Abstract
Very-long-chain acyl-CoA dehydrogenase (VLCAD) is a member of the family of acyl-CoA dehydrogenases (ACADs). Unlike the other ACADs, which are soluble homotetramers, VLCAD is a homodimer associated with the mitochondrial membrane. VLCAD also possesses an additional 180 residues in the C terminus that are not present in the other ACADs. We have determined the crystal structure of VLCAD complexed with myristoyl-CoA, obtained by co-crystallization, to 1.91-Å resolution. The overall fold of the N-terminal ∼400 residues of VLCAD is similar to that of the soluble ACADs including medium-chain acyl-CoA dehydrogenase (MCAD). The novel C-terminal domain forms an α-helical bundle that is positioned perpendicular to the two N-terminal helical domains. The fatty acyl moiety of the bound substrate/product is deeply imbedded inside the protein; however, the adenosine pyrophosphate portion of the C14-CoA ligand is disordered because of partial hydrolysis of the thioester bond and high mobility of the CoA moiety. The location of Glu-422 with respect to the C2-C3 of the bound ligand and FAD confirms Glu-422 to be the catalytic base. In MCAD, Gln-95 and Glu-99 form the base of the substrate binding cavity. In VLCAD, these residues are glycines (Gly-175 and Gly-178), allowing the binding channel to extend for an additional 12Å and permitting substrate acyl chain lengths as long as 24 carbons to bind. VLCAD deficiency is among the more common defects of mitochondrial β-oxidation and, if left undiagnosed, can be fatal. This structure allows us to gain insight into how a variant VLCAD genotype results in a clinical phenotype.
Very-long-chain acyl-CoA dehydrogenase (VLCAD)3 is one of five acyl-CoA dehydrogenases (ACADs) that catalyze the initial, rate-limiting step of mitochondrial fatty acid β-oxidation, with distinct but overlapping fatty acyl chain-length specificities (1, 2). In addition to VLCAD, which has optimal chain length specificity for fatty acyl-CoAs having 16 carbons in length, there are long-, medium-, and short-chain acyl-CoA dehydrogenases (LCAD, MCAD, and SCAD), which are most active with 14, 8, and 4 carbon substrates, respectively (3, 4). In addition, acyl-CoA dehydrogenase 9 (ACAD-9) is most active with unsaturated long-chain acyl-CoAs (5). The ACAD family also includes four members involved in amino acid metabolic pathways: isobutyryl-CoA dehydrogenase (IBD) in valine metabolism, isovaleryl-CoA dehydrogenase (IVD) in leucine metabolism, glutaryl-CoA dehydrogenase (GCAD) in lysine and tryptophan metabolism, and short-branched chain acyl-CoA dehydrogenase (SBCAD) in isoleucine metabolism. Fatty acyl-CoAs are oxidized to the corresponding trans-2,3-enoyl-CoA products with a concurrent reduction of the enzyme-bound FAD cofactor (6). Electron transfer flavoprotein (ETF) reoxidizes the reduced flavin and transfers reducing equivalents to the main mitochondrial respiratory chain through the enzyme ETF-ubiquinone oxidoreductase (7). Unlike other ACADs, which are soluble homotetramers with 45-kDa subunits, mature VLCAD and ACAD-9 are homodimers of 67-kDa subunits bound to the inner mitochondrial membrane (8, 9). VLCAD and ACAD-9 comprise a class of ACADs that have been heretofore structurally uncharacterized. Whereas the tetrameric ACADs share a 30% identity with the first ∼400 amino acids of the two enzymes, VLCAD and ACAD-9 possess an additional 180 residues on the C-terminal end. Mutations in this domain have been shown to affect binding to mitochondrial membranes, implicating it in the normal interaction of VLCAD with the inner mitochondrial membrane (10).
The structures of MCAD (11-13), SCAD (14), IVD (15), and glutaryl-CoA dehydrogenase (GCAD) (16) have been determined (see review in Ref. 17). The subunits of the tetrameric ACADs are arranged as a dimer of dimers. These ACADs have the same polypeptide fold, composed of an N-terminal α-helical domain (α-dom1), a middle β-sheet domain, and a C-terminal α-helical domain (α-dom2). The catalytic base of all known ACADs is a glutamate. In MCAD, this residue is Glu-376 (11, 18) and is the homologous glutamate in SCAD, IBD, and GCAD (14, 16, 19). In LCAD and IVD, however, this residue is not conserved. Instead, Glu-261 (Glu-254 in IVD) in helix G acts as the catalytic residue (15, 20). Sequence alignment of VLCAD and ACAD-9 with the other ACADs indicates that both VLCAD and ACAD-9 possess an MCAD-like catalytic glutamate.
In the peroxisome, β-oxidation is initiated by acyl-CoA oxidases (ACOs). ACOs are flavooxidases with one FAD per subunit and belong to the same superfamily as ACADs (21). In the oxidative half-reaction, however, molecular oxygen rather than ETF reoxidizes the flavin, producing hydrogen peroxide. Like VLCAD and ACAD-9, ACO is a homodimeric protein, which possesses an additional C-terminal domain that is not present in the tetrameric ACADs (22). However, ACOs are soluble enzymes. The N-terminal structure of ACO differs slightly from tetrameric ACADs. The FAD of ACO is more solvent-accessible, allowing for its oxygen reactivity.
The study of VLCAD has been limited because of difficulties with prokaryotic expression. An N-terminally truncated variant of VLCAD was recently reported in GenBank™ (NM_001033859); it lacks exon 3 because of alternative splicing (ΔEx3) (23). This ΔEx3 VLCAD variant is missing 22 amino acids (residues 7-28 of the mature sequence) resulting in an N terminus that more closely resembles that of ACAD-9. Recombinant ΔEx3 VLCAD was shown to be stable with very high specific activity and a substrate specificity profile similar to that previously reported for VLCAD purified from tissue or expressed in mammalian systems (4, 10, 24, 25). Additionally, it was shown that recombinant ΔEx3 VLCAD (hereafter referred to as simply VLCAD) can bind to isolated mitochondrial membranes that have been washed to remove all peripherally associated membrane proteins. Boiling the membranes, which should denature the remaining integral membrane proteins, did not reduce VLCAD binding, suggesting that membrane binding of VLCAD does not require an additional protein (10).
