Abstract
Human immunodeficiency virus, type 1, negative factor (Nef) initiates down-regulation of cell-surface major histocompatibility complex-I (MHC-I) by assembling an Src family kinase (SFK)-ZAP70/Syk-phosphoinositide 3-kinase (PI3K) cascade through the sequential actions of two sites, Nef EEEE65 and PXXP75. The internalized MHC-I molecules are then sequestered in endosomal compartments by a process requiring Nef Met20. How Nef assembles the multikinase cascade to trigger the MHC-I down-regulation pathway is unknown. Here we report that EEEE65-dependent binding to the sorting protein PACS-2 targets Nef to the paranuclear region, enabling PXXP75 to bind and activate a trans-Golgi network (TGN)-localized SFK. This SFK then phosphorylates ZAP-70 to recruit class I PI3K by interaction with the p85 C-terminal Src homology 2 domain. Using splenocytes and embryonic fibroblasts from PACS-2-/- mice, we confirm genetically that Nef requires PACS-2 to localize to the paranuclear region and assemble the multikinase cascade. Moreover, genetic loss of PACS-2 or inhibition of class I PI3K prevents Nef-mediated MHC-I down-regulation, demonstrating that short interfering RNA knockdown of PACS-2 phenocopies the gene knock-out. This PACS-2-dependent targeting pathway is not restricted to Nef, because PACS-2 is also required for trafficking of an endocytosed cation-independent mannose 6-phosphate receptor reporter from early endosomes to the TGN. Together, these results demonstrate PACS-2 is required for Nef action and sorting of itinerant membrane cargo in the TGN/endosomal system.
HIV-12 negative factor (Nef), a 27-kDa N-myristoylated protein, enhances viral replication and virion infectivity, and it is required for the onset of AIDS following HIV-1 infection (1, 2). Nef affects cells in many ways, including altering T-cell activation and maturation (3-5), subverting the apoptotic machinery, and down-regulating CD4 molecules and major histocompatibility complex class I (MHC-I) molecules encoded by the HLA-A and -B loci (2, 6). But the precise mechanism of how Nef mediates these pathways has remained elusive.
Nef diverts cell-surface MHC-I molecules to trans-Golgi network (TGN)-associated endosomal compartments by an endocytic pathway that is stimulated by class I phosphoinositide 3-kinase (PI3K) and dependent on ADP-ribosylation factor-6 (ARF6) (2, 7, 8). This MHC-I down-regulation requires the action of three motifs (1, 2) as follows: an N-proximal α-helical region (residues 7-26) (9) containing a critical methionine (Met20) that promotes association of MHC-I with the heterotetrameric adaptor AP-1 (10, 11); an acidic cluster (EEEE65) required for binding to phosphofurin acidic cluster sorting protein-1 (PACS-1) (12, 13); and an SH3-binding domain formed by a type II polyproline helix (PXXP75) (9, 12) that promotes association of Nef with Src family tyrosine kinases (SFKs) (14-17). The conservation of these three motifs in the pandemic M group HIV-1, which accounts for over 90% of all AIDS cases worldwide, suggests they control an essential pathway required for HIV-1 pathogenesis (18, 19).
The EEEE65 and PXXP75 sites act sequentially to recruit and stimulate class I PI3K by directing the assembly of an SFK—ZAP-70/Syk—PI3K cascade in primary CD4+ T-cells (8). Assembly of this multikinase complex is initiated by the EEEE65-dependent targeting of Nef to the paranuclear region, which enables the PXXP75 SH3 domain-binding motif to recruit a TGN-localized SFK. This Nef-activated SFK then stimulates tyrosine phosphorylation of ZAP-70/Syk, recruiting class I PI3K by an unknown mechanism to increase endocytosis of MHC-I through an ARF6-regulated pathway (7, 8). MHC-I molecules internalized by this signaling pathway are then redistributed to paranuclear endosomal compartments by a process that requires Nef Met20, which mediates interaction with AP-1 (7, 11). This Nef-assembled signaling-based pathway that triggers MHC-I down-regulation was elucidated using PACS and SFK interfering mutants, siRNA knockdown of ZAP-70 or Syk, and isoform-directed inhibitors of class I PI3Ks (8). It also required the utilization of primary cells or cell lines replete for PI3K regulation, including H9 CD4+ cells; the use of transformed cell lines with dysregulated PI3K signaling, such as phosphatase and tensin homologue-deficient Jurkat and U373 cells, which are unresponsive to PI3K inhibitors, had confounded identification of signaling pathways orchestrated by Nef (20, 21).
The acidic cluster-dependent trafficking of Nef is similar to the acidic cluster-dependent trafficking of many cellular and viral proteins, which require the sorting proteins PACS-1 and PACS-2. The PACS proteins integrate secretory pathway traffic with endoplasmic reticulum (ER)-mitochondria communication and apoptotic pathways. They bind to acidic cluster motifs on client proteins to direct their localization to secretory and endosomal compartments or to mitochondria (22-28). The ability of the PACS proteins to mediate these manifold sorting steps is achieved by their interaction with COPI; with sorting adaptors, including AP-1, AP-3, and GGA3; or with regulatory proteins, including 14-3-3 proteins, protein kinase CK2, protein phosphatase 2A, and ubiquitin ligases (24, 26, 29-32).3 Mutations in PACS proteins that interfere with these interactions block protein traffic in a cargo- and compartment-dependent manner (23-26, 30, 32, 33).
The analysis of interfering mutants to dissect the interactions of PACS proteins with these regulatory proteins has provided new insights into how these specific interactions affect protein and membrane traffic. Identifying specific sorting steps mediated by PACS-1 or PACS-2 has, however, required genetic approaches. Accordingly, RNA interference knockdown of Caenorhabditis elegans PACS, which is expressed from a single gene and localizes to presynaptic endosomes, disrupted synaptic transmission and led to paralysis (34). Studies in human cells using antisense or siRNA knockdown reveal that PACS-1 is required for the TGN localization of the cellular proteins furin and CI-MPR, as well as viral envelope glycoproteins, such as human cytomegalovirus gB and the feline RD114 retrovirus envelope glycoprotein, to regulate virus assembly (22-24, 28). PACS-1 has additional roles in late secretory trafficking steps, among them the recycling of internalized cargo from early endosomes to the cell surface (31) and the trafficking of acidic cluster-containing cargo to the primary cilium in epithelial cells (35). Likewise, the recently identified PACS-2 is required for localizing profurin and the calcium-permeable ion channel polycystin-2 to the ER (26, 36). PACS-2 also combines with PACS-1 to mediate the stepwise movement of polycystin-2 from the ER to the TGN and to the cell surface in cell types ranging from epithelial cells to Xenopus oocytes (26).
Although PACS-1 is required for Nef-mediated MHC-I down-regulation and binds Nef in an EEEE65-dependent manner (8, 13), whether Nef requires PACS-1 to form the SFK—ZAP-70/Syk—PI3K cascade has not been determined. Moreover, the roles of PACS-1 in late secretory pathway sorting do not exclude similar roles for PACS-2. Here we report that Nef requires PACS-1 and PACS-2 to down-regulate MHC-I but that their roles in Nef action are distinct. Whereas Nef requires PACS-1 distal to PI3K stimulation, studies using siRNA knockdown and cells from PACS-2-/- mice demonstrate that Nef requires PACS-2 for targeting to the late Golgi/TGN region, for assembly of the multikinase complex, and for down-regulation of MHC-I. Although Nef requires PACS-2 to target to the paranuclear region to assemble the multikinase complex, the itinerant cellular protein CI-MPR requires both PACS-1 and PACS-2 to localize to the TGN and to sort from early endosomal compartments. Together, these results suggest sequential roles for PACS-2 and PACS-1 in Nef-mediated MHC-I down-regulation and explain how the PACS proteins combine to mediate the trafficking of cellular cargo in the TGN/endosomal system.
EXPERIMENTAL PROCEDURES
Cells and Recombinant Virus—H9 CD4+ T-cells, HeLa cells, and HeLa CD8-MPR cells (provided by M. Seaman) were cultured as described (7, 8, 33). Naive CD4+ T-cells were isolated using a MACS CD4+ T-cell isolation kit (Miltenyi Biotec) from freshly drawn blood or from leukapheresed cells donated by healthy volunteers as described (8). Phycoerythrin-conjugated anti-CD4 (mAb Leu-3a, BD Biosciences) was used to verify the purity of the CD4+ T-cell population by FACS. Isolated cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum and supplemented with 2 ng/ml interleukin-7 for 4 days prior to infection. Primary splenocytes were isolated from 3-month-old WT or PACS-2-/- C57BL/6J mice, and red blood cells were removed by hypotonic lysis. The recovered PBMCs were resuspended in RPMI 1640 medium containing 10% fetal bovine serum and used immediately for assays. WT and PACS-2-/- embryo fibroblasts were prepared from 13.5-day littermate embryos by standard methods. Briefly, after removal of head and evisceration, the embryos were minced, incubated with trypsin, and then resuspended in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. The isolated MEFs were cultured in the same medium. A description of the generation and phenotype of PACS-2-/- mice will be presented elsewhere.
Vaccinia virus wild-type (VV:WT) and VV recombinants expressing Nef, FLAG-tagged Nef (Nef/f), the EEEE65→AAAA Nef mutant (NefE4A/f), Hck, or hemagglutinin-tagged PACS-2 (PACS-2ha) were generated and titered using BSC-40 cells as described (8).
