Abstract
Anandamide (AEA) is an endogenous ligand of cannabinoid receptors and a well characterized mediator of many physiological processes including inflammation, pain, and appetite. The biosynthetic pathway(s) for anandamide and its N-acyl ethanolamine (NAE) congeners remain enigmatic. Previously, we proposed an enzymatic route for producing NAEs that involves the double-O-deacylation of N-acyl phosphatidylethanolamines (NAPEs) by α/β-hydrolase 4 (ABDH4 or Abh4) to form glycerophospho (GP)-NAEs, followed by conversion of these intermediates to NAEs by an unidentified phosphodiesterase. Here, we report the detection and measurement of GP-NAEs, including the anandamide precursor glycerophospho-N-arachidonoylethanolamine (GP-NArE), as endogenous constituents of mouse brain tissue. Inhibition of the phosphodiesterase-mediated degradation of GP-NAEs ex vivo resulted in a striking accumulation of these lipids in brain extracts, suggesting a rapid endogenous flux through this pathway. Furthermore, we identify the glycerophosphodiesterase GDE1, also known as MIR16, as a broadly expressed membrane enzyme with robust GP-NAE phosphodiesterase activity. Together, these data provide evidence for a multistep pathway for the production of anandamide in the nervous system by the sequential actions of Abh4 and GDE1.
N-Acylated ethanolamines (NAEs)2 were first isolated from preparations of egg yolk, peanut oils, and soybeans in the 1950s due to their anti-inflammatory and anti-allergenic properties (1). In the ensuing decades, NAEs have been found in nearly all mammalian tissues and ascribed a number of bioactivities (2). Studies by Schmid and colleagues (3) first identified N-acylated phosphatidylethanolamines (NAPEs) as markers of ischemic shock in brain and heart tissues and postulated that these lipids were the metabolic precursors of NAEs. In 1992, anandamide (N-arachidonoyl ethanolamine, NArE, AEA), was identified as an endogenous ligand for the brain (type I) cannabinoid receptor (4). The discovery of AEA as an endocannabinoid sparked renewed interest in NAE biochemistry and a search for pathways responsible for NAE biosynthesis and degradation. The integral membrane protein fatty acid amide hydrolase has been identified as the primary enzyme responsible for NAE degradation in most tissues, including brain (5). Pharmacological (6) or genetic (7) blockade of fatty acid amide hydrolase leads to elevated NAE levels and anti-inflammatory and analgesic effects mediated through cannabinoid receptors. Despite these advances, a detailed understanding of the biosynthetic pathway(s) for NAEs has proven more elusive.
Substantial evidence implicates NAPEs as the key metabolic precursor for NAEs. NAPEs are formed by the calcium-dependent transfer of an acyl chain from the sn-1 position of phosphatidylcholine to the amine of phosphatidylethanolamine by an as-of-yet unidentified enzyme (8). N-Arachidonoyl NAPE, which serves as the AEA precursor, is found at very low levels in brain tissue, consistent with the low abundance of arachidonate in the sn-1 position of phosphatidylcholine (9). Recently, a calcium-independent transacylase capable of generating NAPE from phosphatidylethanolamine and phosphatidylcholine was cloned, however, this protein displays markedly different properties from the brain enzyme that generates NAPEs (10).
Release of NAEs from NAPEs was originally postulated to occur via a calcium-stimulated D-type phospholipase (PLD). In 2004, Ueda and colleagues isolated and identified an NAPE-selective PLD (NAPE-PLD) (11). Mice with a disrupted NAPE-PLD gene (NAPE-PLD-/- mice) were found to exhibit dramatically reduced brain levels of long chain, saturated NAEs. In contrast, polyunsaturated NAEs, including AEA, were unaltered in these animals (12). These results indicate that enzymes distinct from NAPE-PLD are involved in the biosynthesis of AEA in vivo. Consistent with this premise, RNA interference-mediated knockdown of NAPE-PLD in RAW264.7 macrophages failed to alter AEA levels in these cells (13).
That NAPE-PLD does not appear to contribute to AEA biosynthesis, at least in certain cell and tissue types, necessitates the existence of alternative biosynthetic enzymes. Consistent with this premise, substantial rates of conversion of NAPE to NAE were observed in tissue extracts from NAPE-PLD-/- mice (12). Interestingly, this residual activity was not due to another D-type phospholipase, but rather to a multienzyme cascade that involves the initial A/B-type phospholipase-catalyzed double O-deacylation of NAPEs to generate glycerophospho-(GP)-NAEs, followed by phosphodiesterase-catalyzed conversion of these intermediates to NAEs (Fig. 1). This pathway was further found to be inhibited by millimolar concentrations of calcium, which may explain why it had previously gone undetected in assays for NAPE-PLD activity that included high (2-10 mm) levels of exogenous calcium. Conventional protein chromatography coupled with activity-based proteomic analysis identified the hitherto unannotated enzyme α/β-hydrolase 4 (Abh4 or ABHD4) as a B-type NAPE lipase capable of removing both O-acyl chains from NAPE to yield GP-NAE (14). This finding suggested that glycerophospho-N-arachidonoylethanolamine (GP-NArE) may be a physiologically relevant precursor of AEA and pointed to the GP-NAE phosphodiesterase as a critical final step leading to AEA release. However, to our knowledge, GP-NAEs have not previously been identified as natural brain constituents, nor has a phosphodiesterase capable of converting these lipids to NAEs been characterized. Herein, we describe the detection and measurement of GP-NAEs, including the AEA precursor GP-NArE, as endogenous brain constituents. We observe the enzymatic formation of endogenous GP-NAEs in ex vivo brain preparations and, moreover, we identify the integral membrane enzyme glycerophosphodiesterase 1 (GDE1) as a GP-NAE phosphodiesterase enriched in brain that is capable of generating NAEs, including AEA, in vitro.