Although the N-terminal domain of VLCAD (∼400 amino acids) has a high sequence homology, and therefore likely structural homology to the other ACADs, no information on the structure of the C-terminal domain (∼180 amino acids) is available. Here we describe the x-ray crystallographic analysis of human VLCAD. Strikingly, the novel C-terminal domain mimics the other dimer in the tetrameric ACADs.
Preliminary reports of this work were presented at the 2007 Meeting of the American Society for Biochemistry and Molecular Biology (26) and at the American Crystallographic Association 2007 meeting (27).
EXPERIMENTAL PROCEDURES
Protein Purification—Protein was expressed and purified as previously described (10). Briefly, a pET-21a expression plasmid containing ΔEx3 VLCAD was transformed into Escherichia coli C43 (DE3) (Avidis, Saint Beauzire, France). Cells were grown at 37 °C to an absorbance of 0.8-1.0 at 600 nm and induced overnight by the addition of 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. Cells were lysed by sonication in 200 mm potassium phosphate buffer, pH 7.5, with Complete™ protease inhibitor tablets (Roche Applied Science, Manheim, Germany), 1 mm EDTA, and 1 mm dithiothreitol. The mixture was centrifuged at 50,000 × g for 40 min. The supernatant was precipitated with 55% saturated ammonium sulfate and re-centrifuged. The precipitate was resuspended in 50 mm Tris-HCl, pH 7.0, 1 mm EDTA, 1 mm dithiothreitol, 0.1% soy lecithin, and 0.2% Tween-20 and dialyzed against 50 mm Tris-HCl, pH 7.0 and 0.2% Tween-20. Following dialysis, 0.3 mg/ml (final concentration) dodecylmaltoside was added. The sample was then loaded onto a DEAE-Sepharose Fast Flow column (GE Healthcare, Piscataway, NJ) and eluted with a 0-500 mm potassium phosphate gradient. Fractions with VLCAD activity were pooled, diluted 2:1 (water/sample), and loaded again onto a DEAE-Sepharose Fast Flow column. This was eluted with a 0-380 mm NaCl gradient. Fractions with activity were again combined, dialyzed in 25 mm potassium phosphate, pH 6.3, and loaded onto an SP-Sepharose Fast Flow column (GE Healthcare). The sample was eluted with a 0-500 mm KPO4, pH 8.0 gradient. Tween-20 (0.2%) was added to all chromatography buffers. Fractions containing VLCAD were combined, concentrated, flash-frozen in liquid nitrogen, and stored at -80 °C. VLCAD enzyme activity was assayed as previously described (10, 28). Briefly, activity was measured with the anaerobic ETF fluorescence reduction assay, using a LS50B fluorescence spectrophotometer (PerkinElmer Life Sciences). The reaction was started with the addition of the CoA ester substrate to give a final concentration of 25 μm.
Crystallization—Purified protein was crystallized by vapor diffusion using the hanging drop or sitting drop methods (29). VLCAD (5 mg/ml) in 50 mm Tris, pH 7.5 and 5% glycerol, with 3 molar equivalents (per FAD) of myristoyl-CoA, was added in a 1.5:1 ratio with a buffer solution (0.1 m HEPES, pH 7.5, 17.6% polyethylene glycol 2000, 0.047 m MgCl2, and 5% glycerol) and equilibrated against the same buffer solution. When myristoyl-CoA was added, the yellow enzyme solution turned colorless, indicating that the enzyme flavin was reduced by the substrate. Within 1 week, rod-like yellow crystals were obtained (0.25 × 0.08 × 0.04 mm), suggesting that the enzyme was reoxidized during the crystallization. For data collection, crystals were flash-frozen in liquid nitrogen using 0.1 m HEPES, pH 7.5, 20% polyethylene glycol 2000, and 10% glycerol as a cryoprotectant. Two heavy atom derivatives of VLCAD crystals were used for phasing. A mercury derivative was obtained by crystallizing the protein in the presence of 0.6 mm thimerosal. An osmium derivative was obtained by soaking preformed crystals with 6 mm K2OsCl6 dissolved in cryo-protectant for 30 min prior to data collection.
Data Collection and Structure Determination—A complete native dataset was collected in-house using a Rigaku Micro-Max-007 x-ray generator equipped with an R-AXIS IV2+ image plate system and an MSC X-stream cooling system set at -180 °C. All high resolution and anomalous data were collected at the Advanced Photon Source at Argonne National Laboratories. Single wavelength anomalous data were collected with a VLCAD/mercury (thimerosal) co-crystal at the mercury absorbance peak wavelength (1.0083 Å) at the SBC-CAT beamline 19-ID. Diffraction data for osmium (K2OsCl6)-soaked VLCAD crystals were collected at the osmium absorbance peak (1.1405 Å) at the DND-CAT beamline 5-ID. Processing of image data was done using the HKL2000 suite of programs (30) for the native and mercury datasets and X-GEN (31) for the osmium data. Phasing was calculated by MIRAS with the program SOLVE at 50-3.00-Å resolution (32), and phases were extended to 1.91 Å by DM (33), which is a part of the CCP4 program suite (34). Data collection, phasing, and refinement statistics are summarized in Table 1. Initial, automated model building was done with the program RESOLVE (35). Manual density fitting and model building was done with the program Coot (36). Refmac5, also from CCP4, was used for multiple cycles of reciprocal space refinement (37). The program ARP/wARP (38) was used for the addition of waters, with manual adjustment using the program Coot. At a later stage of refinement, water molecules were added or removed on the basis of peak height (3.0σ in the Fo-Fc map) and distance from a potential hydrogen bonding partner (<3.5 Å). The structural model was refined to a final Rwork = 15.9% and Rfree = 21.6%.
TABLE 1.
