Abstract
Transforming growth factorβ (TGF-β) signals through Smad-dependent and Smad-independent pathways. However, Smad signaling is altered by allelic deletion or intragenic mutation of the Smad4 gene in more than half of pancreatic ductal adenocarcinomas. We show here that loss of Smad4-dependent signaling leads to aberrant expression of RON, a phosphotyrosine kinase receptor, and that signaling by RON cooperates with Smad4-independent TGF-β signaling to promote cell motility and invasion. Restoring Smad4 expression in a pancreatic ductal adenocarcinoma cell line that is deficient in Smad4 repressed RON expression. Conversely, small interference RNA knock down of Smad4 or blocking TGF-β signaling with a TGF-β type I receptor kinase inhibitor in Smad4-intact cell lines induced RON expression. TGF-β-induced motility and invasion were inhibited in cells that express Smad4 and that have low levels of RON compared with isogenically matched cells that were deficient in Smad4. Furthermore, knocking down RON expression in Smad4-deficient cells suppressed TGF-β-mediated motility and invasion. We further determined that Smad4-dependent signaling regulated RON expression at the transcriptional level by real-time reverse transcription PCR and RON promoter luciferase reporter assays. Functional inactivation by site-directed mutations of two Smad binding sites on the RON promoter inhibited TGF-β-mediated repression of RON promoter activity. These studies indicate that loss of Smad4 contributes to aberrant RON expression and that cross-talk of Smad4-independent TGF-β signaling and the RON pathway promotes an invasive phenotype.
Transforming growth factor-βs (TGF-βs)2 are multifunctional cytokines that regulate numerous cellular processes, including cell growth, differentiation, and apoptosis. TGF-βs signal downstream of TGF-β receptors through a canonical Smad pathway and through Smad-independent pathways (1). TGF-βs signal by binding to transmembrane serine-threonine kinases termed type I receptor (RI) and type II receptor (RII). A third TGF-β receptor (RIII) is not believed to be involved directly in TGF-β signaling but acts to present TGF-βs to RII (2). TGF-β ligand first binds RII, which then recruits RI to form a functional receptor complex. After this complex is formed, RI is phosphorylated by the constitutively active and autophosphorylated RII. For Smad-dependent signaling, RI directly interacts with and phosphorylates Smads 2 and 3 at a conserved consensus SXS motif; these activated Smads bind Smad4, and subsequently the complex translocates to the nucleus. This Smad complex associates with other cofactors to bind efficiently to specific DNA sequences, and the activated complex promotes transcription or repression of TGF-β-responsive genes (3, 4). TGF-βs also signal through their activated receptors independent of Smads, although these pathways are less fully defined. It is known that TGF-β can activate, independent of Smads, the Ras/ERK, TAK1/p38, RhoA/JNK, and phosphatidylinositol 3-kinase/AKT pathways (1, 5).
TGF-β signaling is tumor-suppressive in normal epithelial cells, but this pathway is often altered in cancer cells, leading to tumor progression (6, 7). A number of mechanisms are attributed to the loss of TGF-β tumor-suppressive activities in cancer cells. These mechanisms include allelic deletion or mutation of Smad4, also designated as DPC4 (deleted in pancreatic carcinoma locus 4) (8); interference of Smad activation by expression of inhibitory Smads (Smad6, Smad7); phosphorylation of the linker region of Smads by ras (9); epigenetic silencing or mutations of TGF-β receptor genes (10, 11); and TGF-β-mediated effects on the microenvironment (12, 13). TGF-β-induced pathways may also cause a different phenotype in tumor cells compared with normal cells by augmenting or cooperating with oncogenic pathways (14–16). Of these mechanisms, loss of Smad4 by allelic deletion or mutation occurs in 55% of pancreatic ductal adenocarcinomas (PDAC) as a relatively late event in tumor progression (8, 17). Pancreatic cancer patients whose tumors are Smad4-negative display a more aggressive cancer and have shorter survival, suggesting that Smad4 has tumorsuppressive properties in PDAC (18, 19).
Studies suggest that TGF-β and phosphotyrosine kinase receptor pathways including erbB2 (16) and epidermal growth factor receptor (20) may cooperate to promote cancer progression. The receptor tyrosine kinase RON (recepteur d'origine nantais), a member of the MET proto-oncogene family, was reported recently to be overexpressed in PDAC (21). Activation of RON by its ligand induces cell signaling through multiple downstream targets, including mitogen-activated protein kinase, phosphatidylinositol 3-kinase/AKT, c-Src, focal adhesion kinase, β-catenin/TCF, and likely other unknown signaling molecules (22). These signals are involved in many cellular processes, including adhesion, motility, proliferation, and apoptotic resistance. Elevated RON kinase expression is reported to correlate with the invasive activity of tumors (22, 23). Separate studies indicate that RON signaling suppresses TGF-β-induced apoptosis (24) and that both TGF-β and RON pathways may promote epithelial to mesenchymal transition (25). These findings prompted us to further examine the interactions of TGF-β and RON pathways in PDAC.