FIGURE 1.
Proposed biosynthetic pathway for NAEs including AEA. Both O-acyl chains are removed from N-arachidonoyl phosphatidylethanolamine (NArPE) by Abh4 yielding GP-NArE. The phosphodiester bond of GP-NArE is then hydrolyzed by a phosphodiesterase (identified herein as GDE1) to release free anandamide and glycerol 3-phosphate.
EXPERIMENTAL PROCEDURES
Materials—1-Oleoyl-2-hydroxy-sn-glycero-3-phosphoethanolamine was purchased from Avanti Polar Lipids (Alabaster, AL). Pentadecenoyl chloride, palmitoyl chloride, oleoyl chloride, arachidonoyl chloride, eicosanoyl chloride, and docosahexaenoyl chloride were purchased from NuCheck Prep (Elysian, MN). [1′-14C]Palmitic acid was purchased from Moravek Biochemicals (Brea, CA). Methoxy arachidonoylfluorophosphonate (MAFP) was purchased from Cayman Chemicals (Ann Arbor, MI).
Synthesis of GP-NAE standards and substrates was performed essentially as described (14). Briefly, acid chlorides were reacted with an excess of 1-oleoyl-2-hydroxy-sn-glycero-3-phosphoethanolamine and allowed to react for 1 h in CH2Cl2 with a catalytic amount of triethylamine. N-Acylated lipids were then purified by silica gel flash column chromatography. The resulting lyso-NAPE was stirred vigorously in CHCl3, MeOH, 2 n LiOH (2:1:1) for 4 h before acidification and purification via silica gel flash column chromatography.
Brain-Lipid Preparation and Tandem MS Analysis of GP-NAE—8-Week-old male C57BL/6 mice were anesthetized with 50% CO2 and killed by decapitation. Brains were removed within 30 s of decapitation and immediately frozen in liquid N2. Frozen brains were placed in a Dounce homogenizer with 8 ml of CHCl3, MeOH, 50 mm Tris, pH 8.0 (2:1:1), and 200 pmol of 1,2-dihydroxy-sn-glycero-3-phospho(N-pentadecenoyl)ethanolamine was added as an internal standard. Mild basification of the aqueous layer was required to prevent the artifactual accumulation of GP-NAEs due to the non-enzymatic (acid-catalyzed) breakdown of sn-1 alkenyl lyso-NAPEs. Brains were immediately homogenized with 8-10 strokes of the pestle and centrifuged for 10 min at 1,400 × g to separate phases. The organic phase was extracted and an additional 4 ml of CHCl3 added to the remaining homogenate, which was vortexed, recentrifuged, and extracted again. The pooled organic phases were evaporated to dryness under N2 yielding ∼20-25 mg of lipid per brain. For the ex vivo analysis of GP-NAEs, brains were homogenized in a Dounce homogenizer in 2 ml of 50 mm Tris, pH 8.0, or 50 mm Tris, pH 8.0, with 10 mm EDTA. 1.4 μl of 28 mm MAFP in Me2SO (for 20 μm final concentration) or Me2SO alone was added to homogenates immediately after homogenization. Samples were either extracted immediately or incubated at room temperature for 4 h with rotation prior to extraction and GP-NAE quantitation as described above.
Lipid extracts were resuspended in 120 μl of CHCl3:MeOH (2:1) and 30 μl of the solution was injected into an Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA) with an Agilent 1100 autosampler. Samples were chromatographed on a Gemini 5-μm C18 column (50 × 4.6 mm, Phenomenex, Torrance, CA) with a 50-min gradient from 0 to 100% buffer B followed by 7 min of isocratic flow with 100% buffer B. Buffer A consisted of MeOH:H2O (5:95) with 0.1% of 28% NH4OH and buffer B consisted of H2O:MeOH:iPrOH (5:35:60) with 0.1% of 28% NH4OH. Analysis was performed with an ESI source in the negative ionization mode with a source voltage of -3.5 kV, flow rate of 0.4 ml/min, and capillary temperature of 325 °C. Care was taken to avoid carryover by running “blank” samples prior to all endogenous measurements. The method included a high-resolution full scan in the Orbitrap (operated at 30,000 resolution) followed by six fragmentation events in which fragment ions were analyzed in the ion trap and/or sent to the Orbitrap for high-resolution mass analysis depending on the experiment. Fragmentation events were chosen to detect palmitoyl-, oleoyl-, arachidonoyl-, eicosanoyl-, docosahexaenoyl-, and pentadecenoyl-GP-NAEs by targeting m/z values of 452, 478, 500, 508, 524, and 436, for MS/MS analysis, respectively. For quantitation, extracted ion chromatograms of the phospho-NAE peaks ([M - H-74]-) were integrated and compared with the internal N-C15:1 GP-NAE standard. Extractions using synthetic standards revealed extraction efficiencies of 3, 25, 45, 8, 18, and 77% for N-C15:1, C20:4, C22:6, C16:0, C18:1, and C20:0 GP-NAEs, respectively, and measurements of endogenous GP-NAEs were corrected accordingly.
