (0.165) | 0.071 (0.184) | 0.065 (0.331) | 0.083 (0.316) | 0.067 (0.322) | 0.125 (0.417) | |
| <I/σI> | 33.7 (16.5) | 39.8 (18.3) | 38.2 (17.7) | 36.7 (15.4) | 18.7 (6.5) | 15.2 (6.2) | 25.6 (3.7) | 11.4 (2.9) | |
| Completeness (%) | 99.8 (97.6) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (99.9) | 99.6 (100.0) | |
| Redundancy | 22.6 (21.4) | 22.7 (22.2) | 22.7 (22.2) | 22.6 (21.3) | 11.3 (10.9) | 10.8 (10.4) | 5.3 (5.0) | 10.9 (10.9) | |
| Refinement | |||||||||
| Resolution (Å) | 44.49-1.75 | 44.39-1.90 | 39.56-1.85 | 38.36-1.90 | |||||
| No. reflections | 39,074 | 30,366 | 32,415 | 30,385 | |||||
| Rwork/Rfree (%) | 18.2/20.2 | 18.5/20.8 | 23.7/27.3 | 19.6/21.7 | |||||
| No. of atoms | |||||||||
| Protein | 2,032 | 2,031 | 2,025 | 2,032 | |||||
| Ligand/ion | 8 | 2 | 2 | 15 | |||||
| Water | 302 | 285 | 285 | 268 | |||||
| B-factors (Å2) | |||||||||
| Protein | 23.1 | 25.3 | 23.6 | 29.0 | |||||
| Ligand/ion | 32.5 | 19.6 | 19.9 | 28.2 | |||||
| Water | 34.9 | 35.9 | 33.1 | 38.9 | |||||
| Root mean square deviations | |||||||||
| Bond lengths (Å) | 0.005 | 0.005 | 0.008 | 0.006 | |||||
| Bond angles (°) | 1.33 | 1.31 | 1.46 | 1.31 | |||||
| Ramachandran analysis | |||||||||
| Most favored regions (%) | 91.7 | 91.3 | 90.4 | 90.4 | |||||
| Additional allowed regions (%) | 7.8 | 8.3 | 9.2 | 9.2 | |||||
| Generously allowed regions (%) | 0.5 | 0 | 0.5 | 0 | |||||
| Disallowed regions (%) | 0 | 0.5 | 0 | 0.5 | |||||
Phase Determination, Model Building, and Refinement—Experimental phases were determined by the multiwavelength anomalous diffraction method (Table 1) (21). Five selenium sites expected for one VACVase molecule were found using the anomalous difference Patterson method as implemented in CNS (22). Heavy atom parameters were refined, and multiwavelength anomalous diffraction phases were calculated and improved by density modification with CNS. The high quality experimental map and the known selenium sites allowed unambiguous tracing of the protein backbone and most of the side chains using O (23). The initial model of selenomethionine VACVase was used as a starting point for the refinement against the native data set using CNS, with the quality of the model cross-validated by using 5% of randomly selected test set. The structures of S122A and D123N mutants and the VACVase-TYC complex were determined by molecular replacement with CNS using the structure of native VACVase as the search model. TYC coordinates, topology, and parameter files were modified from those of l-tyrosine obtained from the HICUP server. The stereochemistry of the final models was analyzed with PROCHECK (24). Refinement statistics are summarized in Table 1.
RESULTS
Substrate Specificity of VACVase—VACVase is synthesized in the cell as a precursor protein with a leader sequence of 20 residues (7, 25). The mature form of human VACVase (residues 21-274) was overexpressed, purified, and used in this study. We first carried out detailed analysis on its substrate specificity. The ability of VACVase to hydrolyze not only valacyclovir and valganciclovir but also the amino acid ester prodrugs of a range of nucleoside analogues prompted us to examine the role of an α-amino group in the substrates. As shown in Table 2, the enzyme efficiently hydrolyzed benzyl esters of l-Phe and l-Ala; however, its activity toward β-amino acid esters was 3 orders of magnitude lower than toward the corresponding α-amino acid esters (barely measurable). Furthermore, the enzyme can neither hydrolyze benzyl esters with corresponding α-hydroxyl acyl moieties (benzyl (S)-(-)-2-hydroxy-3-phenylpropionate and l-lactic acid benzyl ester) nor hydrolyze benzyl esters with aliphatic or aromatic acyl moieties (3-phenylpropionic acid ethyl ester and benzyl propionate). Additionally, the enzyme exhibited no activity for esters with a blocked α-amino group (N-acetyl-l-Phe ethyl ester), further supporting that the α-amino group is required for activity.
TABLE 2.