PI3K Inhibitors, siRNA, and Plasmids—The class I PI3K inhibitor PI-103 (a pyridinylfuranopyrimidine derivative), the inactive PI-103 analogue PIK-112 (8, 37), and LY294002 (Calbiochem) were used as indicated. Control siRNA (scr) and siRNAs specific for human PACS-1 and PACS-2 (Smartpool, Dharmacon) were nucleofected into cells according to Amaxa instructions. In some experiments, two rounds of siRNA nucleofection were performed or cells were co-nucleofected with siRNA and pmaxGFP (Dharmacon) to enrich transfected cell populations by FACS as indicated in the legends. After 48 h cells were processed for flow cytometry or confocal microscopy. pNef-eYFP (8), pGalT-CFP (provided by Dr. J. Lippincott-Schwartz), pUHD10.1HLA-A2.1 (provided by Dr. K. Früh), and plasmids expressing the FLAG- and eGFP-tagged PI3K regulatory subunit p85α (p85α/f), eGFP-p85αR358A/f, eGFP-p85αR649A/f, and eGFP-p85αRARA/f (provided by Dr. L. Cantley) were transfected according to Amaxa instructions.
Flow Cytometry—Flow cytometry was conducted as described (8). Cells were washed and resuspended in FACS buffer (phosphate-buffered saline, pH 7.2, containing 0.5% bovine serum albumin and 0.1% NaN3). Cells were incubated with mAb BB7.1 (anti-HLA-A2.1, 1:400, BD Biosciences) at 4 °C for 1 h using an isotype-matched antibody as a negative control, washed, and then incubated with phycoerythrin-conjugated donkey anti-mouse IgG (1:600; Jackson Immuno-Research) at 4 °C for 30 min. Cells were washed and analyzed by list mode acquisition on a FACSCalibur (BD Biosciences) using CellQuest acquisition/analysis software (BD Biosciences).
In Vitro Binding Assays, Co-immunoprecipitation, and Western Blot—GST, GST-Nef, and GST-NefE4A proteins were prepared as described previously (32). His6-PACS1-FBR (human PACS-1 residues 117-294 corresponding to the cargo binding region) and His6-PACS2-FBR (human PACS-2 residues 38-217 corresponding to the cargo binding region) were purified using nickel-nitrilotriacetic acid-agarose according to the manufacturer's instructions and stored in PACS dialysis buffer (sodium phosphate, pH 7.4, 350 mm NaCl, 5 mm MgCl, 10% glycerol). 5-20 μg of GST, GST-Nef, or GST-NefE4A were mixed with 25 μl of glutathione-Sepharose beads for 1-2 h at 4 °C in binding buffer (20 mm Tris, pH 7.9, 150 mm NaCl, 0.1 mm EDTA, 0.1% Nonidet P-40) and washed with 50-100 volumes of binding buffer to remove unbound protein. Pre-bound GST proteins were incubated with 20 μg of His6-PACS1-FBR or 150 μg of His6-PACS2-FBR in binding buffer or PACS dialysis buffer, respectively, for 1-2 h at 4 °C. Bound proteins were eluted with Laemmli sample buffer, resolved by SDS-PAGE, and analyzed by Western blot using an anti-His6 antibody (Cell Signaling).
For co-immunoprecipitations, cells were harvested in phosphate-buffered saline containing 1% Nonidet P-40, protease inhibitors (0.5 mm phenylmethylsulfonyl fluoride and 0.1 μm each of aprotinin, E-64, and leupeptin), and phosphatase inhibitors (1 mm Na3VO4 and 20 mm NaF). In some experiments, cells were harvested in 20 mm Tris, pH 7.4, 150 mm NaCl, plus 1% Nonidet P-40. FLAG-tagged Nef constructs were immunoprecipitated with mAb M2-agarose (Sigma), washed three times in phosphate-buffered saline containing 1% Nonidet P-40 or (as indicated) five times in 50 mm Tris, pH 7.4, 175 mm NaCl, 1% Nonidet P-40, 0.3% deoxycholate (Nef/f:PACS-1 co-immunoprecipitation) or three times in 50 mm Tris, pH 7.4, 200 mm NaCl, 1% Nonidet P-40, 1% deoxycholate (Nef/f:PACS-2 co-immunoprecipitation), and co-immunoprecipitating proteins were detected by Western blot. The following antibodies were obtained as indicated: anti-FLAG (mAb M2, Sigma); anti-Hck (Santa Cruz Biotechnology); anti-phospho-Tyr292-ZAP-70 (BD Biosciences); anti-p85 (Upstate); anti-phospho-Tyr319Zap70/Tyr352Syk, anti-His6 antibody (Cell Signaling); anti-actin (Chemicon); anti-Nef (AIDS Research and Reference Reagent Program, National Institutes of Health), anti-MHC-I (K455, provided by Dr. K. Früh) and anti-PACS-1 703 (32). Anti-PACS-2 (193) was generated against the PACS-2 peptide DLDEDDEDVGKPKKQRRS.
PI3K Assay—In vitro lipid kinase assays were performed as described (8). Briefly, Nef/f immunoprecipitates from H9 CD4+ cells or primary splenocytes, where indicated, were incubated with 0.2 mg/ml PI (Sigma) and resuspended in assay buffer (20 mm Hepes, pH 7.4, 30 mm MgCl2, and 20 μm ATP) and 10 μCi of [α-32P]ATP for 15 min at room temperature. Phospholipids were extracted with an HCl and chloroform/methanol (1:1) solution, spotted on TLC plates (Fisher), and separated in a solvent mixture composed of CHCl3/MeOH/H2O/NHOOH (45:35:8.5:1.5). [32P]Phosphatidylinositol 1,4,5-trisphosphate was visualized by autoradiography and quantified using Scion Image 1.62.
Immunofluorescence and Confocal Microscopy—H9 CD4+ T-cells, HeLa, or HeLa CD8-CIMPR cells (which express a chimera containing the CD8 lumenal and membrane-spanning domains fused to the CI-MPR cytosolic domain) grown to ∼50% confluency were infected with VV:Nef or transfected with pNef-eYFP alone or in combination with siRNAs as stated in the legends. Cells were fixed and processed for immunofluorescence or confocal microscopy as previously described previously (32). The following primary antibodies were obtained as indicated: mAb W6/32 (anti-MHC-I, 3 μg/ml for antibody uptake studies); mAb anti-CI-MPR (ABR); anti-CI-MPR 8738 (provided by P. Luzio and S. Gary); anti-Golgin 97 (Molecular Probes); anti-mannosidase II (provided by K. Moreman); anti-Rab9 (Calbiochem); anti-TfR (provided by C. Enns); mAb anti-EEA1 (IgG1, BD Biosciences); mAb anti-CD8 (Sigma, 4 μg/ml for antibody uptake studies); and anti-PACS-2 number 604 (24). As indicated in the figure legends, following incubation with species- and subtype-specific fluorescently labeled secondary antisera (Molecular Probes), images were captured at room temperature using a 60× oil immersion objective on an Olympus Fluo-View FV300 confocal laser scanning microscope and processed with the NIH Image J program or a 63× oil immersion objective on a Leica DM-RB microscope and Hamamatsu C4742-95 digital camera. Co-localization of Nef-eYFP, CI-MPR, and CD8-CIMPR with each Golgi/endocytic marker was quantified morphometrically using Metamorph 7.0. A single-bit binary mask for each field of cells was generated based on the fluorescent signal of the specific cargo. The signal from the secretory/endocytic markers outside the mask was then quantified. The percent overlap of staining for the cargo with each marker was determined and presented as the mean. Internalized HLA-A2.1 (W6/32) was quantified morphometrically using NIH Image J. The average pixel intensity per total cell area was determined from the captured confocal images and presented as the mean. Error bars represent standard deviation.
RESULTS
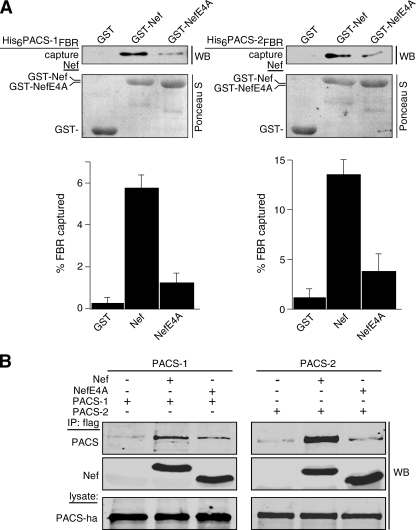
The overlapping specificity of PACS-1 and its recently identified homologue PACS-2 for binding client proteins prompted us to determine whether both PACS proteins bind HIV-1 Nef. Accordingly, we incubated GST-Nef or GST-NefE4A, which contains an EEEE65 → AAAA substitution, with His6-PACS-1fbr or His6-PACS-2fbr. The GST-Nef proteins were captured with glutathione-agarose, and bound His6-PACS proteins were detected by Western blot (Fig. 1A). We found that both His6-PACS FBRs bound GST-Nef more efficiently than they bound GST-NefE4A. To determine whether Nef associates with both PACS-1 and PACS-2 in vivo, we co-expressed FLAG-tagged Nef (Nef/f) or NefE4A/f with hemagglutinin-tagged PACS-1 (PACS-1ha) or PACS-2ha in H9 CD4+ T-cells. The infected cells were harvested, and Nef/f was immunoprecipitated, and co-precipitating PACS proteins were detected by Western blot (Fig. 1B). In agreement with the in vitro protein capture assays, we found that in these cells, PACS-1 and PACS-2 co-immunoprecipitated Nef and that the EEEE65 → AAAA substitution reduced their interaction.