Subcloning and Recombinant Expression of GDE Domain Containing Enzymes—The following expressed sequence tags were purchased from OpenBioSystems (Huntsville, AL) and subcloned into pcDNA3.1-myc/His using conventional techniques: GDE1/MIR16, IMAGE 3601420; GDE2, IMAGE 4224574; GDE3, IMAGE 4457335; GDE4, IMAGE 3602449; and GDE7, IMAGE 3486726. COS-7 cells were grown at 37 °C with 5% CO2 to ∼70% confluence in Dulbecco's modified Eagle's medium containing 10% fetal calf serum in 10-cm dishes and transfected with 6 μg of plasmid DNA using the FuGENE 6 (Roche Applied Science) transfection reagent according to the manufacturer's protocol. After 48 h, the cells were washed twice with phosphate-buffered saline, scraped, resuspended in 50 mm Tris, pH 8.0, and sonicated to lyse. The lysates were then spun at 145,000 × g for 45 min to isolate the insoluble (membrane) fraction, which was sonicated to resuspend in 50 mm Tris, pH 8.0. Cell membranes thus prepared were stored as single-use aliquots at 0.01 mg/ml for enzyme assays or 2 mg/ml for Western blots at -80 °C. For Western blots the cell membranes were treated with (or without) PNGase F (New England Biolabs, Ipswich, MA) according to the manufacturer's protocol prior to SDS-PAGE analysis.
GP-NAE Phosphodiesterase Activity Assay—Enzyme assays were performed in 4-ml glass vials in 50 mm Tris, pH 8.0, with 2 mm MgCl2 except for the following experiments. pH rate profiles were performed in buffers containing 50 mm Tris, 50 mm HEPES, and 50 mm glycine that was adjusted with HCl to the desired pH. The cation-sensitivity assays were performed in 50 mm Tris, pH 8.0, and the indicated cation (or EDTA) at 2 mm final concentration. For all assays, 10 μl of protein (100 μg for cell-membranes or 500 μg for brain samples) was added to 85 μl of the specified buffer and 5 μl of GP-NAE substrate (2 mm in Me2SO or EtOH, 100 μm final concentration). Vials were incubated at 37 °C for 30 min and then quenched with 1.5 ml of CHCl3:MeOH (2:1) and 400 μl of 1% formic acid. For LC-MS assays, d4-N-oleoylethanolamine (10 μl of 100 μm stock in EtOH; 1 nmol total) was added as an internal standard. Phases were separated by centrifugation at 1,400 × g for 3 min and the organic phase transferred to a new vial and evaporated to dryness under a stream of N2. For LC-MS assays the dried reactions were resuspended in 500 μl of MeOH and 50 μl injected into an Agilent 1100 series LC-MS for analysis using an 8-min gradient from 30 to 100% buffer B. Buffer A consisted of MeOH:H2O (5:95) with 0.1% formic acid and buffer B consisted of H2O: MeOH:iPrOH (5:35:60) with 0.1% formic acid. NAE formation was quantified by comparison with the internal d4-N-oleoylethanolamine standard. For radiochromatographic assays, the dried reactions were dissolved in 20 μl of CHCl3:MeOH (2:1), spotted on thin layer silica plates, and developed in CHCl3, MeOH, 28% NH4OH (40:10:1). Distribution of radioactivity on the plate was quantified by a phosphorimaging device (Packard), and products were identified by comparison with 14C radiolabled synthetic standards. NAE formation was calculated from percentage values of the total radioactivity on the TLC plates. All of the assays were conducted with n = 2 or 3, and error bars represent the mean ± S.E.
Generation of Polyclonal Antibodies against mGDE1 and Western Blotting—Polyclonal antibodies that recognize GDE1 were prepared by expressing an N-terminal glutathione S-transferase fusion of the C-terminal 163 amino acids of GDE1 prepared using traditional subcloning techniques. This fusion protein was expressed in BL21-DE3 cells by growing at 37 °C to A600 = 0.4 and inducing expression with 1 mm isopropyl β-d-1-thiogalactopyranoside. After 4 h, cells were harvested and sonicated. Lysate was centrifuged for 30 min at 12,000 × g and the pellet was resuspended in phosphate-buffered saline and visualized by SDS-PAGE and protein staining. The fusion protein was excised from the gel, homogenized, mixed 1:1 with RIBI adjuvant, and injected three times into rabbits at 2-week intervals. One week after the final injection, blood was taken and antibodies affinity purified with the original antigen immobilized on polyvinylidene difluoride membrane. These polyclonal antibodies were used at a 1:200 dilution in TBST (20 mm Tris-HCl, 150 mm NaCl, 0.2% Tween 20) overnight at 4 °C followed by incubation with secondary horseradish peroxidase-conjugated goat anti-rabbit antibody (Bio-Rad Laboratories) at 1:5000 in TBST prior to developing with chemiluminescent ECL substrate (Pierce) and exposing to x-ray film. Specificity of antibodies was tested via immunoblots of mock- and GDE1-transfected COS-7 cell membranes (supplemental Fig. S1).