Substrate specificity of human VACVase
Activity was measured as described under “Experimental Procedures.” Initial rates of hydrolysis were determined by measuring substrate disappearance using HPLC, except for valacyclovir, for which the initial rate was determined by measuring the production of acyclovir.
| Compound | Specific activity |
|---|---|
| Units/mg | |
| Requirement for the α-amino group | |
| l-Phe benzyl ester | 358.3 ± 34.8 |
| 3-Amino-3-phenylpropionic acid ethyl ester | 0.6 ± 0.2 |
| Benzyl (S)-(—)-2-hydroxy-3-phenylpropionatea | −b |
| 3-Phenylpropionic acid ethyl estera | − |
| N-Acetyl-l-Phe ethyl ester | − |
| l-Ala benzyl ester | 1367.7 ± 94.3 |
| β-Ala benzyl ester | 1.8 ± 0.3 |
| l-Lactic acid benzyl ester | − |
| Benzyl propionatea | − |
| Specificity towards the amino acyl group | |
| Valacyclovir | 103.7 ± 2.7 |
| l-Ala benzyl ester | 1367.7 ± 94.3 |
| l-Gly benzyl ester | 849.4 ± 25.1 |
| l-Pro benzyl ester | 5148.4 ± 215.5 |
| l-Leu benzyl ester | 635.0 ± 23.9 |
| l-Val benzyl ester | 156.3 ± 7.4 |
| l-Met methyl ester | 156.2 ± 12.3 |
| l-Phe benzyl ester | 358.3 ± 34.8 |
| l-Tyr benzyl ester | 41.4 ± 0.9 |
| l-Trp methyl ester | 35.3 ± 2.0 |
| l-Ser benzyl ester | 484.3 ± 30.0 |
| l-Asn p-nitrobenzyl ester | − |
| l-Asp benzyl ester | − |
| Specificity towards the leaving group | |
| l-Phe benzyl ester | 358.3 ± 34.8 |
| l-Phe ethyl ester | 75.3 ± 3.1 |
| l-Phe t-butyl ester | − |
Assayed at 2 mm.
−, No detectable activity.
To gain further insight into the specificity of VACVase for the acyl moiety, a range of α-amino acid esters (mostly benzyl esters) were investigated (Table 2). Surprisingly, the enzyme showed fairly broad specificity against the amino acid side chains. Of the amino acid esters examined, the l-Pro ester was most efficiently hydrolyzed followed by the small amino acid esters, l-Gly and l-Ala esters. The hydrophobic and aromatic amino acid esters were also efficiently hydrolyzed by the enzyme. Compared with the l-Phe ester, the activities for the l-Tyr and l-Trp esters were significantly lower. Interestingly, the l-Asn ester is not a substrate for the enzyme, in contrast to the l-Ser ester, which was hydrolyzed very efficiently. Furthermore, the enzyme showed no activity for the ester with a negatively charged amino acid side chain, l-Asp benzyl ester.
Previously, it has been shown that VACVase hydrolyzed the 5′ l-Val ester of floxuridine more efficiently than the 3′ l-Val ester, suggesting that the enzyme prefers a primary alcohol leaving group to a secondary one (17). To gain more insight into the specificity for the alcohol leaving group, three l-Phe esters were tested (Table 2). Compared with the l-Phe benzyl ester, the activity for the l-Phe ethyl ester was significantly lower, whereas the tertiary alcohol ester of l-Phe, l-Phe t-butyl ester, was not a substrate for the enzyme. Thus, the enzyme has a preference for a primary alcohol leaving group but appears to have no major constraints on the nature of the primary alcohol as amino acid esters with various leaving groups ranging from the small methyl group to the larger nucleoside analogues are all efficiently hydrolyzed by the enzyme. These specificity analyses demonstrated that VACVase is a specific α-amino acid ester hydrolase that prefers small, hydrophobic, and aromatic side chains and does not have a stringent requirement for the leaving group other than preferring a primary alcohol.
Overall Structure of VACVase—VACVase, the S122A and D123N mutants, and its complex with the product analogue TYC were all crystallized in the space group P62 with one protein molecule per asymmetric unit. The native structure was determined to 1.75 Å by the multiwavelength anomalous diffraction method (Table 1), and the other structures were determined by molecular replacement. These various structures have essentially the same conformation, with their Cα atoms superposing to within 0.3 Å.
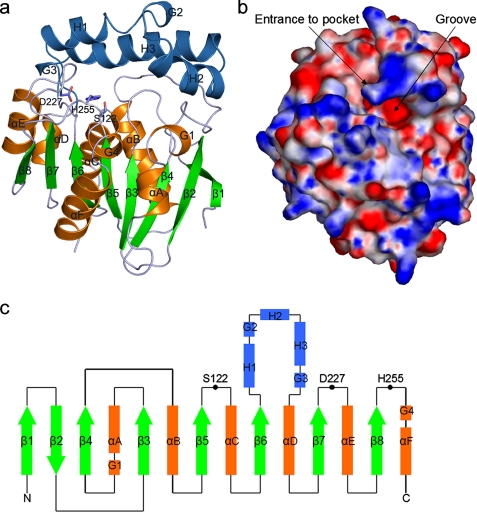
VACVase contains 8 β-strands and 13 α-helices, which form two domains, an α/β hydrolase domain and a cap domain (Fig. 2, a-c). The α/β hydrolase domain consists of a central eight-stranded, twisted β sheet flanked by 5 α-helices. The cap domain is inserted between β6 and αD, and “sits” on top of the loop regions of the α/β hydrolase domain, mimicking a cap. It consists of 3 helices, with helices H1 and H3 in antiparallel orientation and helix H2 oriented ∼90° to H1 and H3. Between the two domains lies a cavity system containing a cave-like pocket with a single opening and a large open groove extending from the pocket mouth (Fig. 2b). Residues in the α/β hydrolase domain, especially those forming the central β sheet, exhibit lower B-factors compared with the cap domain, consistent with the function of this domain in providing a stable structural frame for the catalytic machinery. The relatively high B-factors of the cap domain suggest that it is more flexible, which is required for its function in substrate recognition as observed in other α/β hydrolases (18, 26).
