Abstract
The 2′,3′-dideoxy-3′-thiacytidine drug-resistant M184I HIV-1 reverse transcriptase (RT) has been shown to synthesize DNA with decreased processivity compared with the wild-type RT. M184A displays an even more severe processivity defect. However, the basis of this decreased processivity has been unclear, and both primer-template binding and dNTP interaction defects have been proposed to account for it. In this study, we show that the altered properties of the M184I and M184A RT mutants that we have measured, including decreased processivity, a slower rate of primer extension, and increased strand transfer activity, can all be explained by a defect in dNTP utilization. These alterations are observed only at low dNTP concentration and vanish as the dNTP concentration is raised. The mutant RTs exhibit a normal dissociation rate from a DNA primer-RNA template while paused during synthesis. Slower than normal synthesis at physiological dNTP concentration, coupled with normal dissociation from the primer-template, results in the lowered processivity. The mutant RTs exhibit normal DNA 3′-end-directed and RNA 5′-end-directed ribonuclease H activity. The reduced rate of DNA synthesis causes an increase in the ratio of ribonuclease H to polymerase activity thereby promoting increased strand transfer. These latter results are consistent with an observed higher rate of recombination by HIV-1 strains with Met-184 mutations.
HIV-13 reverse transcriptase (RT) is a heterodimer consisting of a 66-kDa subunit (p66) and a 51-kDa subunit (p51). The p51 and p66 subunits share a common N terminus; the p51 subunit lacks 120 amino acids from the C terminus of the p66 subunit. Active sites for polymerase and ribonuclease H (RNase H) reside in p66. The highly conserved motif YXDD in the polymerase active site of HIV-1 and other retroviral RTs is essential for polymerase activity (1). In HIV-1 RT, Met-184 is the second amino acid in this conserved YXDD motif (2). Two mutations, M184I and M184V, arise in HIV-1 RT after treatment with the RT inhibitor 2′,3′-dideoxy-3′-thiacytidine, or 3TC (3–5), and confer high level resistance to 3TC. 3TC is a nucleoside analog that lacks the 3-OH of the ribose ring; incorporation of 3TC blocks viral DNA synthesis. It is the l-pseudo sugar version of 3TC that is used to treat patients. The M184I/V mutations cause steric hindrance between the oxathiolane ring of 3TCTP and the β-branched side chain of valine or isoleucine, which hinders incorporation of 3TCTP (6–8). Wild-type MLV RT has valine in the second position of the YXDD motif (Val-223), and wild-type MLV is resistant to 3TC (9). Replacing the valine at position X in the YXDD motif of the WT MLV RT with methionine (V223M) makes the virus more susceptible to 3TC, and the MLV RT more sensitive to 3TCTP (10). Both HIV-1 and MLV RTs with an alanine substitution at the X position are moderately resistant to 3TC in vitro, even though alanine is not a β-branched amino acid (10, 11). This suggests that other factors, such as altered positioning of the template primer in the RT active site, also contribute to 3TC resistance (10). Mutation of the HIV-1 RT in response to 3TC therapy produces drug resistance, but also alters other properties of the RT. New properties might be helpful, harmful, or neutral with respect to the welfare of the infected patient. For this reason, it is important to understand the biochemical characteristics of 3TC-resistant forms of HIV-1 RT. This has been a goal of previous and current work.
Most polymerases exhibit processive synthesis, adding a number of nt each time they bind a primer-template. Processivity is the quantitative parameter of processive synthesis, indicating the average number of nucleotides added each time the RT engages the primer (12). The M184V mutant of HIV-1 RT has, relative to the wild-type enzyme, reduced processivity (13–16). The M184I RT mutant is even less processive (15–17). It has been suggested that the reduced processivity of M184I contributes to the reduced fitness of viruses carrying this mutation and to the selection, in vivo, of M184V (8). HIV-1 RT structure determinations show that the YMDD motif lies on the surface of the palm domain and is part of the polymerase active site. A type II′ turn connects β7 to β8 and allows the side chains of these four amino acids to interact with the primer-template, metal cofactor and the dNTP (8). Previously, x-ray structural comparison analyses of complexes of the wild-type and M184I mutant RTs showed differences in the positioning of the primer-template (8). Specifically the last few nt of the primer are distorted by the mutant RT. The 3′-OH is moved 1.5 Å out of place. There are also changes on the template position and the β12–β13 “primer grip” loop of the RT. These observations suggest that the repositioning of the primer-template accounts for the decreased processivity (8).
Mutations at position 184 have also been shown to increase the Michaelis constant (Km) for dNTP utilization by HIV-1 RT (18). This is indicative of a dNTP binding defect. The decreased ability to bind dNTPs would be expected to reduce the primer elongation rate, affecting the measured processivity value. Moreover, the reduced processivity of Met-184 mutants was exacerbated by limiting dNTP concentrations (16, 17). It was unclear from these results whether the effects of the mutations on dNTP utilization were a major determinant of the measured processivity defect. An alternative is that the mutant RT forms have a defect that causes premature dissociation even if dNTP concentrations were raised to a point where the mutant and wild-type RTs elongate primers at the same rate.
Additional evidence suggests that the M184I mutation also influences strand transfer in HIV (19). HIV-1 packages two copies of its viral genome in the virion. Strand transfer occurs when the RT begins DNA synthesis on one template and switches to the other. Two strand transfers are required for the synthesis of linear viral DNA: minus strand strong stop transfer and plus strand transfer (20). Additional stand transfers occur during viral DNA synthesis; these can cause genetic recombination if the two RNA genomes in the virion differ in sequence. Such recombination events promote viral diversity and contribute to the development of drug resistance (20–23). An RNase H-minus mutant of RT does not make products that can participate in transfer (24, 25), indicating that degradation of the RNA template is necessary for strand transfer.
The ratio of polymerization and RNase H activities determines the efficiency of strand transfer (26). Slow polymerization provides an opportunity for the RNase H to carry out additional cleavages of the RNA template, increasing the number of strand transfers; reducing the RNase H activity results in fewer strand transfers (26). A mutation that reduces the rate of DNA polymerization without affecting RNase H activity would be expected to promote strand transfer in vitro and recombination in vivo. Nikolenko and colleagues recently demonstrated that HIV-1 containing the drug-resistant M184I RT had an increased recombination frequency compared with the wild-type RT (19). This observation supports an expectation that the M184I mutation promotes strand transfer in vitro.
In this study, we have attempted to understand the consequences of mutations at position 184 in HIV-1 RT. We found that the M184I and M184A mutant HIV-1 RTs, relative to wild-type, display reduced processivity and primer elongation rate, and increased strand transfer. However, these altered properties can all be explained by the effects of the mutations on the decreased utilization of dNTPs.
