Abstract
BI-1 (Bax inhibitor-1) is an evolutionarily conserved multitransmembrane protein that resides in the endoplasmic reticulum (ER) and that has documented cytoprotective functions in both animals and plants. Recent studies indicate that BI-1 shares in common with Bcl-2/Bax family proteins the ability to regulate the amounts of Ca2+ that can be released from the ER by agents, such as the ER-Ca2+-ATPase (SERCA) inhibitor thapsigargin (TG). Using an ER-targeted, Ca2+ indicator (cameleon), with characteristics optimized for measuring ER Ca2+ ([Ca2+]er), we studied the effects of BI-1 on [Ca2+]er in resting and TG-treated cells. Similar to cells overexpressing antiapoptotic Bcl-2 or Bcl-XL, overexpression of BI-1 resulted in lower resting [Ca2+]er, with concomitantly less Ca2+ released into the cytosol upon stimulation by TG and with a higher rate of Ca2+ leakage from the ER. Co-expression of SERCA restored levels of [Ca2+]er to normal, showing opposing actions of the ER-Ca2+ATPase and BI-1 on ER Ca2+ homeostasis. Conversely, cells with deficient BI-1 have increased [Ca2+]er, and release more Ca2+ into the cytosol when challenged with TG. In BI-1-deficient cells, Bcl-XL fails to reduce [Ca2+]er, indicating that BI-1 functions downstream of Bcl-XL. In bax-/-bak-/- double knock-out cells, both BI-1 and Bcl-XL retained their ability to reduce [Ca2+]er, suggesting that BI-1 and Bcl-XL operate downstream of or parallel to Bax/Bak. The findings reveal a hierarchy of functional interactions of BI-1 with Bcl-2/Bax family proteins in regulating ER Ca2+ homeostasis.
Cell death is essential for development of most multicellular organisms, and its dysregulation contributes to many diseases characterized by inappropriate cell loss or cell accumulation. Bcl-2/Bax family proteins are central regulators of cell death in animals. Many Bcl-2/Bax family members reside in the membranes of mitochondria, where they play well defined roles in controlling release of apoptosis-inducing proteins from these organelles and regulating other aspects of mitochondrial function (1).
Besides mitochondria, some members of the Bcl-2/Bax family also localize to membranes of the ER, where their functions are less clear at present. Overexpression of antiapoptotic Bcl-2 or Bcl-XL or ablation of proapoptotic Bax and Bak has been variably reported to result in lower basal concentrations of free Ca2+ in the ER ([Ca2+]er) and to reduce the amount of Ca2+ released from this organelle in response to SERCA inhibitors or agonists of inositol triphosphate-regulated Ca2+ channels (IP3Rs)4 (2-8). Conversely, overexpression of proapoptotic Bax protein or knocking down the level of Bcl-2 by antisense increases ([Ca2+]er), and augments the amount of releasable ER Ca2+ (9, 10). The molecular mechanism by which Bcl-2/Bax family proteins modulate ER Ca2+ is unknown but could be due either to the intrinsic channel-forming properties of these proteins (11-13) or to interactions with ER Ca2+ channel proteins, such as SERCA (14-16) or IP3R1 (10, 17-20).
BI-1 (Bax inhibitor 1), also known as TEGT (21), was identified as a suppressor of Bax-induced cell death in a yeast-based screen for mammalian cytoprotective proteins (22). BI-1 is an evolutionarily conserved, multitransmembrane protein that resides in the ER in both animal and plant cells (22-25) and that displays cytoprotective activity when overexpressed in animal or plant cells (23, 26-31). Antisense-mediated knockdown of BI-1 expression increases sensitivity of animal and plant cells to certain stress-induced forms of cell death (22, 26). Targeted ablation of the bi-1 gene in mice results in increased sensitivity to cell death induced by ischemia-reperfusion injury and pharmacological inducers of ER stress (32, 33), whereas ablation of the gene encoding BI-1 in plants increases sensitivity to several biotic and abiotic stimulators of cell death (34). Similar to Bcl-2/Bax family proteins, a role for BI-1 in regulating the amounts of Ca2+ released from ER by SERCA inhibitors, such as TG, was recently demonstrated using BI-1-overexpressing and bi-1-/- cells (32, 35) and subsequently confirmed in plant cells (25). Moreover, BI-1 has been reported to associate with Bcl-2 and Bcl-XL, based on co-immunoprecipitation experiments (22), suggesting the possibility of collaborations among these proteins. Using genetically engineered cell lines, we show that BI-1 functions downstream of Bcl-2 family proteins in a hierarchical pathway regulating ER Ca2+ homeostasis.
EXPERIMENTAL PROCEDURES
Plasmids—The plasmids, pcDNA3 BI-1 HA, pcDNA3 myc-Bcl-XL, pcDNA3 SERCA2b, ER-cameleon, and pMito-YC2, have been described (8, 32, 36-38). To generate retroviral expression construct pCLXSN/BI-1-HA, pcDNA3-BI-1-HA was digested with XhoI to release the BI-1-HA fragment, which was then filled in with Klenow, digested with EcoRI, and inserted into the pCLXSN vector (Imgenex, 10041P), using standard molecular cloning methods.
Cell Culture—Early passage HeLa cells (P5-P15) were obtained from ATCC (Manassas, VA) and cultured in Dulbecco's modified Eagle's medium low glucose (1 g/liter; Invitrogen) supplemented with 10% fetal bovine serum, 1 mm l-glutamine, and antibiotics. HeLa cells were transfected by Fugene 6 (Roche Applied Science). Mouse embryo fibroblast (MEF) cells were cultured in Dulbecco's modified Eagle's medium high glucose (4.5 g/liter) (Irvine Scientific) supplemented with 10% fetal bovine serum, 1 mm l-glutamine, and antibiotics. Immortalized bax-/-bak-/- double knock-out MEFs were provided by S. J. Korsmeyer (6). Immortalized bi-1-/- and bi-1+/+ MEFs (32) were generated by infecting cells with SV40 T antigen-producing retrovirus in the presence of 8 μg/ml Polybrene for 3 h and then diluting the Polybrene to 2 μg/ml and culturing for an additional 3 days. The infected cells were then split at 1:20 and selected for single colonies in medium containing 1 mg/ml G418 for 10 days. The MEF cells were then transfected by Fugene 6 (Roche Applied Science) or infected with protein-encoding retroviruses (see below) for imaging studies.