FIGURE 2.
Overall structure of VACVase. a, ribbon representation of VACVase. Insertions (cap domain) relative to the α/β hydrolase fold are colored blue. The catalytic triad residues are shown as sticks. b, surface representation of VACVase colored by electrostatic potential. The continuum electrostatic potential (ranging from -10 to + 10 kT), where k is the Boltzman constant and T is the temperature, was calculated with APBS (43). c, topology diagram of VACVase. Helices are represented as rectangles, strands as arrows, and coils as lines. The eight strands labeled as β1-β8 and six helices labeled as αA-αF constitute the α/β hydrolase fold. Helices H1-H3 constitute the cap domain. Four short helices are labeled from G1 to G4. Black circles represent the catalytic triad residues.
A DALI search (27) revealed dozens of other α/β hydrolases as structural homologues, of which 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase (BphD) from Rhodococcus sp. (28) is the closest, with a root mean square deviation of 2.8 Å over 230 aligned Cα atoms. BphD is involved in the degradation of aromatic compounds in the meta-cleavage pathway (28) and is capable of breaking a C-C bond. Another interesting homologue is the tricorn-interacting aminopeptidase F1 (29) with a root mean square deviation of 3.1 Å over 193 aligned Cα atoms. The α/β hydrolase domains among the three enzymes are strikingly similar; however, their cap domains exhibit high structural variation (supplemental Fig. 1). A further DALI search with the Cα atoms of the VACVase cap domain did not result in any significant structural homologue, suggesting that the cap domain has a novel fold.
The Crystallographic Dimer and Metal Ions—A notable feature of VACVase is its oligomeric state and the associated metal ions observed in the crystal structure. The enzyme appears to be a dimer formed between two crystallographic symmetry-related protomers and is held together by close symmetric interactions of residues from helices G3, αD, αB, and the loops between helices H3 and G3 and helices G3 and αD (supplemental Fig. 2a). The dimer interface buries a surface area of 1046 Å2 (5% of the total surface of the dimer) and is not contiguous. In fact, the interface interactions are mostly polar, involving hydrogen bonds and van der Waals contacts, with the cavities of the interface filled with water molecules (supplemental Fig. 2, b and c).
An interesting feature of the dimer interface is a metal ion located on the 2-fold axis. This ion is shared between the two monomers and is coordinated by two symmetry-related Asp residues (Asp-204) from each monomer. It is further coordinated by four water molecules clearly observed in the electron density map (supplemental Fig. 2b). Initially the metal ion in the dimer interface was built as a manganese ion since it is a component of the crystallization solution. Upon refinement, however, Mn2+ displayed a negative difference electron density peak. Replacing the metal to Mg2+ eliminated the negative peak. Although we cannot rule out the possibility of a Mn2+ cation with fractional occupancy, several lines of evidence suggest that a Mg2+ cation is more reasonable interpretation. The refined bond lengths between the ion and its coordinating oxygen atoms (2.09, 2.04, and 2.05 Å) are similar to the average values calculated from crystal structures of magnesium-containing proteins in the Protein Data Bank and small molecules in the Cambridge Structural Data base (30, 31). Most importantly, the presence of a Mg2+ here is supported by metal analysis (see the supplemental methods), which showed that Mg2+ is the major metal species in the enzyme with a stoichiometry of one Mg2+ per two polypeptide chains (supplemental Table 1).
A question arises of whether the dimer observed in the crystal is representative of the species found in solution. Gel filtration analysis (see the supplemental methods), in contrast to the crystallography and metal analysis results, suggested that VACVase is a monomer in the presence of 100 mm NaCl in 50 mm Tris (pH 7.2). The enzyme still behaved as a monomer in gel filtration analysis in the presence of 1 mm Mg2+.It is possible that there exists an equilibrium of monomeric and dimeric species of VACVase in solution and that the condition of the gel filtration analysis favors the monomeric form.
In addition to the Mg2+ cation at the dimer interface, each VACVase monomer contains an Mn2+ cation, which is bound by His-35, Glu-57, Asp-78, and His-84 from the α/β hydrolase domain and one water molecule (supplemental Fig. 2d). This metal is 18 Å from the O-γ of the catalytic Ser-122 and is further distant from the substrate binding pocket, suggesting that it is not directly involved in the catalytic activity. Indeed, the addition of Mn2+ or Mg2+ to the assay mixture at concentrations ranging from 10 μm to2mm or EDTA at the concentration up to 2 mm has no effect on enzyme activity (data not shown), further suggesting that the metal ions are not involved in catalysis. Although Mg2+ plays a structural role, Mn2+ is more likely a crystallographic artifact as crystallization requires the presence of this metal ion.
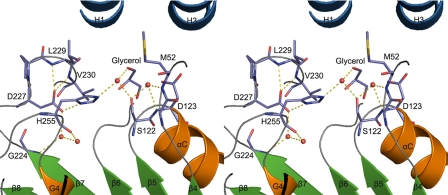
Catalytic Residues of VACVase—The putative catalytic triad resides at the base of the pocket (Figs. 2c and 3). Ser-122 is located on the nucleophile elbow that is the most conserved structural feature of the α/β hydrolase fold (Fig. 3) (26) and adopts an energetically strained main chain conformation with ϕ and ψ angles of 56.5° and -121.8°, respectively. Such unique conformation of Ser-122 allows easy access on one side by His-255 and on the other side by the substrate. The mechanism of serine hydrolases relies on flexibility in the active site to allow for shift of the acid-His pair relative to the nucleophile. The acid of the triad, Asp-227, is fixed by a hydrogen bonding network in which Oδ1 of Asp-227 is anchored via two hydrogen bonds with the main chain amides of the residues two and three amino acids downstream in the sequence, Leu-229 and Val-230 (Fig. 3). The strong hydrogen bond between Asp-227 Oδ2 and His-255 Nδ1 further ensures conformational rigidity of this Asp-His pair.
