Abstract
FGF10, a heparan sulfate (HS)-binding growth factor, is required for branching morphogenesis of mouse submandibular glands (SMGs). HS increases the affinity of FGF10 for FGFR2b, which forms an FGF10·FGFR2b·HS ternary signaling complex, and results in diverse biological outcomes, including proliferation and epithelial morphogenesis. Defining the HS structures involved in specific FGF10-mediated events is critical to understand how HS modulates growth factor signaling in specific developmental contexts. We used HS-deficient BaF3/FGFR2b cells, which require exogenous HS to proliferate, to investigate the HS requirements for FGF10-mediated proliferation and primary SMG epithelia to investigate the structural requirements of HS for FGF10-mediated epithelial morphogenesis. In BaF3/FGFR2b cells, heparin with at least 10 saccharides and 6-O-, 2-O-, and N-sulfates were required for maximal proliferation. During FGF10-mediated SMG epithelial morphogenesis, HS increased proliferation and end bud expansion. Defined heparin decasaccharide libraries showed that 2-O-sulfation with either an N-or 6-O-sulfate induced end bud expansion, whereas decasaccharides with 6-O-sulfation alone induced duct elongation. End bud expansion resulted from increased FGFR1b signaling, with increased FGFR1b, Fgf1, and Spry1 as well as increased Aqp5 expression, a marker of end bud differentiation. Duct elongation was associated with expression of Cp2L1, a marker of developing ducts. Collectively, these findings show that the size and sulfate patterns of HS modulate specific FGF10-mediated events, such as proliferation, duct elongation, end bud expansion, and differentiation, and provide mechanistic insight as to how the developmental localization of specific HS structures in tissues influences FGF10-mediated morphogenesis and differentiation.
The diverse biological activities of FGFs2 are mediated by FGFRs that undergo alternative splicing, resulting in cell-, tissue-, and stage-specific expression and alteration in the ligand binding properties (1-6). The spatial and temporal expression of FGFR and FGF isoforms is further complicated by the diversity of heparan sulfate (HS) structures (7), which also play a critical role in developmental processes (8). HS increases FGFR signaling by stabilizing FGF·FGFR complexes, binding both to ligand and receptor molecules, and by increasing local ligand concentrations at the cell surface and in the extracellular matrix (ECM) (2, 9, 10). HS chains are polymers of repeating disaccharides, which undergo enzymatic modifications that include N-sulfation, N-acetylation, and O-sulfation (7). Assembly of the FGF·FGFR·HS complex requires simultaneous binding by the FGF and FGFR to distinct sulfation patterns within the HS chain, implying that tissue-specific HS may regulate FGF signaling (11).
FGF10 and its receptor FGFR2b are required for mouse SMG development (12-14). FGF10 heterozygous mice are viable and fertile but display SMG and lacrimal gland hypoplasia (15, 16), suggesting that their development is particularly sensitive to levels of FGF10. We have shown that FGFR2b signaling regulates SMG proliferation and morphogenesis, which are dependent on laminin-511 and β1 integrin signaling, which reciprocally regulate FGFR and Fgf1 expression (17, 18). Both FGF10 and FGF7 bind FGFR2b, but FGF10 mitogenic activity is more heparin-dependent than FGF7 (3, 19). FGF10 is highly expressed in the condensing SMG mesenchyme, whereas FGFR2b is in the initial epithelial bud that forms on the end of a single primary duct (20). Investigating the mechanisms that control end bud expansion, clefting, duct formation, and differentiation is fundamental to understanding branching morphogenesis.
HS protects FGFs from proteolytic degradation; participates in FGF internalization, processing, and nuclear localization; and provides an extracellular reservoir from which FGFs can be released (21-23). We reported that heparanase, an endoglycosidase that degrades heparan sulfate, modulates the biological activity of FGF10 in the SMG (24). Heparanase releases FGF10 from perlecan HS in the basement membrane, increasing mitogen-activated protein kinase signaling, epithelial clefting, and lateral branch formation, which results in increased branching morphogenesis. We proposed that heparanase-derived HS fragments might influence both the bioavailability and the bioactivity of FGF10, resulting in distinct morphogenic outcomes.
Efforts to identify HS structures that bind specific FGFs or FGFRs using heparin-derived oligosaccharide libraries give different results, depending on the FGF, cell type, size of heparin, and type of assay used. To summarize, HS is both a positive and a negative regulator of FGF signaling (25, 26), the size and sulfation patterns influence FGFR binding and biological activity (27), many interactions depend more on the overall organization of HS domains than on their fine structure (28), and the stability of the HS·FGF·FGFR complex correlates with overall O-sulfation rather than the precise distribution of sulfates (29). Taken together, binding studies suggest that specific structural features of HS influence specific FGF-receptor interactions in different biological contexts. Our goal is to define the structural requirements of HS that modulate FGF10-mediated biological processes during SMG morphogenesis.
Here, we define the HS structure in terms of size and sulfation patterns that modulate FGF10-mediated proliferation and morphogenesis using two complementary model systems: BaF3 cells expressing specific FGFRs and isolated SMG epithelial organ culture, which lacks the endogenous mesenchyme-derived FGF10 and HS. We demonstrate that differences in HS sulfate patterns, and not just charge density, influence morphogenesis and cellular differentiation, resulting in both duct elongation and end bud expansion. The end bud expansion is due to increased FGFR1b signaling, which also results in cell differentiation, suggesting that specific HS structures modulate diverse FGF10 functions during SMG branching morphogenesis.
EXPERIMENTAL PROCEDURES
Materials—Human recombinant FGF1 FGF2, FGF7, and FGF10 were purchased from R&D Systems. For BaF3 cell culture, recombinant mouse FGF10 was expressed in a bacterial expression vector, purified by a heparin-Sepharose column, followed by Mono S cation exchange chromatography (30, 31). Heparan sulfate, (Sigma), heparin (Celsus, OH), de-N-sulfated N-acetylated heparin, completely desulfated N-acetylated heparin, and completely desulfated N-sulfated heparin were prepared as previously described (32) or purchased from Neoparin Inc. (San Leandro, CA). Sized heparin fragments (a gift from Dr. A. Marolewski, RepliGen Corp.) were prepared as previously described (33) either by alkaline depolymerization or following partial heparinase cleavage of low molecular weight heparin. Fragments obtained following alkaline depolymerization of heparin (average mass ∼5 kDa; Enzyme Research Laboratories, South Bend, IN) were fractionated on a Sephadex G50 column using ammonium bicarbonate buffer. Fractions were dried and weighed, and size was determined by gradient electrophoresis by comparison with known standards. Following partial heparinase cleavage, the saccharides were separated by high resolution gel filtration as described (34).
BaF3 Cells and ex Vivo Submandibular Gland Organ Culture—The pre-B lymphocyte cell line BaF3, expressing FGFRs, was grown in RPMI 1640 supplemented with 10% fetal calf serum and 10% interleukin-3-conditioned medium from WEHI-3B cells. The cells expressing the FGFRs were also grown in RPMI 1640 plus 10 ng/ml recombinant FGF1 and 5 μg/ml heparin as previously described (27).