Reverse Transcription-PCR—cDNA was prepared using the SuperScript III reverse transcriptase kit (Invitrogen) according to the manufacturer's protocol. PCR was performed using 30 cycles with cycling times of 30 s of melting at 95 °C, 30 s of annealing at 58 °C, and 30 s of extension at 72 °C with Taq DNA polymerase using the following primers: GDE1 oligonucleotides, 5′-CCGACTATGTGATCTGACATTTGAACAAGTTAGG-3′ and 5′-CCATACTGTTATTGTACAGCTGTGGAAATTCCG-3′; and glyceraldehyde-3-phosphate dehydrogenase oligonucleotides, 5′-TGTCTTCACCACCATGGAGAAGGC-3′ and 5′-TGGCAGTGATGGCATGGAACTGTGG-3′. The products were visualized on a 2% agarose gel.
RESULTS
Measurement of Endogenous GP-NAE—Initial attempts to detect endogenous GP-NAE in mouse brain were frustrated by the presence of abundant and isobaric lyso-phosphatidylethanolamine lipids that displayed overlapping chromatographic behavior. To accurately detect and measure GP-NAEs, we employed LC-MS/MS to identify characteristic fragment ion peaks (Fig. 2) (15). First, synthetic GP-NAEs were prepared and their chromatographic behavior and fragmentation studied. Fragmentation of GP-NAEs in the negative mode results in three dominant ions: a phospho-NAE peak at [M - H - 74]- and two peaks corresponding to glycer-phosphate and its dehydrate at 171 and 153, respectively (Fig. 2, A and D). The phospho-NAE peak was selected for identification and quantification by multiple reaction monitoring, as this peak also verified the N-acyl chain of the parent GP-NAE species. Thus, by scanning for the [M - H]- > [M - H - 74]- transition, several endogenous GP-NAEs, including C16:0, C18:1, and C20:4 GP-NAEs, could be detected that displayed identical retention and fragmentation to synthetic standards (Table 1 and Fig. 2). Parent and fragment mass signals were strongest for the more abundant GP-NAEs (C16:0, C18:1) (Fig. 2), but could be detected for all GP-NAEs listed in Table 1. Furthermore, we employed a hybrid ion trap/Orbitrap instrument, which enabled high-resolution mass measurements of parent and fragment ions. This instrument yielded measurements of endogenous GP-NAEs and key fragments that were within ∼2 ppm of predicted masses thereby confirming the identity of these NAE precursors (supplemental Table 1). Quantification of endogenous GP-NAEs was performed by comparison to an internal unnatural (N-C15:1) GP-NAE standard. Endogenous GP-NAEs were present at levels somewhat lower than endogenous NAEs (Table 1) (12).
FIGURE 2.
Fragmentation and chromatography of endogenous GP-NAEs in mouse brain via LC-MS/MS. Fragmentation and chromatographic behavior are shown for endogenous N-palmitoyl (A-C) and N-oleoyl (D-F) GP-NAEs. Three major fragment ions are produced, a diagnostic NAE-phosphate fragment (378 and 404 for palmitoyl- and oleoyl-GP-NAEs, respectively) and two fragments corresponding to glycerol 3-phosphate and its dehydrate at 171 and 153, respectively (A and D). B and E, representative multiple reaction monitoring chromatographs of synthetic and endogenous GP-NAEs. C and F, tandem MS/MS fragmentation spectra of synthetic and endogenous GP-NAEs.
TABLE 1.
Endogenous levels of GP-NAEs in mouse brain as measured by LC-MS/MS
C15:1 GP-NAE was included as an internal standard. Measurements were corrected for extraction efficiency as described under “Experimental Procedures” and represent average values ± S.E. (n = 4 per group).
| Wet tissue | |
|---|---|
| pmol/g | |
| C16:0 GP-NAE | 13.17 ± 4.02 |
| C18:1 GP-NAE | 3.18 ± 1.09 |
| C20:0 GP-NAE | 0.07 ± 0.03 |
| C20:4 GP-NAE | 0.16 ± 0.07 |
| C22:6 GP-NAE | 0.54 ± 0.19 |
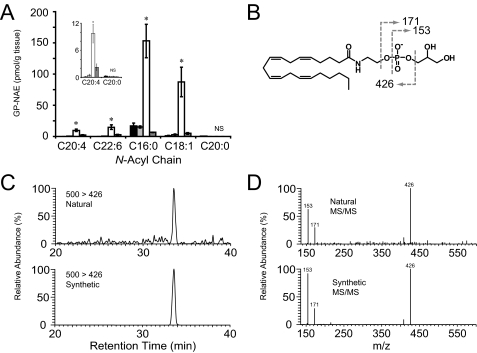
Due to the low levels of endogenous GP-NAEs, we sought additional evidence that these lipids represented potential intermediates in the enzymatic biosynthesis of NAE. To measure enzymatic turnover of GP-NAEs we employed an ex vivo assay. Previous studies had determined that brain GP-NAE phosphodiesterase activity was inhibited by EDTA (14). Here, we found that treatment of brain extracts with EDTA resulted in dramatic accumulation of endogenous GP-NAEs in brain homogenates over time (Fig. 3A). MAFP is a general inhibitor of serine hydrolases including the NAPE-lipase Abh4 (14). The accumulation of GP-NAEs was completely blocked by addition of MAFP thereby confirming that these lipids originated from the enzymatic sn-1/sn-2-deacylation of NAPE precursors. Notably, long chain saturated GP-NAEs (e.g. C20:0) did not accumulate suggesting that this pathway may only be active on GP-NAEs bearing select N-acyl chains (see “Discussion”). The rapid enzymatic turnover of GP-NAEs in brain extracts, combined with the low endogenous levels of these precursors, suggests that they represent transient intermediates in the biosynthesis of NAEs. The significant accumulation of the AEA precursor C20:4 GP-NAE in EDTA-treated brain extracts permitted acquisition of much cleaner MS profiles for this lipid, which matched precisely those observed with a synthetic standard (Fig. 3, C and D).