FIGURE 3.
Stereoview of the active site residues of VACVase. The red spheres represent water molecules.
It is worth noting that a glycerol molecule is observed in the active site of native VACVase, although it does not make any direct contact with the catalytic residues. Possibly because of the presence of this glycerol, Oγ of Ser-122 is not directly hydrogen-bonded with His-255. Instead, it forms hydrogen bonds with two water molecules, one of which occupies the oxyanion hole, whereas the other one is further hydrogen-bonded with Nε2 of His-255. Thus, the Oγ of Ser-122 is not activated by the Asp-His pair, and the three residues represent a catalytic triad in a latent state (Fig. 3). Interestingly, a water molecule hydrogen-bonded to His-255 resembles the nucleophilic water during the deacylation step.
The roles of the catalytic triad residues were examined by site-directed mutagenesis (supplemental Table 2). The H255A mutant completely lost activity. Mutation of Ser-122 to Ala or Asp-227 to Asn essentially eliminated activity (<0.1% of wild-type activity). Furthermore, the S122A mutant structure (Table 1) shows no appreciable conformational changes in the active site residues compared with the native enzyme, indicating its inactivity is only due to the absence of the nucleophilic hydroxyl group. Thus, these results confirmed that Ser-122—His-255—Asp-227 constitutes a functional catalytic triad. In an attempt to engineer a modified catalytic triad into the VACVase structural frame, the nucleophilic hydroxyl group was replaced by a sulfhydryl group in S122C (supplemental Table 2). The resulting mutant, however, did not exhibit significant activity above the background (<0.1% of wild-type activity). This result is similar to the results observed in other α/β hydrolases, such as the S203C mutant of human acetylcholinesterase (32), the S114C mutant of Vibrio harveyi myristoyl-ACP thioesterase (33), and the S232C mutant of mouse cytosolic type I acyl-CoA thioesterase (34).
The transient oxyanion that forms during the acylation and deacylation steps is stabilized in a positively charged oxyanion hole occupied by a water molecule in the native VACVase. It is formed by the main chain amides of the residue after the nucleophile, Asp-123, and a residue located at the loop between β3 and αA, Met-52 (Fig. 3).
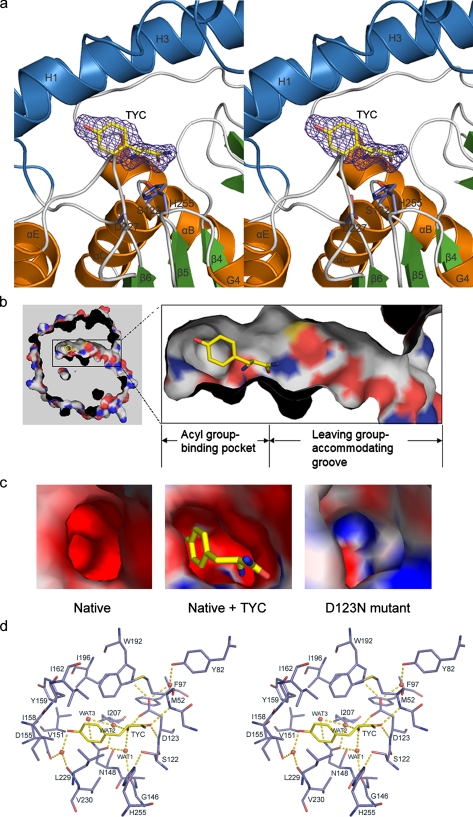
Binding Mode of TYC—The molecular details of the cavity system and substrate binding are best revealed by the structure of VACVase complexed with the product analog TYC (Fig. 4a) whose amide bond is not cleavable by the enzyme (data not shown). The cavity can be divided into an acyl group binding pocket and a leaving group-accommodating groove, with the Oγ of Ser-122 located at the bottom of the pocket opening (Fig. 4b). TYC is bound within the pseudo-tubular pocket which has a depth of ∼12 Å and a maximum diameter of ∼10 Å. Notably, the pocket walls are mostly hydrophobic (Fig. 4b) and are formed by hydrophobic and aromatic side chains from both the α/β hydrolase (Met-52, Phe-97, Leu-229, and Val-230) and cap (Val-151, Ile-158, Tyr-159, Ile-162, Trp-192, Ile-196, and Ile-207) domains. The bottom of the pocket, however, contains a few hydrophilic elements contributed by the side chains from Asp-123, Asp-155, and Asn-148 as well as the main chain carbonyl oxygens of Gly-146 and Leu-229 (Fig. 4d).
FIGURE 4.