ICR mice were used to obtain mesenchyme-free SMG epithelium as previously described (17, 35). Isolated epithelium were cultured in 15 μl of laminin-111 (1 mg/ml; Trevigen, MD) on track-etched polycarbonate filters (13 mm, 0.1-μm pore size) floated on top of 200 μl of Dulbecco's modified Eagle's medium/F-12 in 50-mm glass bottom microwell dishes (Mat-Tek). The medium was supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 150 μg/ml vitamin C, 50 μg/ml transferrin, and 200 ng/ml FGF10. Heparan sulfate or modified heparin fragments were also added directly to the medium. Up to six gland rudiments were cultured on each filter at 37 °C in a humidified 5% CO2, 95% air atmosphere. Glands were photographed at 44 h, and the number, width of end buds (measured in pixels and expressed as arbitrary units (AU), and length (AU) of the ducts were measured using MetaMorph software. The number of end buds, width, and length were also multiplied to give a morphogenic index (AU × 103), which is a measure of the complexity and amount of branching morphogenesis. Each experiment was repeated at least three times.
Detection of Cell Proliferation—BaF3 cells were washed three times and seeded into 96-well microtiter plates (20,000 cells/well) in RPMI 1640 containing 10% fetal calf serum and the indicated concentration of growth factors, heparin, and oligosaccharides. Fresh growth factors and other supplements were added every other day, and the viable cells were counted on day 5 of incubation. Each data point was preformed in duplicate, and each experiment was repeated at least twice. A representative graph is shown with the means and the range.
SMG epithelial proliferation was detected as described (17), using a bromodeoxyuridine labeling kit (Roche Applied Science). The SMGs were blocked using a mouse on mouse kit (M.O.M. reagents; Dako) and anti-bromodeoxyuridine and anti-α3 integrin rabbit polyclonal antiserum (a gift from Dr. M. DiPersio) were incubated for 2 h at 37 °C in the M.O.M. incubation buffer, and the appropriate secondary antibodies and SYBR green were incubated for 1 h at 37 °C. Images were obtained using a Zeiss LSM 510 confocal microscope.
Inhibition of HS-induced End Bud Expansion—Epithelia were cultured as described above with FGF10, HS oligosaccharide, and FGFR inhibitor, SU5402 (1 and 2.5 μm), all added at the beginning of the experiment with Me2SO as a carrier control. SMG epithelia were cultured with recombinant FGFR1b chimera (10 and 15 μg/ml or 125 and 187 nm) (R&D Inc.). Gene expression was measured by RT-PCR after 48 h of culture, and the epithelial morphology was measured as described above.
Real Time PCR—DNase-free RNA was prepared from at least six epithelial rudiments using an RNAqueous-4PCR kit and DNase removal reagent (Ambion, Inc., Austin, TX). Real time PCR was performed using primers designed with similar properties by Beacon Designer software (sequences available on request), SYBR-green PCR Master Mix, and a MyIQ real time PCR thermocycler (all from Bio-Rad). Each cDNA (0.5-1 ng) was amplified with 40 cycles of 95 °C for 10 s and 62 °C for 30 s. Gene expression was normalized to 29 S. The reactions were run in triplicate, and the experiment was repeated three times.
In Situ Hybridization—Digoxigenin-11-UTP-labeled single-stranded RNA probes were prepared using the DIG RNA labeling kit (Roche Applied Science) according to the manufacturer's instructions. Plasmids for FGFR1b and FGFR2b/PGEM-5Zf (+) were provided by Dr. Rosana Dono (Campus de Luminy, Marseille, France), and in situ hybridization was performed as previously described (36).
RESULTS
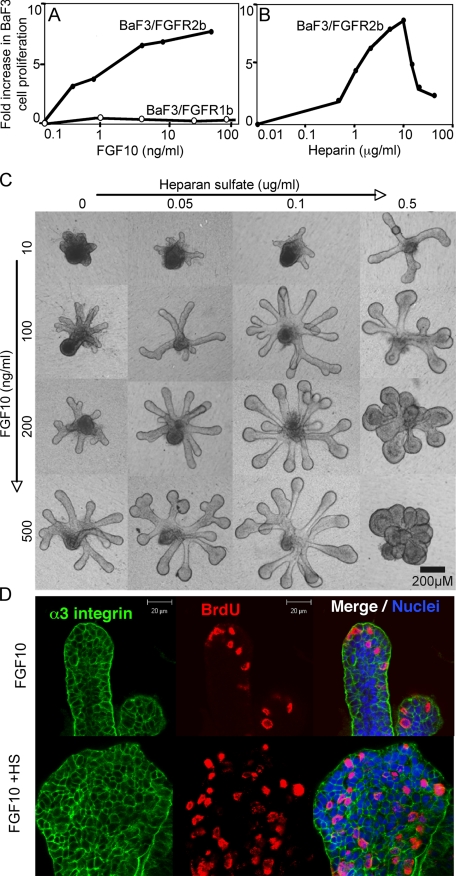
FGF10 Induces Heparin-dependent Proliferation of HS-deficient BaF3/FGFR2b Cells—The proliferation of BaF3 cells overexpressing either FGFR2b (BaF3/FGFR2b) or FGFR1b (BaF3/FGFR1b) in response to FGFs is dependent on exogenous heparin or HS species (37). FGF10 induced a dose-dependent increase in proliferation of BaF3/FGFR2b cells (Fig. 1A) in the presence of 5 μg/ml heparin. However, FGF10 did not induce proliferation in BaF3/FGFR1b cells with 5 μg/ml of heparin (Fig. 1A), suggesting that FGF10 is not a physiological ligand for FGFR1b proliferation. FGF1 increased BaF3/FGFR1b cell proliferation (data not shown), indicating that the cells were functional. Additionally, there was a dose-dependent increase in proliferation with increasing heparin (Fig. 1B) and 1 ng/ml of FGF10, which was an ∼50% maximal response in Fig. 1A. BaF3/FGFR2b cell proliferation was not observed with FGF10 in the absence of heparin. There was a maximal proliferative response with 5-10 μg/ml heparin, but with increasing heparin the proliferation decreased, suggesting that excess heparin bound competitively to FGF10, reducing its effective binding to FGFR and thus proliferation.
FIGURE 1.
FGF10 induces heparin-dependent proliferation of BaF3 cells expressing FGFR2b, and FGF10-dependent SMG epithelial morphogenesis and proliferation are modulated by heparan sulfate. A, BaF3/FGFR1b cells (open circles) or FGFR2b (filled circles) were cultured with FGF10 and 5 μg/ml heparin. FGF10 increases BaF3/FGFR2b cell proliferation in a dose-dependent manner but does not induce BaF3/FGFR1b cells to proliferate. B, BaF3/FGFR2b cells were cultured with 1 ng/ml FGF10 and increasing concentrations of heparin. C, E13 SMG epithelia were cultured with increasing doses of FGF10 and HS. Increasing concentrations of FGF10 induce duct elongation, whereas HS induces end bud expansion. D, heparan sulfate (0.5 μg/ml) increases FGF10-mediated (200 ng/ml) proliferation throughout the epithelium resulting in end bud expansion. Images are single confocal slices through the end bud with bromodeoxyuridine labeling (red), α3 integrin (green), and nuclei (blue).