FIGURE 1.
Nef EEEE65-dependent binding to PACS-1 and PACS-2. A, purified His6PACS-1FBR (residues 117-294) or His6PACS-2FBR (residues 38-217) was incubated with GST, GST-Nef, or GST-NefE4A pre-bound to glutathione-Sepharose. Top, bound proteins were analyzed by Western blot (WB) with anti-His6 antibody showing a 10% input. Bottom, Ponceau S stain of input proteins. Quantification showed that GST-Nef captured 5.8-fold more His6PACS-1FBR and 4.7-fold more His6PACS-2FBR than did GST-NefE4A. Data are representative of three independent experiments. B, H9 CD4+ T-cells were co-infected with VV:PACS-1 (m.o.i. = 3, 8 h) or VV:PACS-2 (m.o.i. = 5,8h) and either VV:WT, VV:Nef/f, or VV:NefE4A/f (m.o.i. = 5, 8 h). FLAG-tagged Nef proteins were immunoprecipitated (IP), and co-precipitating PACS proteins were analyzed by Western blot with antibody 17703 (PACS-1) or antibody 18193 (PACS-2). Quantification showed that Nef/f co-precipitated 3.7-fold more PACS-1 and 6.3-fold more PACS-2 than did NefE4A/f.
Next, we asked whether PACS-1, PACS-2, or both proteins are required for Nef-mediated MHC-I down-regulation. Primary human CD4+ T-cells isolated from healthy donors were nucleofected (Amaxa) with a control siRNA (scr) or with siRNAs that deplete PACS-1 or PACS-2 (Fig. 2A). The cells were then infected with wild-type vaccinia virus or with a vaccinia recombinant expressing HIV-1 Nef. The amount of cell-surface MHC-I was determined by flow cytometry. We used recombinant vaccinia virus in these studies because it expresses Nef in H9 CD4+ cells at levels similar to HIV-1 and down-regulates cell-surface MHC-I at a similar efficiency, but it infects nearly 100% of the cell population (8). We found that depleting either PACS-1 or PACS-2 inhibited Nef from efficiently down-regulating MHC-I in primary CD4+ T-cells.
FIGURE 2.
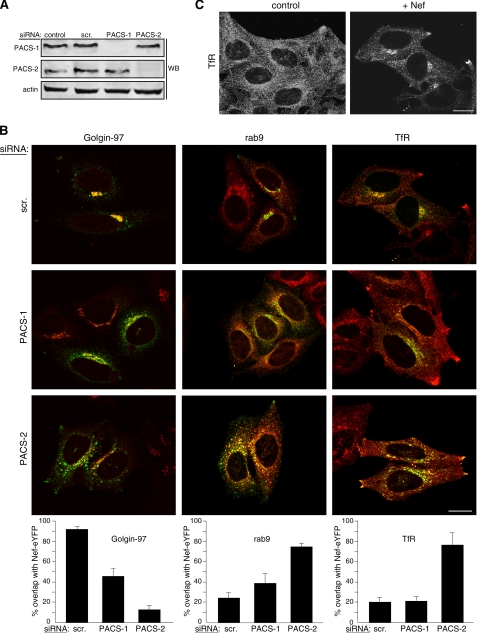
Nef-mediated MHC-I down-regulation in primary CD4+ T-cells requires PACS-1 and PACS-2. A, primary human CD4+ T-cells isolated from healthy donors were nucleofected (Amaxa) with a control siRNA (scr) or siRNAs specific for PACS-1 or PACS-2. At 72 h post-transfection, the cells were either harvested for Western blot (WB) of PACS proteins (bottom) or infected with VV:WT or VV:Nef (m.o.i. = 10, overnight), and cell-surface HLA-A2.1 was analyzed by flow cytometry using mAb BB7.1 (top). The expression of Nef is shown in the insets for each graph. B, top, H9 cells were nucleofected (Amaxa) with pmaxGFP together with a control siRNA or siRNAs specific for PACS-1 or PACS-2. At 72 h post-transfection, the GFP+ cells were enriched by FACS and then infected with VV:WT or VV:Nef (m.o.i. = 10, 5 h) and then fixed and stained with W6/32 to visualize MHC-I down-regulation. Scale bar = 20 μm. Bottom, Western blot of lysates from parallel samples of nucleofected cells.
Next, replicate plates of H9 CD4+ T-cells treated with siRNAs that specifically depleted PACS-1 or PACS-2 were infected with a recombinant virus expressing HIV-1 Nef and the subcellular distribution of MHC-I was monitored by immunofluorescence microscopy (Fig. 2B). Under these conditions, the Nef-induced redistribution of MHC-I is caused by down-regulation of cell-surface molecules and not by altering MHC-I stability or biosynthetic transport (7, 8). In cells treated with the control siRNA, Nef induced the redistribution of cell-surface MHC-I to the paranuclear region. By contrast, siRNA depletion of either PACS-1 or PACS-2 inhibited MHC-I down-regulation.
The EEEE65-dependent binding of Nef to PACS-1 and PACS-2, together with the requirement of both proteins for Nef to down-regulate MHC-I, led us to ask whether one or both PACS proteins were also required for localizing Nef to the late Golgi/TGN region, a step required for Nef to assemble the SFK—ZAP-70/Syk—PI3K cascade that triggers the MHC-I down-regulation pathway (8). As T-cell lines are not ideal for morphologic analyses, we used HeLa cells, which support the same Nef-triggered multikinase cascade used to down-regulate cell-surface MHC-I in HIV-1 permissive cells (8). Replicate plates of HeLa cells were treated with siRNAs that specifically depleted PACS-1 or PACS-2 (Fig. 3A) and then transfected with a plasmid expressing Nef-eYFP. The subcellular localization of Nef-eYFP was determined by confocal microscopy (Fig. 3B). In control cells, Nef-eYFP concentrated in the paranuclear region, where it co-localized with Golgin-97, a late Golgi/TGN marker. Morphometric analysis showed a greater than 90% overlap of Nef-eYFP with Golgin-97 (Fig. 3B).
FIGURE 3.
Paranuclear localization of Nef-eYFP requires PACS-2. A, Western blot (WB) of extracts from HeLa cells nucleofected (Amaxa) with a control siRNA (scr) or siRNAs specific for PACS-1 or PACS-2. B, cells in A were nucleofected a second time with pNef-eYFP (green). At 16 h post-transfection the cells were fixed, stained with antibodies for Golgin 97, Rab9, and TfR followed by incubation with fluorescently labeled secondary antibodies (red), and analyzed by confocal microscopy. Morphometric analysis was performed as described under “Experimental Procedures.” Error bars represent mean and S.D. from 20 cells per marker in five independent experiments. Scale bar = 20 μm. C, untreated HeLa cells or HeLa cells expressing Nef-eYFP were fixed, stained with anti-TfR, and analyzed by confocal microscopy. Scale bar = 20 μm.
In agreement with others (38), Nef-eYFP also co-localized with a Nef-induced, paranuclear-restricted subpopulation of endosomal compartments containing Rab9, a late endosomal marker, and transferrin receptor (TfR), an early/recycling endosome marker (see also Fig. 3C). As in previous studies (13), the Nef-eYFP staining pattern was slightly dispersed in PACS-1-depleted cells, resulting in a more limited overlap with Golgin-97. The dispersed Nef-eYFP overlapped with a slightly distended population of Rab9- and TfR-containing endosomal compartments. Morphometric analysis showed that siRNA depletion of PACS-1 reduced the overlap of Nef-eYFP with Golgin-97 to 45% and correspondingly increased the overlap with Rab9 from 24 to 38%. Depletion of PACS-2, however, caused the Nef-eYFP staining pattern to be markedly dispersed throughout the cytosol, reducing the overlap with Golgin-97 to 13%. Moreover, depletion of PACS-2 prevented the Nef-eYFP-induced paranuclear restriction of early/recycling endosomes. Instead, Nef-eYFP accumulated in dispersed Rab9 and TfR-containing compartments, including a population of TfR-containing endosomal compartments concentrated at the tips of cells. Morphometric analysis revealed that the percentage of Rab9- and TfR-containing endosomal compartments that overlapped with Nef-eYFP each increased to 75% in PACS-2-depleted cells. Together, these results suggest a major role for PACS-2, and to a lesser extent, PACS-1, in localizing Nef-eYFP to the late Golgi/TGN and associated endosomal compartments, which concentrate in the paranuclear region in a Nef/PACS-dependent manner.