Data collection and refinement statistics
| VLCAD + C14-CoA | VLCAD + C14-CoA + Hg co-crystal | VLCAD + C14-CoA + Os crystal soak | |
|---|---|---|---|
| Data collection | |||
| Resolution (Å) | 30 (2.06)a -1.91 | 30 (2.31)-2.20 | 30 (2.21)-2.10 |
| Space group | C2221 | C2221 | C2221 |
| a, b, c (Å) | 74.7, 107.9, 149.9 | 74.6, 108.1, 150.0 | 74.6, 108.2, 150.1 |
| Total reflections | 354462 | 169028 | 145187 |
| Unique reflections | 46776 | 44481 | 43731 |
| Completeness (%) | 98.8 (98.4) | 91.9 (98.3) | 91.5 (97.9) |
| I/σ(I) | 35.5 (3.9) | 16.7 (4.4) | 14.6 (5.1) |
| Rsym (%) | 7.9 (53.2) | 9.9 (37.2) | 6.11 (13.3) |
| Mosaicity (°) | 0.55 | 0.85 | 0.52 |
| X-ray source | In-house | APS: 19-ID | APS: 5-ID |
| Wavelength (Å) | 1.5419 | 1.0083 | 1.1405 |
| Phasing method | MIRAS | MIRAS | |
| Resolution (Å) | 50.0-3.0 | 50.0-3.0 | |
| Number of sites | 2 | 1 | |
| Figure of merit | 0.52 (combined) | ||
| Refinement | |||
| Resolution range (Å) | 30-1.91 | ||
| Number of reflections | 47377 | ||
| Rwork/Rfree (%) | 15.9/21.6 | ||
| R.m.s.d from ideality | |||
| Bond lengths (Å)/Angles (°) | 0.014/1.35 | ||
| Ramachandran plot | |||
| Most favored | 95.4% | ||
| Additional allowed | 4.4% | ||
| Generously allowed | 0.2% | ||
| Num. of atoms/avg. B-value (Å2) | |||
| Protein | 4277/22.3 | ||
| Main chain | 2217/20.9 | ||
| Side chain | 2060/24.0 | ||
| FAD | 53/19.3 | ||
| C14-CoA | 23/53.9 | ||
| Water | 461/32.2 |
Numbers in parentheses are values for the highest resolution shells.
RESULTS AND DISCUSSION
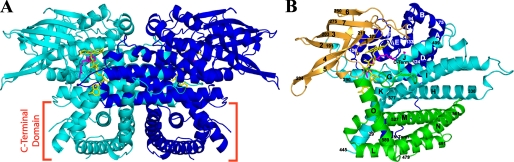
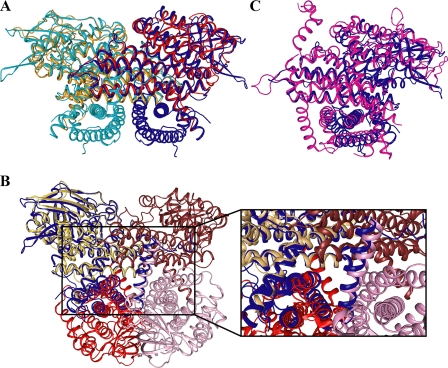
Overall Structure of VLCAD—VLCAD forms a dimer of identical monomers along a crystallographic 2-fold axis (Fig. 1A), confirming previous gel filtration analysis (8). In the presence of myristoyl-CoA, the enzyme crystallized in the space group C2221 with one monomer per asymmetric unit. The electron density map is generally well ordered for the entire polypeptide chain with the exception of the first seven N-terminal amino acids and residues 446-478 (using the full-length mature VLCAD numbering scheme), which, therefore, were not included in the structural model. The N-terminal 400 residues of VLCAD have the same overall fold as MCAD and the other tetrameric ACADs (17). There is an N-terminal α-helical domain (α-dom1), followed by a β-sheet domain and another α-helical domain (α-dom2). The C-terminal 180 amino acids of VLCAD form an α-helical bundle (α-dom3) that is positioned perpendicular to the N-terminal helical domains. Fig. 1B presents the secondary structure labeling convention used for MCAD and extended for VLCAD that will be used in the present description. When the monomer structure was compared with the dimer structure, it is immediately noticeable that the final helix (Helix O) in one monomer is swapped with the same helix from the other monomer. It is unclear what function, if any, there is for this helix swapping. The swapped helices may simply function to stabilize the dimer. Fig. 2A shows an MCAD dimer (PDB 3MDE) overlaid onto the VLCAD dimer. The two structures align well throughout most of the MCAD structure with an r.m.s.d. of 1.4 Å over 346 α-carbons. VLCAD has the same dimer interface as MCAD. Surprisingly, when the VLCAD monomer is aligned with one monomer of an MCAD tetramer (PDB 3MDE), both the N-terminal and C-terminal domains of the VLCAD align with portions of all four monomers of the MCAD tetramer (2.0 Å r.m.s.d. over 428 α-carbons; Fig. 2B). Thus, the C-terminal 180 residues of VLCAD interact with the N-terminal residues in a similar manner as the dimer-dimer interaction of the tetrameric ACADs. Furthermore, residues in the α-dom3 domain of VLCAD must then correspond structurally to those in the N terminus (α-dom2). A structural alignment of the C-terminal 180 residues with the N terminus shows that this is the case (1.7 Å r.m.s.d. over 114 α-carbons). The C-terminal residues show a 14% sequence identity with those residues in the N terminus with which they align structurally. Thus, the C-terminal amino acids most likely originated from a partial gene duplication. When aligned with VLCAD (Fig. 2C) and with the exception of the first ∼30 residues and the last ∼30 residues, ACO (PDB 2DDH) (22) shows a structural similarity with both the N-terminal and the C-terminal domains (2.7 Å r.m.s.d. over 413 α-carbons). One notable difference, however, is that the ACO monomers do not swap helices during dimer formation.
FIGURE 1.
A, ribbon diagram of the overall fold of the human VLCAD dimer. The monomers are represented in cyan and blue. The FADs are shown in yellow, and a partial model of C14-CoA is shown in magenta. The C-terminal domain is marked near the bottom of the figure. B, overall polypeptide fold of a VLCAD monomer showing the N-terminal α-dom1 (blue), the β-sheet domain (gold), α-dom2 (cyan), and the C-terminal α-dom3 (green). The FAD cofactor and the partially hydrolyzed model of substrate/product (C14-CoA) are shown with sticks in yellow and pink, respectively. α-Helices are labeled alphabetically, and β-strands are numbered consecutively from the N to C terminus. The numbers in a smaller font are residue numbers. Unless otherwise noted, all figures were generated using Pymol (52).
FIGURE 2.
Superposition of: A, VLCAD dimer (blue and cyan) on an MCAD dimer (red and orange; PDB 3MDE); B, VLCAD monomer (blue) on an MCAD tetramer (shades of brown and red; PDB 3MDE) with a close-up of the overlay of the C-terminal domain with the other monomers; C, VLCAD monomer (blue) on an ACO monomer (magenta; PDB 2DDH).