HS Increases FGF10-dependent Morphogenesis and Proliferation of SMG Epithelium—Changes in SMG epithelial morphogenesis were measured in response to increasing concentrations of FGF10 with either HS or heparin. The results observed with HS (shown) were similar to those with heparin (data not shown). FGF10 alone (100-500 ng/ml) resulted in a dose-dependent increase in duct length (Fig. 1C), with narrow end buds. Further increases in FGF10 up to 1500 ng/ml resulted in longer ducts, as previously reported (17). HS alone does not support epithelial growth (not shown). However, HS (0.05-0.5 μg/ml) in combination with FGF10 resulted in more complex epithelial morphogenesis, and with 200 ng/ml FGF10 and 0.1 μg/ml HS or with 100 ng/ml FGF10 and 0.5 μg/ml HS, the epithelium had both elongating ducts and enlarged end buds. However, as concentrations of either HS or FGF10 increased, there was less morphogenesis with large end buds with no ducts (see 500 ng/ml FGF10 and 0.5 μg/ml HS), and with further increases in HS and FGF10, there was complete loss of morphogenesis, and the epithelia formed an amorphous clump of cells (not shown). Importantly, bromodeoxyuridine labeling (Fig. 1D) showed that the end bud expansion with exogenous HS was due to an increased area of proliferation. Epithelia treated with FGF10 alone proliferated mainly at the tips, whereas exogenous HS or heparin (not shown) resulted in more generalized proliferation throughout the end buds (Fig. 1D).
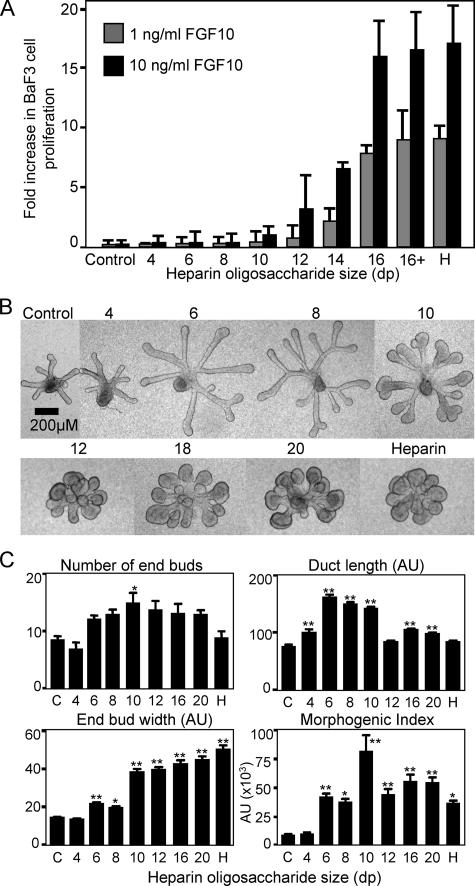
FGF10-dependent BaF3/FGFR2b Proliferation and SMG Epithelial Morphogenesis Are Dependent on the Size of Heparin—We used heparin oligosaccharides ranging from dp4 to dp16, to define the minimal size of heparin required for FGF10-mediated (either 1 or 10 ng/ml) BaF3/FGFR2b cell proliferation (Fig. 2A). Heparin fragments shorter than dp10 did not induce FGF10-dependent proliferation when added at 5-50 μg/ml (Fig. 2A) (data not shown); however, there was a 2-fold increase in cell number with dp10 in the presence of 10 ng/ml of FGF10. Longer oligosaccharides activated FGF10 at both growth factor concentrations, and the potency of heparin oligosaccharides longer than dp16 was comparable with full-length heparin.
FIGURE 2.
Heparin oligosaccharides larger than dp10 increase BaF3/FGFR2b cell proliferation and SMG epithelial end bud expansion and morphogenesis. A, FGF10 (1 and 10 ng/ml) was added to BaF3/FGFR2b cells with 5 μg/ml sized heparin fragments. BaF3/FGFR2b cell proliferation begins to increase (2-fold) with 10 ng/ml FGF10 and dp10 heparin fragments and continues to increase as the saccharide size increases up to dp16, which is similar to heparin (H). B, SMG epithelia were cultured for 48 h with 200 ng/ml FGF10 (control) and 0.5 μg/ml sized heparin oligosaccharides. C, the number of buds, width of the end buds, and duct length were measured and combined to give a morphogenic index. At least five epithelia/condition were measured, and experiments were repeated three times (one-way analysis of variance compared with control: **, p < 0.01; *, p < 0.05).
FGF10-mediated SMG morphogenesis was also dependent on the length of heparin-derived oligosaccharides (Fig. 2B). Morphometric analysis (Fig. 2C) involved counting the number of end buds, measuring the length of the ducts (AU) and the width of the end buds (AU), and then multiplying all three parameters to obtain an overall morphogenic index (×103 AU). Increasing the length of the heparin oligosaccharides resulted in a gradual change in epithelial morphogenesis with 200 ng/ml FGF10 (Fig. 2, B and C). Epithelium treated with dp4 heparin fragments appeared similar to the epithelial rudiments cultured with FGF10 alone. However, dp6 and dp8 heparin fragments increased duct elongation, and dp10 fragments also caused end bud expansion and clefting. The increase in duct length with dp6 and dp8 fragments may be due to activation of autocrine FGF1-dependent mechanism (17), since FGF1 can activate all FGFRs with oligosaccharides shorter than 8 sugar units (32). The combined morphogenesis graph (Fig. 2C) shows that fragments greater than dp6 have a significant effect on morphogenesis, with dp10 giving the greatest increase, and fragments greater than dp12 appeared similar to heparin.
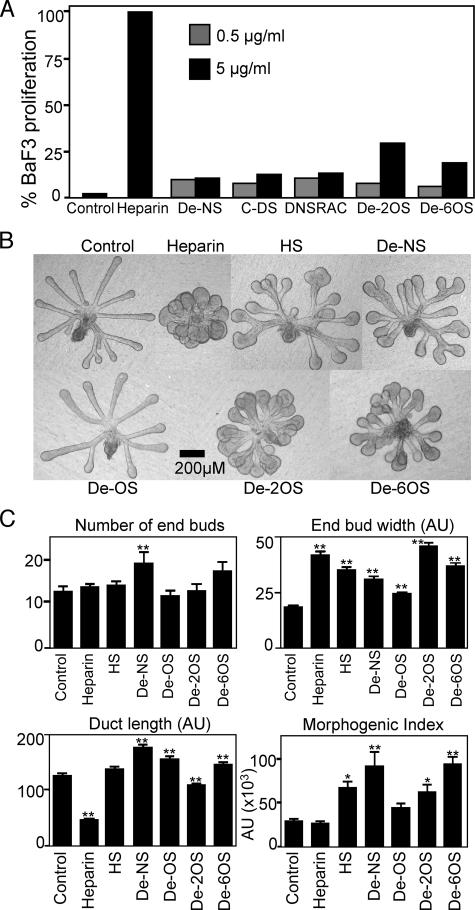
Heparin Sulfation Influences BaF3 Proliferation and SMG Epithelial Morphogenesis—We used selectively desulfated heparins to broadly identify the structural characteristics of heparin required for FGF10 activity. In BaF3/FGFR2b assays, removal of 2-O- or 6-O-sulfates (De-2OS and De-6OS) from heparin (Fig. 3A) reduced proliferation by 75% compared with heparin, whereas removal of N-sulfates (De-NS), de-N-sulfated and reacetylated heparin (DeNSRAC), and completely desulfated heparin (C-DS) reduced proliferation by 92% when compared with heparin (Fig. 3A). Therefore, any reduction in sulfate content reduced FGF10-mediated proliferation of BaF3/FGFR2b cells.