In the present study, we linked the loss of Smad4-dependent TGF-β signaling with an increase in expression of RON receptor tyrosine kinase in human pancreatic cancer cells. We further established that Smad4-dependent signaling regulates RON expression, at least in part, by suppressing RON promoter activity. Interestingly, aberrant RON expression enhanced TGF-β-mediated motility and invasion in Smad4-deficient cells. These findings suggest that the tumor-suppressing function of Smad4 may be mediated in part by regulating the expression of RON. The cooperation of loss of Smad4 and up-regulation of RON may contribute to the switch of TGF-β signaling from being tumor-suppressive to an altered pathway that promotes tumor progression in pancreatic cancer cells.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatment—Human PDAC cell lines BxPC-3, Capan-2, MIA PaCa-2, CFPAC-1, Panc-1, AsPC-1 were from American Type Culture Collection (ATCC, Rockville, MD); UK Pan-1 was established in our laboratory (26). BxPC-3 cells were maintained in RPMI 1640 medium, and other cell lines were grown in Dulbecco's modified Eagle's medium; each medium was supplemented with 10% fetal bovine serum. Human recombinant TGF-β1 was purchased from R&D Systems (Minneapolis, MN). The TGF-β receptor type I kinase inhibitor (RIKI) and the proteasome inhibitor MG132 were purchased from Calbiochem. The Hsp90 inhibitor 17-allylamino-17-demethoxygeldanamycin (17-AAG) was from Sigma-Aldrich, Inc.
Expression of Smad4, siRNA-Smad4, and siRNA-RON—pB-abe/Smad4 (provided by Bernard E. Weissman, University of North Carolina), pSuper/siRNA-Smad4 (provided by LuZhe Sun, The University of Texas Health Science Center at San Antonio, TX), or pSuper/siRNA-RON plasmids were transfected into human embryonic kidney 293T amphotrophic packaging cells (ATCC) using FuGENE 6 (Roche Applied Science) according to the manufacturer's protocol. Cells were then infected with retroviral medium from the packaging cells 48 h after transfection. The stable clones were selected with 1 μg/ml puromycin. The expression levels of Smad4 and RON in these clones were determined by Western blotting analysis.
3TP-Luciferase Reporter Assay and MTT Assay—Responsiveness of cells to TGF-β was determined by transfection of a TGF-β-responsive reporter construct, p3TP-Lux (27), and a control plasmid, CMV-Renilla. 48 h after transfection, 3TP-luciferase activity was measured using a Dual-Luciferase assay kit (Promega) according to the manufacturer's instruction and normalized by Renilla luciferase activity. To determine cell proliferation rate, 1000 cells/well were seeded in 96-well plates and treated with vehicle or 10 ng/ml TGF-β1. At the indicated time points, 0.5 mg/ml MTT (3-(4,5 dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (Sigma) was added and the cells were incubated for another 4 h. MTT assay was performed as described previously (28).
Western Blotting Analysis—Whole cell extracts were obtained in radioimmune precipitation buffer. Western blotting analysis was performed using standard methods. Primary antibodies against RON-β, Smad4 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-phospho-Smad2 was purchased from Chemicon International (Temecula, CA), and anti-Hsp90 was from Cell Signaling (Beverly, MA).
Cell Motility and Invasion Assay—The migratory and invasive behavior of the cells was analyzed by in vitro Transwell motility and Matrigel invasion assays according to the manufacturers' protocols. Briefly, 3 × 104 cells/well were plated in a 24-well Transwell insert with 8-μm pore membrane or Matrigel invasion chambers (BD Biosciences) in 0.3 ml of serum-free medium. The outer chambers contained 0.7 ml of 10% fetal bovine serum medium. The cells were treated with 5 ng/ml TGF-β1. After 48 h of incubation, the cells on the top surface of the membrane were gently removed with cotton swabs. The cells migrating or invading to the undersurface of the membrane were fixed in 70% ethanol and stained with crystal violet. The migration and invasion values were determined by eluting crystal violet in 10% acetic acid, and the absorbance was taken using a Fluostar Optima Plate Reader at 584 nm (29).
Quantitative Reverse Transcription PCR—Total RNA was isolated from cells with the indicated treatments using TRIzol reagent (Invitrogen). 100 ng of total RNA was subjected to realtime reverse transcription PCR reaction with SYBR Green realtime PCR mix (Applied Biosystems) according to the manufacturer's instruction. The primer sequences for RON are described elsewhere (30). β-Actin mRNA was amplified simultaneously for an endogenous control. The primer sequences for β-actin were described previously (31).
RON Promoter Luciferase Reporter Assay—The 1.2-kb human RON gene promoter-CAT reporter vector was kindly provided by Dr. Richard Breathnach. This Ron promoter region was subcloned into pGL3 basic luciferase reporter plasmid. The predicted transcription factor binding sites of RON promoter were analyzed by the MatInspector algorithm (Genomatix). Smad binding element mutant constructs (SBE-mut) of RON promoter were generated using the QuikChange site-directed mutagenesis kit (Stratagene). The primer sequences for the mutants were: SBE-A (–83 ∼–87), wild type, 5′-ATTTGGCCCAGTCCAGACCTC GAGTCGGGCCCCCAG-3′ and mutant, 5′-ATTTGG CCCAGTCTACATCTCGAGTCGGGCCCCCAG-3′; SBE-B (–184 ∼ –188), wild type, 5′-GCAGGCGTCAGGTGCTCAGACCCGAGGGCCGGGAAG-3′ and mutant, 5′-GCAGGCGTCAGGTGCTTACATCCGA GGGCCGGGAAG-3′. Underlined nucleotides represent Smad binding sites. Bold lettering represents the location of wild type and corresponding mutated nucleotides. Cells were transfected with the wild type and SBE mutant of RON promoter luciferase constructs using FuGENE HD (for BxPC3 cells) or FuGENE 6 (for UK Pan-1 cells) transfection reagents (Roche Applied Science). 24 h after transfection, cells were pretreated with vehicle or with 1 μm RIKI for 6 h and then treated with 5 ng/ml TGF-β1 for another 24 h. The RON promoter activities were assayed using the Dual-Luciferase assay kit (Promega) and normalized by CMV-Renilla luciferase activity. All experiments were performed at least three times.
Statistics Analysis—Data are presented as mean ± S.D. of at least two independent triplicate experiments. Statistical analysis was performed using analysis of variance (followed by Bonferroni multiple comparison test to determine the significance among groups).