The Active Site—When myristoyl-CoA was added to the enzyme for crystallization set-up, the solution became colorless, indicating that the enzyme was completely reduced, and that the substrate was oxidized to form the product. However, after a few days, pale yellow crystals started to grow, suggesting that the enzyme was slowly re-oxidized by the molecular oxygen to either fully oxidized or semiquinone form. Because the stoichiometry of the enzyme (active site) (substrate was 1:3, and the flavin was nearly completely reduced upon addition of the substrate) it is reasonable to assume that the fatty acyl ligand in the crystalline protein is primarily the product, trans(2)-tetradecenoyl-CoA. However, the exact proportions of the different enzyme and the bound-ligand species in each crystal could not be determined. Therefore, we refer to the bound fatty acyl ligand as C14-CoA, even though the predominant form is the product.
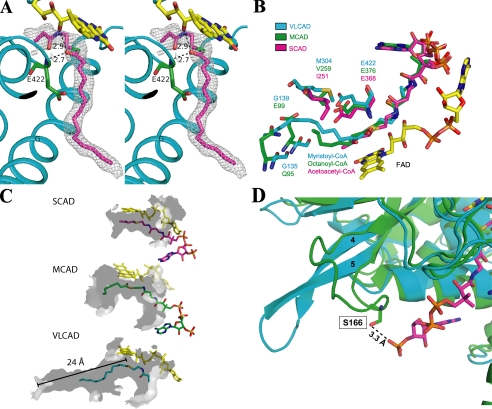
The Fo-Fc difference Fourier map near the isoalloxazine ring of the bound FAD exhibits a residual electron density, which could be fitted with a partial model of C14-CoA (Fig. 3A). Although the electron density for the myristate portion was very clear, the density in the area near where the CoA moiety is expected to bind was weak. Therefore, a partial CoA model showing a truncation in the pantothenic acid region was modeled into the binding cavity. The weak density in this region is most likely caused by a combination of a non-enzymatic partial hydrolysis of substrate/product at the thioester bond and high mobility of the CoA moiety. Although the crystallization medium contained substrate (myristoyl-CoA), at the relatively high pH of the solution (pH 7.5) and long crystallization period, it is not unusual to find the thioester hydrolysis product in the active site of a crystalline protein. To further support this contention, diffraction data were collected for co-crystals soaked for 24 h in the well buffer that did not contain substrate. Strong density was still observed for the acyl chain, but no interpretable density was observed for the entire CoA moiety (data not shown). These observations are a further indication: 1) that substrate was bound to and catalyzed by the enzyme to form the product, and 2) that after product was formed, non-enzymatic hydrolysis of the thioester bond has occurred. As a result, the acyl moiety was retained because of tight binding to the active site through two hydrogen bonds provided by the carbonyl group of the acyl moiety and the extensive hydrophobic interactions of the hydrocarbon chain with the polypeptide. The loosely bound CoA portion of the thioester substrate must have then dissociated from the active site after the hydrolysis. Thus, the binding mode of the partial C14-CoA modeled into the substrate cavity is the same as that of the acyl chain of the full C14-CoA bound to the enzyme. A similar situation was observed in the structure of the complex of ACO with dodecenoic acid (39). When crystals of the binary complex were obtained by co-crystallization with dodecanoyl-CoA, only dodecenoic acid was observed in the crystal structure. Alternatively, weak electron density in the region expected to bind the CoA moiety may be explained by a combination of partial hydrolysis of the thioester bond and higher mobility of the CoA moiety as compared with other ACAD structures. In support of this possibility, some residual electron density is observed beyond the substrate thioester bond.
FIGURE 3.
A, stereo diagram of a 2Fo-Fc electron density map (1.0σ) fitted with a partial model of C14-CoA. The catalytic base, Glu-422 (green), is located in a position analogous to the catalytic residues of MCAD and SCAD. As with MCAD and SCAD, substrate position is stabilized by hydrogen bonds between the thioester carbonyl and both the amide nitrogen of the catalytic glutamate (2.7 Å) and the ribityl 2′-hydroxyl of FAD (2.9 Å). Helices are labeled. Part of the pantothenic acid and adenosine pyrophosphate moiety of CoA are not visible. The thioester sulfur is shown as green for clarity. B, overlay of the binding site residues of VLCAD (blue), MCAD (green; ID: 3MDE), and SCAD (magenta; ID: 1JQI). Gln-95 and Glu-99 in MCAD form the bottom of its binding cavity. In VLCAD, the corresponding residues are glycines (Gly-135 and Gly-138). This effectively extends the cavity and allows for much longer substrates to bind. C, surface renderings (inside, light gray; outside, dark gray) of the binding cavities of SCAD, MCAD, and VLCAD illustrate the basis of their substrate chain length specificities. The cavity depths of SCAD, MCAD, and VLCAD, measured from the substrate thioester carbonyls (shown for VLCAD), are 8, 12, and 24 Å. Only a partial model of the CoA moiety of C14-CoA is shown. D, overlay of the substrate binding sites of VLCAD (cyan) and MCAD (green; ID: 3MDE). In MCAD, residue Ser-166 on the loop between β-strands 4 and 5 hydrogen bonds with the 3′-phosphate on the CoA moiety of octanoyl-CoA. In VLCAD, the β-sheet formed by strands 4 and 5 extends much further away from the substrate binding site, thereby precluding an analogous hydrogen bond, and resulting in a wider opening of the substrate binding cavity.
In VLCAD, the acyl moiety is buried deep inside the molecule at the re-face of the isoalloxazine ring of FAD. Glu-422 is located in a position analogous to the catalytic residues of MCAD and SCAD, near the C2-C3 atoms of the acyl-CoA substrate. Thus, the structure of VLCAD confirms the results of mutagenesis studies that indicate Glu-422 as the catalytic base (40). As in the other ACAD structures, the arrangement of the isoalloxazine ring of FAD, the C-2 and C-3 atoms of substrate, and the carboxylate of Glu-422 (Fig. 3A) is ideally suited for the abstraction of the pro-R hydrogen as a proton from the C-2 atom and transfer of the C-3 pro-R hydrogen to the N-5 atom of FAD as a hydride ion (17, 41). The thioester carbonyl oxygen of the partially hydrolyzed substrate/product makes hydrogen bonds to the 2′-hydroxyl of the FAD ribityl chain (2.9 Å) and the amide nitrogen of Glu-422 (2.7 Å), as observed in other ACAD structures whose entire CoA derivatives were observed. These interactions are not only responsible for proper positioning and orientation of the substrate, they are also crucial for polarization of the substrate and the lowering of the pKa of the C-2 proton for abstraction by the catalytic base.