FIGURE 3.
Chemical desulfation of heparin decreases FGF10-mediated BaF3/FGFR2b cell proliferation and influences SMG epithelial morphogenesis. A, BaF3/FGFR2b cells were cultured with FGF10 (10 ng/ml) and 0.5 or 5 μg/ml de-N-sulfated (De-NS), completely desulfated (C-DS), N-desulfated and reacetylated (DNSRAC), de-2-O-sulfated (De-2OS), or de-6-O-sulfated (De-6OS) heparin. Results are expressed as a percentage of cell proliferation compared with heparin. B, SMG epithelia were cultured with FGF10 (Control) and 0.5 μg/ml of heparin, chemically modified heparins, and HS. C, the number of buds, width of the end buds, and duct length were measured and combined to give a morphogenic index. Removal of any sulfates from heparin increases the morphogenic index, similar to heparan sulfate. Removal of all O-sulfates (De-OS) results in a morphogenic index similar to control. At least five epithelia/condition were measured, and experiments were repeated three times (one-way analysis of variance compared with control: **, p < 0.01; *, p < 0.05.
SMG epithelial morphogenesis was also influenced by the removal of sulfates from heparin. Epithelia cultured with FGF10 (200 ng/ml) and 0.5 μg/ml heparin underwent extensive budding but no duct elongation (Fig. 3, B and C), and the morphogenic index was similar to epithelium cultured with no heparin, whereas HS, which has a lower overall and more heterogeneous pattern of sulfation, increased duct length and end bud width, resulting in increased morphogenesis compared with heparin. Removal of any sulfates from heparin, de-N-sulfated heparin, 2-O-sulfate, or 6-O-sulfate all resulted in increased morphogenesis compared with heparin, suggesting that overall sulfation does not correlate with morphogenesis. The epithelial morphology with selective desulfation appeared more similar to HS than heparin treatment (Fig. 3, B and C). However, heparin with all O-sulfation removed appeared similar to FGF10 alone, suggesting that O-sulfation was important for FGF10-dependent end bud expansion.
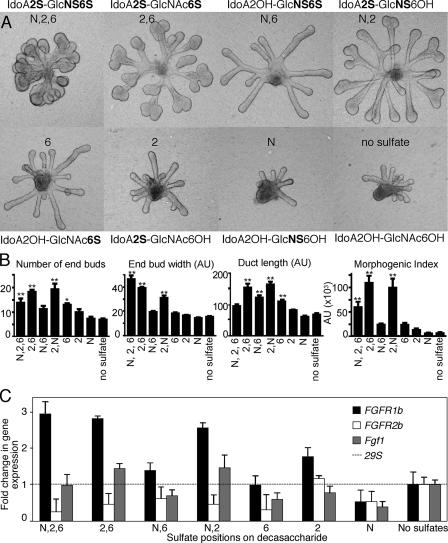
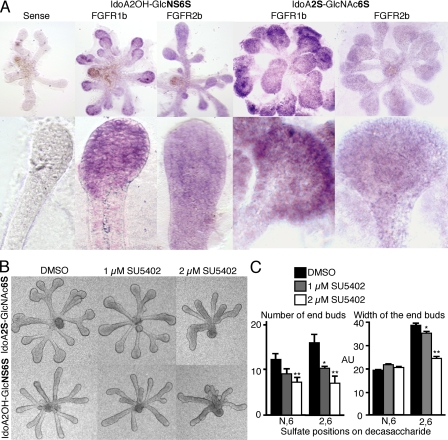
Sulfate Patterns Influence Epithelial Morphogenesis and FGFR1b and Fgf1 Expression—Heparin decasaccharide (dp10) libraries with defined sulfate groups have previously been shown to influence FGFR signaling (26). SMG epithelia were cultured with FGF10 (200 ng/ml) and 0.5 mg/ml of the decasaccharides, and the morphogenesis was measured (Fig. 4, A and B). Decasaccharides containing three sulfate groups per disaccharide unit (IdoA2S-GlcNS6S) resulted in a budding epithelial morphology (Fig. 4A), similar to that observed with 0.5 mg/ml heparin and 200 ng/ml FGF10 (Fig. 2B). Decasaccharides with two sulfates per disaccharide unit (IdoA2S-GlcNS6OH, IdoA2OH-GlcNS6S, and IdoA2S-GlcNAc6S) all increased duct length, but IdoA2OH-GlcNS6S did not induce end bud expansion. This result is important, because it implies that specific sulfate patterns, and not just sulfate density, are important for end bud expansion. Interestingly, decasaccharides containing a 2-O-sulfate with any other sulfate group (IdoA2S-GlcNS6OH and IdoA2S-GlcNAc6S) grew more robustly than those with other decasaccharides and increased the number of buds, the width of the end buds, and the duct length (Fig. 4, A and B). Notably, epithelium treated with decasaccharides containing only 6-O-sulfate (IdoA2OH-GlcNAc6S) had longer ducts than the other decasaccharides with single sulfates/disaccharide unit and were a similar length to N- and 6-O-sulfated decasaccharide (IdoA2OH-GlcNS6S). Epithelium cultured with decasaccharides with only 2-O-sulfation caused a slight, but not significant, increase in duct length, and decasaccharides with only N-sulfation resembled those with no sulfation (Fig. 4, A and B).
FIGURE 4.
Oligosaccharide libraries with defined sulfation show that the position of sulfates increases end bud morphogenesis and FGFR1b expression. A, oligosaccharide libraries (dp10, 0.5 mg/ml) with defined sulfate groups were added to SMG epithelial cultures with FGF10 (200 ng/ml). B, the number and width of the end buds and length of the ducts were measured and combined to give a morphogenic index. C, oligosaccharides that increase end bud width also have increased FGFR1b expression, and the SMGs with the greatest combined morphogenesis also have increased Fgf1 expression. Gene expression was normalized to 29S and expressed as a -fold increase compared with the expression in the Ido2AOH-GlcNAc6OH-treated group. At least five epithelia/condition were measured, and the experiment was repeated three times (one-way analysis of variance compared with no sulfate group: **, p < 0.01; *, p < 0.05).