Stable HeLa cell lines with BI-1 knockdown (by short hairpin RNA (shRNA)) were generated using retrovirus infection. 293T cells were first transfected (using Lipofectamine 2000) with 2 μg of VSVG, 4 μg of cytomegalovirus-gag polymerase, and 5 μg of either BI-1 shRNA (Open Biosystems, V2SH_153887) or a control shRNA to produce viruses that can express the corresponding shRNA. Virus-containing medium was collected, filtered, and used to infect early passage HeLa cells. The infected cells were then diluted and selected for single colonies in medium containing 1 μg/ml puromycin for 15 days. BI-1 knockdown was confirmed by reverse transcription (RT)-PCR. Retroviruses were generated by the same method as for infecting MEFs.
Ca2+ Imaging—HeLa cells (4 × 105) were plated on 35-mm glass-bottom dishes (MatTek, Ashland, MA) and transfected the next day using Fugene 6 (Roche Applied Science) with 0.08 μg of ER-cameleon-encoding plasmid and either 1 μg of pcDNA3 control (“neo”), 1 μg of pcDNA3-BI-1-HA, or 0.5 μg of pcDNA3-BI-1-HA and 0.5 μg of pcDNA3-SERCA2b. To obtain mitochondrial Ca2+, HeLa cells were transfected with 0.3 μg of Mito-YC2-encoding plasmid and 1.7 μg of pcDNA3 control (“vector”) or 1.7 μg of pcDNA3-BI-1-HA. MEF cells (8 × 104 to 1.6 × 105) were plated and transfected with 0.4 μg of ER-cameleon-encoding plasmid and 1.6 μg of pcDNA3 control (“vector”), 1.6 μg of pcDNA3-BI-1-HA or pcDNA3-myc-Bcl-XL.
Cells were imaged 2-3 days after transfection using either a Zeiss Axiovert microscope with a cooled CCD camera, controlled by MetaFluor version 6.1 software (Universal Imaging, Downington, PA), or an inverted Olympus IX81 fluorescence microscope equipped with a thermo-controlled stage (Warner Instruments Inc., Hamden, CT) with oil immersion objective UAPO ×40/340, numerical aperture 1.35-0.65, fitted with a cooled Cascade 512B camera (Photometrics, Inc., Tucson, AZ) and employing MetaFluor software, version 7.1.4 (Molecular Devices, Downingtown, PA). The cells were imaged either in Ca2+-containing or Ca2+-free Hanks' balanced salt solution (HBSS) (Invitrogen), containing 20 mm HEPES (pH 7.4), 1-2 g/liter d-glucose (osmolarity ∼300). Dual emission ratio imaging of the cameleons was accomplished with the Zeiss system using a 436DF20 excitation filter, a 450DRLP dichroic mirror, and two emission filters (475DF30 for ECFP, 535DF25 for EYFP) controlled by a Lambda 10-2 filter wheel (Sutter Instrument Co., Novato, CA). With the Olympus system, we used ultra-high speed wavelength switcher DG4 (Sutter Instrument Co.) with the D436/20× excitation filter, with 455 long pass dichroic mirror, and D480/40m and D535/30m emission filters for cyan-emitting fluorescent protein (CFP) and yellow-emitting fluorescent protein (YFP), respectively, installed in a Sutter Lambda 10B filter wheel. All filters were obtained from Chroma Technology Co. (Rockingham, VT). For cameleon imaging, the yellow and cyan intensities were measured 5-7 times every 15 s, and the average ratios were calculated after background subtraction for each individual cell. Mito-YC2 cells were imaged using the same parameters as the ER-cameleon cells. The ER-cameleon calibration was performed as described (8). To deduce [Ca2+]er, Rmin was obtained by treating the cells with 3 mm EGTA and 2 μm ionomycin, and Rmax was determined by treating the cells with 25 μm digitonin, followed by 5-10 mm Ca2+, 1 mm ATP, and 1 mm Mg2+. The Rmin and Rmax values were then used to convert cell data to [Ca2+]er as described previously (8).
Fura-2-based imaging of cytosolic Ca2+ was performed as described previously (8). In brief, the cells were incubated in HBSS containing 4 μm Fura-2/AM (Molecular Probes, Inc., Eugene, OR) with 0.04% Pluronic F-127 for 30 min at room temperature in the dark. Cells were then washed with HBSS and incubated for 15 min before changing to fresh HBSS for imaging. Excitation ratio imaging for Fura-2 was accomplished by using 350- and 380-nm excitation filters (both 10-nm bandwidths), and (for the Zeiss system) a 450-nm dichroic mirror with a 535/45 emission filter or (for the Olympus system) a 400 long pass dichroic mirror with a D340X filter. To calibrate Fura-2, Rmin was obtained by treating the cells with 8 μm ionomycin and 10 mm EGTA in Ca2+-deficient HBSS, and Rmax was obtained by treating the cells with 2 μm ionomycin and 20 mm Ca2+. The 350/380 nm ratio was then converted to [Ca2+]i, as described (39).
Immunoblotting—Total protein was isolated from HeLa cells homogenized in detergent-containing buffer, normalized for protein content (50 μg/sample), and analyzed by SDS-PAGE (8-16% gels) and immunoblotting using the following primary antibodies: anti-HA (3F10; Roche Applied Science), anti-Bax (40), anti-Bcl-2, anti-Bcl-XL (41), anti-Grp94 (SPA-851; Stressgen Biotechnologies), anti-tubulin (catalog number 317; Santa Cruz Biotechnology), anti-calreticulin (GTX20004; GeneTex), anti-SERCA2 (GTX30342; GeneTex), and anti-IP3R1, -2, and -3 (sc-6093/7277/7278; Santa Cruz Biotechnology, Inc., Santa Cruz, CA). Antibody detection was accomplished by using horseradish peroxidase-conjugated secondary antibodies (anti-mouse IgG or anti-rabbit IgG; Amersham Biosciences) and an enhanced chemiluminescence method (Amersham Biosciences).
RT-PCR—Total RNAs were isolated from cells using the RNeasy kit (74104; Qiagen), reverse-transcribed using the Superscript RT-PCR kit (10928-034; Invitrogen), following the manufacturer's instructions. cDNAs were then used as template for PCR amplification using a BI-1-specific primer pair as well as a CPH-specific primer pair as control.