EXPERIMENTAL PROCEDURES
Materials—HIV-1 nucleocapsid protein NCp7-(1–72) was chemically synthesized as described (27). DNA oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA). Oligo(dT)-poly(rA) was purchased from Amersham Biosciences. The [γ-32P]ATP (6000 Ci/mmol) was purchased from PerkinElmer Life Sciences. All other enzymes and dNTP solutions were obtained from Roche Applied Science.
Preparation of HIV-1 RTs—Wild-type HIV-1 reverse transcriptase and the Met-184 mutants, M184I and M184A, were prepared as p66/p51 heterodimers as described previously (6, 10). The relative amounts of each RT preparation were normalized by their relative specific activities of polymerization. The specific activities of all of the RTs were determined by measuring the rate of dTTP incorporation using oligo(dT)-poly(rA) as the primer-template. The reaction was performed in 50 mm Tris-HCl, 50 mm NaCl, 1 mm EDTA, 1 mm DTT, 98.75 μm dTTP, 1.25 μm [α-32P]dTTP, and 300 fmol of oligo(dT)-poly(rA) duplex in each 10-μl reaction. The ratio of oligo(dT)∼200 to poly(rA)12–18 is ∼1:1. Three different amounts of each enzyme were tested to make sure that the oligo(dT)-poly(rA) was in considerable excess over the amount of enzyme used. 2 μl of each reaction mixture was spotted onto DE-81 paper, which was washed by 0.1 m sodium phosphate (dibasic) five times to remove unincorporated radioactivity, and counts per minute were determined by scintillation counting. One unit is defined as the amount of enzyme required to incorporate 1 nmol of dTTP on this template in a 10-min reaction mixture at 37 °C. The specific activities of the wild-type, M184I, and M184A RT were 56,480, 48,240, and 45,430 units/mg, respectively.
Preparation of the RNA Templates—The D520 RNA template, which contains the first 520 nt from the 5′-end of the HIV-1 NL4-3 genome, used for the processivity and rate of polymerization assays, was generated as previously described (28). The weakly structured substrates used for the transfer activity assay, donor RNA template Dpol, 184 nt in length, and acceptor RNA template Apol, 227 nt in length, were generated as previously described (29). The sequence of this and the following templates is provided in the cited references. The 41-nt RNA template (30) used for the dissociation rate (koff) and intrinsic RNase H assays was purchased from Integrated DNA Technologies. All RNA templates were purified using polyacrylamide/urea gel electrophoresis. RNAs were quantitated by UV absorption using a GeneQuant II from Amersham Biosciences.
Preparation of Substrates—To 5′-end label the RNA templates, purified RNA was first treated with calf intestinal phosphatase to remove the 5′-phosphate using the manufacturer's protocol. Then the RNA templates or DNA primers were 5′-end-labeled using [γ-32P]ATP and polynucleotide kinase. Unincorporated ATP was removed using a Bio-Rad P-30 Micro BioSpin size-exclusion column. The RNA template and DNA primer were mixed in a 1:2 ratio and annealed by incubation at 95 °C for 5 min, followed by slow cooling to room temperature.
Processivity Assays—Processivity of the RTs was measured using the D520 RNA template annealed with the 5′-end 32P-labeled DNA primer lg31 (5′-GCCTTCTGATGTCTCTAAAAGGCCAGGA). RTs were pre-bound to the substrates at room temperature for 5 min prior to the initiation of the reaction by addition of MgCl2, dNTPs, and the oligo(dT)-poly(rA) trap. The final 12-μl reaction mixture contained 4 nm primer, 2 nm donor RNA, 2 units of RT (25 nm WT, 28 nm M184I or 30 nm M184A), 5 μg (∼1000-fold excess over the substrates) oligo(dT)-poly(rA), 50 mm Tris-HCl (pH 8.0), 6 mm MgCl2, 50 mm KCl, 1 mm dithiothreitol, 1 mm EDTA and dNTP concentrations as described. The reactions were performed at 5 μm, 50 μm, 100 μm, and 200 μm dNTPs and sampled at 20 min. To verify that the oligo(dT)-poly(rA) can trap RT effectively, a control experiment was performed in which the oligo(dT)-poly(rA) was preincubated with substrates prior to RT addition. A second control reaction was performed in the absence of the oligo(dT)-poly(rA) allowing multiple rounds of extension. Products were resolved by 6% electrophoresis on a polyacrylamide/urea gel and visualized using a Storm PhosphorImager (GE Healthcare).
Determination of koff (Dissociation Rate Constants) for the Primer-Template—The value of koff was determined by preincubating HIV-1 RT with the substrates that the 41-nt RNA template (30) was annealed to the 5′-end 32P-labeled 24-nt DNA primer (5′-TAGAGGATCCCCGGGTACCGAGCT). Oligo(dT)-poly(rA) was added, and the incubation was continued for various times in the absence of dNTPs so that any RT that dissociated from the substrate would be trapped by the oligo(dT)-poly(rA). RT that remained bound to substrate at various times was quantitated by primer extension. The assay was performed as previously described with slight modifications (31). After incubation with the trap mix, poly(rA)-oligo(dT), for 0–80 s, MgCl2, dCTP, dGTP, and ddATP were added to initiate the reaction. The ddATP limits synthesis to 3 nt. The final 12-μl reaction mixture contained 2 nm substrate, 2 units of RT (25 nm WT, 28 nm M184I, or 30 nm M184A), 1.5 μg (∼1000-fold excess over the substrates) of poly(rA)-oligo(dT), 50 mm Tris-HCl (pH 8.0), 6 mm MgCl2, 50 mm KCl, 1 mm dithiothreitol, 1 mm EDTA, and 1 μm of each dCTP, dGTP, and ddATP. Products were resolved on a 10% polyacrylamide/urea gel, visualized using a Storm PhosphorImager (GE Healthcare) and quantitated with ImageQuaNT software v. 1.2 (GE Healthcare). The dissociation rate was determined by graphing the total remaining capacity for incorporation versus time. Results were fitted to an equation for single-exponential decay (Y = e-kofft), where Y is incorporation at time t relative to incorporation at time 0.