RESULTS
Generation of Isogenic Cell Lines That Differ in Smad4 Expression—TGF-β signaling through Smads is known to have tumor-suppressive activity. However, during tumor progression >50% of PDAC cells lose expression of Smad4 due to allelic deletion or intragenic mutations (32). To investigate whether Smad4 plays a role in the malignant behavior of PDAC cells, we generated two sets of isogenically matched pancreatic cancer cell lines that differ only in their expression levels of Smad4. The two cell lines chosen for these studies were BxPC3 and UK Pan-1. BxPC3 cells have biallelic deletion of Smad4 (8), and UK Pan-1 cells express Smad4 (26). Smad4 was restored in BxPC3 cells by infecting cells with a retroviral vector that expresses the Smad4 cDNA. These Smad4-expressing cells were referred to as Bx/Smad4 (Fig. 1A, left). Smad4 expression was knocked down in UK Pan-1 cells by stably expressing an siRNA directed against Smad4, and these cells were referred to as UK/siSmad4. UK/siSmad4 cells showed a dramatic reduction of Smad4 expression compared with vector control cells (Fig. 1A, right). TGF-β/Smad transcriptional responses were determined for the two isogenic cell sets using a luciferase readout of a TGF-β response element (3TP-Luc). Restoring Smad4 expression in BxPC3 cells induced a Smad-dependent response; conversely, knock down of Smad4 in UK Pan-1 cells decreased the Smad-dependent response (Fig. 1B). TGF-β inhibited growth of the Smad4-intact Bx/Smad4 and UK Pan-1 cells, suggesting that the autonomous TGF-β tumor-suppressive effects were present in these cells (Fig. 1C).
FIGURE 1.
Generation of isogenic cell lines that differ in Smad4 expression. A, Smad4 was stably expressed in BxPC3 cells (Bx/Smad4) or knocked down in UK Pan-1 cells (UK/siSmad4) by retroviral infection. These cells were compared with parental cells infected with an empty vector. Efficacy of restoration or knock down of Smad4 was determined by Western blotting. B, TGF-β/Smad transcriptional responses were determined by 3TP-luc assays. Cells were transfected with 0.5 μg of p3TP-luciferase reporter plasmid along with CMV-Renilla plasmid in 12-well plates. 24 h after transfection, cells were treated with 5 ng/ml TGF-β1 or vehicle control for 24 h. 3TP-luciferase activities were measured and normalized by the Renilla luciferase activities. The data represent mean ± S.D. from experiments performed in triplicates. C, BxPC3, Bx/Smad4, UK Pan-1, and UK/siSmad4 cells were seeded (1000 cells/well) in 96-well plates and treated with 5 ng/ml TGF-β1 or vehicle control. 3-(4,5 Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) proliferation assays were performed at the indicated time points. The data are mean ± S.D. from experiments performed in six replicates. **, p < 0.01 compared with control.
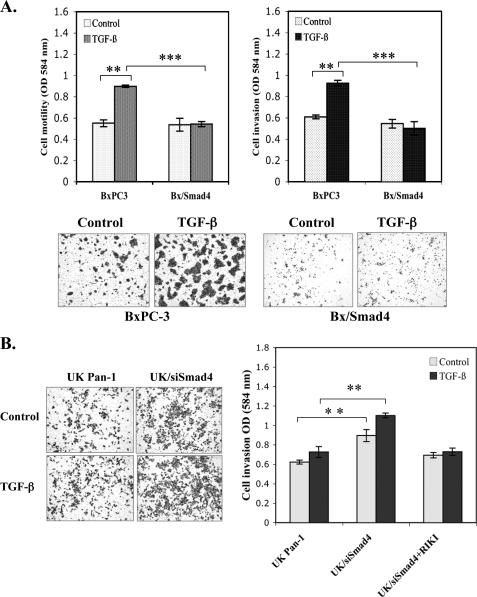
Smad4 Suppresses TGF-β-mediated Motility and Invasion—Studies indicate that TGF-β can induce motility and invasion in some late stage tumors (13). It is not clear whether TGF-β-mediated invasion is through Smad-dependent or Smad-independent pathways, although current studies suggest that TGF-β signaling can be altered in tumor cells and may cooperate with oncogenic signaling pathways to promote tumor progression (14, 15). We examined the role of Smad4 in regulating TGF-β signaling effects on motility and invasion using the two isogenic matched PDAC cell models that differ only in their level of Smad4 expression. TGF-β induced motility and invasion in BxPC3 cells but not in BxPC3/Smad4 cells (Fig. 2A). Consistent with the premise that signaling through Smad4 reduces TGF-β-induced invasiveness, knock down of Smad4 in UK Pan-1 cells (UK/siSmad4) increased TGF-β-induced invasion (Fig. 2B) The role of autocrine TGF-β in promoting invasion in UK/siSmad4 was confirmed because treatment of these cells with TGF-β RIKI reduced TGF-β-mediated invasion (Fig. 2B, right bars).
FIGURE 2.
Smad4 expression suppresses TGF-β-mediated cell invasiveness. A, BxPC3 and Bx/Smad4 cells (3 × 104) were seeded in the Transwell inserts (upper left) or Matrigel invasion chambers (upper right) and treated with 5 ng/ml TGF-β1 alone or vehicle control for 48 h. Cells on the top surface of the chamber were removed. The cells migrating to the under surface were fixed and stained with crystal violet. The motility and invasion value were measured by eluting the dye from the cells on the undersurface of inserts. The data represent mean ± S.D. from experiments performed in triplicate. **, p < 0.01; ***, p < 0.001. The lower panels are the representatives of light microscopy photos (×4 objective) of the invasion assay. B, Matrigel invasion assays were performed in UK Pan-1 and UK/siSmad4 cells. Left, the representatives of light microscopy photos (×4 objective). Right, quantification data representing the mean ± S.D. of three separate invasion assays; **, p < 0.01.