Cell Death Assay—HeLa and MEF cell lines were treated with 2 μm TG or 1.0 μg/ml TRAIL in Dulbecco's modified Eagle's medium low glucose (1 g/liter; Invitrogen) supplemented with 10% fetal bovine serum, 1 mm l-glutamine, and antibiotics for 12 h to induce apoptosis. Both floating and attached cells were then collected and labeled using the annexin V-fluorescein isothiocyanate/propidium iodide apoptosis detection kit (catalog number K101-100; BioVision), following a protocol provided by the manufacturer. 104 cells from each condition were analyzed by fluorescence-activated cell sorting to identify cells into four groups: annexin-negative/PI-negative, annexin-negative/PI-positive, annexin-positive/PI-negative, and annexin-positive/PI-positive. Aliquots of the collected cells were also suspended at 106 cells/ml in PBS with 2 μg/ml Hoechst dye, and the percentage of cells with apoptotic nuclear morphology was determined by UV microscopy.
RESULTS
BI-1 Alters Steady-state ER Ca2+ Levels—Our previous investigations showed that mammalian BI-1 regulates a cell death pathway linked to ER stress and modulates the levels of releasable Ca2+ from ER, as measured by a Ca2+-sensitive ratiometric dye (Fura-2) that reports cytosolic free Ca2+ concentrations, in experiments where cells were treated with SERCA inhibitor, TG, to release Ca2+ into cytosol (32). Westphalen et al. (35) also measured ER [Ca2+] in BI-1-overexpressing cells using first generation ER-targeted Ca2+-sensitive fluorescent proteins that report [Ca2+] via fluorescence resonance energy transfer. To lay a foundation for our studies on the relation of BI-1 to ER [Ca2+], we compared the effects of BI-1 overexpression and BI-1 knockdown, achieved with shRNAs on steady-state Ca2+ concentrations in the ER ([Ca2+]er).
To directly measure [Ca2+]er, we utilized an improved ER-targeted, Ca2+-sensitive fluorescent protein, termed D1ER-cameleon, that consists of tandem fusions of CFP and YFP, separated by Ca2+-responsive elements (mutant calmodulin and a complementary mutant version of the calmodulin-binding peptide M13) (8). Binding of Ca2+ to the calmodulin moiety induces binding to M13, bringing CFP and YFP into a new orientation that enhances fluorescence resonance energy transfer. The characteristics of the ER-targeted cameleon used here have been reported in detail (8). The cameleon was targeted to ER by the addition of an N-terminal calreticulin signal sequence and C-terminal ER retention sequence (KDEL), and its localization to ER was confirmed by microscopy (supplemental Fig. S1). This improved ER-targeted cameleon has been used to study the effects of Bcl-2 and Bcl-XL on ER Ca2+ dynamics (8), confirming and extending prior studies.
We first tested the effects of BI-1 overexpression on [Ca2+]er using a gene transfer approach, where HeLa cells were co-transfected with plasmids encoding ER-targeted cameleon and either control vector or BI-1-encoding plasmid, measuring fluorescence resonance energy transfer and [Ca2+]er in individual cells by previously established ratiometric methods that normalize for differences in levels of protein production (8, 42, 43). Immunoblotting confirmed expression of the epitope-tagged BI-1 protein in cells transfected with BI-1-HA but not in control-transfected cells (Fig. 1A). Although individual cells varied, on average, BI-1-overexpressing cells contained significantly lower resting [Ca2+]er, as represented by the YFP/CFP ratio (Fig. 1B) (BI-1-HA = 1.85 ± 0.02, n = 24; vector = 2.05 ± 0.02, n = 24, p < 0.01 by Student's t test). For some experiments, we also converted the YFP/CFP ratio to an estimated [Ca2+]er by a calibration method, as described previously (8). Recognizing that accurate calibrations of the maximum YFP/CFP ratio (Rmax) are difficult to obtain due to the toxic effects of permeabilizing cells in the presence of high extracellular Ca2+ (reviewed in Ref. 44), our values of resting ER [Ca2+] in control HeLa cells nevertheless agree well with prior reports (supplemental Fig. S2), based on methods using ER-targeted green fluorescent protein-based cameleons or aequorin (8, 35, 45). Also, it bears noting that whereas the absolute YFP/CFP ratio values varied, depending on which Ca2+ imaging system (microscope) was used, the relative differences in ER-targeted cameleon fluorescence seen for these genetically engineered cells were reproducible.
FIGURE 1.
BI-1 modulates basal ER [Ca2+]. A and B, BI-1-overexpressing cells have lower resting [Ca2+]er compared with control cells. HeLa cells were co-transfected with either pcDNA3-BI-1-HA or control vector pcDNA3 and ER-cameleon-encoding plasmid at a 12:1 ratio. A, lysates were prepared from cells after imaging, normalized for total protein content, and analyzed using antibodies directed against various proteins as indicated. B, cameleon-expressing cells were imaged 2-3 days after transfection to determine the yellow/cyan ratio. Data for individual cells are represented by triangles. YFP/CFP ratios (mean ± S.E.) for BI-1 and control cells were 1.85 ± 0.02 and 2.0 ± 0.02, respectively (n = 24; p < 0.01 by Student's t test). Data are representative of three independent experiments. C and D, BI-1-deficient cells have increased resting [Ca2+]er compared with wild-type cells. HeLa cells were transfected with plasmids encoding BI-1 shRNA or control vector, together with ER-cameleon-encoding plasmid. C, cells were lysed for RNA recovery, and relative levels of BI-1 and CPH-1 (control) mRNA were evaluated by RT-PCR. D, cells were imaged 2-3 days after transfection to determine yellow/cyan ratio. Data for individual cells are represented by triangles. YFP/CFP ratios (mean ± S.E.) for BI-1 knockdown and control cells were 2.16 ± 0.02 and 2.04 ± 0.02, respectively (n = 24; p < 0.01 by Student's t test). Data are representative of three independent experiments.
Next, we knocked down BI-1 expression in HeLa cells using shRNA targeting BI-1 mRNA and confirmed reduced BI-1 mRNA by reverse transcription-PCR (Fig. 1C). (Note that antibodies that detect endogenous BI-1 have not been attainable to date, despite considerable effort, thus precluding analysis of endogenous BI-1 protein.) We used three different shRNA sequences to target BI-1, all three of which were comparably effective (not shown). Comparison of resting [Ca2+]er in control- and shRNA-transfected HeLa cells revealed that average resting [Ca2+]er was significantly higher in BI-1-deficient cells, with BI-1 knockdown cells having an average YFP/CFP ratio of 2.16 ± 0.02 (n = 24) compared with control-transfected cells with an average YFP/CFP ratio of 2.04 ± 0.02 (n = 24) (p < 0.01 by unpaired t test) (Fig. 1D). Additional immunoblotting studies demonstrated that BI-1 overexpression and BI-1 knockdown do not alter the levels of proteins known to modulate [Ca2+]er, including SERCA2, Bax, Bcl-2, Bcl-XL, Grp94, calreticulin, and IP3Rs in HeLa cells (Fig. 2).