DNA Polymerization and Strand Transfer Assay—For DNA polymerization assays, the D520 template and lg31 primer described earlier, and the Dpol template and the MB22 primer (29) were used. Transfer assays used the Dpol template, with weak pausing, and the MB22 primer (29). In all cases the donor template and primer were annealed as described above and preincubated with RT at room temperature for 5 min prior to the initiation of the reactions by addition of MgCl2 and dNTPs. The final reaction mixtures contained 2 nm substrates, 50 mm Tris-HCl (pH 8.0), 6 mm MgCl2, 50 mm KCl, 1 mm dithiothreitol, 1 mm EDTA, 2 units of RT (25 nm wild-type, 28 nm M184I, or 30 nm M184A), and varying dNTPs concentrations as described. For the RT proteins, the rate of synthesis was obtained by dividing the maximum number of nt incorporated by the reaction time. For strand transfer assays, the acceptor RNA template Apol was used at a final concentration of 8 nm. Transfer reactions and DNA polymerization using Dpol template and MB22 primer were performed in the presence of NC. Sufficient NC to provide 200% coverage of the RNA templates (2 NC per 7 nt) was added to the mixture prior to the addition of RT and incubated at room temperature for 5 min. All reactions were incubated at 37 °C and terminated by addition of 2× termination dye mix (20 mm EDTA (pH 8.0), 90% formamide, and 0.1% each of bromphenol blue and xylene cyanole). Reaction products were separated on a 6% polyacrylamide/urea gel and visualized and quantitated as described above. The Two-Sample t-Test was used to compare the transfer efficiency of the wild-type RT and M184I mutant, and the wild-type RT and M184A RT mutant. The significance level of the test (denoted α) was set to 0.05.
RNase H Assays—3′-End DNA-directed RNase H assays and 5′-end RNA-directed RNase H assays were performed as previously described with slight modifications (30). The ratio of the 5′-end 32P-labeled 41-nt RNA template to the 24-nt DNA primer or the 77-nt DNA primer (30) was 1:2. The reactions were performed using identical reaction conditions as the DNA polymerization assay described above, except 0.1 unit of RT was prebound with the substrates, and the reaction was initiated by addition of 6 mm MgCl2. Reaction products were separated on a 10% polyacrylamide/urea gel and visualized and quantitated as described above.
RESULTS
The drug-resistant mutant of HIV-1 RT M184I, which is usually the first resistance mutation to appear in patients treated with 3TC, displays a moderate decrease in processivity compared with the WT RT (15–17). Another mutant engineered at the same position, M184A, is even less processive and is partially resistant to 3TC (10, 11). Our initial goal was to determine the biochemical deficiency that causes this reduction in processivity.
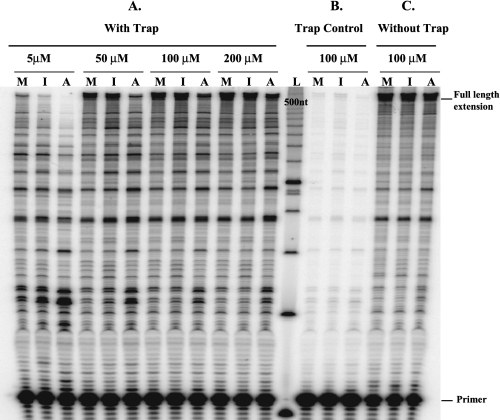
The Processivity Defect of the Met-184 RT Mutants Is Minimized at High dNTP Concentrations—To compare the processivity of the WT and mutant RTs, DNA synthesis was carried out in reactions that contained a polymer trap, which sequesters any RT that dissociates from the substrate. We used a 520-nt RNA template derived from the 5′-end region of the HIV-1 genome. A 5′-end-labeled DNA primer was annealed to the 3′-end of the template (see “Materials and Methods”). Equivalent amounts of wild-type and mutant RT proteins, as determined from specific activity measurements on a oligo(dT)-poly(rA) template with 100 μm dTTP, were used in each experiment. This equalized any preparation-related differences in polymerization. The RT was bound to the primertemplate, and DNA synthesis was initiated by the addition of magnesium, dNTPs, and excess oligo(dT)-poly(rA) trap (Fig. 1A). The size of the DNA products increased with time until all of the RT had fallen off the primer-template and had been sequestered by the trap (data not shown). The final products then represent the distribution of primer extension lengths from one round of RT binding and DNA synthesis (Fig. 1A). The distribution of product lengths on this long RNA template is a very sensitive indicator of processivity.
FIGURE 1.
The dNTP concentration dependence of the processivity of M184I and M184A HIV-1 RTs. A, the processivity assays of the RT proteins were performed at dNTP concentrations of 5, 50, 100, and 200 μm each. The substrates used for the processivity assays were made by annealing a 5′-end-labeled DNA primer to a 520-nt RNA template (see “Experimental Procedures”). Equivalent amounts of each RT protein normalized by specific activity were prebound to the substrate, and the reaction was initiated by addition of Mg2+, dNTPs, and the oligo(dT)-poly(rA) trap. Each reaction was performed in duplicate and terminated at 20 min. B, for the trap control, the reaction conditions were the same as for the processivity assay except the oligo(dT)-poly(rA) trap was added before RT addition. Only the reaction at 100 μm of each dNTP is shown. C, the extension reaction was also performed without the trap mix under the same reaction conditions. Only the reaction at 100 μm of each dNTP is shown. L, 25-bp DNA ladder.
Assays performed with 5 μm of each dNTP showed that, relative to wild-type RT, M184I has a substantially lower processivity, whereas the M184A mutant had an even lower processivity than M184I. At 50 μm of each dNTP, the processivity of M184I and wild-type RT were similar, whereas M184A still showed a somewhat decreased processivity. At 100 and 200 μm of each dNTP some of the products are of full length, but 80% or more are still a distribution of partial extensions. Based on the distribution of product sizes, all three proteins displayed similar processivity. The effectiveness of the trap was verified by preincubating RT with trap before the addition of substrate, magnesium, and dNTPs. Little to no primer extension was observed under these conditions (Fig. 1B), showing that the trap efficiently sequestered the RT, preventing it from re-binding the labeled substrate. A reaction performed in the absence of a trap showed that the wild-type and mutant RTs synthesized more fully extended products than they did in the presence of a trap. The reaction with 100 μm of each dNTP is shown (Fig. 1C).