TGF-β Negatively Regulates RON Expression in a Smad4-dependent Manner—Previous findings indicate that RON is overexpressed in PDAC and aberrant expression of RON is correlated with tumor invasiveness in cancer cell lines (22, 33, 34). RON and TGF-β both induce epithelial to mesenchymal transition, and RON signaling can suppress TGF-β-mediated apoptosis (24, 25). These findings led us to examine the interaction of RON and TGF-β signaling in PDAC. Surprisingly, we found that RON expression was inversely correlated with the expression of Smad4. Cells that were deficient of Smad4 expressed dramatically higher levels of RON compared with Smad4-intact PDAC cell lines (Fig. 3A). To further examine whether Smad4-dependent TGF-β signaling might regulate the expression of RON, we compared two Smad4 null cell lines, Capan-2 and BxPC3, and two Smad4 intact cell lines, Panc-1 and UK Pan-1. Treatment of these cells with exogenous TGF-β further decreased the low level of RON expression in Smad4-intact cells but did not have an effect on the expression level of RON in cells deficient of Smad4 (Fig. 3B). Moreover, knock down of Smad4 in UK Pan 1 cells dramatically increased the expression level of RON (Fig. 3B, right panel). We further determined in Smad4-intact cells (Bx/Smad4, UK Pan-1, and Panc-1) that blocking TGF-β signaling, using an RIKI, caused an increase in the steady state level of RON (Fig. 3, C and D). Thus, these data suggest that RON expression is suppressed by TGF-β signaling in a Smad4-dependent manner.
FIGURE 3.
TGF-β negatively regulates RON kinase expression in a Smad4-dependent manner. Western blotting analyses were performed for the expression of Smad4 and RON-β of whole cell lysates (50 μg/lane) from indicated treatments. β-Actin was used for protein loading control. A, cell lysates were isolated from exponential growing cells, and the expression levels of RON-β and Smad4 were inversely correlated in PDAC cell lines. B, Panc-1, UK Pan-1, CF PAC-1, and BxPC3 cells were treated with 5 ng/ml TGF-β1 or vehicle for 48 h. C, BxPC3 and Bx/Smad4 cells were pretreated with 1 μm RIKI for 6 h, followed by 5 ng/ml TGF-β1 or vehicle for 48 h. The efficacy of RIKI to block TGF-β-induced phosphorylation of Smad2 is indicated (middle panel). D, RIKI reversed TGF-β-mediated down-regulation of RON-β expression in UK Pan-1 and Panc-1 cells.
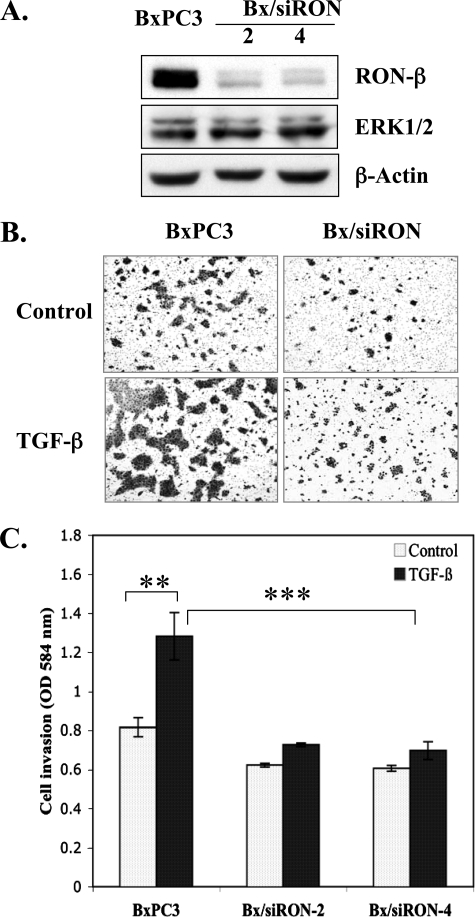
RON Is Required for TGF-β-mediated Invasion in Samd4-deficient Cells—Our data indicate that signaling through Smad4 inhibited invasion and suppressed RON expression in PDAC. We hypothesized that RON activity was required for TGF-β-induced invasion. To test this hypothesis we compared the capacity of TGF-β to induce invasion in vector control BxPC3 cells with BxPC3 cells where RON expression was knocked down by stably expressing RON siRNA. Two cell clones where RON was dramatically knocked down were compared with BxPC3 vector control cells for invasion through Matrigel (Fig. 4). TGF-β induced invasion in the control BxPC3 cells; however, knock down of RON by siRNA suppressed TGF-β-induced invasion as shown for two separate cell clones (Fig. 4, B and C). These results confirm the cooperativity of RON for TGF-β-induced invasion in BxPC3 cells and mimic the effect of Smad4-intact PDAC cells lines that express low levels of RON (Fig. 2A).
FIGURE 4.
RON is required for TGF-β-mediated invasion in Smad4-deficient cells. A, BxPC3 cells were stably infected with siRNA-RON (Bx/siRON). Expression levels of RON-β were determined by Western blotting analysis as described in Fig. 3. Expression levels of ERK1/2 were determined for the specificity of siRNA-RON. B, a representative of light microscopy photos (×4 objective) of Matrigel invasion assays. Quantification of the Matrigel invasion assays showing the inhibition of TGF-β-mediated cell invasion by knocking down RON. C, cell invasion was measured as described in Fig. 2. The data are mean ± S.D. from experiments performed in triplicate. **, p < 0.01; ***, p < 0.001.