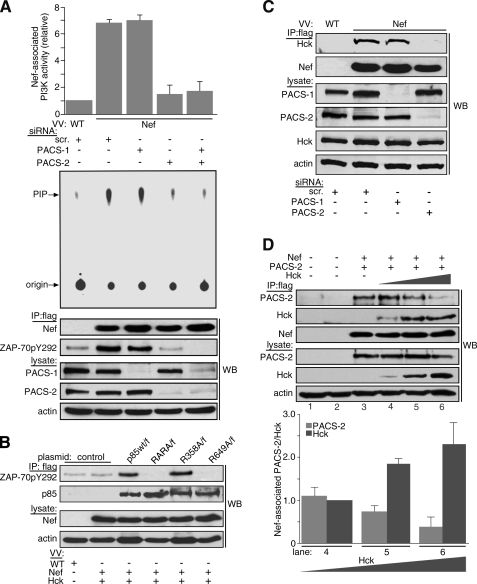
The PACS-dependent localization of Nef-eYFP to the TGN region led us to ask whether PACS-2 or PACS-1 was required for Nef to assemble the SFK—ZAP-70/Syk—PI3K cascade that triggers MHC-I down-regulation. To test this possibility, we expressed Nef/f in H9 CD4+ T-cells that were treated with siRNAs to deplete PACS-1, PACS-2, or both proteins. Nef/f was immunoprecipitated, and the amount of co-immunoprecipitating class I PI3K activity was determined using an in vitro lipid kinase assay (Fig. 4A, top). We found that siRNA depletion of PACS-2 but not PACS-1 blocked the ability of Nef to stimulate PI3K. We then asked at which point along the SFK—ZAP-70/Syk—PI3K cascade Nef requires PACS-2. To associate with PI3K in CD4+ T-cells, Nef must recruit and trigger activation of ZAP-70 (8). We found that siRNA depletion of PACS-2, but not PACS-1, blocked the ability of Tyr292-phosphorylated ZAP-70 to associate with Nef/f (Fig. 4A, bottom). However, depletion of both PACS-2 and PACS-1 further reduced Tyr292-phosphorylated ZAP-70, suggesting PACS-1 may augment the PACS-2-dependent phosphorylation of ZAP-70.
FIGURE 4.
PACS-2 is required for Nef to assemble the SFK—ZAP-70/Syk—PI3K cascade. A, H9 CD4+ cells were nucleofected (Amaxa) with a control (scr) siRNA or siRNAs specific for PACS-1 or PACS-2 on day 1 and again on day 3. On day 5, cells were infected with either VV:WT or VV:Nef/f (m.o.i. = 10, 8 h). Nef was immunoprecipitated (IP), and co-immunoprecipitating Tyr(P)292Zap70 was detected by Western blot (WB). The amount of Nef-associated PI3K activity was quantified as described under “Experimental Procedures.” Bottom, Western blot showing expression of PACS-1, PACS-2, and actin. Error bars represent the mean ± S.D. of three independent experiments. B, H9 cells were transfected on day 1 with plasmids expressing p85α/f, p85αR358A/f, p85αR649A/f, or p85αRARA/f. On day 3, the cells were infected with VV:WT (m.o.i. = 10, 8 h) or co-infected with VV:Nef (untagged, m.o.i. = 10, 8 h) and VV:Hck (m.o.i. = 2, 8 h). Cells were harvested, and FLAG-tagged p85α was immunoprecipitated with mAb M2 and co-precipitating Tyr(P)292ZAP-70 detected by Western blot. Data are representative of two independent experiments. C, H9 cells were transfected with siRNAs as described in A. On day 5, the cells were co-infected with VV:WT or VV:Nef/f (m.o.i. = 10, 8 h) and VV:Hck (m.o.i. = 2, 8 h). Nef was immunoprecipitated, and co-immunoprecipitating Hck was detected by Western blot. Data are representative of three independent experiments. D, parallel plates of H9 CD4+ cells were infected or not with VV:Nef/f (m.o.i. = 6 each) and VV:PACS-2ha (m.o.i. = 5 each) and with decreasing amounts of VV:WT (m.o.i. in lane 2 = 14, lane 3 = 3, lane 4 = 2, and lane 5 = 1) or VV:Hck (m.o.i. in lane 4 = 1, lane 5 = 2, lane 6 = 3; total m.o.i. = 14, 18 h) and harvested as indicated under “Experimental Procedures.” Nef/f was immunoprecipitated with mAb M2, and co-precipitating Hck and PACS-2ha were detected by Western blot. Error bars represent mean and S.D. from three independent experiments.
Next, we asked how Nef-SFK-ZAP-70 recruits class I PI3K. One possibility was suggested by a recent study showing that Tyr-phosphorylated ZAP-70 recruits PI3K by binding to p85 regulatory subunit SH2 domains (39). We therefore tested whether Nef directs formation of phospho-ZAP-70 to recruit PI3K by interacting with one of these domains. We transfected H9 CD4+ T-cells with plasmids expressing either FLAG-tagged p85α (p85α/f) or p85α constructs containing Arg→Ala mutations that block Tyr(P) binding in the N-terminal SH2 domain (p85αR358A/f), the C-terminal SH2 domain (p85αR649A/f), or both SH2 domains (p85αRARA/f). After 48 h the transfected cells were infected with vaccinia recombinants co-expressing untagged Nef and Hck, which is an SFK that is activated by binding to the Nef PXXP75 motif (15). The cells were harvested, and FLAG-tagged p85α constructs were immunoprecipitated, and co-precipitating ZAP-70 was detected with a Tyr(P)292-specific antibody (Fig. 4B). We found that p85α/f and p85αR358A/f but not p85αR649A/f or p85αRARA/f associated with Nef/Hck-activated ZAP-70. These findings suggest that Nef recruits PI3K by directing the ligation of phospho-ZAP-70 to the C-terminal SH2 domain in the p85α regulatory subunit.
Unlike the classic mechanism of PI3K stimulation described for cell-surface receptors, our studies suggest Nef directs phosphorylation of ZAP-70 at Tyr292 by first recruiting an SFK at the TGN region (8). To test whether Nef required PACS-2 to bind a TGN-localized SFK, we treated H9 CD4+ cells with siRNAs specific for PACS-1 or PACS-2. We then co-expressed Nef/f and Hck, which localizes to the TGN where it binds Nef and stimulates it to recruit PI3K (8). Nef/f was immunoprecipitated, and co-precipitating Hck was detected by Western blot (Fig. 4C). We found that siRNA depletion of PACS-2 but not PACS-1 inhibited the ability of Nef to co-immunoprecipitate Hck. Together, these siRNA-based experiments suggest Nef requires PACS-2 to target to the late Golgi/TGN, thereby triggering formation of the multikinase cascade, and requires PACS-1 at a subsequent step in the MHC-I down-regulation pathway.
PACS-2 binds Nef and is required for Nef to target to the TGN region, where it binds an SFK to assemble the SFK—ZAP-70/Syk—PI3K cascade. However, we found that PACS-2 did not co-immunoprecipitate with the multikinase complex (data not shown). We therefore asked if PACS-2 molecules bound to Nef were exchanged for an SFK, because this would explain the absence of PACS-2 in the multikinase complex. To test this possibility, we co-expressed a constant amount of PACS-2ha and Nef/f with increasing amounts of Hck in replicate plates of H9 CD4+ cells. Nef/f was immunoprecipitated, and co-precipitating PACS-2ha and Hck were detected by Western blot (Fig. 4D). We found that increased expression of Hck reduced the amount of PACS-2 that co-immunoprecipitated with Nef, which was replaced by a corresponding increase in the amount of co-precipitating Hck.
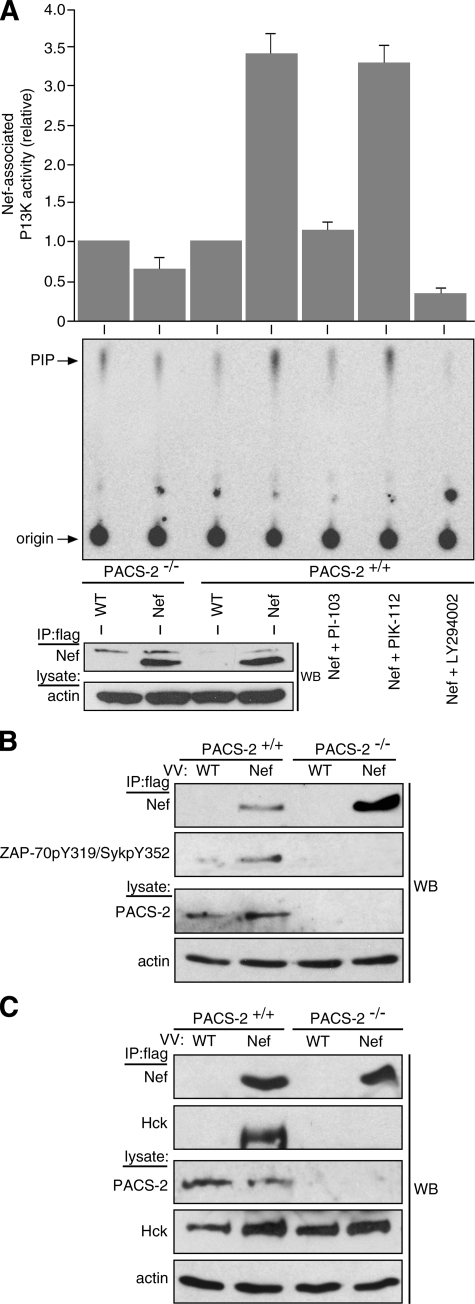
To determine unequivocally whether PACS-2 is required for Nef to assemble the SFK—ZAP-70/Syk—PI3K cascade, we tested the ability of Nef to stimulate class I PI3K in cells isolated from WT or PACS-2-/- mice. Accordingly, splenocytes were isolated from C57Bl/6 WT or PACS-2-/- mice and then infected with a VV recombinant expressing Nef/f. At 8 h post-infection, Nef/f was immunoprecipitated, and co-precipitating PI3K activity was detected using the in vitro lipid kinase assay (Fig. 5A). In agreement with studies of Nef expressed in human cells (8), we found that Nef/f recruited class I PI3K only in splenocytes from WT mice but not from PACS-2-/- mice. Also in agreement with the findings in human cells, the Nef/f-associated PI3K was sensitive to the PI3K inhibitor LY294002.