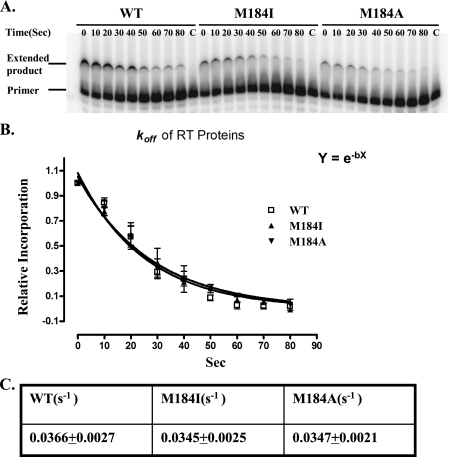
Both Met-184 RT Mutants Exhibit the Same Dissociation Rate from the Primer-Template as Wild-type RT—One possible explanation for the decreased processivity of the mutant RTs is that they dissociate more rapidly from the primer-template than wild-type RT. We measured the dissociation rate (koff) of RT bound to a primer-template. koff was determined by preincubating the RT-primer-template complex with excess oligo(dT)-poly(rA) for progressively longer times before magnesium and dNTPs were added to initiate the polymerase reaction. Because the dissociation of the enzyme from the DNA-RNA substrate was slow relative to the rate of polymerization, the amount of extended product was proportional to the amount of substrate-bound RT remaining over time (Fig. 2A). The dissociation rate was determined by constructing a graph of the total extension as the time of incubation with trap increased (Fig. 2B). Values of koff were calculated after fitting the data to a single exponential decay equation. All three of the RTs showed similar koff values (Fig. 2C). This suggests that the observed decrease in processive synthesis is not the result of premature dissociation of the mutant RTs from the primer-template.
FIGURE 2.
Determination of the dissociation rate constants of the HIV-1 RTs. A, a typical dissociate rate experiment is shown. The substrate used was a 24-nt DNA primer annealed to a 41-nt RNA template. Experiments were performed in the presence of excess trap, oligo(dT)-poly(rA). The time axis indicates how long after the oligo(dT)-poly(rA) trap was added until the Mg2+ and dNTPs were added to initiate DNA synthesis. The amount of the extended products was proportional to the amount of RT remaining bound at each time point. Lane C represents a control reaction in which the trap was added before the RT. B, the amount of primer extension seen at increasing times of incubation with the trap is shown relative to the amount of primer extension when there was no preincubation with the trap. Nonlinear least-square fits of the data from three independent experiments to an equation for single-exponential decay are shown. C, the dissociation rate constants were calculated for the three HIV-1 RTs.
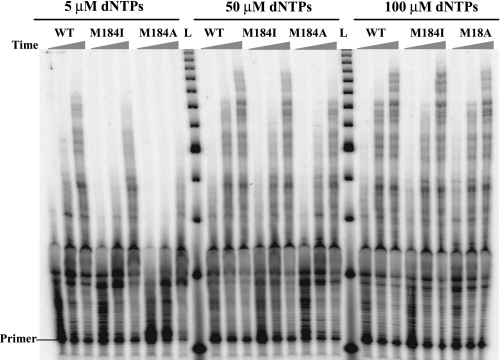
Met-184 RT Mutants Exhibit a Reduced Rate of Primer Elongation at Low dNTP Concentrations—An alternative explanation for the reduced processivity is that the mutant RTs extend the primer more slowly. This would decrease the number of nt added even if the RT is bound to the primer-template for the same length of time. To measure the maximum rate of primer elongation at specific dNTP concentrations, we performed DNA extension reactions for 15, 30, and 60 s using the 520-nt RNA template (Fig. 3). RT molecules were in ∼10-fold excess over the substrate so that any RTs that dissociated from the primer terminus during synthesis would be rapidly replaced. Under these conditions, nt addition instead of RT dissociation should be rate-limiting. Extension reactions were carried out at different dNTP concentrations. The rate of polymerization was calculated as the number of nt added to the substrate divided by the reaction time. The longest primers made at 60 s were used to calculate the polymerization rate. At 5 μm of each dNTP the rate of synthesis for wild-type RT was 2.55 nt/s, and for M184I and M184A the polymerization rates were 2.03 and 1.28 nt/s, respectively. When reactions were performed at 50 μm of each dNTP, M184I and WT showed similar polymerization rates of 3.6 and 3.7 nt/s, respectively. However, M184A still showed a slower rate of polymerization, 2.5 nt/s. At 100 μm of each dNTP all three enzymes had similar polymerization rates of ∼3.7 nt/s. These data are consistent with a previous report showing that the Km values for incorporation by dNTPs are much higher for M184A RT than for wild-type RT (18). The M184V RT had an intermediate values for Km (18). Although Km is not a direct measure of dNTP binding, the higher Km values suggest that the mutations reduced the dNTP binding affinity.
FIGURE 3.
Effect of dNTP concentration on DNA synthesis by the HIV-1 RTs. A DNA primer labeled at the 5′-end was annealed to a 520-nt RNA template, and extension assays were performed with excess wild-type, M184I, and M184A RTs at dNTP concentrations of 5, 50, and 100 μm each. The reactions were stopped at 15, 30, and 60 s. L, 25-bp DNA ladder.
The calculated polymerization rates of the RTs at more than 50 μm of each dNTP are ∼3.7 nt/s, which are consistent with our previous report of 3.3 nt/s maximum polymerization rate for the WT HIV-1 RT on a native RNA template (32). This is slow compared with the rates of polymerization usually detected in pre-steady-state single-turnover experiments (40 ∼ 70 nt/s) (33, 34). The likely reason is that the RT pauses at many locations during long distance synthesis, greatly lowering the average rate of primer extension.
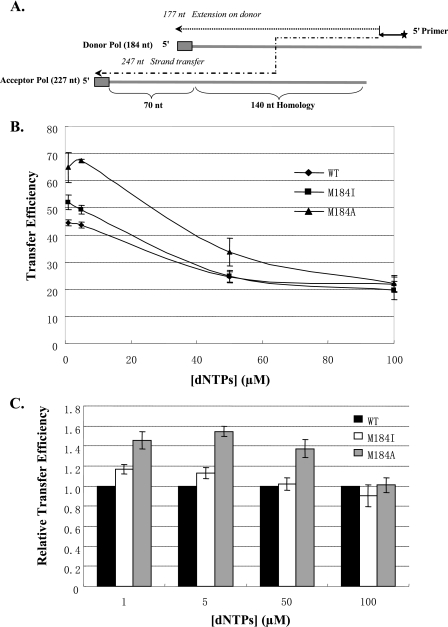
M184I and M184A Display Higher Strand Transfer Efficiency Only at Low dNTP Concentrations—An important characteristic of HIV-1 RT is the ability to generate the substrates for strand transfer. Transfer requires both the polymerization and RNase H activities of the RT. For strand transfer to occur, RT must degrade the RNA template of a DNA-RNA duplex so that the newly synthesized DNA primer can anneal to another RNA template. The “dynamic copy choice” model of strand transfer asserts that transfer is favored by a high ratio of RNase H cleavage to polymerase activity (26). In this model, conditions and mutations that selectively reduce the polymerization rate of the RT, without affecting the RNase H activity, should enhance transfer. To measure the efficiency of strand transfer, we used a previously described transfer system with donor and acceptor templates derived from the HIV-1 pol coding region (29), illustrated in Fig. 4A. A DNA primer is extended on the 184-nt RNA donor template making a 177-nt donor extension product (DE). In the presence of the 227-nt acceptor RNA template, which shares 140 nt of homology with the donor template, strand transfer leads to the synthesis of a 247-nt transfer product (TP). Based on their specific activity, equivalent amounts of RT were used.