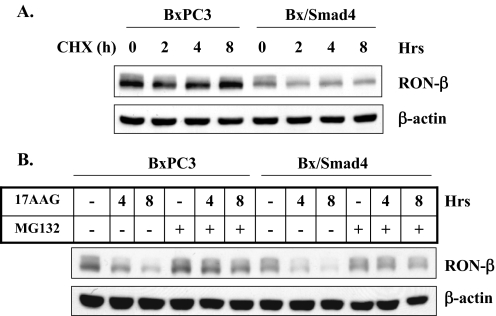
Smad4-dependent Signaling Regulates RON Transcription—To determine the level at which Smad4 signaling regulates RON expression, RON protein stability and the steady state levels of RON mRNA were determined. BxPC3 and Bx/Smad4 cells were treated with cycloheximide for up to 8 h to inhibit de novo protein synthesis. Only a slight level of degradation of RON protein was observed in both cell lines, with the only difference being that Bx/Smad4 cells show lower basal level of RON protein compared with the parental BxPC3 cells which are deficient in Smad4. This finding suggests that RON protein has a relatively long half-life in both Smad4-intact and Smad4-deficient cells. A recent study (35) showed that degradation of RON protein is through an ubiquitin-proteasome pathway and that heat shock protein 90 (Hsp90), a chaperone protein, stabilizes the RON from proteasome degradation. To examine whether Smad4 might accelerate RON degradation by this mechanism, BxPC3 and Bx/Smad4 cells were treated with an Hsp90 inhibitor, 17-allylamino-17-demethoxy-geldanamycin (17-AAG), and a proteasome inhibitor, MG132. Both cell lines displayed similar patterns for RON degradation by inhibiting Hsp90. Inhibiting proteasome activity with MG132 prevented 17-AAG-mediated degradation of RON (Fig. 5A), suggesting that RON protein turnover is caused, at least in part, through a ubiquitin-proteasome pathway and that Hsp90 may protect RON from this degradation. However, these studies suggest that Smad4 signaling does not down-regulate RON expression by altering these processes.
FIGURE 5.
RON protein stability is not regulated by Smad4. A, BxPC3 and Bx/Smad4 cells were treated with 5 ng/ml TGF-β1 for 24 h followed by adding 20 μg/ml cycloheximide (CHX) for the indicated time periods. B, BxPC3 and Bx/Smad4 cells were treated with vehicle (–) or Hsp90 inhibitor 17AAG (1 μm) in the presence or absence of the proteasome inhibitor MG132 (10 μm) for 6 h. Cell lysates were analyzed by Western blotting using antibody to RON-β. β-Actin was used as protein loading control.
The mRNA level of RON was next determined by real-time reverse transcription PCR. TGF-β treatment caused a decrease in the steady state levels of mRNA in both Smad4-restored BxPC3 and Smad4-intact UK Pan-1 cells, but not in Smad4 null BxPC3 and in UK/siSmad4 cells. Blocking TGF-β signaling in Smad4-positive cells by RIKI prevented the decrease of RON mRNA by TGF-β (Fig. 6, A and C). To further determine whether Smad4-dependent TGF-β signaling regulates RON at transcription level, we performed RON promoter reporter assays. Consistent with RON mRNA levels, TGF-β treatment decreased RON promoter activities in Smad4-positive cells, but not in Smad4 null cells or in cells where Smad4 was knocked down (Fig. 6, B and D).
FIGURE 6.
Smad4-dependent TGF-β signaling RON expression at the transcriptional level. A, BxPC3 and Bx/Smad4 cells were pretreated with RIKI (1 μm) for 6 h followed by vehicle or TGF-β1 (5 ng/ml) in the presence or absence of RIKI for another 24 h. 100 ng of total RNA was subjected to reverse transcription and real-time PCR reaction with SYBR Green mix. The relative mRNA levels were presented as compared with untreated BxPC3 control. B, BxPC3 and Bx/Smad4 cells were transfected with full-length of RON promoter luciferase reporter construct and received the same treatment as above. Luciferase activities were measured 48 h after transfection. The data were presented as -fold changes of luciferase activities compared with untreated control. C and D, real-time reverse transcription PCR and RON promoter luciferase reporter assays performed in UK Pan-1 and UK/siSmad4 cells. Data for A–C are presented as the mean ± S.D. of three separate experiments.
An examination of the DNA sequence of the RON promoter reveals that it lacks a TATA box and that it possesses several putative binding sites for Sp1 and STAT family of transcription factors (Fig. 7A). In addition, the RON promoter has three putative Smad binding elements (SBEs) located at –83, –184, and –484 bp relative to the transcription start site (Fig. 7A). The two SBE sites, termed site SBE-A and SBE-B, most proximal to the transcription start site were examined for their role in TGF-β/Smad4-mediated repression of RON promoter activity. Functional inactivation by site-directed mutation of either SBE-A or SBE-B sites of the RON promoter attenuated the TGF-β-mediated suppression of RON promoter activity in Smad4-expressing cells (Fig. 7B). TGF-β treatment reduced RON promoter activity to 35.6% of control for UK Pan-1 and 33.6% for Bx/Smad4 cells (Fig. 7B). Mutation of SBE-A or SBE-B sites reduced the level of TGF-β-mediated inhibition of RON promoter activity to 87.3% (SBE-A) and 79.2% (SBE-B) of control in UK Pan-1 cells and 74.1% (SBE-A) and 64.1% (SBE-B) in Bx/Smad4 cells (Fig. 7B). These findings imply that TGF-β/Smad4-dependent signaling represses RON promoter activity and that the SBE sites may serve as repressor elements.
FIGURE 7.