FIGURE 2.
BI-1 does not alter expression of ER proteins. Total protein was isolated from HeLa cells (from left to right: control shRNA, BI-1 shRNA, pcDNA3 vector, pcDNA3-BI-1-HA) homogenized in detergent-containing buffer, normalized for protein content (50 μg/sample), and analyzed by SDS-PAGE (8-16% gels) and immunoblotting using various antibodies as indicated. Protein levels were assessed by scanning densitometry, normalized using tubulin as internal control. N/A, not applicable.
The shRNA-mediated reduction in BI-1 expression rendered HeLa cells more sensitive to TG-induced cell killing (Fig. 3), as expected (32). Similar conclusions were reached whether measuring the percentage of annexin V-positive/propidium iodide-negative cells by fluorescence-activated cell sorting (Fig. 3), the percentage of dead cells failing to exclude propidium iodide dye (not shown), or the percentage of cells with apoptotic morphology based on microscopic examination of Hoechst dye-stained cells (not shown). Also, consistent with our prior report (29), BI-1-deficient MEF cells exhibited greater sensitivity to apoptosis induction by TG but not TRAIL (Fig. 3), showing selective vulnerability to ER stress-induced cell death.
FIGURE 3.
BI-1 protects cell from apoptosis under ER stress. HeLa and MEF cell lines were treated with 2 μm TG or 1.0 μg/ml TRAIL for 12 h to induce apoptosis. Cells were then collected and labeled using annexin V-fluorescein isothiocyanate and propidium iodide. 104 cells from each condition were analyzed by fluorescence-activated cell sorting, and the percentage of annexin-positive/propidium iodide-negative cells was calculated. BI-1-deficient cells show a significantly higher percentage of apoptosis compared with the control cells under ER stress (TG) but not following challenge with TRAIL (p < 0.05 by unpaired t test). Data shown are the average ± S.D. of three independent assays. UT, untreated control; TG, thapsigargin.
BI-1 Overexpression Increases Ca2+ Leakage Rate of ER—The altered [Ca2+]er seen in BI-1-overexpressing cells could be due to either decreased uptake of Ca2+ into ER or increased passive Ca2+ leakage from ER. Bcl-2/Bax family proteins have been shown to regulate the Ca2+ leakage rate (8, 35, 45). If a functional relation exists between BI-1 and Bcl-2/Bax family proteins with respect to [Ca2+]er regulation, therefore, we predict that BI-1 also should increase the leakage rate.
To answer this question, we used TG to inhibit SERCA-mediated uptake of Ca2+ into the ER, comparing the rate of Ca2+ loss from this organelle in BI-1-overexpressing and control-transfected cells, based on cameleon fluorescence measurements (Fig. 4). Measurements were performed for individual cells in Ca2+-containing versus Ca2+-deficient medium, the latter intended to exclude the effects of capacitative Ca2+ entry, which refills ER Ca2+ from import of extracellular Ca2+ (2, 5). In both control and BI-1-overexpressing cells, TG induced loss of Ca2+ from ER. Although BI-1-overexpressing cells started with approximately half the [Ca2+]er of control cells, TG induced emptying of ER Ca2+ to similar extents, resulting in similar final [Ca2+]er in control and BI-1-overexpressing cells in both Ca2+-containing and Ca2+-deficient medium (Fig. 4 and supplemental Fig. S3).
FIGURE 4.
Effects of TG on [Ca2+]er in BI-1 overexpressing cells. HeLa cells were transfected with 0.08 μg of ER-cameleon-encoding plasmid and either 1 μg of pcDNA3 control (neo), 1 μg of pcDNA3-BI-1-HA, or 0.5 μg of pcDNA3-BI-1-HA and 0.5 μg of pcDNA3-SERCA2b. After 2-3 days, cells were imaged in either Ca2+-containing (A and B) or Ca2+-free (C and D) HBSS and treated with 2 μm (A and B) or 4 μm (C and D) TG (t0 = time of TG addition). Once [Ca2+]er reached a plateau, 3 mm EGTA and 2 μm ionomycin was added to obtain the Rmin for each individual cell (A and B). Data present yellow/cyan ratios (mean ± S.E.; n > 6 cells) versus time (A and C) or calculated mean [Ca2+]er (B and D) versus time, based on calibration methods as described (8).
As a control, we also performed experiments in which SERCA was co-expressed with BI-1, reasoning that enhanced SERCA-mediated pumping of Ca2+ into the ER should overcome the effects of BI-1. Indeed, resting [Ca2+]er levels in cells co-expressing SERCA and BI-1 were restored to the concentrations found in control-transfected cells (Fig. 4, A and B, and supplemental Fig. S3), excluding nonspecific effects of BI-1 overexpression on the cameleon reporter system.
Inhibiting SERCA with TG causes depletion of [Ca2+]er due to passive leak of Ca2+ from ER (37). Thus, when cells are placed into Ca2+-deficient medium to exclude uptake from extracellular sources, the rate of Ca2+ leakage from the ER can be calculated from the first order derivative of the [Ca2+]er change over time (d[Ca2+]/dt) following TG treatment. Accordingly, we compared the leakage rates of vector control and BI-1-overexpressing cells, using the ER-targeted cameleon reporter method as in Fig. 4, C and D. BI-1-overexpressing cells were thus determined to have a higher ER Ca2+ leakage rate compared with vector control cells (Fig. 5), suggesting that the lower resting [Ca2+]er observed in BI-1-overexpressing cells is due at least in part to increased leakage of Ca2+ from ER, as reported recently for Bcl-2/Bax family proteins (2, 5, 10, 46). Although we cannot exclude the possibility that reduced import of Ca2+ into ER also plays a role in the reduced [Ca2+]er of BI-1-overexpressing cells seen under steady-state conditions when cells are cultured in Ca2+-containing medium, these data indicate that BI-1 shares with Bcl-2/Bcl-XL the ability to increase the passive leakage rate of Ca2+ from ER.
FIGURE 5.