FIGURE 3.
Ex vivo accumulation of GP-NAEs in brain tissue via an MAFP-sensitive and EDTA-dependent enzymatic pathway. A, GP-NAE levels in brain tissue homogenized in buffer (50 mm Tris, pH 8.0) alone (black and light gray bars), buffer with 10 mm EDTA (open bars) or 10 mm EDTA and 20 μm MAFP (dark gray bars). Homogenates were incubated for 0 h (black bars) or 4 h (all other bars) and then subjected to organic extraction and LC-MS/MS analysis. The inset shows a magnification of low abundance GP-NAEs. Student's t test compared EDTA-treated to samples treated with EDTA and MAFP; asterisk indicates p < 0.05; NS, not significant (n = 4 per group). B, fragmentation of N-arachidonoyl-GP-NAE produces a diagnostic NAE-phosphate fragment (m/z 426) and two fragments corresponding to glycerol 3-phosphate and its dehydrate at 171 and 153, respectively. Representative chromatographs (C) and tandem MS fragmentation (D) are shown for synthetic GP-NArE and natural GP-NArE, which accumulates in the presence of EDTA.
Identification of GDE1 as the GP-NAE Phosphodiesterase—Having confirmed that GP-NAEs are endogenous constituents of mouse brain, we next attempted to identify the enzyme(s) that catalyze hydrolysis of the phosphodiester bond of these lipids to release NAEs. One candidate enzyme class is the glycerophosphodiester phosphodiesterases (GDEs), which hydrolyze the phosphodiester bond of glycerophosphodiesters to yield glycerol phosphate and an alcohol (16). Enzymes containing this GDE domain have been shown to play roles in glycerophospholipid metabolism in yeast and bacteria (17). Searches of mammalian genomes revealed 7 genes encoding proteins containing GDE domains. The GP-NAE phosphodiesterase activity in brain tissue was found exclusively in the membrane fraction (Fig. 5B) and treatment with NaCl or Na2CO3 did not solubilize the activity (not shown), indicating that it derives from an integral membrane protein. Of the seven genes encoding mammalian GDE domain containing proteins, six are predicted to have transmembrane domains and of these, only five are expressed in brain, as determined by expressed sequence tag counts from the UniGene data base and reports from the literature (18). These five predicted GDEs were heterologously expressed in COS-7 cells as epitope-tagged proteins. Following confirmation of expression by Western blotting, membrane fractions from these cells were tested for GP-NAE phosphodiesterase activity using an LC-MS-based substrate assay with synthetic N-C16:0 GP-NAE. One enzyme, GDE1, displayed robust GP-NAE phosphodiesterase activity, whereas the other four GDEs were indistinguishable from a mock-transfected control (Fig. 4, A and B).
FIGURE 5.
GDE1 and GP-NAE phosphodiesterase activity reside in the membrane. A, Western blot of mouse brain GDE1 showing that it resides exclusively in the membrane fraction and is a glycoprotein as evidenced by a ∼5-kDa shift in migration upon treatment with PNGase F. B, GP-NAE phosphodiesterase activity resides exclusively in the membrane fraction of mouse brain.
FIGURE 4.
Expression and GP-NAE phosphodiesterase activity of GDE domain containing enzymes. A, Myc-tagged GDE-domain containing enzymes were overexpressed in COS-7 cells. Membrane fractions were treated with PNGase F and visualized by SDS-PAGE and Western blotting with an anti-Myc antibody (Cell Signaling). B, GP-NAE phosphodiesterase activity of membrane fractions of COS-7 cells expressing GDE domain-containing proteins was measured by monitoring release of N-C16:0 NAE via LC-MS with 100 μm N-C16:0 GP-NAE as substrate.
Brain GP-NAE Phosphodiesterase and Recombinant GDE1 Exhibit Similar Biochemical Properties—We next compared the activity of GDE1 to the GP-NAE phosphodiesterase activity detected in mouse brain. First, we confirmed by Western blotting that GDE1 (Fig. 5A), like the GP-NAE phosphodiesterase activity, resides in brain membrane fractions (Fig. 5B). Treatment with the glycosidase PNGase F reduced the apparent molecular mass of GDE1 by ∼5 kDa, indicating that, as previously reported, the enzyme is an N-linked glycoprotein (19). These data are consistent with hydropathy plot analyses that predict the enzyme is an integral membrane protein with the majority of the protein sequence, including the catalytic domain, facing the luminal/extracellular compartments of the cell. We next compared the sensitivity of GDE1 and brain GP-NAE phosphodiesterase to various divalent cations and found that both activities were inhibited by EDTA, calcium chloride, and zinc chloride, and markedly enhanced by magnesium chloride (Fig. 6B). Recombinant GDE1 and brain GP-NAE phosphodiesterase activity also displayed equivalent pH rate profiles, showing a slightly alkaline pH optimum (Fig. 6A). Finally, GDE1 and GP-NAE phosphodiesterase activity were found to exhibit nearly identical tissue distributions, with highest levels being observed in the brain and spinal cord, followed by kidney, liver, and testis (Fig. 7). In contrast, little or no GP-NAE phosphodiesterase activity or GDE1 expression was detected in the heart or spleen. Interestingly, this tissue distribution resembles that of other enzymes that participate in NAE metabolism, including fatty acid amide hydrolase (5) and Abh4 (14). These data collectively suggest that GDE1 is a principal GP-NAE phosphodiesterase in mammalian tissues.