Functional inactivation of the SBEs inhibits TGF-β-mediated repression of RON promoter activity. A, a schematic diagram representing the putative Smad binding sites and other transcriptional regulators of RON promoter gene (45). B, comparison of activities of the wild type of RON promoter with the mutants of the putative SBEs, SBE-A and SBE-B. Wild type (WT) and SBE-A and SBE-B site mutants (SBE-A mut and SBE-B mut) of RON promoter constructs were transiently transfected into cells. 24 h after transfection, cells were treated with vehicle or TGFβ1 (5 ng/ml) for 24 h and promoter activities were measured and normalized by Renilla luciferase activities. Data are presented as the mean ± S.D. of a representative experiment performed in triplicate.
DISCUSSION
In this study, we investigated the role of Smad4 in mediating the paradoxical functions of TGF-β signaling of tumor suppression and tumor promotion. In the course of these studies we made the anecdotal observation that pancreatic cancer cells that express Smad4 have lower levels of RON receptor compared with Smad4-deficient cells. These events were linked by expressing Smad4 in Smad4-deficient cells and conversely by knocking down Smad4 expression in Smad4-intact cells. These studies revealed that Smad4-dependent TGF-β signaling was a negative regulator of RON expression and that this Smad4-dependent suppression of RON expression was mediated at least in part at the transcriptional level. Moreover, functional inactivation of SBEs on the RON promoter inhibited TGF-β-mediated suppression of RON promoter activity. This finding suggests that these SBEs are required for TGF-β-mediated suppression of RON promoter activity. More importantly, TGF-β-induced motility and invasion found in PDAC cell lines were reduced in cells that expressed Smad4 and that showed low levels of RON or in Smad4-deficient cells where RON expression was knocked down. Taken together, these studies imply that down-regulating the expression of RON by Smad signaling contributes to TGF-β-mediated tumor suppression and that the tumor-promoting effects of TGF-β in Smad4-deficient cells are dependent on cross-talk of Smad4-independent TGF-β signaling and RON pathways.
It is well recognized that Smad4 is a tumor suppressor in normal cells and in early stages of neoplasia. However, a number of studies indicate that Smad signaling mediates a more invasive phenotype in late stage cancers (36, 37). These studies suggest that Smad signaling is altered in these tumors, perhaps by altering the composition of coactivators/corepressors of Smad complexes, thereby changing TGF-β-induced gene expression, or by cooperating effects of TGF-β signaling (cross-talk) with oncogenic pathways. Examples of this include a report by Deckers et al. (38) that Smad4 enhances bone metastasis in a mammary cancer model, and a study by Arteaga and co-workers (16) indicates that cross-talk of TGF-β/Smad and erbB-2 induces TGF-β-mediated cell motility and cancer progression. In contrast, other studies indicate that intact TGF-β signaling may have tumor-suppressive activities, at least in some cancer models. Bardeesy et al. (39) showed, using a transgenic mouse model of PDAC, that loss of Smad4 caused rapid tumor progression. Moreover, studies by Moses and co-workers (40) showed in a murine mammary tumor virus transgenic model that restoring an intact TGF-β signaling pathway repressed disease progression. In our study, knock down of Smad4 in a Smad4-intact PDAC cell line, UK Pan-1, significantly enhanced TGF-β-induced invasion. Conversely, expressing Smad4 in the Smad4-deficient cell line BxPC3 inhibited TGF-β-mediated motility and invasion. Thus, Smad4 signaling reduces the level of invasiveness caused by TGF-β in these cell line models. Our findings are also in agreement with the clinical observations that patients with PDAC who are deficient in Smad4 have increased metastatic progression and have shorter survival times (18, 19).
Invasion by cancer cells is a complex process involving a variety of factors. Altered expression of receptor tyrosine kinases contributes significantly to invasive growth of cancers (41, 42). Overexpression of receptor tyrosine kinases such as erbB family members is frequently found in pancreatic cancers and pancreatic cancer cell lines (17, 43). In addition, recent studies show that the RON receptor, a member of the Met family of receptor tyrosine kinases, is overexpressed in PDAC (33, 34). We found that RON receptor tyrosine kinase expression was elevated in pancreatic cancer cell lines that are deficient of Smad4 compared with cell lines that possess intact components of the Smad signaling pathway. Re-expression of Smad4 in BxPC3 cells down-regulated RON expression. TGF-β1 treatment further decreased the levels of RON in Smad4-positive cell lines. We did not find that TGF-β caused any appreciable down-regulation in the expression of erbB family of receptor tyrosine kinases (not shown). Our findings are the first to indicate that Smad4-dependent TGF-β signaling is a negative regulator of RON. Thus, loss of Smad4 likely contributes to the overexpression of RON observed in some pancreatic cancers. Our studies further indicate that TGF-β/Smad-independent signaling mediates motility and invasiveness and that optimal TGF-β-mediated motility and invasion require RON signaling. It is known that TGF-β can activate, independent of Smads, the Ras/ERK, TAK1/p38, RhoA/JNK, and phosphatidylinositol 3-kinase/AKT pathways (1). Our studies imply that cross-talk between RON and Smad4-independent TGF-β pathways cooperates in TGF-β-induced invasiveness.
Although RON is reported to be overexpressed in PDAC tumors (33), the mechanism causing this overexpression has not been investigated. Analysis of the RON promoter revealed three consensus SBEs. Smad complexes associate with other cofactors to efficiently bind SBEs and promote transcription or repression of TGF-β-responsive genes (3, 4). Activated Smads induce the expression of cyclin-dependent kinase inhibitors, including p21WAF1/CIP1, p27kip1, and p15INK4B, but repress the transcription of c-Myc (4, 44). Our findings suggest a role of Smad4-containing complexes as transcriptional repressors of the RON promoter. The importance of these SBEs in repressing RON promoter activity is supported by site-directed mutation studies. TGF-β-mediated repression of RON promoter activity was inhibited by functional inactivation of either of the two SBE sites that are most proximal to the transcriptional start site. These studies imply that Smads act as transcriptional repressors of RON, perhaps in a manner similar to that seen for c-Myc. Studies by others (35) show that RON protein turnover is mediated through ubiquitylation/proteasome pathway. Our studies indicate that in the absence of exogenous RON ligand RON protein is relatively long lived in both Smad4-intact and Smad4-deficient cells. Our studies indicate that TGF-β regulates RON expression through transcriptional repression. However, these studies do not rule out the possibility that TGF-β could partially regulate RON expression through post-translational mechanisms.