BI-1 increases ER Ca2+ leakage rate. HeLa cells transfected with control (“vector”) (black circles) or BI-1-encoding (white circles) plasmids, together with ER-cameleon plasmid (12:1 ratio), were imaged in Ca2+-free HBSS. TG was added, the yellow/cyan ratio was determined for individual cells, and then data were converted to [Ca2+]er by calibration. The leak rate (d[Ca2+]/dt) was plotted against [Ca2+]er.
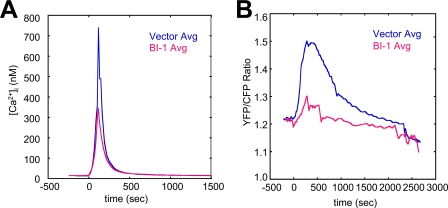
BI-1 reduces Ca2+ uptake by mitochondria. The reduced [Ca2+]er of BI-1-overexpressing cells would be predicted to result in less Ca2+ efflux into the cytosol, and consequently less uptake into mitochondria, when cells are treated with TG. Indeed, in experiments in which Fura-2 was used for simultaneously measuring cytosolic free Ca2+ concentrations ([Ca2+]i), along with cameleon-based measurements of [Ca2+]er, we observed that the TG-induced maximum [Ca2+]i was typically ∼50% lower in BI-1-overexpressing compared with control cells (Fig. 6A). Note that the time course of TG-induced Ca2+ efflux into cytosol showed a quick “spike” (∼200 s) after TG stimulation, whereas it takes about ∼1200 s for the TG-induced Ca2+ leak from ER to reach steady-state, probably due to the efflux of cytosolic Ca2+ out of cells or its sequestration into non-ER storage pools, such as mitochondria.
FIGURE 6.
BI-1 reduces TG-induced entry of Ca2+ into cytosol and mitochondria. A, HeLa cells transfected with control (vector) (blue) or BI-1-encoding (red) plasmids, together with ER-cameleon plasmid (12:1 ratio), were loaded with Fura-2 and treated with 4 μm TG (at time = 0 s), and cameleon-expressing cells were imaged in Ca2+-free HBSS. The Fura-2 350/380 ratio was then converted to [Ca2+]i after standard calibration (8). B, HeLa cells were transfected with pcDNA3 (Vector) or BI-1-encoding plasmids, in combination with mitochondria-targeted yellow cameleon-encoding plasmid (pMito-YC2) at a 6:1 ratio. Cameleon-expressing cells were imaged 3 days after transfection. The YFP/CFP ratio was monitored after treatment with 2 μm TG (t0 = time of TG addition). Data represent mean (n > 8 cells imaged).
Next, we compared TG-induced increases in mitochondrial Ca2+ using an alternative cameleon protein (pMito-YC2) that was designed for measuring mitochondrial Ca2+ concentrations and that was targeted to the mitochondrial matrix using duplicated mitochondrial targeting sequence of subunit VIII of human COX (38) (supplemental Fig. S1). Ratiometric determinations demonstrated that BI-1 overexpression reduces the accumulation of Ca2+ in mitochondria (Fig. 6B). These findings thus independently corroborate prior measurements of mitochondrial Ca2+ obtained using mitochondria-targeted aequorin protein (35).
BI-1 Regulates [Ca2+]er Downstream of Bcl-XL—Since BI-1, Bcl-2, and Bcl-XL all regulate basal [Ca2+]er, and all increase ER Ca2+ leakage rate, we studied the functional relations between BI-1 and Bcl-XL using genetically engineered cells. First, we examined the ability of overexpressed Bcl-XL to regulate [Ca2+]er in HeLa cells in which BI-1 was knocked down using shRNA. Using the ER-targeted, Ca2+-sensitive cameleon reporter, we observed that Bcl-XL reduced [Ca2+]er in control-transfected but not in BI-1-deficient HeLa cells (Fig. 7A). Second, we used mouse fibroblasts from bi-1-/- versus bi-1+/+ embryos (MEFs), transfecting Bcl-XL into these cells along with the ER-targeted cameleon reporter. In wild-type MEFs, Bcl-XL significantly reduced [Ca2+]er (Fig. 7B, columns 1 and 2: YFP/CFP ratio Bcl-XL = 2.00 ± 0.02 versus vector = 2.07 ± 0.02; n = 30; p < 0.01 by unpaired t test). In contrast, in BI-1-deficient MEFs, Bcl-XL failed to reduce [Ca2+]er and actually appeared to produce an increase rather than a decrease in [Ca2+]er (Fig. 7B). Immunoblot analysis confirmed equivalent levels of Bcl-XL protein production in transfected bi-1-/- and bi-1+/+ cells (data not shown).
FIGURE 7.
BI-1 is required for Bcl-XL-mediated modulation of [Ca2+]er. A, stable HeLa cell lines containing BI-1 shRNA or control shRNA were transiently transfected with plasmids encoding Bcl-XL or a control plasmid, as indicated, together with ER-cameleon plasmid. After 2 days, cells were imaged to determine the YFP/CFP ratio. Ratios (mean ± S.E.) for columns 1-4 are 2.04 ± 0.02, 1.94 ± 0.02, 2.18 ± 0.02, and 2, 16 ± 0.02, respectively (n = 30). The difference between column 1 and 2 is significant (p < 0.01 by unpaired t test). B, bi-1+/+ or bi-1-/- MEFs were co-transfected with ER-cameleon plasmid and either pcDNA3 or Bcl-XL-encoding plasmid. After 2 days, cells were imaged to determine the YFP/CFP ratio. Ratios (mean ± S.E.) for columns 1-4 are 2.07 ± 0.02, 2.00 ± 0.02, 2.04 ± 0.02, and 2.17 ± 0.02, respectively (n = 30). The difference between columns 1 and 2 is significant (p < 0.01 by unpaired t test).