FIGURE 4.
Strand transfer activity of wild-type and mutant HIV-1 RTs. A, schematic of the substrate is shown. For donor extension reactions, a 5′-end-labeled DNA primer was annealed to a 184-nt donor RNA. A 177-nt donor extension product (DE) was formed when the DNA primer synthesized to the 5′-end of the donor (small dotted line). For transfer reactions, a second 227-nt RNA template acceptor, which shares 140 nt of homology with the donor, was added to the reaction. A 247-nt transfer product (TP) was produced when the DNA transferred from donor template to the acceptor (thick dotted line). End transfers are prevented by a 16-nt non-viral sequence at the 5′-end of the donor template (gray box). B, the transfer efficiencies of wild-type and mutant HIV-1 RTs are presented at varied dNTP concentrations. Strand transfer reactions were performed with 1, 5, 50, and 100 μm of each dNTP in the presence of NC, added to a concentration sufficient to give 200% coating (2 NC molecules per 7 nt), with wild-type, M184I, and M184A RTs. All reactions were terminated after 30 min. The transfer efficiency was calculated using the equation (TE = (TP/(TP + DE)) × 100%). Values were averaged from at least three experiments. C, the relative transfer efficiencies of mutant HIV-1 RTs to wild-type RT were indicated.
Transfer efficiency (TE) is a quantitative measure of how frequently the primer terminus switches from the donor to the acceptor template and was calculated as TE = (TP/(TP + DE)) × 100%. As previously observed with the wild-type RT (25), transfer efficiency of all of the RTs rose as the concentration of dNTPs was lowered. This is the result of a lowering of the polymerization/RNase H ratio as the primer elongation rate decreased. At 1 and 5 μm dNTPs, the transfer efficiencies of the Met-184 mutant RTs were higher than that of the wild-type RT. The greatest observed difference was, as anticipated, with the more severe M184A mutation (Fig. 4B). Compared with wild-type RT, the transfer efficiency of M184A was 46% higher (p = 0.003) at 1 μm dNTPs and 55% higher (p < 0.001) at 5 μm (Fig. 4C). More interesting, M184I also showed significant difference compared with wild-type RT at low dNTPs concentration, 17% higher (p = 0.012) at 1 μm dNTPs and 13% higher (p = 0.007) at 5 μm (Fig. 4C). At 50 μm dNTPs, the transfer efficiencies of the wild-type and M184I mutant RTs were similar, but the value for M184A RT was 37% higher (p = 0.045) than that of wild-type RT (Fig. 4C). By 100 μm dNTPs, all differences disappeared. The general outcome is that the M184I RT transfers more efficiently than the wild-type RT, and the M184A RT transfers considerably more efficiently than either the wild-type or M184I RT. However, the differences in transfer efficiency disappear at high dNTP concentration. At low dNTP concentration the polymerase/RNase H ratio must be considerably lower for the mutant compared with the wild-type RTs. This outcome would be expected if the sole defect of the mutant RTs was in dNTP binding.
Met-184 RT Mutants Have the Same Intrinsic RNase H Activities as the Wild-type RT—In view of the increased strand transfer efficiency and decreased rate of primer elongation observed with the mutant RTs, the dynamic copy choice model would predict that the mutations have little or no effect on the RNase H activity. Both the DNA 3′-end-directed cleavages produced by polymerizing RT, and RNA 5′-end directed cleavage produced by non-polymerizing RTs were examined.
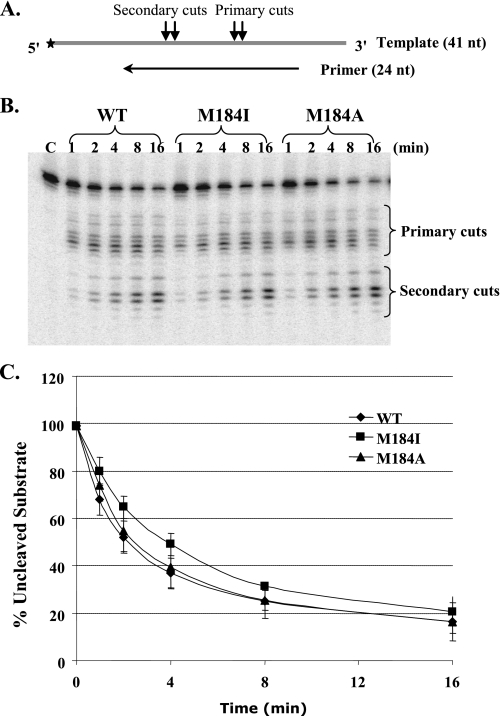
The 3′-end-directed RNase H activity was tested using a 5′-end-labeled 41-nt RNA annealed to an unlabeled 24-nt DNA primer. In the duplex the 3′-end of the DNA was recessed relative to the 5′-end of the RNA (Fig. 5A). Equivalent amounts of RT activity were used based on the polymerase-specific activity. Magnesium was added to initiate the reaction in the absence of dNTPs. All of the RTs made two sets of cleavages, primary cuts which were clustered around 18 nt from the 3′-end of the DNA, and secondary cuts, appearing ∼8 nt from the 3′-end of the DNA (Fig. 5B). To compare the RNase H cleavage efficiency of the WT and the Met-184 RT mutants, the percentage of uncleaved RNA substrates (Fig. 5C) and the percentage of the secondary cut products (data not shown) were measured, and the amounts of these RNAs were plotted over time for each enzyme. The Met-184 RT mutants and WT enzyme showed similar DNA 3′-end-directed RNase H activity. Note that at 100 μm dTTP used for equalizing the RTs by specific activity, the mutant RTs do not display a polymerization defect. Therefore, observed equivalence of RNase H activities indicates that the mutants display normal RNase H activity.
FIGURE 5.
DNA 3′-end-directed RNase H activity of HIV-1 RT proteins. A, the substrates were made by annealing a 24-nt DNA primer to a 5′-end-labeled 41-nt RNA template. The two sequential RNase H cleavages that occur are indicated as primary and secondary cuts. B, the reactions were performed using equivalent amounts of polymerase activity of the wild-type, M184I, and M184A in the absence of dNTPs. Reactions were sampled at different time points, as indicated above the gel. Lane C is a control reaction performed in the absence of RT. C, quantitation of the amount of the uncleaved substrate over time. The percentage of the uncleaved substrate for each time point was calculated as ((uncleaved substrate/(primary cuts + secondary cuts + uncleaved substrate)) × 100%). Results represent the average of three independent experiments.