How TGF-β signaling switches from tumor-suppressive to tumor-promoting activity remains an intriguing question for cancer biologists. We show here that one critical and novel mechanism involves cross-talk between TGF-β and RON signaling pathways and that the presence or absence of intact Smad4-dependent signaling regulates TGF-β-induced invasive properties in PDAC. This TGF-β/RON axis may be exploited for cancer therapy, and the status of Smad4-dependent signaling may impact response to therapy by TGF-β RI kinase inhibitors, which are now entering clinical trials.
This work was supported by National Institutes of Health Grant CA96122 and a Veterans Affairs merit award (to J. F.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TGF-β, transforming growth factor β; ERK, extracellular signal-regulated kinase; JNK, c-Jun N-terminal kinase; PDAC, pancreatic ductal adenocarcinoma; RIKI, receptor type I kinase inhibitor; siRNA, small interference RNA; SBE, Smad binding element.
References
- 1.Derynck, R., and Zhang, Y. E. (2003) Nature 425 577–584 [DOI] [PubMed] [Google Scholar]
- 2.Lopez-Casillas, F., Wrana, J. L., and Massague, J. (1993) Cell 73 1435–1444 [DOI] [PubMed] [Google Scholar]
- 3.Attisano, L., and Wrana, J. L. (2002) Science 296 1646–1647 [DOI] [PubMed] [Google Scholar]
- 4.Feng, X. H., and Derynck, R. (2005) Annu. Rev. Cell Dev. Biol. 21 659–693 [DOI] [PubMed] [Google Scholar]
- 5.Yu, L., Hebert, M. C., and Zhang, Y. E. (2002) EMBO J. 21 3749–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott, R. L., and Blobe, G. C. (2005) J. Clin. Oncol. 23 2078–2093 [DOI] [PubMed] [Google Scholar]
- 7.Massague, J., Blain, S. W., and Lo, R. S. (2000) Cell 103 295–309 [DOI] [PubMed] [Google Scholar]
- 8.Schutte, M., Hruban, R. H., Hedrick, L., Cho, K. R., Nadasdy, G. M., Weinstein, C. L., Bova, G. S., Isaacs, W. B., Cairns, P., Nawroz, H., Sidransky, D., Casero, R. A., Jr., Meltzer, P. S., Hahn, S. A., and Kern, S. E. (1996) Cancer Res. 56 2527–2530 [PubMed] [Google Scholar]
- 9.Kretzschmar, M., Doody, J., Timokhina, I., and Massague, J. (1999) Genes Dev. 13 804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Markowitz, S., Wang, J., Myeroff, L., Parsons, R., Sun, L., Lutterbaugh, J., Fan, R. S., Zborowska, E., Kinzler, K. W., Vogelstein, B., Brattain, M., and Willson, J. K. V. (1995) Science 268 1336–1338 [DOI] [PubMed] [Google Scholar]
- 11.Zhao, S., Venkatasubbarao, K., Li, S., and Freeman, J. W. (2003) Cancer Res. 63 2624–2630 [PubMed] [Google Scholar]
- 12.Siegel, P. M., and Massague, J. (2003) Nat. Rev. Cancer 3 807–821 [DOI] [PubMed] [Google Scholar]
- 13.Jakowlew, S. B. (2006) Cancer Metastasis Rev. 25 435–457 [DOI] [PubMed] [Google Scholar]
- 14.Oft, M., Peli, J., Rudaz, C., Schwarz, H., Beug, H., and Reichmann, E. (1996) Genes Dev. 10 2462–2477 [DOI] [PubMed] [Google Scholar]
- 15.Seton-Rogers, S. E., Lu, Y., Hines, L. M., Koundinya, M., LaBaer, J., Muthuswamy, S. K., and Brugge, J. S. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 1257–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueda, Y., Wang, S., Dumont, N., Yi, J. Y., Koh, Y., and Arteaga, C. L. (2004) J. Biol. Chem. 279 24505–24513 [DOI] [PubMed] [Google Scholar]
- 17.Bardeesy, N., and DePinho, R. A. (2002) Nat. Rev. Cancer 2 897–909 [DOI] [PubMed] [Google Scholar]
- 18.Hua, Z., Zhang, Y. C., Hu, X. M., and Jia, Z. G. (2003) World J. Gastroenterol. 9 2764–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilentz, R. E., Iacobuzio-Donahue, C. A., Argani, P., McCarthy, D. M., Parsons, J. L., Yeo, C. J., Kern, S. E., and Hruban, R. H. (2000) Cancer Res. 60 2002–2006 [PubMed] [Google Scholar]
- 20.Wilkins-Port, C. E., Higgins, C. E., Freytag, J., Higgins, S. P., Carlson, J. A., and Higgins, P. J. (2007) J. Biomed. Biotechnol. 2007 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ronsin, C., Muscatelli, F., Mattei, M. G., and Breathnach, R. (1993) Oncogene 8 1195–1202 [PubMed] [Google Scholar]
- 22.Camp, E. R., Liu, W., Fan, F., Yang, A., Somcio, R., and Ellis, L. M. (2005) Ann. Surg. Oncol. 12 273–281 [DOI] [PubMed] [Google Scholar]