BI-1 deficiency renders both HeLa and MEF cells more sensitive to ER stress-induced apoptosis (Fig. 3). Note that BI-1 deficiency in MEFs appears to have only a slight effect on resting [Ca2+]er, as measured using the ER-targeted cameleon reporter (YFP/CFP ratio = 2.04 ± 0.02 for bi-1-/- versus 2.07 + 0.02 for bi-1+/+; n = 30), although prior cytosolic Ca2+ imaging studies detected reduced TG-releasable intracellular Ca2+ pools in these cells (32), perhaps reflecting a sensitivity difference in these methods or implying that BI-1 makes less of a contribution to regulating basal [Ca2+]er in these cells compared with HeLa cells, at least under nonstressed conditions. Attempts to restore bi-1 expression in bi-1-/- MEFs using either BI-1 protein-encoding plasmid or recombinant retrovirus were only modestly successful due to poor levels of expression (supplemental Fig. S4), making it difficult to contrast the effects of Bcl-XL in control and reconstituted bi-1-/- MEFs. Nevertheless, the observation that Bcl-XL fails to reduce [Ca2+]er in both HeLa and MEFs with BI-1 deficiency demonstrates that this Bcl-2 family member requires BI-1 for regulating [Ca2+]er.
Bax and Bak Are Not Required for BI-1-mediated [Ca2+]er Regulation—The effects of BI-1 and Bcl-XL were examined in cells deficient in both Bax and Bak, since prior data have suggested that these proapoptotic proteins operate downstream of Bcl-2 and Bcl-XL (10) and because simultaneous deficiency of Bax and Bak has been reported to lower [Ca2+]er (6). Using ER-targeted cameleon, we confirmed that [Ca2+]er is slightly lower in bax-/-bak-/- double knock-out cells compared with wild-type cells (average YFP/CFP ratio = 2.03 ± 0.01 for wild-type versus 1.96 ± 0.02 for double knock-out; p < 0.01 by unpaired t test), corroborating data obtained by alternative Ca2+ measurement methods (6). Despite Bax/Bak deficiency, both BI-1 and Bcl-XL retained the ability to lower average [Ca2+]er (Fig. 8), demonstrating that Bax and Bak are not required for the ER Ca2+-modulating activity of either BI-1 or Bcl-XL and implying that these antiapoptotic proteins operate downstream of or parallel to Bax and Bak, at least with respect to [Ca2+]er regulation.
FIGURE 8.
BI-1 and Bcl-XL lower [Ca2+]er in Bax/Bak double knock-out cells. bax-/-bak-/- double knock-out (DKO) cells co-transfected with ER-cameleon plasmid and pcDNA3 (vector) or plasmids encoding BI-1 or Bcl-XL as indicated. Cells were imaged 2 days later to determine yellow/cyan emission ratio. YFP/CFP ratios (mean ± S.E., n = 40) were 1.97 ± 0.02, 1.82 ± 0.02, and 1.82 ± 0.02, respectively. Differences between vector control and BI-1 or Bcl-XL were significant (p < 0.01 by unpaired t test).
DISCUSSION
Using an ER-targeted Ca2+-sensitive fluorescent protein (cameleon), we extended recent studies (32, 35) about the [Ca2+]er-regulating activity of BI-1, demonstrating here that BI-1 is similar to antiapoptotic Bcl-2 family proteins in its ability to regulate [Ca2+]er. In common with Bcl-2 and Bcl-XL, BI-1 overexpression lowers resting [Ca2+]er and increases the rate of passive Ca2+ leakage from ER. Conversely, as shown here, BI-1 deficiency causes higher resting [Ca2+]er. One consequence of lower [Ca2+]er caused by BI-1 overexpression is reduced entry of Ca2+ into the cytosol upon discharge from the ER and reduced accumulation of Ca2+ in mitochondria, compared with control cells. Since mitochondrial Ca2+ accumulation can precipitate mitochondrial permeability transition (MPT) (reviewed in Refs. 47 and 48), resulting in release of apoptogenic mitochondrial proteins, such as cytochrome c and consequent caspase activation, we speculate that the effects of BI-1 on Ca2+ dynamics could account at least in part for the previously reported ability of BI-1 to inhibit downstream activation of the mitochondrial pathway for cell death following exposure of cells to TG and other inducers of ER stress (32). It should be noted, however, that [Ca2+]er affects many other events directly and indirectly, besides its role in regulating MPT, including the function of ER-resident chaperones (Grp78 and Grp94) that control signaling by several ER proteins (PERK, Ire1, and ATF6) (reviewed in Ref. 49). Indeed, tissues from bi-1-/- mice show evidence of increased signaling via the Ire1 and ATF6 pathways (33), which results in changes in the activities of stress kinases and several transcription factors (reviewed in Ref. 49). Also, with respect to MPT, because Bcl-2-mediated apoptosis regulation has been dissociated from MPT based on experiments using cyclophilin D knock-outs (50, 51), it is also possible that BI-1 regulates cell death through MPT-independent mechanisms.
In this regard, though BI-1-mediated changes in [Ca2+]er correlate with previously demonstrated alterations in resistance to certain types of cell death stimuli (32), it remains to be determined whether the ability of BI-1 to modulate [Ca2+]er accounts for the cytoprotective activity of this protein. Bcl-2 family proteins also regulate [Ca2+]er, correlating with their effects on cell death, but direct cause and effect data linking [Ca2+]er regulation to cell death control are presently lacking. However, consistent with an important role for [Ca2+]er, experimental manipulations that independently regulate [Ca2+]er, such as overexpression of SERCA to increase [Ca2+]er and expression of calreticulin to sequester luminal ER Ca2+ and thereby lower [Ca2+]er, have been shown to alter cellular sensitivity to several types of cell death stimuli and to mimic or negate the actions of Bcl-2/Bax family proteins (4, 52-54). Still, the mechanisms connecting altered [Ca2+]er to cell death responses are unclear and may be multifactorial.
By using cells genetically engineered to produced BI-1 deficiency, we determined that BI-1 is required for the [Ca2+]er-lowering activity of Bcl-XL, suggesting that BI-1 operates downstream of Bcl-XL. Conversely, Bcl-XL retained its ability to lower [Ca2+]er in cells lacking both Bax and Bak, consistent with a recent report (10, 17-20). Taken together, these findings suggest a hierarchical pathway for regulation of [Ca2+]er in which Bax and Bak operate upstream of or parallel to Bcl-XL and in which Bcl-XL is upstream of BI-1. The placing of BI-1 as a distal component of this proposed hierarchical pathway is consistent with data indicating a more ancient function for BI-1 as a protector against cellular stress in both plants and animals, compared with Bcl-2/Bax family proteins, which exist only in animal species. Since plants contain no recognizable homologs of Bcl-2/Bax family proteins based on sequence comparisons, it is possible that they possess functional analogs that regulate BI-1 without sharing overt sequence homology.