TYC binding in VACVase. a, stereoview of the Fo - Fc-simulated annealing omit electron density of TYC. The map is shown at a 3.8 σ contour level. TYC and the catalytic triad residues are shown as sticks. b, cutaway view of the surface showing the acyl group binding pocket and the leaving group-accommodating groove. TYC is bound in the cave-like pocket. c, the local surface electrostatic potential of the acyl group binding pockets in native, TYC-bound VACVase, and the D123N mutant. The continuum electrostatic potential (ranging from -10 to + 10 kT), where k is the Boltzman constant and T is the temperature, was calculated with APBS (43). TYC is shown as sticks. d, the binding mode of TYC in stereoview. The red spheres represent water molecules.
Upon TYC binding, the Oγ of Ser-122 switches toward the Nε2 of His-255 resulting in a hydrogen bond of 2.8 Å between Oγ and Nε2 (Fig. 4d). Thus, this structure captures the catalytic triad in a productive configuration. As expected, the carbonyl oxygen of TYC occupies the oxyanion hole, whereas the amide nitrogen points toward the solvent and makes no contact with the protein. The aromatic side chain of TYC is surrounded by the hydrophobic pocket walls and forms van der Waals contacts with the side chains of Ile-158, Tyr-159, Ile-162, Trp-192, Leu-229, and Val-230. The phenolic OH is hydrogen-bonded to a water molecule which is fixed by hydrogen bonding to the side chain of Asp-155 and the carbonyl oxygen of Leu-229. The α-amino group of TYC is anchored by a hydrogen-bonding network in which the free amine is apparently in a protonated state and forms ionic interaction with the side chain of Asp-123 and hydrogen bonds with two water molecules. One of the water molecules is further hydrogen-bonded to the side chain of Asn-148 and the carbonyl oxygen of Gly-146, whereas the other one also forms hydrogen bonds to a third water and the side chain of Asn-148.
Compared with the native structure, minor main chain conformational changes are observed in the cap domain, including residues in helix H1 and the C-terminal half segment of helix H3. The main chains of these residues show movements (Cα shifts less than 1.4 Å) in a direction pointing away from the pocket center, leading to an expansion of the pocket. Direct conformational changes in the pocket forming residues are observed with Tyr-159, Ile-162, and Ile-196 (supplemental Fig. 3). As a result, the volume of the acyl group binding pocket in the complex structure (72.7 Å3) increases by 3-fold compared with that of the apoenzyme (24.3 Å3), indicating that the pocket is highly flexible. Such flexibility and the mostly hydrophobic nature of the pocket walls also account for the ability of VACVase to hydrolyze a wide spectrum of mostly nonpolar amino acid esters ranging from the small glycyl to the bulky tryptophanyl ester (Table 2).
Electrostatic Properties of VACVase—As a basic protein, VACVase displays mostly positive electrostatic potential on the surface (Fig. 2b). However, except for the oxyanion hole, the electrostatic potential in the acyl group binding pocket is strongly negative (Fig. 2b and 4c). Further examination of the pocket-lining residues suggested that the side chains of Asp-123 and Asp-155 as well as the main chain carbonyl oxygens of Gly-146 and Leu-229 may all contribute to the electrostatic potential in the pocket (Fig. 4d). In particular, Asp-123, the residue immediately after the nucleophile, is isolated from other hydrophilic elements and forms a hydrophilic patch in a hydrophobic wall. As the only acidic residue that is close to and makes direct contact with the α-amino group of TYC, Asp-123 is examined for its role in substrate recognition.
As shown in supplemental Table 3, both D123A and D123N mutants completely lost activity for α-amino acid benzyl esters and showed no activity for the ester with an aliphatic acyl moiety (benzyl propionate). Remarkably, the D123N mutant exhibited low activity for the hydrolysis of α-hydroxyl acid esters (l-lactic acid benzyl ester and benzyl (S)-(-)-2-hydroxy-3-phenylpropionate), whereas neither the wild-type nor the D123A enzyme hydrolyzed these two esters. Thus, the salt bridge between the α-amino group and the Asp observed in the native enzyme is substituted by a hydrogen bond between the α-hydroxyl group and the Asn in the D123N mutant. The importance of Asp-123 is also supported by the structure of the D123N mutant in which the α-amino group binding region shows positive electrostatic potential in contrast to the negative potential observed in the native enzyme (Fig. 4c). Furthermore, the side chain carboxylate of Asp-123 is fixed by two hydrogen bonds that ensure its conformational rigidity, one with the side chain Nε1 of Trp-192, which may increase the acidity of Asp-123, and the other with a water molecule which is further hydrogen-bonded to Y82 (Fig. 4d). Therefore, these observations indicate that this acidic residue functions as the single most important specificity determinant by (i) playing a critical role in generating the local negative electrostatic potential and (ii) forming a crucial electrostatic interaction with the substrate α-amino group.
DISCUSSION
VACVase was previously suggested as a nucleoside prodrug activating enzyme (7, 17). The present biochemical and structural studies demonstrated that the specificity of this enzyme resides mainly in the acyl moiety and to a less extent in the alcohol moiety; thus, the enzyme is better defined as an α-amino acid ester prodrug-activating enzyme. The conclusion of VACVase as an α-amino acid ester hydrolase here is also supported by a previous report of a VACVase isolated from rat liver (35), which showed that the α-amino group in the substrate is important for activity. Moreover, neither rat nor human VACVase exhibits peptidase activity (17, 35), further supporting its function as an esterase. Notably, the wide specificity spectrum of VACVase toward the nonpolar aminoacyl moiety resembles that of its structural homologue tricorn-interacting aminopeptidase F1 (29, 36), which differs from VACVase in that it hydrolyzes peptides, whereas VACVase does not. Interestingly, in addition to F1, there exist tricorn-interacting amin-opeptidases F2 and F3 that exhibit preferences for polar and charged residues in the P1 position of peptide substrates (37), whereas no such α-amino acid ester hydrolases have been found.