- 23.Zhou, Y. Q., He, C., Chen, Y. Q., Wang, D., and Wang, M. H. (2003) Oncogene 22 186–197 [DOI] [PubMed] [Google Scholar]
- 24.Wang, D., Shen, Q., Xu, X. M., Chen, Y. Q., and Wang, M. H. (2005) Carcinogenesis 26 27–36 [DOI] [PubMed] [Google Scholar]
- 25.Wang, D., Shen, Q., Chen, Y. Q., and Wang, M. H. (2004) Oncogene 23 1668–1680 [DOI] [PubMed] [Google Scholar]
- 26.Fralix, K. D., Ahmed, M. M., Mattingly, C., Swiderski, C., McGrath, P. C., Venkatasubbarao, K., Kamada, N., Mohiuddin, M., Strodel, W. E., and Freeman, J. W. (2000) Cancer 88 2010–2021 [DOI] [PubMed] [Google Scholar]
- 27.Ahmed, M. M., Alcock, R. A., Chendil, D., Dey, S., Das, A., Venkatasubbarao, K., Mohiuddin, M., Sun, L., Strodel, W. E., and Freeman, J. W. (2002) J. Biol. Chem. 277 2234–2246 [DOI] [PubMed] [Google Scholar]
- 28.Venkatasubbarao, K., Choudary, A., and Freeman, J. W. (2005) Cancer Res. 65 2861–2871 [DOI] [PubMed] [Google Scholar]
- 29.Sieg, D. J., Hauck, C. R., and Schlaepfer, D. D. (1999) J. Cell Sci. 112 Pt. 16, 2677–2691 [DOI] [PubMed] [Google Scholar]
- 30.Matsuzaki, S., Canis, M., Vaurs-Barriere, C., Boespflug-Tanguy, O., Dastugue, B., and Mage, G. (2005) Fertil. Steril. 84 Suppl. 2, 1180–1190 [DOI] [PubMed] [Google Scholar]
- 31.Venkatasubbarao, K., Ahmed, M. M., Swiderski, C., Harp, C., Lee, E. Y., McGrath, P., Mohiuddin, M., Strodel, W., and Freeman, J. W. (1998) Genes Chromosomes Cancer 22 138–144 [DOI] [PubMed] [Google Scholar]
- 32.Hahn, S. A., Schutte, M., Hoque, A. T., Moskaluk, C. A., da Costa, L. T., Rozenblum, E., Weinstein, C. L., Fischer, A., Yeo, C. J., Hruban, R. H., and Kern, S. E. (1996) Science 271 350–353 [DOI] [PubMed] [Google Scholar]
- 33.Thomas, R. M., Toney, K., Fenoglio-Preiser, C., Revelo-Penafiel, M. P., Hingorani, S. R., Tuveson, D. A., Waltz, S. E., and Lowy, A. M. (2007) Cancer Res. 67 6075–6082 [DOI] [PubMed] [Google Scholar]
- 34.Camp, E. R., Yang, A., Gray, M. J., Fan, F., Hamilton, S. R., Evans, D. B., Hooper, A. T., Pereira, D. S., Hicklin, D. J., and Ellis, L. M. (2007) Cancer 109 1030–1039 [DOI] [PubMed] [Google Scholar]
- 35.Germano, S., Barberis, D., Santoro, M. M., Penengo, L., Citri, A., Yarden, Y., and Gaudino, G. (2006) J. Biol. Chem. 281 21710–21719 [DOI] [PubMed] [Google Scholar]
- 36.Kang, Y., He, W., Tulley, S., Gupta, G. P., Serganova, I., Chen, C. R., Manova-Todorova, K., Blasberg, R., Gerald, W. L., and Massague, J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 13909–13914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian, F., DaCosta Byfield, S., Parks, W. T., Yoo, S., Felici, A., Tang, B., Piek, E., Wakefield, L. M., and Roberts, A. B. (2003) Cancer Res. 63 8284–8292 [PubMed] [Google Scholar]
- 38.Deckers, M., van Dinther, M., Buijs, J., Que, I., Lowik, C., van der Pluijm, G., and ten Dijke, P. (2006) Cancer Res. 66 2202–2209 [DOI] [PubMed] [Google Scholar]
- 39.Bardeesy, N., Cheng, K. H., Berger, J. H., Chu, G. C., Pahler, J., Olson, P., Hezel, A. F., Horner, J., Lauwers, G. Y., Hanahan, D., and DePinho, R. A. (2006) Genes Dev. 20 3130–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pierce, D. F., Jr., Gorska, A. E., Chytil, A., Meise, K. S., Page, D. L., Coffey, R. J., Jr., and Moses, H. L. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 4254–4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holbro, T., Civenni, G., and Hynes, N. E. (2003) Exp. Cell Res. 284 99–110 [DOI] [PubMed] [Google Scholar]
- 42.Ozawa, F., Friess, H., Tempia-Caliera, A., Kleeff, J., and Buchler, M. W. (2001) Teratog. Carcinog. Mutagen. 21 27–44 [DOI] [PubMed] [Google Scholar]
- 43.Freeman, J. W., DeArmond, D., Lake, M., Huang, W., Venkatasubbarao, K., and Zhao, S. (2004) Front. Biosci. 9 1889–1898 [DOI] [PubMed] [Google Scholar]
- 44.Frederick, J. P., Liberati, N. T., Waddell, D. S., Shi, Y., and Wang, X. F. (2004) Mol. Cell. Biol. 24 2546–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Del Gatto, F., Gilbert, E., Ronsin, C., and Breathnach, R. (1995) Biochim. Biophys. Acta 1263 93–95 [DOI] [PubMed] [Google Scholar]