Of particular interest in the ACAD family is the nature of chain length specificity. In SCAD, Ile-251 on Helix G extends partially into the binding site (Fig. 3B), limiting the depth of the substrate binding cavity (14). In MCAD, two prolines (Pro-257 and Pro-258) N-terminal to the analogous residue (Val-259) alter the trajectory of Helix G, widening the binding cavity. In VLCAD, Arg-313 on Helix G forms a salt bridge with Glu-354 on Helix H, widening the binding cavity. Gln-95 and Glu-99 in MCAD form the base of the binding cavity. In VLCAD, these residues are glycines (Gly-135 and Gly-139), which effectively open up and deepen the binding pocket. Beyond these residues, the channel extends for an additional 12 Å, allowing for substrate acyl chain lengths as long as 24 carbons. Fig. 3C shows renderings of the hydrophobic substrate binding cavities of SCAD, MCAD, and VLCAD. Measured from the substrate thioester carbonyls, the cavity depths of SCAD, MCAD, and VLCAD are 8, 12, and 24 Å, respectively.
The depth of the substrate binding cavity only accounts for part of the story for chain length specificity. VLCAD shows little activity for substrates with chain lengths of less than 12 carbons (4). Fig. 3D shows an overlay of the structures of MCAD and VLCAD. Ser-166 on the loop between β-strands 4 and 5 in MCAD (Ser-190 in IVH) makes a hydrogen bond with the 3′-phosphate on the CoA moiety of octanoyl-CoA. In VLCAD, the β-sheet formed by strands 4 and 5 extends further away from the substrate binding site, thereby precluding an analogous hydrogen bond. This loss in binding energy is offset by additional hydrophobic interactions made by longer acyl chains. Therefore, as demonstrated in the literature, VLCAD not only allows longer chain-length substrates to bind, but it prefers them (4). It has been shown for MCAD that substrate/product binding energy increases linearly with chain length (390 cal/CH2 group) (42). For ACADs, product release is the rate-limiting step (43). This means that the binding energy from the acyl moiety of the substrate/product in VLCAD would be considerably higher than those in other ACADs. Therefore, the lack of this hydrogen bond to the loop between β-strands 4 and 5 in VLCAD offsets the tighter binding of the fatty acyl chain and allows for longer chain length products to be more easily released, resulting in a more efficient catalytic turnover. Indeed, it has been shown with MCAD that the weaker binding 5-hydroxydecanoyl-CoA actually has a higher Vmax than the tighter binding decanoyl-CoA (44). This absence of a hydrogen bond (between the CoA moiety and the polypeptide) in VLCAD may also be the cause of higher mobility of the CoA moiety, which results in the weak electron density observed in this region. In the other ACADs, the loop between β-strands 4 and 5 serves to make the opening of the substrate binding cavity much narrower resulting in a tighter binding of the CoA moiety relative to the acyl chain as compared with VLCAD.
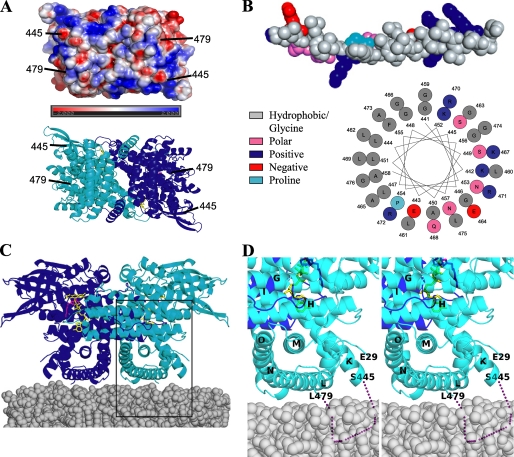
Membrane Binding—VLCAD is a monotopic membrane protein. The C-terminal domain of VLCAD has been shown to be responsible for binding to the matrix side of the inner mitochondrial membrane (8, 10). In vitro membrane binding studies with human clinical mutants A450P and L462P, both of which reside in the C-terminal domain, showed that while these two variants are active and stable, they have a greatly reduced ability to bind the membrane. Assuming each monomer binds the membrane equally well, VLCAD must interact with the membrane on a plane that is perpendicular to the 2-fold axis. An electrostatic representation of a surface in the C-terminal domain that fits this criterion is shown in Fig. 4A. There is no clear hydrophobic patch visible that is capable of interacting with the membrane in a symmetrical manner. However, the residues 446-478 are disordered in the VLCAD structure. Because of the proximity of residues both 445 and 479 to the surface (Fig. 4A), it is expected that the disordered residues also pack at the surface of the molecule. Fig. 4A shows a helix model built from residues 441-476, which include the disordered residues. The helix shows a periodicity that is strikingly amphipathic in nature, with six positively charged residues, which would interact strongly with negatively charged lipid head groups. This motif is completely conserved in mammals and well conserved in zebrafish and pufferfish. Prostaglandin H2 synthase-1 (PGHS-1) (45, 46) and squalene cyclase (47) are two monotopic membrane proteins that anchor to one leaflet of the membrane through amphipathic α-helices. Thus, it is not unreasonable to assume that residues 441-476 are responsible for anchoring VLCAD to the membrane in a similar manner. It is possible that these residues may only become ordered upon interaction with the membrane. Additionally, the human clinical mutations A450P and L462P inhibit membrane binding, and these mutation sites are within this region, further lending credence to this supposition (10). It is possible that the proline mutations disrupt the putative amphipathic helix in such a way as to inhibit membrane binding. Further studies are needed to unequivocally identify membrane binding regions.
FIGURE 4.
Putative membrane binding site of VLCAD. A, an electrostatic representation (top) and a ribbon representation of the membrane-binding surface are shown (90° rotation from Fig. 1 along the x-axis). B, a helix model built from residues 441-481, which include the disordered residues. The helix shows a periodicity that is strikingly amphipathic in nature and may be capable of interacting with the mitochondrial membrane. C, putative VLCAD membrane orientation. It is proposed that residues 445-479 are responsible for anchoring VLCAD to the membrane in the orientation pictured. D, magnification and stereo view of the rectangular region of Fig. 4C. The dotted purple line represents the disordered region.