As the driving force for the entry and binding of a positively charged α-amino acid ester substrate, the localized negative electrostatic potential of VACVase resembles that of the active site gauge of acetylcholine esterase (38, 39) in which electrostatic steering guides the entry of positively charged acetylcholine. In fact, such electrostatic focusing within a protein cleft is also a general phenomenon in biology and often plays an important role in the function of proteins (40).
The most unique structural feature of VACVase, perhaps, is the surprisingly important role of Asp-123 in defining the specificity. Fixation of the substrate α-amino group by Asp-123 and the carbonyl oxygen by the oxyanion hole would bring the carbonyl carbon in close proximity to the nucleophile and, thus, present the substrate in an optimal orientation for catalysis. Apparently, binding of esters without an α-amino group would leave a buried negative charge in Asp-123, which is energetically unfavorable. Interestingly, although VACVase and tricon-interacting aminopeptidase F1 possess similar specificity for the acyl moiety, they differ significantly in the binding mode of the substrate α-amino group. In F1, the nucleophile-following residue (Tyr-106) points its side chain away from the active site pocket and does not participate in substrate binding. Instead, the substrate α-amino group is coordinated in F1 by a hydrogen-bonding network of two glutamates, one tyrosine, and the carbonyl oxygen of a glycine (29). To our knowledge, the observation with Asp-123 here is the first time that a nucleophile-following residue has been observed to be directly involved in substrate binding and discrimination in serine hydrolases.
Although VACVase has a stringent requirement for an aminoacyl moiety, it can accommodate a variety of primary and secondary alcohol leaving groups but not tertiary alcohols. The relative promiscuity toward the leaving group may be attributed to the wide and open groove of the enzyme (∼16Åin length) extending from the pocket (Fig. 4b). In fact, molecular docking of valacyclovir and amino acid benzyl esters on this enzyme revealed that the leaving groups are exposed to the solvent and make no specific interaction with the groove-lining residues (data not shown), indicting that catalysis does not require tight binding of the leaving group.
The inactivity of VACVase toward esters with a tertiary alcohol leaving group is probed by modeling l-tyrosine t-butyl ester in the liganded structure. With the l-tyrosinyl moiety fixed in the same conformation as in the TYC liganded structure, the t-butyl group clashes with the pocket mouth-lining residues when rotated along the ester C-O bond (data not shown). Hence, the narrow pocket mouth functions as a bottleneck to preclude an ester formed by a tertiary alcohol from binding the enzyme in a catalytically competent conformation.
Bioinformatic searches revealed that homologous genes displaying significant identity to human VACVase are not present in prokaryotes, fungi, or plants, i.e. VACVases exist only in metazoa, indicating a physiological function exclusive in metazoa for this esterase activity. The enzyme also shows low sequence similarity (<25% identity) to two known subfamilies of α/β hydrolases, the bacterial BphD enzymes (25, 41) and the archaeal tricon-interacting amin-opeptidases F1 (36). Both enzymes are also the closest structural homologues of VACVase, implying that they may have closer evolutionary relationship to VACVase than other α/β hydrolases. Nevertheless, they have been subjected to considerable evolutionary changes especially in the cap domains to perform different functions.
To classify the enzyme within the α/β hydrolase-fold superfamily, phylogenetic analysis was performed on the currently available VACVase homologous sequences distributed in 18 species ranging from Nematoda to primates (supplemental Fig. 4). Clustering of these sequences is consistent with taxonomic classification. Notably, these sequences cluster into two major groups. The Heterodera glycine, Caenorhabditis elegans, and Brugia malayi sequences are clustered in a distinct clade, temporarily assigned as Group I, whereas the other sequences are in a larger clade, temporarily assigned as Group II. The functional relevance of this two-group bifurcation is not known. However, examination of the aligned sequences revealed that Group I members have a Glu residue in the position corresponding to the α-amino group binding residue Asp-123, whereas all Group II members display an Asp residue in this position (Fig. 1b). We speculate that this could be one major difference distinguishing the two clades of VACVases. Certainly, biochemical studies on VACVases from various sources are needed to support these computational results. Based on the biochemical, structural, and phylogenetic studies, we propose that human VACVase and its homologues from metazoa constitute a new family of eukaryotic α-amino acid ester hydrolases.
Although the present study has established unequivocally that VACVase is an α-amino acid ester hydrolase, its physiological function remains unknown. Previous studies have shown that the enzyme is widely expressed in a variety of human tissues with highest levels in liver and kidney (25). Such broad distribution/abundance may indicate an important physiological role for its activity. In addition, the sequence features of the VACVase encoding gene BPHL promoter region conform to those of a typical housekeeping (constitutively expressed) gene (42), further suggesting the importance of this enzyme.