FIGURE 5.
Pacs-2 gene knock-out prevents Nef assembly of the SFK—ZAP-70/Syk—PI3K cascade in mouse splenocytes. A, splenocytes isolated from WT C57Bl/6 or PACS-2-/- C57Bl/6 mice were infected with VV:WT or VV:Nef/f (m.o.i. = 25, 8 h). Nef was immunoprecipitated (IP), and PI3K activity was measured as described under “Experimental Procedures.” Western blot (WB) depicts immunoprecipitated Nef/f. A replicate immunoprecipitate from Nef/f-expressing WT C57Bl/6 splenocytes was aliquoted and treated with 0.1 μm PI-103, PIK-112, or 5 μm LY294002, and Nef-associated PI3K activity was measured. Error bars represent the mean ± S.D. of five independent experiments (10 mice total). B, primary splenocytes isolated from WT or PACS-2-/- C57Bl/6 mice were infected as described in A. Nef/f was immunoprecipitated and co-immunoprecipitating Tyr(P)319Zap-70/Tyr(P)352Syk was detected by Western blot. C, primary splenocytes isolated from three paired sets of WT or PACS-2-/- C57Bl/6 mice were co-infected with VV:WT or VV:Nef/f (m.o.i. = 10, 18 h) and VV:Hck (m.o.i. = 2, 18 h). Nef was immunoprecipitated, and co-immunoprecipitating Hck was detected by Western blot. The lower m.o.i. in the VV:WT-infected samples does not affect the result (see Fig. 4D).
As LY294002 inhibits all classes of PI3K (40), we asked if Nef/f expressed in mouse splenocytes associated specifically with a class I PI3K. We treated the immunoprecipitates with the class I PI3K inhibitor PI-103, which blocks Nef-associated class I PI3K in vitro and represses MHC-I down-regulation in primary human CD4+ T-cells and H9-CD4+ cells (8). Treatment of the Nef-associated PI3K activity from mouse splenocytes with PI-103, but not its inactive analogue PIK-112, similarly blocked the Nef-associated PI3K activity. Moreover, Nef/f expressed in mouse cells appeared to recruit and stimulate class I PI3K by the same signaling cascade we identified in human cells; only Nef/f expressed in WT splenocytes co-precipitated tyrosine-phosphorylated (activated) ZAP-70/Syk (Fig. 5B).
To test whether Nef requires PACS-2 to bind an SFK, we co-expressed Nef/f and Hck, which localizes to the TGN region (8, 41), in WT or PACS-2-/- splenocytes. The Nef/f was immunoprecipitated, and co-precipitating Hck was detected by Western blot (Fig. 5C). We found that Nef associated with Hck in WT splenocytes but not in PACS-2-/- splenocytes. Together, these studies confirm genetically that Nef requires PACS-2 to assemble the SFK—ZAP-70/Syk—PI3K cascade and that our findings from studies using siRNA knockdown in human primary cells and cell lines recapitulate those in PACS-2-/- mice.
As PACS-2 was initially found to localize membrane cargo to the ER (26), our determination that PACS-2 was required for localization of Nef-eYFP to the late Golgi/TGN region led us to ask whether in addition to PACS-1, PACS-2 may also regulate the sorting of cellular cargo in the late secretory pathway. One candidate protein is CI-MPR, an itinerant cellular cargo protein that contains in its cytosolic domain a CK2-phosphorylatable acidic cluster, DDpSDED2487, required for PACS binding (32). To test this possibility, we first determined the localization of endogenous CI-MPR in cells depleted of PACS-1 or PACS-2. We found that siRNA depletion of either PACS-1 or PACS-2 caused CI-MPR to mislocalize from the TGN to a population of Rab9- and TfR-containing endosomal compartments (supplemental Fig. S1).
The requirement for PACS-1 and PACS-2 to maintain the steady-state localization of CI-MPR to the TGN raised the possibilities that PACS-1 and PACS-2 mediate solely the localization of CI-MPR to the TGN or that one or both sorting proteins are required for routing of endocytosed CI-MPR to the TGN. To test these possibilities, we measured the fate of an internalized CI-MPR reporter construct, CD8-CIMPR, by antibody uptake in HeLa:CD8-CIMPR cells depleted of PACS-1 or PACS-2. Replicate plates of HeLa:CD8-CIMPR cells were treated with a control siRNA or with siRNAs specific for PACS-1 or PACS-2 (Fig. 6, bottom) and then incubated with anti-CD8 for 1 h and processed for confocal microscopy. In control cells or in PACS-1 siRNA cells, internalized CD8-CIMPR concentrated in the paranuclear region where it overlapped with Golgin-97 but not with TfR (Fig. 6, top, and data not shown).
FIGURE 6.
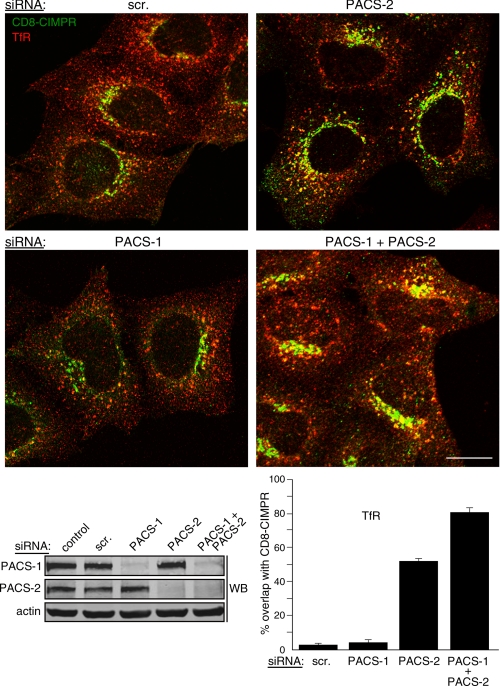
PACS-1 and PACS-2 mediate the sorting of endocytosed CD8-CIMPR. HeLa-CD8CIMPR cells were nucleofected (Amaxa) with a control siRNA (scr) or siRNAs specific for PACS-1 or PACS-2. After 48 h, anti-CD8 (mAb UCHT-4, green) was added to the medium for 1 h at 37 °C. The media were removed and replaced with fresh culture medium for 30 min, and the cells were fixed and stained with anti-TfR (red) followed by isoform-specific fluorescent secondary antibodies. The cells were then visualized by confocal microscopy. Morphometric analysis was performed as described under “Experimental Procedures.” Error bars represent mean ± S.D. from 20 cells per marker in eight independent experiments. Bottom, Western blot (WB) of lysates from parallel samples of nucleofected cells. Scale bar = 20 μm.
These findings suggested that although PACS-1 mediates the steady-state localization of itinerant membrane cargo to the TGN, it is not required for the sorting of endocytosed cargo to the TGN. By contrast, siRNA depletion of PACS-2 inhibited targeting of internalized CD8-CIMPR to the TGN region, causing the internalized CD8-CIMPR to accumulate in TfR-containing endosomes. Morphometric analysis revealed that siRNA knockdown of PACS-2 increased the overlap of endocytosed CD8-CIMPR with TfR from 3 to 52%. Because our previous studies showed that, in addition to maintaining furin and other proteins locally at the TGN, PACS-1 is required for recycling endocytosed furin from endosomes to the cell surface (31), we asked what effect siRNA co-depletion of PACS-2 and PACS-1 would have on the localization of internalized CD8-CIMPR. Accordingly, we found that depletion of both PACS proteins markedly increased the amount of internalized CD8-CIMPR that accumulated in TfR-containing endosomes, increasing the overlap of CD8-CIMPR with TfR to 81% (Fig. 6B). Together, these results suggest both PACS-1 and PACS-2 are required for the local concentration of CI-MPR to the TGN but, whereas PACS-2 is required for the delivery of internalized CD8-CIMPR to the TGN, PACS-1 mediates recycling of the endocytosed cargo to the cell surface (31).