If VLCAD binds the membrane in the orientation pictured in Fig. 4C, then the substrate binding site is somewhat distal to the membrane surface (∼20 Å), which presents a problem if the enzyme obtains substrate directly from the membrane phase. In PGHS-1, a hydrophobic channel allows the substrate to migrate from the membrane to the active site (46). As shown in Fig. 4 (lower panel of A and C), the interface between the two VLCAD monomers forms a central cavity, the opening to which is expected to be proximal to the membrane surface. However, it is unlikely that PGHS-like substrate channeling occurs in VLCAD. The cavity wall is lined with hydrophilic residues and it would take a major structural rearrangement for the substrate to migrate from the central cavity to the active site, which is located at either side of the dimeric molecule. Alternatively, the regions flanking the putative membrane anchoring domain are glycine-rich and, thus, highly flexible. These regions may act like hinges which allow VLCAD the freedom of movement to ensure favorable adaptation to a fluid membrane that is characterized by a large amount of molecular disorder. This freedom of movement may also permit a rocking motion that would alternate in bringing the substrate binding site of one monomer and then the other into closer proximity to the membrane surface. VLCAD, however, may not need to obtain substrates that are free in the membrane. Because of the low concentrations of mitochondrial β-oxidation intermediates (48), it has been suggested that these enzymes are organized into a multienzyme complex (49). Therefore, the substrate may pass directly from carnitine palmitoyl transferase-II (CPT-II) to VLCAD and onto the mitochondrial trifunctional enzyme complex.
Fig. 4D, shows a close-up of the putative VLCAD membrane interface. The N terminus is somewhat proximal to the membrane surface and could possibly play a role in membrane binding. This may also be true of the full-length VLCAD isoform, which possesses an additional 22 residues at the N terminus. However, it has been shown that these 22 residues are not required for membrane binding (10).
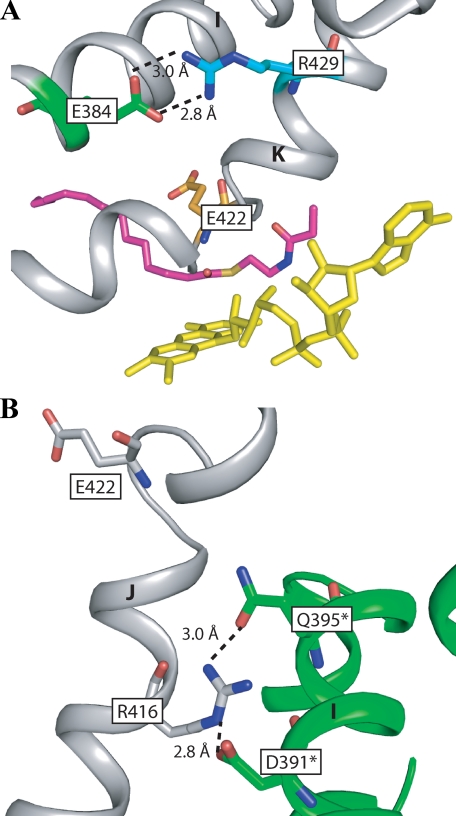
VLCAD Clinical Mutants—The structure of VLCAD affords us the opportunity to analyze what role the VLCAD human mutations might play in causing the diseased state. Three distinct phenotypes of VLCAD deficiency have been described. These range in symptoms from severe neonatal cardiomyopathy and liver failure to a purely myopathic form which develops in adolescence or adulthood (50). About 80 pathogenic (clinical) mutation sites have been identified, and there is a clear correlation between the genotype and the severity of the disease (51). We have examined our structure at the regions of two VLCAD mutations representative of the severe and the mild phenotypes (Fig. 5). The mutation R429W is associated with the severe childhood phenotype. Four patients have been reported with this variant, but it is unknown how the mutation affects the protein function, resulting in the diseased state. Arg-429 is on helix K and makes a salt bridge with Glu-384 on helix I (Fig. 5A). This residue is proximal to the catalytic glutamate (Glu-422) and appears to play a key role in its positioning as well as the overall stability of the protein. Replacing this charged residue with a bulky neutral residue would break the salt bridge and destabilize the enzyme. The mutation R416H is located on helix J, and also is near the catalytic glutamate (Fig. 5B). This variant is associated with the mild, late-onset phenotype (51). In Fig. 5B, it is shown that Arg-416 makes a salt bridge with Asp-391 and hydrogen bonds with Gln-395, both of which are located on the other monomer. A mutation at this site would affect the position of helix J and, thus, the catalytic glutamate. Additionally, because it forms a salt bridge with the opposing monomer, it may have an effect on dimer interaction. The substitution of an arginine with a histidine, however, is a relatively conservative mutation. The histidine is able to make only one of the two bonds with the residues of the other monomer. As a result, the interaction is slightly weaker and a mild phenotype is observed. Both the A450P and L462P mutations have been reported to result in a mild clinical phenotype (24, 51), suggesting that interference with normal enzyme localization might leave enough residual enzyme activity in the cell to support some physiologic function.
FIGURE 5.
A, site of the R429W clinical mutation. Arg-429 (cyan) on helix K forms a salt bridge with Glu-384 (green) of helix I, which positions the catalytic glutamate, Glu-422 (orange). Truncated C14-CoA and FAD are depicted as magenta and yellow, respectively. B, site of the R416H mutation. Arg-416 forms a salt bridge with Asp-391 and a hydrogen bond with Gln-395 on the other monomer, thus stabilizing the dimer. Residues on the opposing monomer are labeled with an asterisk.
After the preliminary report was presented and during preparation of this manuscript, an independently determined set of coordinates for VLCAD cocrystallized with trans 2-palmitenoyl-CoA has been released in the PDB (ID, 2UXW). With the exception of an additional 10 residues in the C-terminal domain, this unpublished structure shows no significant differences from the structure described in this report (r.m.s.d., 0.5 Å).
Conclusion—We have presented here the crystal structure of human VLCAD in complex with C14-CoA. In MCAD, Glu-376 acts as a catalytic base, and a sequence alignment shows this residue is conserved in VLCAD (Glu-422). Mutagenesis studies also implicated that Glu-422 is the catalytic base. The structure of VLCAD confirms that this residue is optimally positioned for catalysis. The substrate binding pocket of VLCAD extends for an additional 12 Å beyond where the pocket of MCAD ends, allowing VLCAD to bind substrates with much longer acyl chain lengths. Additionally, VLCAD is selective for long-chain acyl-CoA substrates: the opening of the binding pocket is much wider than any of the other ACADs, thus providing an optimum binding affinity for catalytic turnover of the product having longer fatty acyl chain.