We hypothesized that the decasaccharides were increasing FGF10/FGFR2b signaling, so we measured expression of FGFR1b, FGFR2b, and Fgf1 (Fig. 4C), which we had previously shown were downstream of FGFR signaling (18). Gene expression was normalized to 29S and expressed as a -fold change in expression relative to the expression with IdoA2OH-GlcNAc6OH, a decasaccharide with no sulfates, which supports minimal growth and duct elongation (Fig. 4A). End bud expansion was associated with an ∼3-fold increase of FGFR1b expression, and the decasaccharides with 2 sulfates/unit (IdoA2S-GlcNS6OH and IdoA2S-GlcNAc6S) also increased Fgf1 ∼1.5-fold (Fig. 4C). Increased Fgf1 expression was similar to that previously reported, and anti-FGF1 antibodies have previously been shown to decrease FGF10-dependent morphogenesis (17). End bud expansion was also associated with decreased FGFR2b expression, suggesting an autocrine negative regulatory loop, similar to our previous findings (17). The decasaccharides resulting in the highest morphogenic index (with IdoA2S-GlcNS6OH and IdoA2S-GlcNAc6S) are associated with both increased FGFR1b and Fgf1 expression.
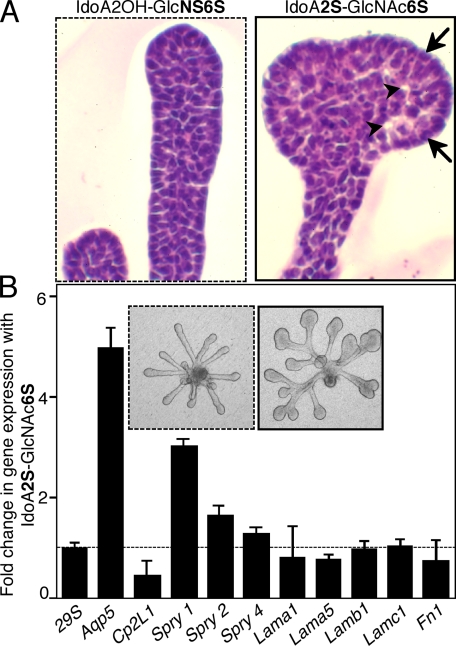
End Bud Expansion Also Involves Cellular Differentiation—We next compared SMG epithelia cultured with either IdoA2OH-GlcNS6S or IdoA2S-GlcNAc6S to investigate their effects on cell differentiation; both have 2 sulfates/disaccharide, but only one induces end bud expansion. Histological analysis of epithelia cultured with IdoA2S-GlcNAc6S shows that end bud cells at the periphery that are in contact with the ECM have basally located nuclei, and lumens are forming in the center of the bud, suggesting the end bud cells may be differentiating (Fig. 5A). Therefore, we measured expression of the water channel, aquaporin 5 (Aqp5), a marker of apical polarity and epithelial differentiation during SMG development (38), which increased ∼5-fold (Fig. 5B) compared with epithelia not forming end buds. We also hypothesized that increased FGFR signaling resulting in end bud expansion would also increase expression of Sprouty genes, which are negative regulators of FGFR signaling; Sprouty 1 (Spry1) expression increases ∼3.5-fold, and Spry2 expression increases ∼1.5-fold with end bud expansion but no change in Spry4. In addition, the expression of the grainyhead-related transcription factor, Cp2L1, a marker of SMG duct development (39, 40), decreased ∼2-fold in epithelia with enlarged end buds compared with those forming ducts. End bud expansion did not result in changes in expression of laminins α1, α5, β1, or γ1 or fibronectin, ECM genes previously associated with SMG branching morphogenesis. Our findings suggest that end bud expansion associated with IdoA2S-GlcNAc6S decasaccharide increases expression of genes associated with FGFR-mediated proliferation (FGFR1b and Fgf1), the regulation of FGFR signaling (Spry1 and Spry2), and epithelial differentiation (Aqp5).
FIGURE 5.
End bud expansion is associated with changes in cellular organization, differentiation, and FGFR signaling. A, histological analysis by hematoxylin/eosin staining of end bud epithelium shows cellular organization with areas of lumen formation (arrowheads) and cells in contact with the ECM that have basally located nuclei (arrows). B, end bud expansion is associated with increased expression of Aqp5, a marker of cell differentiation; Spry1, a regulator of FGFR signaling; and reduced expression of Cp2L1, a marker of ductal differentiation. Gene expression was normalized to 29S and expressed as -fold increase compared with the epithelium treated IdoA2OH-GlcNS6S. Light micrographs of the epithelium are also shown. The experiment was repeated three times with similar results.
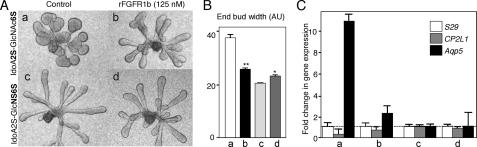
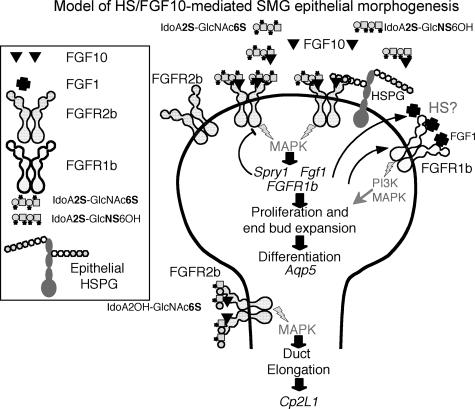
FGF10 Decasaccharide-mediated End Bud Expansion and Differentiation Occurs via Increased FGFR1b Signaling—Whole mount in situ analysis of FGFR1 and FGFR2 confirmed that the increase in FGFR1b expression was localized in the end bud (Fig. 6A), whereas FGFR2 is expressed throughout the epithelium. The sense probe for FGFR1b is shown, which appeared similar to the FGFR2 sense probe (not shown). We also confirmed that the end bud expansion was due to increased FGFR signaling by adding SU5402, a chemical inhibitor of FGFR phosphorylation (Fig. 6B). Low doses of SU5402 decreased end bud expansion with IdoA2S-GlcNAc6S (Fig. 6, B and C) and also decreased the number of end buds with both decasaccharides. Since SU5402 inhibits phosphorylation of all FGFRs, we also used soluble recombinant FGFR1b to confirm a functional role in end bud expansion (Fig. 7). Epithelia cultured with FGF10 and either IdoA2S-GlcNAc6S or IdoA2OH-GlcNS6S were treated with rFGFR1b (125 nm), and the end bud width (Fig. 7B) and gene expression of Cp2L1 (ductal marker) and Aqp5 (an end bud marker) were measured. The rFGFR1b reduced the end bud width of IdoA2S-GlcNAc6S-treated epithelium to a similar width as IdoA2OH-GlcNS6S-treated epithelium. Importantly, the rFGFR1b had little effect on the morphogenesis of IdoA2OH-GlcNS6S-treated epithelium, suggesting that duct elongation was independent of increased FGFR1b signaling (see the model in Fig. 8). In addition, the increased Aqp5 expression measured with IdoA2S-GlcNAc6S treatment was reduced by rFGFR1b, and the Cp2L1 expression was increased with rFGFR1b treatment compared with control, suggesting that the end bud differentiation was downstream of increased FGFR1b signaling (Fig. 8). In our model, we have also included information about mitogen-activated protein kinase and PI3K signaling and the role of FGF1 from our previous work (17, 18, 24); in addition, we propose that specific FGF10-binding HS structures around the end buds bind FGFR2b, increasing both FGFR1b and Fgf1 gene expression. The increased FGFR1b results in increased proliferation and differentiation with increased Aqp5 expression. Additionally, we speculate that there may be epithelial HSPG that plays a role in the epithelial FGFR signaling.