To test unequivocally the requirement of PACS-2 for trafficking both HIV-1 Nef and CI-MPR, we determined the localization of each protein in genetically paired WT and PACS-2-/- MEFs. We first expressed Nef-eYFP in WT and PACS-2-/- MEFs (Fig. 7A). In WT MEFs, Nef-eYFP overlapped with the Golgi marker mannosidase II. As with human cells, Nef-eYFP also caused a subpopulation of early endosomes to collapse toward the Golgi region. By contrast, Nef-eYFP in PACS-2-/- MEFs was dispersed and displayed an increased overlap with EEA1-positive early endosomes, which also failed to collapse to the paranuclear region. We used mannosidase II and EEA1 to identify the Golgi and early endosomes in these experiments as neither the Golgin-97 nor the TfR antibodies stained MEF cells. To determine the effect of genetic loss of PACS-2 on endogenous CI-MPR localization, WT and PACS-2-/- MEFs were transfected with cyan fluorescent protein-tagged galactosyltransferase (GalT-CFP), a late Golgi marker, stained with anti-CI-MPR and then visualized by confocal microscopy (Fig. 7B). In WT MEFs, CI-MPR overlapped with GFP-GalT whereas in PACS-2-/- MEFs CI-MPR displayed a dispersed staining pattern with an increased redistribution to EEA1-positive compartments. These experiments with cells from knock-out mice demonstrate that PACS-2 is required for localization of Nef and cellular cargo and validate genetically the requirement for PACS-2 by Nef to assemble the SFK-ZAP-70-PI3K complex in primary splenocytes.
FIGURE 7.
Nef-eYFP and endogenous CI-MPR are mislocalized in PACS-2-/- MEFs. A, PACS-2+/+ and PACS-2-/- MEFs were transfected (Amaxa) with pNef-eYFP. After 18 h the cells were fixed and stained with anti-mannosidase II or anti-EEA1 (red) and visualized by confocal microscopy. Western blot (WB) in upper right-hand corner of PACS-2 in lysates from PACS-2+/+ and PACS-2-/- MEFs is shown. Inset, Nef-eYFP in dispersed EEA1-postive compartments in PACS-2-/- MEFs. B, PACS-2+/+ and PACS-2-/- MEFs were transfected (Amaxa) or not with pGalT-CFP. After 18 h the cells were fixed, stained with anti-EEA1 (red) or anti-CI-MPR 8738 (red in GalT-expressing cells; green in EEA1 co-stained cells (bottom)), and visualized by confocal microscopy. Inset, CI-MPR in EEA1-positive compartments. Morphometric analysis was performed as described under “Experimental Procedures.” Error bars represent mean ± S.D. from 20 cells per marker in four independent experiments for Nef-eYFP and three independent experiments for CI-MPR. Scale bar = 20 μm.
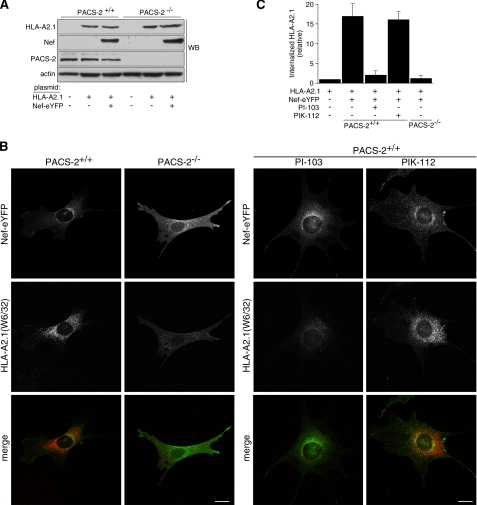
The inability of HIV-1 Nef to localize to the paranuclear region and therefore to assemble the multikinase cascade in PACS-2-/- MEFs suggested that these cells could be used to test genetically the importance of PACS-2 for Nef-mediated down-regulation of MHC-I. Accordingly, we expressed human HLA-A2.1 in replicate plates of WT and PACS-2-/- MEFs alone or together with Nef-eYFP. Western blot analysis showed similar levels of HLA-A2.1 and Nef-eYFP expressed in WT and PACS-2-/- MEFs (Fig. 8A). The cells were then incubated with mAb W6/32 for 1 h and then processed for confocal microscopy (Fig. 8, B and C). We used mAb W6/32 in these experiments because it recognizes fully assembled human MHC-I molecules and we have successfully used this antibody to monitor MHC-I down-regulation by antibody uptake (7, 8, 42). In WT MEFs, we found that Nef-eYFP concentrated in the paranuclear region and induced the internalization of cell-surface HLA-A2.1 to perinuclear endosomal compartments. By contrast, when expressed in PACS-2-/- MEFs, Nef-eYFP failed to concentrate in the paranuclear region or down-regulate cell-surface HLA-A2.1.
FIGURE 8.
PACS-2 and class I PI3K are required for Nef to down-regulate MHC-I in MEFs. A, Western blot (WB) of lysates from PACS-2+/+ and PACS-2-/- MEFs co-transfected (Amaxa) with pUHD10.1HLA-A2.1 and pNef-eYFP. B, after 48 h, anti-MHC-I (mAb W6/32, red) was added to the media for 1 h at 37 °C. Where indicated the cells were pretreated with 1 μm PI-103 or PIK-112 for 1 h prior to antibody uptake, and the inhibitors were maintained in the culture medium for the duration of the antibody uptake. Following uptake, the media were removed and replaced with fresh culture media for 30 min in the absence or presence of inhibitor. The cells were fixed, stained with a fluorescent secondary antibody, and then visualized by confocal microscopy. C, morphometric quantitation of internalized HLA-A2.1 (W6/32). The amount of internalized antibody was the same in WT or PACS-2-/- cells transfected with pUHD10.1HLA-A2.1 alone, and therefore the same value was used for normalization of uptake in lane 1. The error bars represent the mean ± S.D. from 20 cells per marker from three independent experiments. Scale bar = 20 μm.
Next, to test whether the PACS-2-dependent down-regulation of MHC-I by Nef requires PI3K activity, replicate wells of WT MEFs were treated with the class I PI3K inhibitor PI-103 or its inactive analogue PIK-112 during the antibody uptake. We found that PI-103 but not PIK-112 inhibited the ability of Nef to down-regulate HLA-A2.1. Together, these experiments demonstrate using gene knock-out and pharmacologic inhibition of class I PI3K that PACS-2 is required for HIV-1 Nef to assemble the SFK—ZAP-70/Syk—PI3K cascade that triggers down-regulation of cell-surface MHC-I, confirming our determination that siRNA knockdown of PACS-2 and inhibition of class I PI3K in human primary CD4+ T-cells inhibits MHC-I down-regulation (Fig. 2) (8).
DISCUSSION
PACS-2 was initially shown to regulate the ER trafficking of acidic cluster-containing cargo (26, 30, 36). Here, we identified new roles for PACS-2 in regulating protein and membrane traffic in the TGN/endosomal system. PACS-2 is required for localization of HIV-1 Nef to the paranuclear region, which enables Nef to assemble the SFK—ZAP-70/Syk—PI3K cascade that triggers down-regulation of cell-surface MHC-I. We also found that PACS-2 and PACS-1 combine to mediate distinct trafficking steps of cellular cargo in the late secretory pathway, including the steady-state localization of CI-MPR to the TGN and the sorting of endocytosed CD8-CIMPR from early endosomes. Our results using siRNA knockdown in human primary cells and cell lines phenocopy those using cells from PACS-2-/- mice, demonstrating genetically the role of PACS-2 in regulating Nef action and the sorting of itinerant cell cargo.
PACS-1 and PACS-2 appear to mediate parallel pathways maintaining the steady-state localization of CI-MPR to the TGN but appear to have separate roles in regulating the sorting of internalized CD8-CIMPR from early endosomes (Fig. 6 and supplemental Fig. S1). The sorting of itinerant membrane proteins from early endosomes is complex, delivering cargo to several destinations, including recycling to the cell surface, transfer to late endosomes, or sorting to the TGN. Together with previous work (31), our results suggest PACS-2 mediates delivery from early endosomes to the TGN, whereas PACS-1 mediates recycling to the cell surface. These opposing sorting steps mediated by the PACS proteins may explain why PACS-1 interfering mutants block retrieval of endocytosed acidic cluster-containing cargo from early endosomes to the TGN (24, 32, 33). That PACS-1 and PACS-2 regulate sorting from early endosomes is supported by genetic studies demonstrating that C. elegans PACS is localized to presynaptic endosomes and regulates neurotransmission (34). But how mammalian PACS-1- and PACS-2-dependent sorting from early endosomes is controlled and how PACS-2 may cooperate with other sorting molecules that mediate early endosome-to-TGN trafficking of internalized cargo (see Refs. 43-46 and for review see Ref. 47) remains to be determined.
Although PACS-2 is required for delivery of both endocytosed CD8-CIMPR and Nef-eYFP from an early endosome population to the TGN, it appears to do so by different mechanisms. Only Nef-eYFP caused the PACS-2-dependent restriction of a population of early endosomal compartments to the paranuclear region in HeLa cells or MEFs (Figs. 3 and 7). Our studies are in agreement with others who reported Nef collapses early endosomal compartments to the paranuclear region and disrupts transferrin uptake (38). These studies, however, identified a requirement for the Nef DXXXLL165 motif (48), which binds to AP-1 and AP-3 (48-50), but not the EEEE65 motif. As the EEEE65 → AAAA mutation suppressed, but did not block, interaction of Nef with PACS-2 or PACS-1 in vivo (Fig. 1), we suspect that the residual interaction between NefE4A and PACS-2 may be sufficient to affect redistribution of endosomal compartments but not to efficiently target Nef to the Golgi/TGN or enable Nef to stimulate PI3K. Whether EEEE65/PACS-2 and DXXXLL165/AP act independently or together to collapse early endosomal compartments to the paranuclear region and what role this organellar redistribution has on the mechanism of Nef action warrants further investigation.