How BI-1 regulates [Ca2+]er remains to be determined. The multiple membrane-spanning domains of the BI-1 protein are suggestive of intrinsic ion channel activity, but BI-1 may indirectly control [Ca2+]er through interactions with other ER Ca2+ channel proteins, such as IP3Rs, as recently suggested for Bcl-2 family proteins (10). In yeast, however, the ability of BI-1 to protect against Bax-induced cell death was recently reported to be dependent on genes encoding homologs of SERCA (25), indicating a functional link between BI-1 and ER Ca2+ ATPases. If BI-1 does indirectly regulate the activity of ER Ca2+ channels, then we surmise that these channel proteins must be conserved in yeast, plants, and animals, because BI-1 overexpression has been shown to block cell death induced by ectopic expression of Bax in such diverse eukaryotic organisms. However, BI-1 may possess other functions besides [Ca2+]er modulation that account for its cytoprotective activity.
Acknowledgments
We thank T. Pozzan for pMito-YC2 plasmid, J. Lytton for pcDNA3-SERCA2b plasmid, S. Korsmeyer for bax-/-bak-/- cells, M. Thomas for preparing the bi-1-/- MEFs, M. Hanaii and T Siegfried for manuscript preparation, and R. Y. Tsien for comments on the manuscript.
This work was supported by National Institutes of Health Grants F32-NS047855, F32-GM067488, AG15393, and NS27177 and Department of Energy Grant DE-FG-01-ER63276. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S4.
Footnotes
The abbreviations used are: IP3R, inositol triphosphate-regulated Ca2+ channel; TG, thapsigargin; MEF, mouse embryo fibroblast; CFP, cyan-emitting fluorescent protein; YFP, yellow-emitting fluorescent protein; HBSS, Hanks' balanced salt solution; RT, reverse transcription; ER, endoplasmic reticulum; shRNA, short hairpin RNA; HA, hemagglutinin; [Ca2+]er, endoplasmic reticulum Ca2+; MPT, mitochondrial permeability transition.
References
- 1.Reed, J. C., Jürgensmeier, J. M., and Matsuyama, S. (1998) Biochim. Biophys. Acta 1366 127-137 [DOI] [PubMed] [Google Scholar]
- 2.Foyouzi-Youssefi, R., Arnaudeau, S., Borner, C., Kelley, W. L., Tschopp, J., Lew, D. P., Demaurex, N., and Krause, K.-H. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 5723-5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lam, M., Dubyak, G., Chen, L., Nuñez, G., Miesfeld, R. L., and Distelhorst, C. W. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 6569-6573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinton, P., Ferrari, D., Rapizzi, E., Di Virgilio, F., Pozzan, T., and Rizzuto, R. (2001) EMBO J. 20 2690-2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinton, P., Ferrari, D., Magalhaes, P., Schulze-Osthoff, K., Di Virgilio, F., Pozzan, T., and Rizzuto, R. (2000) J. Cell Biol. 148 857-862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scorrano, L., Oakes, S. A., Opferman, J. T., Cheng, E. H., Sorcinelli, M. D., Pozzan, T., and Korsmeyer, S. J. (2003) Science 300 135-139 [DOI] [PubMed] [Google Scholar]
- 7.Zong, W. X., Li, C., Hatzivassiliou, G., Lindsten, T., Yu, Q. C., Yuan, J., and Thompson, C. B. (2003) J. Cell Biol. 162 59-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer, A. E., Jin, C., Reed, J. C., and Tsien, R. Y. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 17404-17409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chami, M., Prandini, A., Campanella, M., Pinton, P., Szabadkai, G., Reed, J. C., and Rizzuto, R. (2004) J. Biol. Chem. 279 54581-54589 [DOI] [PubMed] [Google Scholar]
- 10.Oakes, S. A., Scorrano, L., Opferman, J. T., Bassik, M. C., Nishino, M., Pozzan, T., and Korsmeyer, S. J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 105-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schendel, S. L., Xie, Z., Montal, M. O., Matsuyama, S., Montal, M., and Reed, J. C. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 5113-5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minn, A. J., Velez, P., Schendel, S. L., Liang, H., Muchmore, S. W., Fesik, S. W., Fill, M., and Thompson, C. B. (1997) Nature 385 353-357 [DOI] [PubMed] [Google Scholar]
- 13.Antonsson, B., Montessuit, S., Sanchez, B., and Martinou, J. C. (2001) J. Biol. Chem. 276 11615-11623 [DOI] [PubMed] [Google Scholar]
- 14.Kuo, T. H., Kim, H. R., Zhu, L., Yu, Y., Lin, H. M., and Tsang, W. (1998) Oncogene 17 1903-1910 [DOI] [PubMed] [Google Scholar]
- 15.Dremina, E. S., Sharov, V. S., and Schoneich, C. (2006) Biochemistry 45 175-184 [DOI] [PubMed] [Google Scholar]
- 16.Dremina, E. S., Sharov, V. S., Kumar, K., Zaidi, A., Michaelis, E. K., and Schoneich, C. (2004) Biochem. J. 383 361-370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, R. G., Bui, T., White, C., Madesh, M., Krawczyk, C. M., Lindsten, T., Hawkins, B. J., Kubek, S., Frauwirth, K. A., Wang, Y. L., Conway, S. J., Roderick, H. L., Bootman, M. D., Shen, H., Foskett, J. K., and Thompson, C. B. (2007) Immunity 27 268-280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, C., Wang, X., Vais, H., Thompson, C. B., Foskett, J. K., and White, C. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 12565-12570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White, C., Li, C., Yang, J., Petrenko, N. B., Madesh, M., Thompson, C. B., and Foskett, J. K. (2005) Nat. Cell Biol. 7 1021-1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, R., Valencia, I., Zhong, F., McColl, K. S., Roderick, H. L., Bootman, M. D., Berridge, M. J., Conway, S. J., Holmes, A. B., Mignery, G. A., Velez, P., and Distelhorst, C. W. (2004) J. Cell Biol. 166 193-203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walter, L., Dirks, B., Rothermel, E., Heyens, M., Szpirer, C., Levan, G., and Gunther, E. (1994) Mamm. Genome 5 216-221 [DOI] [PubMed] [Google Scholar]