We have proposed that a region from residues 441 to 476 mediates membrane binding. This region is disordered in the crystal structure. However, it is optimally positioned in the novel C-terminal domain for membrane interaction. Furthermore, clinical variants, A450P and L462P, both of which lie within this region show a reduced ability to bind the membrane, strongly suggesting that this region is involved in membrane binding. Further studies are needed to unequivocally confirm this hypothesis. Also unresolved is the issue of how substrate gets into the binding cavity, which is somewhat distal (∼20 Å) from the membrane surface. It is possible that the mobility of the putative membrane interacting domain allows the binding pocket to move nearer to the membrane surface. It is also possible that VLCAD is part of a multienzyme complex and receives substrate directly from its interacting partners. Further studies will be directed to answer these outstanding questions.
Acknowledgments
We thank Drs. Owen Griffith and Vaughn Jackson for careful reading of the manuscript and Dr. Andrew Howard for assistance in data reduction using X-GEN. We also thank the staff at the APS beamlines SBC 19ID and DND 5ID for excellent assistance in data collection. Use of the Advanced Photon Source was supported by the United States. Department of Energy, Basic Energy Sciences, Office of Science, under Contract W-31-109-Eng-38 and Office of Biological and Environmental Research under Contract DE-AC02-06CH11357.
The atomic coordinates and structure factors (code 3B96) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported in part by National Institutes of Health Grant GM29076 (to J.-J. P. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: VLCAD, very-long-chain acyl-CoA dehydrogenase; ACAD, acyl-CoA dehydrogenase; LCAD, long-chain acyl-CoA dehydrogenase; MCAD, medium-chain acyl-CoA dehydrogenase; SCAD, short-chain acyl-CoA dehydrogenase; IBD, isobutyryl-CoA dehydrogenase; GCAD, glutaryl-CoA dehydrogenase; ETF, electron transfer flavoprotein; ACO, acyl-CoA oxidase; PGHS, prostaglandin H2 synthase; r.m.s.d., root mean square deviation; PDB, protein data bank.
References
- 1.Beinert, H. (1963) in The Enzymes. (Boyer, P. D., Lardy, H., and Myrback, K., eds) Vol. 7, pp. 447-476, Academic Press, New York [Google Scholar]
- 2.Aoyama, T., Souri, M., Ushikubo, S., Kamijo, T., Yamaguchi, S., Kelley, R. I., Rhead, W. J., Uetake, K., Tanaka, K., and Hashimoto, T. (1995) J. Clin. Investig. 95 2465-2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ikeda, Y., Okamura-Ikeda, K., and Tanaka, K. (1985) J. Biol. Chem. 260 1311-1325 [PubMed] [Google Scholar]
- 4.Izai, K., Uchida, Y., Orii, T., Yamamoto, S., and Hashimoto, T. (1992) J. Biol. Chem. 267 1027-1033 [PubMed] [Google Scholar]
- 5.Ensenauer, R., He, M., Willard, J. M., Goetzman, E. S., Corydon, T. J., Vandahl, B. B., Mohsen, A. W., Isaya, G., and Vockley, J. (2005) J. Biol. Chem. 280 32309-32316 [DOI] [PubMed] [Google Scholar]
- 6.Crane, F. L., and Beinert, H. (1956) J. Biol. Chem. 218 717-731 [PubMed] [Google Scholar]
- 7.Ruzicka, F. J., and Beinert, H. (1977) J. Biol. Chem. 252 8440-8445 [PubMed] [Google Scholar]
- 8.Souri, M., Aoyama, T., Hoganson, G., and Hashimoto, T. (1998) FEBS Lett. 426 187-190 [DOI] [PubMed] [Google Scholar]
- 9.Zhang, J., Zhang, W., Zou, D., Chen, G., Wan, T., Zhang, M., and Cao, X. (2002) Biochem. Biophys. Res. Commun. 297 1033-1042 [DOI] [PubMed] [Google Scholar]
- 10.Goetzman, E. S., Wang, Y., He, M., Mohsen, A. W., Ninness, B. K., and Vockley, J. (2007) Mol. Genet. Metab. 91 138-147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, J. J., Wang, M., and Paschke, R. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 7523-7527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, H. J., Wang, M., Paschke, R., Nandy, A., Ghisla, S., and Kim, J. J. (1996) Biochemistry 35 12412-12420 [DOI] [PubMed] [Google Scholar]
- 13.Satoh, A., Nakajima, Y., Miyahara, I., Hirotsu, K., Tanaka, T., Nishina, Y., Shiga, K., Tamaoki, H., Setoyama, C., and Miura, R. (2003) J. Biochem. (Tokyo) 134 297-304 [DOI] [PubMed] [Google Scholar]
- 14.Battaile, K. P., Molin-Case, J., Paschke, R., Wang, M., Bennett, D., Vockley, J., and Kim, J. J. (2002) J. Biol. Chem. 277 12200-12207 [DOI] [PubMed] [Google Scholar]
- 15.Tiffany, K. A., Roberts, D. L., Wang, M., Paschke, R., Mohsen, A. W., Vockley, J., and Kim, J. J. (1997) Biochemistry 36 8455-8464 [DOI] [PubMed] [Google Scholar]
- 16.Fu, Z., Wang, M., Paschke, R., Rao, K. S., Frerman, F. E., and Kim, J. J. (2004) Biochemistry 43 9674-9684 [DOI] [PubMed] [Google Scholar]
- 17.Kim, J. J., and Miura, R. (2004) Eur. J. Biochem. 271 483-493 [DOI] [PubMed] [Google Scholar]
- 18.Bross, P., Engst, S., Strauss, A. W., Kelly, D. P., Rasched, I., and Ghisla, S. (1990) J. Biol. Chem. 265 7116-7119 [PubMed] [Google Scholar]