Together with our recent report (8), the studies here identify the molecular basis for the sequential roles of the Nef EEEE65 and PXXP75 motifs, conserved in pandemic M group HIV-1, in initiating the MHC-I down-regulation pathway. Here we established the requisite role for PACS-2 in mediating the ability of Nef to assemble the SFK—ZAP-70/Syk—PI3K cascade that triggers MHC-I down-regulation using siRNA knockdown, gene knock-out, and pharmacologic inhibition of class I PI3Ks. Our results suggest that Nef EEEE65-dependent binding to PACS-2 (Fig. 1) directs trafficking of Nef and Nef-containing early endosomal compartments to the paranuclear region (Figs. 3 and 7), where the PXXP75 motif then binds and activates a TGN-localized SFK (Figs. 4 and 5) (8). This step directs the exchange of PACS-2 for the SFK, as their association with Nef in vivo appears to be mutually exclusive (Fig. 4D). The absence of PACS-2 in the multikinase complex is further supported by the lack of effect of Nef on the subcellular distribution of endogenous PACS-2 (supplemental Fig. S2).
SFK activation generally requires the C-terminal dephosphorylation of the kinase by plasma membrane-associated tyrosine phosphatase. However, Nef directly activates a subset of SFKs, including Hck, Src, and Lyn (14), pools of which are localized to the Golgi/TGN region (41, 51), by a PXXP75-mediated displacement of the kinase SH3 domain (52). The activated, Nef-bound SFK then phosphorylates ZAP-70 or its homologue Syk in a reservoir-specific manner to recruit and stimulate a class I PI3K by ligating the C-terminal SH2 domain in p85 (Fig. 4B). This ligation is similar to the mechanism used by phagocytic cells to trigger ARF6-mediated Fc receptor internalization, which is stimulated by the binding of class I PI3K to SFK-phosphorylated ZAP-70/Syk (39). The recruited class I PI3K then triggers the ARF6-controlled down-regulation of cell-surface MHC-I to TGN/endosomal compartments, a process that requires Nef Met20 (7), which mediates the interaction of MHC-I with AP-1 (11). In the absence of PACS-2, Nef is unable to bind SFKs, recruit and direct ZAP-70 Tyr292 phosphorylation, or stimulate class I PI3K, thereby preventing MHC-I down-regulation (Figs. 2 and 4).
The requirement for PACS-2 by Nef to recruit class I PI3K in H9 cells was not because of off-target effects of the PACS-2 siRNA, because Nef also failed to recruit and stimulate PI3K in splenocytes from PACS-2-/- mice and to localize to the paranuclear region or down-regulate transfected HLA-A2.1 in PACS-2-/- MEFs (Figs. 5, 7, and 8). Together, these results demonstrate genetically an essential role for PACS-2 in mediating Nef action.
Although PACS-1 has a limited role in Nef trafficking (Fig. 3), it is not required for Nef to recruit PI3K (Fig. 4), suggesting that EEEE65/PACS-1 is required for MHC-I down-regulation subsequent to the PACS-2-dependent formation of the SFK—ZAP-70-Syk—PI3K complex. That Nef induces PACS-1 to interact with MHC-I supports this possibility.4 Studies identifying the step in the Nef-mediated MHC-I down-regulation itinerary requiring PACS-1 are currently underway.
Our results describing the roles of PACS-2 and PACS-1 in mediating Nef action and regulating the sorting of itinerant cell cargo agree with biochemical, RNA interference, and gene knock-out analyses from evolutionally diverse metazoans, identifying the PACS family as bona fide membrane and protein traffic regulators (7, 8, 13, 22-28, 30-36). However, our results disagree with a recent report asserting that Nef-mediated MHC-I down-regulation did not involve PACS proteins and challenged whether the PACS proteins mediate protein traffic (53). We do not know the basis for these conflicting results. As our results show that siRNA knockdown of PACS-2 phenocopies the Pacs-2 gene knock-out in preventing Nef-mediated MHC-I down-regulation, it is possible that the conflict between the two studies may have arisen from differences in the extent of siRNA knockdown of the PACS proteins. These authors also attempted to test the importance of PACS proteins in trafficking cellular cargo using a furin reporter protein that was mutated at EEDE779. These residues, however, are required for basolateral sorting of furin but not for binding PACS proteins, which recognize phosphorylated Ser773 and two proximal residues (28, 54). Unfortunately, mutation of residues not required for PACS binding would confound a meaningful interrogation of the roles of the PACS proteins in mediating protein traffic.
The role of PACS-2 in enabling Nef to assemble the SFK—ZAP-70/Syk—PI3K may have broader implications than triggering the signal-dependent down-regulation of cell-surface MHC-I. For example, binding of Nef to Hck at the Golgi region of macrophages is required for down-regulation of the macrophage colony-stimulating factor receptor Fms (55). In addition, the Nef EEEE65 and PXXP75 motifs and ZAP-70 activation are required for several additional pathways of Nef action, including Nef-mediated PAK2 activation, up-regulation of Fas ligand, and formation of virological synapses (1, 2, 56-58). It will therefore be important to determine to what extent PACS-2 contributes to these various Nef-controlled pathways and whether the multikinase cascade can be targeted to treat AIDS.
Acknowledgments
We thank Drs. L. Cantley, K. Früh, K. Shokat, M. Seaman, P. Luzio, S. Gray, C. Ruby, M. Forte, J. Lippincott-Schwartz, and C. Enns for reagents and helpful discussions.
This work was supported by National Institutes of Health National Research Service Awards DK076343 (to R. T. Y.), T32 NS007381 (to M. J. H.), T32 GM71338 (to F. P.), R01 AI49793, and R01 DK37274. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: HIV-1, human immunodeficiency virus, type 1; MHC-I, major histocompatibility complex class 1; siRNA, short interfering RNA; TGN, trans-Golgi network; SFK, Src family kinase; PACS, phosphofurin acidic cluster sorting protein; CI-MPR, cation-independent mannose 6-phosphate receptor; TfR, transferrin receptor; Nef, negative factor; ZAP-70, ζ-chain associated kinase 70; Syk, spleen tyrosine kinase; Hck, hemopoietic cell kinase; PI3K, phosphoinositide 3-kinase; ER, endoplasmic reticulum; VV, vaccinia virus; MEF, mouse embryo fibroblast; FBR, furin(cargo) binding region; ARF6, ADP ribosylation factor 6; WT, wild type; mAb, monoclonal antibody; GST, glutathione S-transferase; FACS, fluorescence-activated cell sorter; eGFP, enhanced green fluorescent protein; m.o.i., multiplicity of infection; SH, Src homology.