FIGURE 6.
There is increased expression of FGFR1b in the end buds, and the oligosaccharide-mediated increase in number and width of end buds is due to increased FGFR signaling. A, whole mount in situ analysis of both FGFR1 and FGFR2 in the salivary epithelium cultured with IdoA2S-GlcNAc6S or IdoA2OH-GlcNS6S. B, both the number and width of end buds are decreased by low doses of SU5402, an FGFR inhibitor. SMG epithelia were cultured with FGF10 (200 ng/ml) and IdoA2S-GlcNAc6S or IdoA2OH-GlcNS6S (0.5 μg/ml) with Me2SO (DMSO) or SU5402 (1 and 2 mm). C, morphometric analysis of B. The decrease in number and width of the end buds was measured using MetaMorph software (one-way analysis of variance compared with the Me2SO group: **, p < 0.01; *, p < 0.05).
FIGURE 7.
End bud expansion and differentiation are reduced by soluble recombinant FGFR1b. A, epithelia cultured with IdoA2S-GlcNAc6S (a) were treated with rFGFR1b (b), and epithelia cultured with IdoA2OH-GlcNS6S (c) were treated with rFGFR1b (d) as a control. B, morphometric analysis of end bud width showing a significant decrease with IdoA2S-GlcNAc6S control (a) compared with rFGFR1b-treated (b). There was also a slight but significant increase in end bud width with c compared with d. C, gene expression of Aqp5, a marker of end bud differentiation, was decreased by rFGFR1b, and there is a relative increase in Cp2L1, a marker of duct differentiation. Gene expression was normalized to the level of expression with IdoA2OH-GlcNS6S treatment and to 29S expression. rFGFR1b did not change levels of gene expression with IdoA2OH-GlcNS6S (Student's unpaired t test: **, p < 0.001; *, p < 0.01).
FIGURE 8.
Model of FGF10-HS interaction during SMG morphogenesis. End bud expansion involves binding of specific HS fragments with 2-O- and either N-or 6-O-sulfation, either alone or in combination with endogenous epithelial cell HSPGs, to FGFR2b. Increased FGFR2b signaling results in increased gene expression of FGFR1b and Fgf1, leading to proliferation, and increased Aqp5, a marker of end bud epithelial differentiation. There also may be specific HS structures that bind FGFR1b/FGF1. Decreased FGFR2b and increased Sprouty expression, negative regulators of FGFR signaling, balance the increased FGFR2b signaling. HS fragments with 6-O-sulfation alone increase duct elongation and expression of duct markers (e.g. Cp2L1).
DISCUSSION
We analyzed the structural modifications of heparin that influence the biological activity of FGF10 using two complementary experimental systems: 1) BaF3 cells, which do not have endogenous HS or FGFRs and are engineered to express FGFR1b and FGFR2b, and 2) SMG epithelial morphogenesis assays, a more complex three-dimensional organ culture system with endogenous epithelial cell surface HS and HS present in the three-dimensional laminin ECM, which we recently showed contained perlecan (24). We report that in both experimental systems, FGF10-mediated cell proliferation depends on saccharide length and sulfate composition. The amount of FGF10 used in the three-dimensional organ culture is higher than that used in Baf3 cell culture, most likely due to the nature of the assay; the SMGs are embedded within a three-dimensional ECM, the FGFs are added to the serum-free media under the filter, and the SMG cells are within a three-dimensional tissue. In comparison, Baf3 cells are in single-cell suspension surrounded by FGF-containing medium.
Previous studies have produced conflicting results as to the ability of recombinant FGF10 to activate FGFR1b (3, 41). Using the BaF3 model system (27), we show that FGF10 and heparin stimulate proliferation with FGFR2b but not with FGFR1b. We also confirmed that FGF10 did not induce a proliferative response in NIH/3T3 cells at concentrations up to 500 ng/ml (data not shown), which is in agreement with previous studies (3). One reason our Baf3 results may differ from previous reports (41) is that we used different assays. Here, we measured cell proliferation, which measures the ability of FGF10 to allow repeated completion of the cell cycle, whereas they used mitogenic assays, which measures DNA synthesis as cells progress to S-phase. It is plausible that in vivo some FGF10 may bind to FGFR1b with a specific HS structure or neoepitope of HS, which could be produced by heparanase cleavage in a particular developmental context. Our previous work also shows that FGF10-dependent SMG epithelial morphogenesis requires autocrine FGF1/FGFR1b signaling (17).
When FGF10 was initially characterized, its mitogenic activity was distinguished from FGF7 by sensitivity to heparin, and although both bound to FGFR2b, it was suggested that their activity was differentially regulated by components of the ECM (3). Importantly, FGF10 has a higher affinity for heparin and was eluted from a heparin-Sepharose column with 1.0 m NaCl (3), whereas FGF7 has a lower affinity and was eluted with 0.45 m NaCl (42). Our data support the hypothesis that FGF10 bioactivity is more HS-dependent than FGF7.
Optimal FGF10-mediated BaF3/FGFR2b cell proliferation was greatly reduced when any sulfates were removed from heparin (Fig. 3A), which is similar to that reported for FGF7 with FGFR2b and is in contrast to FGF1 with FGFR2b, which was activated by oligosaccharides shorter than 8 sugar units, and both 6-O- and 2-O-desulfated heparin (27, 32). High affinity binding of FGF1 to FGFR2b was activated by disaccharides and both de-2-O- and de-6-O-sulfated heparin (43). In our assays, the HS sulfation requirements for BaF3 cell proliferation and SMG epithelial morphogenesis (Fig. 3) were different. Although any removal in sulfates greatly decreased BaF3 cell proliferation, removal of sulfates increased SMG epithelial morphogenesis compared with heparin. However, there is a major difference in these two assays; the BaF3 cell system is HS-free, whereas the SMG epithelial cells have endogenous HS on their cell surface. The endogenous epithelial HS did not support end bud expansion when FGF10 was added, suggesting that the exogenous HS fragments mimic HS found in the mesenchyme in the in vivo situation. This suggests that the SMG epithelial and mesenchymal cells in the intact gland have different HS or that the epithelium alone is unable to cleave or release specific HS from its cell surface that induces FGF10-mediated proliferation. Our recent report suggests that heparanase, which is more abundant in the mesenchyme, may release FGF10 stored in the ECM with an HS fragment to stimulate epithelial morphogenesis (24).