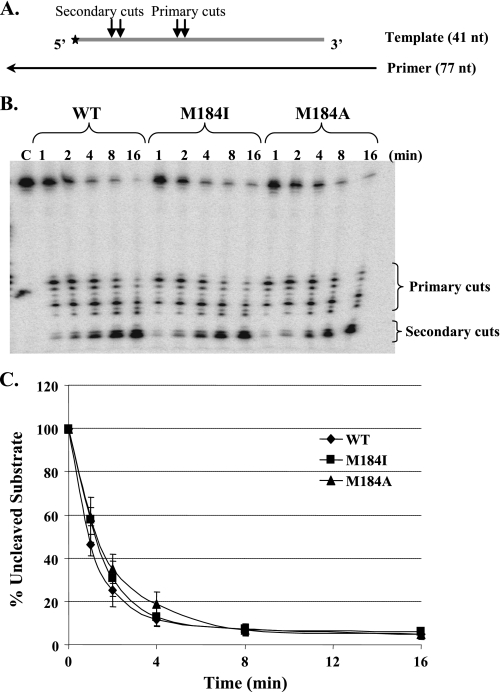
RNA 5′-end-directed RNase H activity was measured using a 5′-end-labeled 41-nt RNA annealed to an unlabeled 77-nt DNA primer. In the duplex the 5′-end of the RNA was recessed relative to the 3′-end of the DNA (Fig. 6A). On this RNA, RT also made two clusters of cuts similar to those seen with the 3′-end-directed cleavage, also referred to as primary and secondary (Fig. 6B). The percentage of uncleaved RNA substrates (Fig. 6C) and the percentage of the secondary cleavage products (data not shown) were again plotted over time. The Met-184 RT mutants and wild-type enzyme showed similar RNA 5′-end-directed RNase H activity. The similar rates, and cleavage patterns, for both 3′-DNA- and 5′-RNA-directed cleavages suggest that the increased transfer efficiency is not the result of an effect of the mutations on RNase H activity or specificity.
FIGURE 6.
RNA 5′-end-directed RNase H activity of HIV-1 RT proteins. A, the substrates used were made by annealing a 77-nt DNA primer to a 5′-end-labeled 41-nt RNA template. Two sequential RNase H cleavages occur that are indicated as the primary and secondary cuts. B, the reactions were performed using equivalent amounts of polymerase activity of the wild-type, M184I, and M184A in the absence of dNTPs. Reactions were sampled at different time points, as indicated above the gel. Lane C is a control reaction in the absence of RT. C, quantitation of the amount of uncleaved substrate over time. The percentage of uncleaved substrate for each time point was calculated as ((uncleaved substrate/(primary cuts + secondary cuts + uncleaved substrate)) × 100%). Results represent the average of three independent experiments.
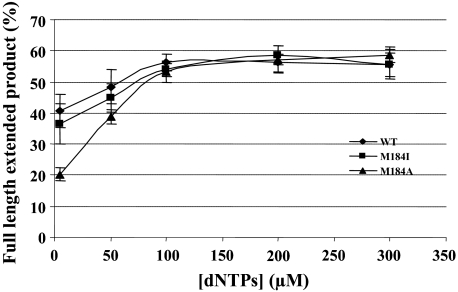
Reduced DNA Synthesis by Met-184 RT Mutants Still Occurs When Template Structure Is Minimized by NC—Lastly, we considered the possibility that at least some of the primer elongation deficiency of the mutant RTs resulted from defects in their ability to synthesize DNA on a structured template. We quantified the amount of the 177-nt full-length donor extension product of the pol template with an amount of NC that should have been sufficient to produce 200% coating at varied dNTP concentrations (Fig. 7). NC is expected to relieve secondary structure in the template, facilitating strand displacement. The pol template is already weakly structured (29) compared with the 520-nt template representing the natural 5′-end region of HIV-1, and the presence of NC should reduce the structure even further. The amount of RNA-dependent DNA polymerization was similar at concentrations of dNTPs over 100 μm each. The amount of fully extended product was ∼55% of the total products for all the enzymes tested. When dNTP concentrations were less than 100 μm dNTPs, the Met-184 mutants carried out increasingly inefficient DNA synthesis. At 50 μm dNTPs, the amount of fully extended product decreased to 48.4% for wild-type RT, 44.7% for M184I, and 38.8% for M184A. This difference was accentuated at 5 μm dNTPs, with just 40.7% for wild-type RT, 36.5% for M184I, and 20.4% for M184A of total products being full-length donor extension. These results represent a similar reduction in primer elongation rate as that measured with the 520-nt template. Apparently, the observed effects of the mutations are related to defects in dNTP utilization, rather than any inability of the mutant RTs to cope with specific template features.
FIGURE 7.
Effect of dNTP concentration on DNA synthesis by HIV-1 RTs in the presence of NC. The graphs show the percentage of extension products that are fully synthesized to the end of the donor template at different dNTP concentrations. Reactions were performed with the weakly structured substrate in the presence of sufficient NC to produce 200% coating of the template. Values are from the average of at least three experiments.
DISCUSSION
We previously showed that M184I HIV-1 RT had a decreased processivity and M184A was even less processive (15). A mutation in RT can decrease processivity by slowing the rate of polymerization without altering the rate of dissociation of the RT from the primer-template. Alternatively, the mutation could increase the rate of dissociation without altering the rate of polymerization, or the mutation could affect both processes. In the current work we explored the basis of the altered processivity seen in the Met-184 mutations.
Prasad and colleagues have suggested that processivity is influenced by the interaction of the enzyme and the primer-template. They reported several mutations in HIV-1 RT that affect the dissociation of RT from the primer-template (35). These include FE20 and FE103, which are insertions of a short series of amino acids into the β3–β4 fingers subdomain of HIV-1 RT. These mutant RTs displayed normal polymerization activity but were 25–30% more processive than the wild-type RT, linking structural changes affecting primer-template interactions with processivity. Our previous analysis of a complex of the mutant M184I RT and a primer-template suggested that repositioning of the primer-template accounts for the decreased processivity (8). However, it was also known that Met-184 mutants of HIV-1 displayed a higher Km value for dNTP utilization than the wild-type RT (18). Therefore, mutations at the Met-184 position have the potential to affect both dNTP utilization and the interactions of RT with the primer-template, and either or both of these defects could decrease processivity.