M. J. Harriff, unpublished results.
M. J. Harriff and G. Thomas, unpublished results.
References
- 1.Das, S. R., and Jameel, S. (2005) Indian J. Med. Res. 121 315-332 [PubMed] [Google Scholar]
- 2.Peterlin, B. M., and Trono, D. (2003) Nat. Rev. Immunol. 3 97-107 [DOI] [PubMed] [Google Scholar]
- 3.Stevenson, M. (2003) Nat. Med. 9 853-860 [DOI] [PubMed] [Google Scholar]
- 4.Stove, V., Naessens, E., Stove, C., Swigut, T., Plum, J., and Verhasselt, B. (2003) Blood 102 2925-2932 [DOI] [PubMed] [Google Scholar]
- 5.Thoulouze, M. I., Sol-Foulon, N., Blanchet, F., Dautry-Varsat, A., Schwartz, O., and Alcover, A. (2006) Immunity 24 547-561 [DOI] [PubMed] [Google Scholar]
- 6.Foster, J. L., and Garcia, J. V. (2007) Adv. Pharmacol. 55 389-409 [DOI] [PubMed] [Google Scholar]
- 7.Blagoveshchenskaya, A. D., Thomas, L., Feliciangeli, S. F., Hung, C. H., and Thomas, G. (2002) Cell 111 853-866 [DOI] [PubMed] [Google Scholar]
- 8.Hung, C. H., Thomas, L., Ruby, C. E., Atkins, K. M., Morris, N. P., Knight, Z. A., Scholz, I., Barklis, E., Weinberg, A. D., Shokat, K. M., and Thomas, G. (2007) Cell Host & Microbe 1 121-133 [DOI] [PubMed] [Google Scholar]
- 9.Mangasarian, A., Piguet, V., Wang, J. K., Chen, Y. L., and Trono, D. (1999) J. Virol. 73 1964-1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akari, H., Arold, S., Fukumori, T., Okazaki, T., Strebel, K., and Adachi, A. (2000) J. Virol. 74 2907-2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roeth, J. F., Williams, M., Kasper, M. R., Filzen, T. M., and Collins, K. L. (2004) J. Cell Biol. 167 903-913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenberg, M. E., Iafrate, A. J., and Skowronski, J. (1998) EMBO J. 17 2777-2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piguet, V., Wan, L., Borel, C., Mangasarian, A., Demaurex, N., Thomas, G., and Trono, D. (2000) Nat. Cell Biol. 2 163-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trible, R. P., Emert-Sedlak, L., and Smithgall, T. E. (2006) J. Biol. Chem. 281 27029-27038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, C. H., Leung, B., Lemmon, M. A., Zheng, J., Cowburn, D., Kuriyan, J., and Saksela, K. (1995) EMBO J. 14 5006-5015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marsh, J. W. (1999) Arch. Biochem. Biophys. 365 192-198 [DOI] [PubMed] [Google Scholar]
- 17.Renkema, G. H., and Saksela, K. (2000) Front. Biosci. 5 D268-D283 [DOI] [PubMed] [Google Scholar]
- 18.Keele, B. F., Van Heuverswyn, F., Li, Y., Bailes, E., Takehisa, J., Santiago, M. L., Bibollet-Ruche, F., Chen, Y., Wain, L. V., Liegeois, F., Loul, S., Mpoudi Ngole, E., Bienvenue, Y., Delaporte, E., Brookfield, J. F., Sharp, P. M., Shaw, G. M., Peeters, M., and Hahn, B. H. (2006) Science 313 523-526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agopian, K., Wei, B. L., Garcia, J. V., and Gabuzda, D. (2007) Virology 358 119-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsen, J. E., Massol, R. H., Nieland, T. J., and Kirchhausen, T. (2004) Mol. Biol. Cell 15 323-331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kasper, M. R., and Collins, K. L. (2003) J. Virol. 77 3041-3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bouard, D., Sandrin, V., Boson, B., Negre, D., Thomas, G., Granier, C., and Cosset, F. L. (2007) Traffic 8 835-847 [DOI] [PubMed] [Google Scholar]
- 23.Crump, C. M., Hung, C. H., Thomas, L., Wan, L., and Thomas, G. (2003) J. Virol. 77 11105-11113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crump, C. M., Xiang, Y., Thomas, L., Gu, F., Austin, C., Tooze, S. A., and Thomas, G. (2001) EMBO J. 20 2191-2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinners, I., Wendler, F., Fei, H., Thomas, L., Thomas, G., and Tooze, S. A. (2003) EMBO Rep. 4 1182-1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kottgen, M., Benzing, T., Simmen, T., Tauber, R., Buchholz, B., Feliciangeli, S., Huber, T. B., Schermer, B., Kramer-Zucker, A., Hopker, K., Simmen, K. C., Tschucke, C. C., Sandford, R., Kim, E., Thomas, G., and Walz, G. (2005) EMBO J. 24 705-716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simmen, T., Aslan, J. E., Blagoveshchenskaya, A. D., Thomas, L., Wan, L., Xiang, Y., Feliciangeli, S. F., Hung, C. H., Crump, C. M., and Thomas, G. (2005) EMBO J. 24 717-729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wan, L., Molloy, S. S., Thomas, L., Liu, G., Xiang, Y., Rybak, S. L., and Thomas, G. (1998) Cell 94 205-216 [DOI] [PubMed] [Google Scholar]
- 29.Benzinger, A., Muster, N., Koch, H. B., Yates, J. R., III, and Hermeking, H. (2005) Mol. Cell. Proteomics 4 785-795 [DOI] [PubMed] [Google Scholar]
- 30.Mansouri, M., Douglas, J., Rose, P. P., Gouveia, K., Thomas, G., Means, R. E., Moses, A. V., and Fruh, K. (2006) Blood 108 1932-1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molloy, S. S., Thomas, L., Kamibayashi, C., Mumby, M. C., and Thomas, G. (1998) J. Cell Biol. 142 1399-1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott, G. K., Fei, H., Thomas, L., Medigeshi, G. R., and Thomas, G. (2006) EMBO J. 25 4423-4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott, G. K., Gu, F., Crump, C. M., Thomas, L., Wan, L., Xiang, Y., and Thomas, G. (2003) EMBO J. 22 6234-6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sieburth, D., Ch'ng, Q., Dybbs, M., Tavazoie, M., Kennedy, S., Wang, D., Dupuy, D., Rual, J. F., Hill, D. E., Vidal, M., Ruvkun, G., and Kaplan, J. M. (2005) Nature 436 510-517 [DOI] [PubMed] [Google Scholar]
- 35.Schermer, B., Hopker, K., Omran, H., Ghenoiu, C., Fliegauf, M., Fekete, A., Horvath, J., Kottgen, M., Hackl, M., Zschiedrich, S., Huber, T. B., Kramer-Zucker, A., Zentgraf, H., Blaukat, A., Walz, G., and Benzing, T. (2005) EMBO J. 24 4415-4424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feliciangeli, S. F., Thomas, L., Scott, G. K., Subbian, E., Hung, C. H., Molloy, S. S., Jean, F., Shinde, U., and Thomas, G. (2006) J. Biol. Chem. 281 16108-16116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knight, Z. A., Gonzalez, B., Feldman, M. E., Zunder, E. R., Goldenberg, D. D., Williams, O., Loewith, R., Stokoe, D., Balla, A., Toth, B., Balla, T., Weiss, W. A., Williams, R. L., and Shokat, K. M. (2006) Cell 125 733-747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madrid, R., Janvier, K., Hitchin, D., Day, J., Coleman, S., Noviello, C., Bouchet, J., Benmerah, A., Guatelli, J., and Benichou, S. (2005) J. Biol. Chem. 280 5032-5044 [DOI] [PubMed] [Google Scholar]
- 39.Moon, K. D., Post, C. B., Durden, D. L., Zhou, Q., De, P., Harrison, M. L., and Geahlen, R. L. (2005) J. Biol. Chem. 280 1543-1551 [DOI] [PubMed] [Google Scholar]
- 40.Ward, S., Sotsios, Y., Dowden, J., Bruce, I., and Finan, P. (2003) Chem. Biol. 10 207-213 [DOI] [PubMed] [Google Scholar]
- 41.Carreno, S., Gouze, M. E., Schaak, S., Emorine, L. J., and Maridonneau-Parini, I. (2000) J. Biol. Chem. 275 36223-36229 [DOI] [PubMed] [Google Scholar]
- 42.Barnstable, C. J., Bodmer, W. F., Brown, G., Galfre, G., Milstein, C., Williams, A. F., and Ziegler, A. (1978) Cell 14 9-20 [DOI] [PubMed] [Google Scholar]
- 43.Arighi, C. N., Hartnell, L. M., Aguilar, R. C., Haft, C. R., and Bonifacino, J. S. (2004) J. Cell Biol. 165 123-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carlton, J., Bujny, M., Peter, B. J., Oorschot, V. M., Rutherford, A., Mellor, H., Klumperman, J., McMahon, H. T., and Cullen, P. J. (2004) Curr. Biol. 14 1791-1800 [DOI] [PubMed] [Google Scholar]
- 45.Popoff, V., Mardones, G. A., Tenza, D., Rojas, R., Lamaze, C., Bonifacino, J. S., Raposo, G., and Johannes, L. (2007) J. Cell Sci. 120 2022-2031 [DOI] [PubMed] [Google Scholar]
- 46.Seaman, M. N. (2004) J. Cell Biol. 165 111-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bonifacino, J. S., and Rojas, R. (2006) Nat. Rev. Mol. Cell Biol. 7 568-579 [DOI] [PubMed] [Google Scholar]
- 48.Coleman, S. H., Madrid, R., Van Damme, N., Mitchell, R. S., Bouchet, J., Servant, C., Pillai, S., Benichou, S., and Guatelli, J. C. (2006) J. Virol. 80 1837-1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaudhuri, R., Lindwasser, O. W., Smith, W. J., Hurley, J. H., and Bonifacino, J. S. (2007) J. Virol. 81 3877-3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doray, B., Lee, I., Knisely, J., Bu, G., and Kornfeld, S. (2007) Mol. Biol. Cell 18 1887-1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bard, F., Patel, U., Levy, J. B., Jurdic, P., Horne, W. C., and Baron, R. (2002) Eur. J. Cell Biol. 81 26-35 [DOI] [PubMed] [Google Scholar]
- 52.Lerner, E. C., and Smithgall, T. E. (2002) Nat. Struct. Biol. 9 365-369 [DOI] [PubMed] [Google Scholar]
- 53.Lubben, N. B., Sahlender, D. A., Motley, A. M., Lehner, P. J., Benaroch, P., and Robinson, M. S. (2007) Mol. Biol. Cell 18 3351-3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simmen, T., Nobile, M., Bonifacino, J. S., and Hunziker, W. (1999) Mol. Cell. Biol. 19 3136-3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hiyoshi, M., Suzu, S., Yoshidomi, Y., Hassan, R., Harada, H., Sakashita, N., Akari, H., Motoyoshi, K., and Okada, S. (2008) Blood 111 243-250 [DOI] [PubMed] [Google Scholar]
- 56.Zauli, G., Gibellini, D., Secchiero, P., Dutartre, H., Olive, D., Capitani, S., and Collette, Y. (1999) Blood 93 1000-1010 [PubMed] [Google Scholar]
- 57.Stumptner-Cuvelette, P., Jouve, M., Helft, J., Dugast, M., Glouzman, A. S., Jooss, K., Raposo, G., and Benaroch, P. (2003) Mol. Biol. Cell 14 4857-4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sol-Foulon, N., Sourisseau, M., Porrot, F., Thoulouze, M. I., Trouillet, C., Nobile, C., Blanchet, F., di Bartolo, V., Noraz, N., Taylor, N., Alcover, A., Hivroz, C., and Schwartz, O. (2007) EMBO J. 26 516-526 [DOI] [PMC free article] [PubMed] [Google Scholar]