The localization of SMG epithelial proliferation at the tips of the ducts (Fig. 1D) suggests that in the absence of exogenous HS, there is an HSPG expressed at the tip of the elongating duct that binds FGF10 to form a ternary signaling complex with FGFR2b. We have previously shown that FGFR2b protein is localized throughout the epithelium (17), and we plan to identify the proteoglycan at the proliferating epithelial tip. Exogenous HS resulted in generalized proliferation and end bud enlargement, but there must have been spatial regulation of the proliferation, because the duct structure was maintained although FGFR2b is present on the duct, which led us to analyze the expression of Sprouty genes (Fig. 5), negative regulators of FGF signaling. It remains to be determined whether increased expression of a particular Sprouty is present in the duct.
Importantly, we showed that FGF10-dependent SMG epithelial morphogenesis is modulated by the sulfation pattern and not just sulfate density. Heparin decasaccharides with 2-O- sulfation and either an N-or 6-O-sulfate induced end bud expansion, whereas decasaccharides with 6-O- and N-sulfation or 6-O-sulfation alone induced duct elongation (Fig. 4). Others have shown that FGF10 requires 6-O-sulfates to bind an octasaccharide affinity column (44), and interestingly, the enzyme 6-O-sulfotransferase was localized in the distal lung epithelium and mesenchyme at sites where FGF10 was expressed (45). Thus, the saccharides with 6-O-sulfation may bind FGF10 and activate FGFR2b signaling, resulting in duct elongation. However, end bud expansion and clefting required a 2-O-sulfate and another sulfate on the other sugar ring, either an N-or 6-O-sulfate. These results show that specific HS structures may modulate FGF10-dependent biological processes resulting in proliferation, morphogenesis, and differentiation (see the model in Fig. 8), suggesting that although there is some specificity in the location, there is also some flexibility in the patterns of sulfation that induce end bud formation. Specific patterns of HS modification may provide an important mechanism whereby ligands with similar FGFR binding properties elicit different biological responses in vivo, and there is probably some redundancy built into the system.
A “two-end” model was proposed from the crystal structure of an FGFR·FGF complex and a heparin fragment, and in this model of the ternary signaling complex, the heparin-binding sites of FGF and FGFR bind two HS chains. The differences between the heparin-binding sites of FGFs and FGFRs, together with ligand-induced changes in FGFR conformation, result in unique HS binding sites specific for individual FGF·FGFR complexes that bind HS, which may be expressed in a cell- or tissue-specific or a developmentally regulated manner (45, 46). Potentially, the HS in the ternary FGF·FGFR signaling complex could be attached to an HSPG on the same cell (in cis), localized on another cell or in the ECM (in trans), a cleavage product of an endogenous HSPG, or a combination of any of these. The decasaccharides in our assays may have influenced both FGF10 and FGF1 binding to FGFR2b and also FGF1 binding to FGFR1b (see the model in Fig. 8). It is not possible to distinguish the effects of the decasaccharides on either of these FGFs or on other FGFs that may be produced by the epithelium.
In conclusion, the biological activities of FGF10 can be modulated by sulfated HS structures. We speculate that the regulation of the bioactivity of FGF10 is influenced by the expression of HSPGs and the enzymes that modify their HS structures as well as the temporal expression patterns of these enzymes during development, which result in specific HS structures in localized cell environments. The data presented here and our previous report (24) suggest that the epithelium and the mesenchyme in the intact SMG may contain different HS structures. Proteoglycans in the ECM, such as perlecan, would thus have HS chains with distinct sulfation patterns and GAG chain length, depending on where they were synthesized, and their modification by heparanase may also alter their function. Future HS analysis of proteoglycans from both the epithelium and the mesenchyme is required. Understanding how HS structures modulate FGF10 binding, storage in the ECM, and formation of ternary signaling complexes with FGFR2b may assist in future design of HS mimetics to modulate FGF10 function for therapeutic applications.
Acknowledgments
We thank Sarah Knox and Ivan Rebustini for critical reading of the manuscript and Dr. John T. Gallagher for the gift of HS reagents.
This work was supported by the Intramural Research Program of the NIDCR, National Institutes of Health, and by grants from the DKFZ, Israel Science Foundation, and Ministry of Health (to D. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: FGF, fibroblast growth factor; FGFR, FGF receptor; HS, heparan sulfate; ECM, extracellular matrix; AU, arbitrary units; SMG, submandibular gland.
References
- 1.Martin, G. R. (1998) Genes Dev. 12 1571-1586 [DOI] [PubMed] [Google Scholar]
- 2.Ornitz, D. M. (2000) BioEssays 22 108-112 [DOI] [PubMed] [Google Scholar]
- 3.Igarashi, M., Finch, P. W., and Aaronson, S. A. (1998) J. Biol. Chem. 273 13230-13235 [DOI] [PubMed] [Google Scholar]
- 4.Yan, G., Fukabori, Y., McBride, G., Nikolaropolous, S., and McKeehan, W. L. (1993) Mol. Cell. Biol. 13 4513-4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miki, T., Fleming, T. P., Bottaro, D. P., Rubin, J. S., Ron, D., and Aaronson, S. A. (1991) Science 251 72-75 [DOI] [PubMed] [Google Scholar]
- 6.Avivi, A., Yayon, A., and Givol, D. (1993) FEBS Lett. 330 249-252 [DOI] [PubMed] [Google Scholar]
- 7.Lindahl, U., Kusche-Gullberg, M., and Kjellen, L. (1998) J. Biol. Chem. 273 24979-24982 [DOI] [PubMed] [Google Scholar]
- 8.Perrimon, N., and Bernfield, M. (2000) Nature 404 725-728 [DOI] [PubMed] [Google Scholar]
- 9.Schlessinger, J., Plotnikov, A. N., Ibrahimi, O. A., Eliseenkova, A. V., Yeh, B. K., Yayon, A., Linhardt, R. J., and Mohammadi, M. (2000) Mol. Cell 6 743-750 [DOI] [PubMed] [Google Scholar]