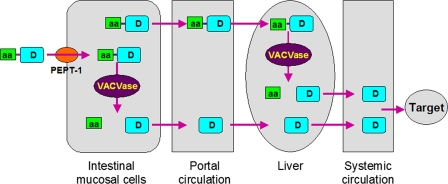
Despite its unknown physiological function, the promiscuity of VACVase for the leaving group suggests a broader application for it as a prodrug activating enzyme, i.e. VACVase-targeted prodrug design can be applied to other classes of drugs in addition to nucleoside analogues. Based on the molecular properties and tissue distribution of VACVase, the pharmacokinetic profiles (10, 11, 16), transport (3, 14), and activation mechanism of valacyclovir, a complete model for a molecular biology-based drug delivery system, is presented in Fig. 5. Drugs that are polar and poorly absorbed (due to low membrane permeability) can be developed as amino acid ester prodrugs that can be transported across intestinal epithelium membrane by the H+-coupled oligopeptide transporter 1 (PEPT1) and then activated by VACVase in the intestine and the liver. As a result, the oral bioavailability of the drug will be significantly improved. This amino acid ester prodrug approach represents a novel drug delivery strategy mechanistically exploiting human physiology and biology and can significantly expand the number of therapeutic candidates for oral systemic delivery.
FIGURE 5.
Model for an amino acid ester prodrug-based drug delivery strategy targeted to intestinal peptide transporters and VACVase. aa represents an amino acid; D represents a drug molecule; PEPT-1, oligopeptide transporter 1.
In summary, this report reveals that human VACVase is a member of a new family of eukaryotic α-amino acid ester hydrolase. The enzyme contains a unique active site characterized by a flexible and mostly hydrophobic acyl pocket, a localized negative electrostatic potential, a large open leaving group-accommodating groove, and most importantly, a pivotal acidic residue, Asp-123. This work provides the molecular basis for prodrug activation by VACVase and establishes this enzyme as an essential physiological component of an amino acid ester prodrug-based oral drug delivery system.
Supplementary Material
Acknowledgments
We thank J. Brunzelle and M. Becker of Advanced Photon Source for help with data collection and the Hauptman-Woodward Medical Research Institute (Buffalo, NY) for performing high throughput crystallization screening. We acknowledge T. Huston of the Department of Geology (University of Michigan) for performing the metal analysis studies.
The atomic coordinates and structure factors (codes 2OCG, 2OCI, 2OCK, and 2OCL) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported in part by National Institutes of Health Grant GM 37188 (to G. L. A.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1-S3 and Figs. S1-S4.
Footnotes
The abbreviations used are: VACVase, valacyclovirase; BphD, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase; TYC, l-tyrosine amide; MOPS, 4-morpholinepropanesulfonic acid; HPLC, high performance liquid chromatography.
References
- 1.Ettmayer, P., Amidon, G. L., Clement, B., and Testa, B. (2004) J. Med. Chem. 47 2393-2404 [DOI] [PubMed] [Google Scholar]
- 2.Stella, V. J., Borchardt, R. T., Hageman, M. J., Oliyai, R., Maag, H., and Tilley, J. W. (2007) Prodrugs: Challenges and Rewards, Springer-Verlag New York Inc., New York
- 3.Han, H., de Vrueh, R. L., Rhie, J. K., Covitz, K. M., Smith, P. L., Lee, C. P., Oh, D. M., Sadee, W., and Amidon, G. L. (1998) Pharmacol. Res. 15 1154-1159 [DOI] [PubMed] [Google Scholar]
- 4.Liederer, B. M., and Borchardt, R. T. (2006) J. Pharmacol. Sci. 95 1177-1195 [DOI] [PubMed] [Google Scholar]
- 5.Satoh, T., and Hosokawa, M. (1998) Annu. Rev. Pharmacol. Toxicol. 38 257-288 [DOI] [PubMed] [Google Scholar]
- 6.Imai, T. (2006) Drug Metab. Pharmacokinet. 21 173-185 [DOI] [PubMed] [Google Scholar]
- 7.Kim, I., Chu, X. Y., Kim, S., Provoda, C. J., Lee, K. D., and Amidon, G. L. (2003) J. Biol. Chem. 278 25348-25356 [DOI] [PubMed] [Google Scholar]
- 8.de Miranda, P., and Blum, M. R. (1983) J. Antimicrob. Chemother. 12 Suppl. B, 29-37 [DOI] [PubMed] [Google Scholar]
- 9.Curran, M., and Noble, S. (2001) Drugs 61 1145-1150 [DOI] [PubMed] [Google Scholar]
- 10.Weller, S., Blum, M. R., Doucette, M., Burnette, T., Cederberg, D. M., Demiranda, P., and Smiley, M. L. (1993) Clin. Pharmacol. Ther. 54 595-605 [DOI] [PubMed] [Google Scholar]
- 11.Soul-Lawton, J., Seaber, E., On, N., Wootton, R., Rolan, P., and Posner, J. (1995) Antimicrob. Agents Chemother. 39 2759-2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung, D., and Dorr, A. (1999) J. Clin. Pharmacol. 39 800-804 [DOI] [PubMed] [Google Scholar]