FIGURE 6.
Mouse brain GP-NAE phosphodiesterase and GDE1 display similar biochemical properties. A, pH rate profile of brain membrane GP-NAE phosphodiesterase (open diamonds) and membranes from COS-7 cells transfected with GDE1 (filled squares) showing equivalent pH optima. Assays were performed in buffer containing 50 mm Tris, 50 mm HEPES, and 50 mm glycine and adjusted to the desired pH with HCl/NaOH. B, mouse brain GP-NAE phosphodiesterase and GDE1 display similar cation sensitivity profiles. Mouse brain membranes (open bars) and GDE1-transfected COS-7 cell membranes (filled bars) were treated with 2 mm CaCl2, ZnCl2, MgCl2, or EDTA. Assays were performed with 100 μm [1′-14C]N-C16:0 GP-NAE substrate, where the release of [1′-14C]N-C16:0 NAE was measured by thin layer radiochromatography.
FIGURE 7.
Tissue distribution of GP-NAE phosphodiesterase activity and GDE1. A, GP-NAE phosphodiesterase activity in membrane fractions of mouse tissues. Assays were performed with 100 μm [1′-14C]N-C16:0 GP-NAE substrate, where the release of [1′-14C]N-C16:0 NAE was measured by thin layer radiochromatography. B, Western blot of GDE1 expression in membrane fractions of mouse tissues following treatment with PNGase F showing strong correlation with GP-NAE phosphodiesterase activity. C, reverse transcription-PCR analysis of GDE1 mRNA expression correlates with GDE1 protein levels and GP-NAE phosphodiesterase activity. The ubiquitously expressed enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a control to confirm integrity of tissue cDNA. Br, brain; Spc, spinal cord; Ht, heart; Lv, liver; Kd, kidney; Spln, spleen; Ts, testis.
Finally, we measured the ability of GDE1 to hydrolyze GP-NAEs with various N-acyl chains. GDE1 was able to generate NAEs from synthetic GP-NAEs bearing multiple types of N-acyl chains, showing the highest activity with C16:0, C18:1, and C20:4 substrates, and lower activity with C20:0 and C22:6 substrates (Fig. 8).
FIGURE 8.
N-Acyl chain selectivity of GDE1. Membrane fractions from GDE1-transfected COS-7 cells were tested for activity against a panel of GP-NAE substrates bearing saturated (C16:0, C20:0), monounsaturated (C18:1), and polyunsaturated (C20:4, C22:6) N-acyl chains. Assays were performed with 100 μm of each substrate using LC-MS to measure release of NAE.
DISCUSSION
Despite over 20 years of study, the biosynthesis of AEA and other NAEs remains poorly understood (20). It is generally accepted that NAPEs are the precursors for NAEs, but the precise enzymatic steps leading to release of NAEs from NAPEs are unclear. Four different enzymatic routes have been proposed. 1) Direct hydrolytic cleavage of the phosphodiester bond of NAPE to release NAE, a reaction that is catalyzed by the NAPE-PLD enzyme (3). Although NAPE-PLD-/- mice possess reduced brain levels of long-chain saturated NAEs, these animals do not display significant alterations in polyunsaturated NAEs in the nervous system (including AEA). These findings invoke the existence of additional enzymatic routes for AEA formation in the brain. 2) Phospholipase C-type cleavage of the phosphodiester bond releasing phospho-NAE, which is rapidly converted to NAE by a lipid phosphatase (11). An enzyme displaying NAPE-phospholipase C-type activity remains to be identified, but a candidate phospho-NAE phosphatase, PTPN22, was cloned from RAW293 immune cells and is also expressed at low levels in the brain (13). However, PTPN22-/- mice possess essentially wild-type levels of AEA in the brain (21). 3) Deacylation of NAPE to generate lyso-NAPE, which is then converted to NAE by a lysophospholipase D. Evidence for this pathway includes the identification of a secreted phospholipase, sPLA2 IB, that singly deacylates NAPE (22). Lyso-NAPE generated by this enzyme was converted by brain extracts to NAE in a reaction presumed to be catalyzed by a D-type lysophospholipase. More recent data suggest that this lyso-NAPE to NAE transformation actually reflects a two-step conversion (see enzymatic route 4) (14). 4) Double deacylation of NAPE to yield glycerophospho-(GP)-NAE, which is then converted to NAE by a metal-dependent phosphodiesterase. This pathway is supported by two principal observations. First, conversion of NAPE or (lyso)NAPE to NAE in NAPE-PLD-/- brains is blocked by MAFP, an inhibitor of serine hydrolases, including many A/B-type phospholipases. Similarly, an NAPE analog with non-hydrolyzable ether bonds in place of natural esters was not converted to NAE in NAPE-PLD-/- brain extracts, demonstrating the absolute requirement for NAPE deacylation (14). Collectively, these observations suggest that double-O-deacylation, as would be performed by an MAFP-sensitive hydrolase, is required for the PLD-independent conversion of NAPE to NAE. This model was bolstered by the identification and molecular characterization of Abh4 as a B-type phospholipase selective for NAPEs over other phospholipid substrates. The resulting GP-NAE products are then rapidly converted by brain homogenates to NAEs by a metal-dependent phosphodiesterase. If this fourth enzymatic pathway is responsible for converting NAPEs to NAEs in brain, then this tissue should contain both the GP-NAE intermediates and a GP-NAE phosphodiesterase.