We report here that the processivity defect of the Met-184 mutant RTs appears to derive completely from a reduction in their ability to utilize dNTPs. This is because increased dNTP concentration restored nearly normal processivity to the mutant RTs (Fig. 1). We also show that the dissociation rates of the mutant RTs from the primer-template (koff) in the absence of DNA synthesis were similar to that of the wild-type RT (Fig. 2). This result suggests that the processivity defect is not at all a consequence of premature dissociation of the RT. The decreased dNTP utilization by the mutant RTs results in a reduction in the primer elongation rate at limiting dNTP concentrations (Fig. 3). Because the primer elongation rate is slower, and the dissociation from the primer-template is the same, the RT dissociates from the primer-template after fewer nt have been incorporated, producing the processivity defect. The primer elongation rate of the mutants also approached that of the wild-type RT as the dNTP concentration was increased (Fig. 3). A processivity or elongation defect caused by a problem with binding of the primer-template would not be fully corrected by increasing dNTP concentration. Taken together, these observations are consistent with the interpretation that the processivity and synthesis defects are caused solely by the dNTP utilization defect.
The accompanying report (36) includes experiments that probe the basis of the dNTP utilization defect in a patient-derived M184I RT. Pre-steady state measurements show that the binding affinity (Kd) for each of the four dNTPs is ∼10-fold higher for M184I. However, the kpol values, representing the conformational change and catalysis steps of synthesis, are unaltered by the mutation. These results indicate that dNTP utilization of M184I RT is impaired specifically at low dNTP concentrations, because the entry of dNTP to the active site of the M184I RT becomes rate-limiting at dNTP concentrations at which dNTP binding to the wild-type RT is still efficient.
We recently showed that weak dNTP binding by the HIV-1 RT mutants V148I and Q151N promoted an increase in strand transfer (37). This observation agrees with the predictions of the dynamic copy choice model for recombination proposed by Pathak and colleagues. In that model, the efficiency of strand transfer is promoted by a higher ratio of RNase H cleavage over nt addition (26). The dNTP binding defect caused by the V148I and Q151N mutations decreased the polymerization rate on a donor RNA template but did not affect RNase H activity. The reduced rate of polymerization led to a relative increase in the RNase H cleavages in the donor template, which increased the likelihood of an interaction with acceptor RNA leading to transfer.
We suspected that observed increases in strand transfer by Met-184 mutant RTs occurred by a similar mechanism. Assays that measured both the DNA 3′-end-directed and the RNA 5′-end-directed RNase H cleavages showed that the levels of RNase H activity of the M184I and M184A mutants were similar to that of wild-type RT (Figs. 5 and 6). As with the dNTP-binding RT mutants V148I and Q151N, strand transfer efficiency of both of the Met-184 mutants approached that of wild-type RT at saturating dNTP concentrations. Taken together these data indicate that the M184I and M184A mutations increase transfer efficiency by decreasing the rate of primer elongation. The differences in this rate are eliminated at high dNTP concentrations, demonstrating that the dNTP utilization defect is the sole cause of the change in transfer efficiency.
Our results are consistent with a previous report that HIV-1 containing the drug-resistant M184I RT has an increased recombination frequency, compared with the wild-type virus, in the human embryonic kidney cell line 293T (26.6% versus 13.5%) (19). It is likely that these cells have a dNTP concentration in the range of 5 μm each, similar to that found for most established transformed lines such as HeLa cells (38). The cellular dNTP concentrations of the primary HIV-1 target cells, activated CD4+ T cells and terminally differentiated macrophage, are in the range of 0.05 μm to 4 μm (39). The levels of dNTPs found in these cells are sufficiently low that Met-184 mutant viruses would be expected to either fail at infection, or have a reduced rate of viral DNA synthesis, which would lead to a higher rate of recombination than the wild-type virus. This suggests that drug resistance mutations that alter dNTP utilization can directly influence the ability of the virus to recombine. Therefore, RT properties that may be desirable or undesirable for the infected patient are a consequence of the mutations that cause drug resistance. The dNTP utilization defect appears to make the virus less fit for replication, but more effective at recombination which could promote viral diversity.
Consistent with the prediction that HIV-1 with wild-type versus M184I RT will respond differently at low concentration cellular dNTPs we investigated the relative infectivity of HIV-1 vectors in human macrophage in the accompanying report (36). The macrophages were found to support infection by vector expressing wild-type but not M184I RT.
NC, which relieves template structure, did not change the dNTP concentration-dependent reduction primer elongation observed with the M184I or M184A mutant RTs (Fig. 7). Our data indicate that, at the normal cellular dNTP concentrations, the M184I RT would be expected to have decreased DNA synthesis activity (Fig. 7) compared with the wild-type RT either in the presence or the absence of NC. This indicates that template structure does not have a major impact on the slowing of synthesis by the mutant RTs at low dNTP concentration.
The fact that MI84V HIV-1 RT has a less severe processivity defect than the M184I is thought to account for its selection in patients (8). The equilibrium dissociation constant (Kd) for dCTP interaction with HIV-1 M184V RT was found to be about three times higher than with WT RT (40). The Kd value for dCTP binding to M184I RT was shown to be ∼9-fold higher than for WT RT in the accompanying report (36). These observations suggest that the biochemical properties of M184V also result from a dNTP utilization defect, but one that is not as severe as that observed with M184I or M184A RTs.
In summary, the tested mutations at the Met-184 position displayed defects in primer elongation rate, lower processivity, and increased strand transfer. We show that all of these effects can be attributed to defective dNTP utilization. Increased recombination seen with HIV-1 M184I in cell culture (19) is consistent with this interpretation.
Acknowledgments
We thank Dr. Vandana Purohit for critical reading of the manuscript. We also thank Dr. Vandana Purohit, Min Song, and Sean Rigby for helpful discussions.
This work was supported by National Institutes of Health grant GM 49573 to R.A.B. and by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HIV-1, human immunodeficiency virus-1; RT, reverse transcriptase; NC, nucleocapsid protein; DE, donor extension product; FB, foldback product; TP, transfer product; TE, transfer efficiency; 3TC, 2′,3′-dideoxy-3′-thiacytidine; nt, nucleotide(s); WT, wild-type; MLV, Moloney leukemia virus; DTT, dithiothreitol; RNase H, ribonuclease H; 3TCTP, 2′,3′-dideoxy-3′-thiacytidine triphosphate.