- 22.Xu, Q., and Reed, J. C. (1998) Mol. Cell 1 337-346 [DOI] [PubMed] [Google Scholar]
- 23.Bolduc, N., Ouellet, M., Pitre, F., and Brisson, L. F. (2003) Planta 216 377-386 [DOI] [PubMed] [Google Scholar]
- 24.Kawai-Yamada, M., Jin, L., Yoshinaga, K., Hirata, A., and Uchimiya, H. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 12295-12300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ihara-Ohori, Y., Nagano, M., Muto, S., Uchimiya, H., and Kawai-Yamada, M. (2007) Plant Physiol. 143 650-660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolduc, N., and Brisson, L. F. (2002) FEBS Lett. 532 111-114 [DOI] [PubMed] [Google Scholar]
- 27.Kawai, M., Pan, L., Reed, J. C., and Uchimiya, H. (1999) FEBS Lett. 464 143-147 [DOI] [PubMed] [Google Scholar]
- 28.Kawai-Yamada, M., Ohori, Y., and Uchimiya, H. (2004) Plant Cell 16 21-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsumura, H., Nirasawa, S., Kiba, A., Urasaki, N., Saitoh, H., Ito, M., Kawai-Yamada, M., Uchimiya, H., and Terauchi, R. (2003) Plant J. 33 425-434 [DOI] [PubMed] [Google Scholar]
- 30.Sanchez, P., de Torres, M., and Grant, M. (2000) Plant J. 21 393-399 [DOI] [PubMed] [Google Scholar]
- 31.Chae, H.-J., Ke, N., Chen, S., Kim, H.-R., Godzik, A., Dickman, M., and Reed, J. C. (2003) Gene (Amst.) 323 101-113 [DOI] [PubMed] [Google Scholar]
- 32.Chae, H. J., Kim, H. R., Bailly-Maitre, B., Zhu, X., Ke, N., Krajewska, M., Krajewski, S., Cui, J., Digicaylioglu, M., Thomas, M., Kress, C. L., Babendure, J., Tsien, R. Y., Lipton, S. A., and Reed, J. C. (2004) Mol. Cell 15 355-366 [DOI] [PubMed] [Google Scholar]
- 33.Bailly-Maitre, B., Fondevila, C., Kaldas, F., Droin, N., Luciano, F., Ricci, J. E., Croxton, R., Krajewska, M., Zapata, J. M., Kupiec-Weglinski, J. W., Farmer, D., and Reed, J. C. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 2809-2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe, N., and Lam, E. (2006) Plant J. 45 884-894 [DOI] [PubMed] [Google Scholar]
- 35.Westphalen, B. C., Wessig, J., Leypoldt, F., Arnold, S., and Methner, A. (2005) Cell Death Differ. 12 304-306 [DOI] [PubMed] [Google Scholar]
- 36.Zhang, H., Xu, Q., Krajewski, S., Krajewska, M., Xie, Z., Fuess, S., Kitada, S., Pawloski, K., Godzik, A., and Reed, J. C. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 2597-2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lytton, J., Westlin, M., and Hanley, M. R. (1991) J. Biol. Chem. 266 17067-17071 [PubMed] [Google Scholar]
- 38.Filippin, L., Abad, M. C., Gastaldello, S., Magalhaes, P. J., Sandona, D., and Pozzan, T. (2005) Cell Calcium 37 129-136 [DOI] [PubMed] [Google Scholar]
- 39.Grynkiewicz, G., Poenie, M., and Tsien, R. Y. (1985) J. Biol. Chem. 260 3440-3450 [PubMed] [Google Scholar]
- 40.Krajewski, S., Krajewska, M., Shabaik, A., Miyashita, T., Wang, H.-G., and Reed, J. C. (1994) Am. J. Pathol. 145 1323-1333 [PMC free article] [PubMed] [Google Scholar]
- 41.Krajewski, S., Krajewska, M., Shabaik, A., Wang, H.-G., Irie, S., Fong, L., and Reed, J. C. (1994) Cancer Res. 54 5501-5507 [PubMed] [Google Scholar]
- 42.Miyawaki, A., Griesbeck, O., Heim, R., and Tsien, R. Y. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 2135-2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyawaki, A., Llopis, J., Heim, R., McCaffery, J. M., Adams, J. A., Ikura, M., and Tsien, R. Y. (1997) Nature 388 882-887 [DOI] [PubMed] [Google Scholar]
- 44.Palmer, A. E., and Tsien, R. Y. (2006) Nat. Protoc. 1 1057-1065 [DOI] [PubMed] [Google Scholar]
- 45.Bassik, M. C., Scorrano, L., Oakes, S. A., Pozzan, T., and Korsmeyer, S. J. (2004) EMBO J. 23 1207-1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanden Abeele, F., Skryma, R., Shuba, Y., Van Coppenolle, F., Slomianny, C., Roudbaraki, M., Mauroy, B., Wuytack, F., and Prevarskaya, N. (2002) Cancer Cell 1 169-179 [DOI] [PubMed] [Google Scholar]
- 47.Bernardi, P. (1999) Physiol. Rev. 79 1127-1155 [DOI] [PubMed] [Google Scholar]
- 48.Kroemer, G., and Reed, J. C. (2000) Nat. Med. 6 513-519 [DOI] [PubMed] [Google Scholar]
- 49.Xu, C., Bailly-Maitre, B., and Reed, J. C. (2005) J. Clin. Invest. 115 2656-2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baines, C. P., Kaiser, R. A., Purcell, N. H., Blair, N. S., Osinska, H., Hambleton, M. A., Brunskill, E. W., Sayen, M. R., Gottlieb, R. A., Dorn, G. W., Robbins, J., and Molkentin, J. D. (2005) Nature 434 658-662 [DOI] [PubMed] [Google Scholar]
- 51.Nakagawa, T., Shimizu, S., Watanabe, T., Yamaguchi, O., Otsu, K., Yamagata, H., Inohara, H., Kubo, T., and Tsujimoto, Y. (2005) Nature 434 652-658 [DOI] [PubMed] [Google Scholar]
- 52.Liu, H., Bowes, R. C., 3rd, van de Water, B., Sillence, C., Nagelkerke, J. F., and Stevens, J. L. (1997) J. Biol. Chem. 272 21751-21759 [DOI] [PubMed] [Google Scholar]
- 53.Nakamura, K., Bossy-Wetzel, E., Burns, K., Fadel, M. P., Lozyk, M., Goping, I. S., Opas, M., Bleackley, R. C., Green, D. R., and Michalak, M. (2000) J. Cell Biol. 150 731-740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arnaudeau, S., Frieden, M., Nakamura, K., Castelbou, C., Michalak, M., and Demaurex, N. (2002) J. Biol. Chem. 277 46696-46705 [DOI] [PubMed] [Google Scholar]