Here, we have identified the integral membrane enzyme GDE1 as a GP-NAE phosphodiesterase. GDE1 belongs to a larger family of enzymes that hydrolyze the phosphodiester bond of glycerol-phosphodiesters releasing glycerol-phosphate and a free alcohol. GDE domain-containing enzymes are found in bacteria, yeast, plants, and mammals and are ascribed functions that include membrane remodeling, phospholipid turnover, and phosphate scavenging. To date, GDEs have been primarily characterized in the context of glycerophosphoethanolamine, glycerophosphocholine, or glycerophosphoinositol metabolism (23-26).
To determine whether any of the mammalian GDE domain containing enzymes possess GP-NAE phosphodiesterase activity, we expressed these enzymes in COS-7 cells. Among the five GDEs tested, one enzyme, GDE1, exhibited robust activity with GP-NAE. GDE1 was originally identified in a yeast two-hybrid screen for proteins that interact with RGS16, a regulator of G-protein signaling (19). Based on its interactions with RGS16, GDE1 is also referred to as MIR16 (for membrane interacting protein of RGS16). Subsequently GDE1 has been shown to cleave glycerolphosphoinositol substrates in vitro, although the endogenous substrate(s) for this enzyme remain unknown (27). The Farquhar group (27) went on to demonstrate that G-protein coupled receptor agonists could modulate the activity of GDE1 suggesting a provocative bidirectional interaction between G-protein coupled receptor signaling and GDE1 activity.
Several additional lines of biochemical evidence support GDE1 as the principal GP-NAE phosphodiesterase in brain tissue. First, GDE1 displayed a pH optimum and cation sensitivity nearly identical to those observed for brain GP-NAE phosphodiesterase activity. Both activities displayed slightly alkaline pH optima, inhibition by EDTA, calcium, and zinc, and activation by magnesium. Second, GDE1 is an integral membrane protein, which matches the subcellular distribution of the brain GP-NAE phosphodiesterase activity. Finally, the tissue distribution of GDE1 matched closely that of GP-NAE phosphodiesterase activity, as well as other enzymes involved in NAE metabolism, being highly expressed in brain and spinal cord, intermediately expressed in testis, kidney, and liver, and only weakly expressed in heart and spleen.
We have also reported herein the detection and quantification of GP-NAEs as endogenous brain constituents. To our knowledge, these lipids have not previously been reported as natural products in mammalian systems. Using liquid chromatography coupled with high-resolution tandem mass spectrometry we detected multiple GP-NAE species, including the proposed AEA precursor GP-NArE, in normal mouse brain. These endogenous metabolites were detected at levels ∼10-fold lower than NAEs suggesting a rapid enzymatic flux through this pathway. Consistent with this premise, substantial accumulation of endogenous GP-NAEs was observed upon incubation of brain lysates with the GP-NAE phosphodiesterase inhibitor EDTA. This accumulation was also blocked by MAFP, which presumably prevents the serine hydrolase (e.g. Abh4)-mediated sn-1/sn-2 deacylation of NAPE precursors. It is interesting to note that the EDTA-dependent accumulation of GP-NAEs was acylchain dependent. Whereas significant increases were observed in long chain polyunsaturated (e.g. C20:4, C22:6), as well as shorter chain saturated and monounsaturated (e.g. C16:0, C18:1) GP-NAEs, no accumulation of long chain saturated GP-NAEs (e.g. C20:0) could be detected. In addition to the N-C20:0 GP-NAE, we detected a number of other long chain saturated GP-NAEs that also did not accumulate upon incubation with EDTA (not shown). This difference presents a striking foil to the levels of NAEs in NAPE-PLD-/- mice, where significant decreases were observed exclusively in the long chain (>C20) saturated/monounsaturated members of this lipid class (12). Together, these data suggest that NAEs may be biosynthesized in vivo by a bifurcated pathway dependent on the identity of their acyl chains.
In summary, this work, in conjunction with previous studies (12, 14), supports a model for AEA biosynthesis that involves double O-deacylation by Abh4 to yield GP-NArE followed by GDE1-mediated conversion of this lipid intermediated to AEA. Validation of this route for AEA biosynthesis through, for example, the creation of mice lacking Abh4 or GDE1, would designate these enzymes as principal regulators of endocannabinoid signaling in vivo and potential therapeutic targets for a range of human disorders, including obesity, smoking cessation, and cancer (28).
This work was supported by National Institutes of Health Grants DA015197 and DA017259 and the ARCS Foundation and a Koshland Graduate Fellowship in Enzyme Biochemistry (to G. M. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
Footnotes
The abbreviations used are: NAE, N-acyl ethanolamine; Abh4, α/β-hydrolase 4; AEA, N-arachidonoyl ethanolamine; GDE1, glycerophosphodiesterase 1; GP-NAE, glycerophospho-N-acyl ethanolamine; GP-NArE, glycerophospho-N-arachidonoylethanolamine; LC, liquid chromatography; MAFP, methoxy arachidonoylfluorophosphonate; MIR16, membrane interacting protein of RGS16; MS, mass spectrometry; NAPE, N-acyl phosphatidylethanolamine; PLD, phospholipase D; PNGase F, peptide:N-glycosidase F.