References
- 1.Poch, O., Sauvaget, I., Delarue, M., and Tordo, N. (1989) EMBO J. 8 3867-3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larder, B. A., Purifoy, D. J., Powell, K. L., and Darby, G. (1987) Nature 327 716-717 [DOI] [PubMed] [Google Scholar]
- 3.Tisdale, M., Kemp, S. D., Parry, N. R., and Larder, B. A. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 5653-5656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao, Q., Gu, Z., Parniak, M. A., Cameron, J., Cammack, N., Boucher, C., and Wainberg, M. A. (1993) Antimicrob. Agents Chemother. 37 1390-1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boucher, C. A., Cammack, N., Schipper, P., Schuurman, R., Rouse, P., Wainberg, M. A., and Cameron, J. M. (1993) Antimicrob. Agents Chemother. 37 2231-2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao, H. Q., Boyer, P. L., Sarafianos, S. G., Arnold, E., and Hughes, S. H. (2000) J. Mol. Biol. 300 403-418 [DOI] [PubMed] [Google Scholar]
- 7.Huang, H., Chopra, R., Verdine, G. L., and Harrison, S. C. (1998) Science 282 1669-1675 [DOI] [PubMed] [Google Scholar]
- 8.Sarafianos, S. G., Das, K., Clark, A. D., Jr., Ding, J., Boyer, P. L., Hughes, S. H., and Arnold, E. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 10027-10032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halvas, E. K., Svarovskaia, E. S., Freed, E. O., and Pathak, V. K. (2000) J. Virol. 74 6669-6674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyer, P. L., Gao, H. Q., Clark, P. K., Sarafianos, S. G., Arnold, E., and Hughes, S. H. (2001) J. Virol. 75 6321-6328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Julias, J. G., Boyer, P. L., McWilliams, M. J., Alvord, W. G., and Hughes, S. H. (2004) Virology 322 13-21 [DOI] [PubMed] [Google Scholar]
- 12.Bambara, R. A., Uyemura, D., and Choi, T. (1978) J. Biol. Chem. 253 413-423 [PubMed] [Google Scholar]
- 13.Sharma, P. L., and Crumpacker, C. S. (1999) J. Virol. 73 8448-8456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, H. Q., Boyer, P. L., Arnold, E., and Hughes, S. H. (1998) J. Mol. Biol. 277 559-572 [DOI] [PubMed] [Google Scholar]
- 15.Boyer, P. L., and Hughes, S. H. (1995) Antimicrob. Agents Chemother. 39 1624-1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Back, N. K., Nijhuis, M., Keulen, W., Boucher, C. A., Oude Essink, B. O., van Kuilenburg, A. B., van Gennip, A. H., and Berkhout, B. (1996) EMBO J. 15 4040-4049 [PMC free article] [PubMed] [Google Scholar]
- 17.Back, N. K., and Berkhout, B. (1997) Antimicrob. Agents Chemother. 41 2484-2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson, J. E., Aulabaugh, A., Caligan, B., McPherson, S., Wakefield, J. K., Jablonski, S., Morrow, C. D., Reardon, J. E., and Furman, P. A. (1996) J. Biol. Chem. 271 13656-13662 [DOI] [PubMed] [Google Scholar]
- 19.Nikolenko, G. N., Svarovskaia, E. S., Delviks, K. A., and Pathak, V. K. (2004) J. Virol. 78 8761-8770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Telesnitsky, A., and Goff, S. P. (1997) in Retroviruses (Coffin, J. M., Hughes, S. H., and Varmus, H. E., eds) pp. 121-160, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 21.Hu, W. S., and Temin, H. M. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 1556-1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodrich, D. W., and Duesberg, P. H. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 2052-2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clavel, F., Hoggan, M. D., Willey, R. L., Strebel, K., Martin, M. A., and Repaske, R. (1989) J. Virol. 63 1455-1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanese, N., Telesnitsky, A., and Goff, S. P. (1991) J. Virol. 65 4387-4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeStefano, J. J., Mallaber, L. M., Rodriguez-Rodriguez, L., Fay, P. J., and Bambara, R. A. (1992) J. Virol. 66 6370-6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svarovskaia, E. S., Delviks, K. A., Hwang, C. K., and Pathak, V. K. (2000) J. Virol. 74 7171-7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Rocquigny, H., Ficheux, D., Gabus, C., Fournie-Zaluski, M. C., Darlix, J. L., and Roques, B. P. (1991) Biochem. Biophys. Res. Commun. 180 1010-1018 [DOI] [PubMed] [Google Scholar]
- 28.Song, M., Balakrishnan, M., Chen, Y., Roques, B. P., and Bambara, R. A. (2006) J. Biol. Chem. 281 24227-24235 [DOI] [PubMed] [Google Scholar]
- 29.Balakrishnan, M., Fay, P. J., and Bambara, R. A. (2001) J. Biol. Chem. 276 36482-36492 [DOI] [PubMed] [Google Scholar]
- 30.Wisniewski, M., Balakrishnan, M., Palaniappan, C., Fay, P. J., and Bambara, R. A. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 11978-11983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeStefano, J. J., Bambara, R. A., and Fay, P. J. (1993) Biochemistry 32 6908-6915 [DOI] [PubMed] [Google Scholar]
- 32.Fuentes, G. M., Rodriguez-Rodriguez, L., Palaniappan, C., Fay, P. J., and Bambara, R. A. (1996) J. Biol. Chem. 271 1966-1971 [DOI] [PubMed] [Google Scholar]
- 33.Skasko, M., Weiss, K. K., Reynolds, H. M., Jamburuthugoda, V., Lee, K., and Kim, B. (2005) J. Biol. Chem. 280 12190-12200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng, J. Y., and Anderson, K. S. (1999) Biochemistry 38 9440-9448 [DOI] [PubMed] [Google Scholar]
- 35.Kew, Y., Olsen, L. R., Japour, A. J., and Prasad, V. R. (1998) J. Biol. Chem. 273 7529-7537 [DOI] [PubMed] [Google Scholar]
- 36.Jamburuthugoda, V. K., Santos-Velazquez, J. M., Skasko, M., Operario, D. J., Purohit, V., Chugh, P., Szymanski, E. A., Wedekind, J. E., Bambara, R. A., and Kim, B. (2008) J. Biol. Chem. 283 9206-9216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Operario, D. J., Balakrishnan, M., Bambara, R. A., and Kim, B. (2006) J. Biol. Chem. 281 32113-32121 [DOI] [PubMed] [Google Scholar]
- 38.Traut, T. W. (1994) Mol. Cell Biochem. 140 1-22 [DOI] [PubMed] [Google Scholar]
- 39.Diamond, T. L., Roshal, M., Jamburuthugoda, V. K., Reynolds, H. M., Merriam, A. R., Lee, K. Y., Balakrishnan, M., Bambara, R. A., Planelles, V., Dewhurst, S., and Kim, B. (2004) J. Biol. Chem. 279 51545-51553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ray, A. S., Murakami, E., Peterson, C. N., Shi, J., Schinazi, R. F., and Anderson, K. S. (2002) Antiviral Res. 56 189-205 [DOI] [PubMed] [Google Scholar]