- 10.Pellegrini, L., Burke, D. F., von Delft, F., Mulloy, B., and Blundell, T. L. (2000) Nature 407 1029-1034 [DOI] [PubMed] [Google Scholar]
- 11.Ford-Perriss, M., Guimond, S. E., Greferath, U., Kita, M., Grobe, K., Habuchi, H., Kimata, K., Esko, J. D., Murphy, M., and Turnbull, J. E. (2002) Glycobiology 12 721-727 [DOI] [PubMed] [Google Scholar]
- 12.Min, H., Danilenko, D. M., Scully, S. A., Bolon, B., Ring, B. D., Tarpley, J. E., DeRose, M., and Simonet, W. S. (1998) Genes Dev. 12 3156-3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sekine, K., Ohuchi, H., Fujiwara, M., Yamasaki, M., Yoshizawa, T., Sato, T., Yagishita, N., Matsui, D., Koga, Y., Itoh, N., and Kato, S. (1999) Nat. Genet. 21 138-141 [DOI] [PubMed] [Google Scholar]
- 14.Ohuchi, H., Hori, Y., Yamasaki, M., Harada, H., Sekine, K., Kato, S., and Itoh, N. (2000) Biochem. Biophys. Res. Commun. 277 643-649 [DOI] [PubMed] [Google Scholar]
- 15.Entesarian, M., Matsson, H., Klar, J., Bergendal, B., Olson, L., Arakaki, R., Hayashi, Y., Ohuchi, H., Falahat, B., Bolstad, A. I., Jonsson, R., Wahren-Herlenius, M., and Dahl, N. (2005) Nat. Genet. 37 125-127 [DOI] [PubMed] [Google Scholar]
- 16.Jaskoll, T., Abichaker, G., Witcher, D., Sala, F. G., Bellusci, S., Hajihosseini, M. K., and Melnick, M. (2005) BMC Dev. Biol. 5 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinberg, Z., Myers, C., Heim, V. M., Lathrop, C. A., Rebustini, I. T., Stewart, J. S., Larsen, M., and Hoffman, M. P. (2005) Development (Camb.) 132 1223-1234 [DOI] [PubMed] [Google Scholar]
- 18.Rebustini, I. T., Patel, V. N., Stewart, J. S., Layvey, A., Georges-Labouesse, E., Miner, J. H., and Hoffman, M. P. (2007) Dev. Biol. 308 15-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeh, B. K., Igarashi, M., Eliseenkova, A. V., Plotnikov, A. N., Sher, I., Ron, D., Aaronson, S. A., and Mohammadi, M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 2266-2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel, V. N., Rebustini, I. T., and Hoffman, M. P. (2006) Differentiation 74 349-364 [DOI] [PubMed] [Google Scholar]
- 21.Vlodavsky, I., Miao, H. Q., Medalion, B., Danagher, P., and Ron, D. (1996) Cancer Metastasis Rev. 15 177-186 [DOI] [PubMed] [Google Scholar]
- 22.Nugent, M. A., and Iozzo, R. V. (2000) Int. J. Biochem. Cell Biol. 32 115-120 [DOI] [PubMed] [Google Scholar]
- 23.Sperinde, G. V., and Nugent, M. A. (1998) Biochemistry 37 13153-13164 [DOI] [PubMed] [Google Scholar]
- 24.Patel, V. N., Knox, S. M., Likar, K. M., Lathrop, C. A., Hossain, R., Eftekhari, S., Whitelock, J. M., Elkin, M., Vlodavsky, I., and Hoffman, M. P. (2007) Development (Cambr.) 134 4177-4186 [DOI] [PubMed] [Google Scholar]
- 25.Yates, E. A., Guimond, S. E., and Turnbull, J. E. (2004) J. Med. Chem. 47 277-280 [DOI] [PubMed] [Google Scholar]
- 26.Guimond, S. E., and Turnbull, J. E. (1999) Curr. Biol. 9 1343-1346 [DOI] [PubMed] [Google Scholar]
- 27.Berman, B., Ostrovsky, O., Shlissel, M., Lang, T., Regan, D., Vlodavsky, I., Ishai-Michaeli, R., and Ron, D. (1999) J. Biol. Chem. 274 36132-36138 [DOI] [PubMed] [Google Scholar]
- 28.Kreuger, J., Spillmann, D., Li, J. P., and Lindahl, U. (2006) J. Cell Biol. 174 323-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jastrebova, N., Vanwildemeersch, M., Rapraeger, A. C., Gimenez-Gallego, G., Lindahl, U., and Spillmann, D. (2006) J. Biol. Chem. 281 26884-26892 [DOI] [PubMed] [Google Scholar]
- 30.Gitay-Goren, H., Soker, S., Vlodavsky, I., and Neufeld, G. (1992) J. Biol. Chem. 267 6093-6098 [PubMed] [Google Scholar]
- 31.Reich-Slotky, R., Shaoul, E., Berman, B., Graziani, G., and Ron, D. (1995) J. Biol. Chem. 270 29813-29818 [DOI] [PubMed] [Google Scholar]
- 32.Ostrovsky, O., Berman, B., Gallagher, J., Mulloy, B., Fernig, D. G., Delehedde, M., and Ron, D. (2002) J. Biol. Chem. 277 2444-2453 [DOI] [PubMed] [Google Scholar]
- 33.Hoffman, M. P., Engbring, J. A., Nielsen, P. K., Vargas, J., Steinberg, Z., Karmand, A. J., Nomizu, M., Yamada, Y., and Kleinman, H. K. (2001) J. Biol. Chem. 276 22077-22085 [DOI] [PubMed] [Google Scholar]
- 34.Merry, C. L., Lyon, M., Deakin, J. A., Hopwood, J. J., and Gallagher, J. T. (1999) J. Biol. Chem. 274 18455-18462 [DOI] [PubMed] [Google Scholar]
- 35.Hoffman, M. P., Kidder, B. L., Steinberg, Z. L., Lakhani, S., Ho, S., Kleinman, H. K., and Larsen, M. (2002) Development (Cambr.) 129 5767-5778 [DOI] [PubMed] [Google Scholar]
- 36.de Vega, S., Iwamoto, T., Nakamura, T., Hozumi, K., McKnight, D. A., Fisher, L. W., Fukumoto, S., and Yamada, Y. (2007) J. Biol. Chem. 282 30878-30888 [DOI] [PubMed] [Google Scholar]
- 37.Rapraeger, A. C., Guimond, S., Krufka, A., and Olwin, B. B. (1994) Methods Enzymol. 245 219-240 [DOI] [PubMed] [Google Scholar]
- 38.Wei, C., Larsen, M., Hoffman, M. P., and Yamada, K. M. (2007) Tissue Eng. 13 721-735 [DOI] [PubMed] [Google Scholar]
- 39.Yamaguchi, Y., Ogura, S., Ishida, M., Karasawa, M., and Takada, S. (2005) Dev. Dyn. 233 484-495 [DOI] [PubMed] [Google Scholar]
- 40.Yamaguchi, Y., Yonemura, S., and Takada, S. (2006) Development (Cambr.) 133 4737-4748 [DOI] [PubMed] [Google Scholar]
- 41.Zhang, X., Ibrahimi, O. A., Olsen, S. K., Umemori, H., Mohammadi, M., and Ornitz, D. M. (2006) J. Biol. Chem. 281 15694-15700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ron, D., Bottaro, D. P., Finch, P. W., Morris, D., Rubin, J. S., and Aaronson, S. A. (1993) J. Biol. Chem. 268 2984-2988 [PubMed] [Google Scholar]
- 43.Ornitz, D. M., Xu, J., Colvin, J. S., McEwen, D. G., MacArthur, C. A., Coulier, F., Gao, G., and Goldfarb, M. (1996) J. Biol. Chem. 271 15292-15297 [DOI] [PubMed] [Google Scholar]
- 44.Ashikari-Hada, S., Habuchi, H., Kariya, Y., Itoh, N., Reddi, A. H., and Kimata, K. (2004) J. Biol. Chem. 279 12346-12354 [DOI] [PubMed] [Google Scholar]
- 45.Izvolsky, K. I., Shoykhet, D., Yang, Y., Yu, Q., Nugent, M. A., and Cardoso, W. V. (2003) Dev. Biol. 258 185-200 [DOI] [PubMed] [Google Scholar]
- 46.Mohammadi, M., Olsen, S. K., and Goetz, R. (2005) Curr. Opin. Struct. Biol. 15 506-516 [DOI] [PubMed] [Google Scholar]