References
- 1.Kuehl, F. A., Jacob, T. A., Ganley, O. H., Ormond, R. E., and Meisinger, M. A. P. (1957) J. Am. Chem. Soc. 79 5577-5578 [Google Scholar]
- 2.Bachur, N. R., Masek, K., Melmon, K. L., and Udenfriend, S. (1965) J. Biol. Chem. 240 1019-1024 [PubMed] [Google Scholar]
- 3.Natarajan, V., Reddy, P. V., Schmid, P. C., and Schmid, H. H. (1982) Biochim. Biophys. Acta 712 342-355 [DOI] [PubMed] [Google Scholar]
- 4.Devane, W. A., Hanus, L., Breuer, A., Pertwee, R. G., Stevenson, L. A., Griffin, G., Gibson, D., Mandelbaum, A., Etinger, A., and Mechoulam, R. (1992) Science 258 1946-1949 [DOI] [PubMed] [Google Scholar]
- 5.Cravatt, B. F., Giang, D. K., Mayfield, S. P., Boger, D. L., Lerner, R. A., and Gilula, N. B. (1996) Nature 384 83-87 [DOI] [PubMed] [Google Scholar]
- 6.Kathuria, S., Gaetani, S., Fegley, D., Valino, F., Duranti, A., Tontini, A., Mor, M., Tarzia, G., La Rana, G., Calignano, A., Giustino, A., Tattoli, M., Palmery, M., Cuomo, V., and Piomelli, D. (2003) Nat. Med. 9 76-81 [DOI] [PubMed] [Google Scholar]
- 7.Cravatt, B. F., Demarest, K., Patricelli, M. P., Bracey, M. H., Giang, D. K., Martin, B. R., and Lichtman, A. H. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 9371-9376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cadas, H., di Tomaso, E., and Piomelli, D. (1997) J. Neurosci. 17 1226-1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugiura, T., Kondo, S., Sukagawa, A., Tonegawa, T., Nakane, S., Yamashita, A., and Waku, K. (1996) Biochem. Biophys. Res. Commun. 218 113-117 [DOI] [PubMed] [Google Scholar]
- 10.Jin, X. H., Okamoto, Y., Morishita, J., Tsuboi, K., Tonai, T., and Ueda, N. (2007) J. Biol. Chem. 282 3614-3623 [DOI] [PubMed] [Google Scholar]
- 11.Okamoto, Y., Morishita, J., Tsuboi, K., Tonai, T., and Ueda, N. (2004) J. Biol. Chem. 279 5298-5305 [DOI] [PubMed] [Google Scholar]
- 12.Leung, A., Saghatelian, A., Simon, G. M., and Cravatt, B. F. (2006) Biochemistry 45 4720-4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, J., Wang, L., Harvey-White, J., Osei-Hyiaman, D., Razdan, R., Gong, Q., Chan, A. C., Zhou, Z., Huang, B. X., Kim, H. Y., and Kunos, G. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 13345-13350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simon, G. M., and Cravatt, B. F. (2006) J. Biol. Chem. 281 26465-26472 [DOI] [PubMed] [Google Scholar]
- 15.Hansen, H. H., Hansen, S. H., Schousboe, A., and Hansen, H. S. (2000) J. Neurochem. 75 861-871 [DOI] [PubMed] [Google Scholar]
- 16.Yanaka, N. (2007) Biosci. Biotechnol. Biochem. 71 1811-1818 [DOI] [PubMed] [Google Scholar]
- 17.Patton-Vogt, J. (2007) Biochim. Biophys. Acta 1771 337-342 [DOI] [PubMed] [Google Scholar]
- 18.Nogusa, Y., Fujioka, Y., Komatsu, R., Kato, N., and Yanaka, N. (2004) Gene (Amst.) 337 173-179 [DOI] [PubMed] [Google Scholar]
- 19.Zheng, B., Chen, D., and Farquhar, M. G. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 3999-4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okamoto, Y., Wang, J., Morishita, J., and Ueda, N. (2007) Chem. Biodivers. 4 1842-1857 [DOI] [PubMed] [Google Scholar]
- 21.Liu, J., Wang, L., Harvey-White, J., Huang, B. X., Kim, H.-Y., Luquet, S., Palmiter, R. D., Krystal, G., Rai, R., Mahadevan, A., Razdan, R. K., and Kunos, G. (2008) Neuropharmacology 54 1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun, Y. X., Tsuboi, K., Okamoto, Y., Tonai, T., Murakami, M., Kudo, I., and Ueda, N. (2004) Biochem. J. 380 749-756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tommassen, J., Eiglmeier, K., Cole, S. T., Overduin, P., Larson, T. J., and Boos, W. (1991) Mol. Gen. Genet. 226 321-327 [DOI] [PubMed] [Google Scholar]
- 24.Almaguer, C., Fisher, E., and Patton-Vogt, J. (2006) Curr. Genet. 50 367-375 [DOI] [PubMed] [Google Scholar]
- 25.van der Rest, B., Boisson, A. M., Gout, E., Bligny, R., and Douce, R. (2002) Plant Physiol. 130 244-255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao, M., and Sockanathan, S. (2005) Science 309 2212-2215 [DOI] [PubMed] [Google Scholar]
- 27.Zheng, B., Berrie, C. P., Corda, D., and Farquhar, M. G. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 1745-1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bifulco, M., Grimaldi, C., Gazzerro, P., Pisanti, S., and Santoro, A. (2007) Mol. Pharmacol. 71 1445-1456 [DOI] [PubMed] [Google Scholar]