- 13.Pescovitz, M. D., Rabkin, J., Merion, R. M., Paya, C. V., Pirsch, J., Freeman, R. B., O'Grady, J., Robinson, C., To, Z., Wren, K., Banken, L., Buhles, W., and Brown, F. (2000) Antimicrob. Agents Chemother. 44 2811-2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balimane, P. V., Tamai, I., Guo, A., Nakanishi, T., Kitada, H., Leibach, F. H., Tsuji, A., and Sinko, P. J. (1998) Biochem. Biophys. Res. Commun. 250 246-251 [DOI] [PubMed] [Google Scholar]
- 15.Sugawara, M., Huang, W., Fei, Y. J., Leibach, F. H., Ganapathy, V., and Ganapathy, M. E. (2000) J. Pharmacol. Sci. 89 781-789 [DOI] [PubMed] [Google Scholar]
- 16.Crooks, T. J., and Murray, A. (1994) Antiviral Chem. Chemother. 5 31-37 [Google Scholar]
- 17.Kim, I., Song, X., Vig, B. S., Mittal, S., Shin, H. C., Lorenzi, P. J., and Amidon, G. L. (2004) Mol. Pharmacol. 1 117-127 [DOI] [PubMed] [Google Scholar]
- 18.Nardini, M., and Dijkstra, B. W. (1999) Curr. Opin. Struct. Biol. 9 732-737 [DOI] [PubMed] [Google Scholar]
- 19.Pflugrath, J. W. (1999) Acta Crystallogr. D Biol. Crystallogr. 55 1718-1725 [DOI] [PubMed] [Google Scholar]
- 20.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 21.Hendrickson, W. A., and Ogata, C. M. (1997) Methods Enzymol. 276 494-523 [DOI] [PubMed] [Google Scholar]
- 22.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 23.Jones, T. A., Zou, J. Y., Cowan, S. W., and Kjeldgaard, M. (1991) Acta Crystallogr. Sect. A 47 110-119 [DOI] [PubMed] [Google Scholar]
- 24.Laskowski, R. A., MacArthur, M. W., Moss, D. S., and Thornton, J. M. (1993) J. Appl. Crystallogr. 26 283-291 [Google Scholar]
- 25.Puente, X. S., and Lopez-Otin, C. (1995) J. Biol. Chem. 270 12926-12932 [DOI] [PubMed] [Google Scholar]
- 26.Ollis, D. L., Cheah, E., Cygler, M., Dijkstra, B., Frolow, F., Franken, S. M., Harel, M., Remington, S. J., Silman, I., Schrag, J., Sussman, J. L., Verschueren, K. H. G., and Goldman, A. (1992) Protein Eng. 5 197-211 [DOI] [PubMed] [Google Scholar]
- 27.Holm, L., and Sander, C. (1995) Trends Biochem. Sci. 20 478-480 [DOI] [PubMed] [Google Scholar]
- 28.Nandhagopal, N., Yamada, A., Hatta, T., Masai, E., Fukuda, M., Mitsui, Y., and Senda, T. (2001) J. Mol. Biol. 309 1139-1151 [DOI] [PubMed] [Google Scholar]
- 29.Goettig, P., Groll, M., Kim, J. S., Huber, R., and Brandstetter, H. (2002) EMBO J. 21 5343-5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harding, M. M. (2006) Acta Crystallogr. D Biol. Crystallogr. 62 678-682 [DOI] [PubMed] [Google Scholar]
- 31.Harding, M. M. (2001) Acta Crystallogr D Biol. Crystallogr. 57 401-411 [DOI] [PubMed] [Google Scholar]
- 32.Shafferman, A., Kronman, C., Flashner, Y., Leitner, M., Grosfeld, H., Ordentlich, A., Gozes, Y., Cohen, S., Ariel, N., Barak, D., Harel, M., Silman, I., Sussman, J. L., and Velan, B. (1992) J. Biol. Chem. 267 17640-17648 [PubMed] [Google Scholar]
- 33.Li, J., Szittner, R., Derewenda, Z. S., and Meighen, E. A. (1996) Biochemistry 35 9967-9973 [DOI] [PubMed] [Google Scholar]
- 34.Huhtinen, K., O'Byrne, J., Lindquist, P. J., Contreras, J. A., and Alexson, S. E. (2002) J. Biol. Chem. 277 3424-3432 [DOI] [PubMed] [Google Scholar]
- 35.Burnette, T. C., Harrington, J. A., Reardon, J. E., Merrill, B. M., and de Miranda, P. (1995) J. Biol. Chem. 270 15827-15831 [DOI] [PubMed] [Google Scholar]
- 36.Tamura, T., Tamura, N., Lottspeich, F., and Baumeister, W. (1996) FEBS Lett. 398 101-105 [DOI] [PubMed] [Google Scholar]
- 37.Tamura, N., Lottspeich, F., Baumeister, W., and Tamura, T. (1998) Cell 95 637-648 [DOI] [PubMed] [Google Scholar]
- 38.Ripoll, D. R., Faerman, C. H., Axelsen, P. H., Silman, I., and Sussman, J. L. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 5128-5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan, R. C., Truong, T. N., McCammon, J. A., and Sussman, J. L. (1993) Biochemistry 32 401-403 [DOI] [PubMed] [Google Scholar]
- 40.Honig, B., and Nicholls, A. (1995) Science 268 1144-1149 [DOI] [PubMed] [Google Scholar]
- 41.Hernaez, M. J., Andujar, E., Rios, J. L., Kaschabek, S. R., Reineke, W., and Santero, E. (2000) J. Bacteriol. 182 5448-5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Puente, X. S., Pendas, A. M., and Lopez-Otin, C. (1998) Genomics 51 459-462 [DOI] [PubMed] [Google Scholar]
- 43.Baker, N. A., Sept, D., Joseph, S., Holst, M. J., and McCammon, J. A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 10037-10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.