- 19.Battaile, K. P., Nguyen, T. V., Vockley, J., and Kim, J. J. (2004) J. Biol. Chem. 279 16526-16534 [DOI] [PubMed] [Google Scholar]
- 20.Djordjevic, S., Dong, Y., Paschke, R., Frerman, F. E., Strauss, A. W., and Kim, J. J. (1994) Biochemistry 33 4258-4264 [DOI] [PubMed] [Google Scholar]
- 21.Matsubara, Y., Indo, Y., Naito, E., Ozasa, H., Glassberg, R., Vockley, J., Ikeda, Y., Kraus, J., and Tanaka, K. (1989) J. Biol. Chem. 264 16321-16331 [PubMed] [Google Scholar]
- 22.Nakajima, Y., Miyahara, I., Hirotsu, K., Nishina, Y., Shiga, K., Setoyama, C., Tamaoki, H., and Miura, R. (2002) J. Biochem. (Tokyo) 131 365-374 [DOI] [PubMed] [Google Scholar]
- 23.Aoyama, T., Uchida, Y., Kelley, R. I., Marble, M., Hofman, K., Tonsgard, J. H., Rhead, W. J., and Hashimoto, T. (1993) Biochem. Biophys. Res. Commun. 191 1369-1372 [DOI] [PubMed] [Google Scholar]
- 24.Souri, M., Aoyama, T., Yamaguchi, S., and Hashimoto, T. (1998) Eur. J. Biochem. 257 592-598 [DOI] [PubMed] [Google Scholar]
- 25.Lea, W., Abbas, A. S., Sprecher, H., Vockley, J., and Schulz, H. (2000) Biochim. Biophys. Acta 1485 121-128 [DOI] [PubMed] [Google Scholar]
- 26.McAndrew, R. P., Wang, Y., Mohsen, A.-W., He, M., Vockley, J., and Kim, J.-J. (2007) FASEB J. 21 649 c.3 [Google Scholar]
- 27.McAndrew, R. P., Wang, Y., Mohsen, A.-W., He, M., Vockley, J., and Kim, J.-J. (2007) American Crystallographic Association, pp. 01.04.05, American Crystallographic Association, Inc., Salt Lake City, UT
- 28.Frerman, F. E., and Goodman, S. I. (1985) Biochem. Med. 33 38-44 [DOI] [PubMed] [Google Scholar]
- 29.McPherson, A. (1999) Crystallization of Biological Macromolecules, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 30.Otwinowski, Z., and Minor, A. (1997) in Methods in Enzymology (Carter, J., and Sweet, R. M., eds) Vol. 276, pp. 307-326, Academic Press, New York [DOI] [PubMed] [Google Scholar]
- 31.Howard, A. J. (2000) in Crystallographic Computing (Bourne, P. E., and Watenpaugh, K. D., eds) Vol. 7, Oxford University Press, Oxford
- 32.Terwilliger, T. C., and Berendzen, J. (1999) Acta Crystallogr. Sect. D. 55 849-861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cowtan, K. (1994) Joint CCP4 and ESF-EACBM, Newsletter on Protein Crystallography, 31 34-38 [Google Scholar]
- 34.Collaborative Computational Project, N. (1994) Acta Crystallogr. Sect. D. 50 760-76315299374 [Google Scholar]
- 35.Terwilliger, T. C. (2003) Methods Enzymol. 374 22-37 [DOI] [PubMed] [Google Scholar]
- 36.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 37.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D 53 240-255 [DOI] [PubMed] [Google Scholar]
- 38.Perrakis, A., Morris, R., and Lamzin, V. S. (1999) Nat. Struct. Biol. 6 458-463 [DOI] [PubMed] [Google Scholar]
- 39.Tokuoka, K., Nakajima, Y., Hirotsu, K., Miyahara, I., Nishina, Y., Shiga, K., Tamaoki, H., Setoyama, C., Tojo, H., and Miura, R. (2006) J. Biochem. (Tokyo) 139 789-795 [DOI] [PubMed] [Google Scholar]
- 40.Souri, M., Aoyama, T., Cox, G. F., and Hashimoto, T. (1998) J. Biol. Chem. 273 4227-4231 [DOI] [PubMed] [Google Scholar]
- 41.Ghisla, S., Thorpe, C., and Massey, V. (1984) Biochemistry 23 3154-3161 [DOI] [PubMed] [Google Scholar]
- 42.Trievel, R. C., Wang, R., Anderson, V. E., and Thorpe, C. (1995) Biochemistry 34 8597-8605 [DOI] [PubMed] [Google Scholar]
- 43.Kumar, N. R., Peterson, K. L., and Srivastava, D. K. (1996) in Flavins and Flavoproteins (Stevenson, K. J., Massey, V., and Williams, C. W., eds), pp. 633-636, University of Calgary Press, Calgary, Canada
- 44.Hanley, P. J., Gopalan, K. V., Lareau, R. A., Srivastava, D. K., von Meltzer, M., and Daut, J. (2003) J. Physiol. 547 387-393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malkowski, M. G., Ginell, S. L., Smith, W. L., and Garavito, R. M. (2000) Science 289 1933-1937 [DOI] [PubMed] [Google Scholar]
- 46.Nina, M., Berneche, S., and Roux, B. (2000) Eur. Biophys. J. 29 439-454 [DOI] [PubMed] [Google Scholar]
- 47.Wendt, K. U., Poralla, K., and Schulz, G. E. (1997) Science 277 1811-1815 [DOI] [PubMed] [Google Scholar]
- 48.Garland, P. B., Shepherd, D., and Yates, D. W. (1965) Biochem. J. 97 587-594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greville, G. D., and Tubbs, P. K. (1968) Essays Biochem. 4 155-212 [PubMed] [Google Scholar]
- 50.Gregersen, N., Andresen, B. S., Corydon, M. J., Corydon, T. J., Olsen, R. K., Bolund, L., and Bross, P. (2001) Hum. Mutat. 18 169-189 [DOI] [PubMed] [Google Scholar]
- 51.Andresen, B. S., Olpin, S., Poorthuis, B. J., Scholte, H. R., Vianey-Saban, C., Wanders, R., Ijlst, L., Morris, A., Pourfarzam, M., Bartlett, K., Baumgartner, E. R., deKlerk, J. B., Schroeder, L. D., Corydon, T. J., Lund, H., Winter, V., Bross, P., Bolund, L., and Gregersen, N. (1999) Am. J. Hum. Genet. 64 479-494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeLano, W. L. (2002) The PyMOL Users Manual, DeLano Scientific, Palo Alto, CA