Abstract
Dopamine receptors belong to the superfamily of G-protein-coupled receptors and are subdivided into D1-type (D1 and D5) and D2-type (D2, D3, and D4) receptors. The D4 receptor has a remarkable polymorphism in its third intracellular loop, which is under intensive investigation and which has been associated with, among other conditions, attention deficit hyperactivity disorder. Here, we demonstrate that KLHL12, a BTB-Kelch protein, specifically binds to this polymorphic region of the D4 receptor through its Kelch domain. Moreover, we show that KLHL12 also interacts with Cullin3 and thereby functions as an adaptor to target the D4 receptor to an E3 ubiquitin ligase complex. By ubiquitination assays in eukaryotic cells, we further demonstrate that overexpression of KLHL12 strongly promotes ubiquitination of the D4 receptor. In addition, we show that also other dopamine receptor subtypes undergo basal ubiquitination, but this is not affected by KLHL12. These data are the first to show ubiquitination of dopamine receptors and the first to identify a protein specifically interacting with the D4 polymorphism, thereby building up an E3 ligase complex with substrate specificity toward the D4 receptor.
Dopamine is an important neurotransmitter in mammalian brain that controls many basic functions, such as movement, cognition, emotion, reward, sexual behavior, and endocrine regulation. Malfunction of dopamine signaling has been implicated in many neurological disorders, such as Parkinson disease, attention deficit hyperactivity disorder, and schizophrenia (1, 2). By binding to dopamine receptors, dopamine can give rise to several possible signaling cascades. Dopamine receptors belong to the superfamily of G-protein-coupled receptors (GPCRs)5 or seven-transmembrane receptors and are divided in two subfamilies: the D1-like subfamily (D1 and D5 receptors) that signal through Gs to activate adenylyl cyclase and the D2-like subfamily (D2, D3, and D4 receptors) that signal through Gi/o to inhibit adenylyl cyclase. The D4 receptor has an important polymorphism in its third intracellular loop (IC3), consisting of a 2–11-fold repeat of 16 amino acids, denoted as D4.2 to D4.11 receptors (3, 4). Possible associations between this polymorphism and attention deficit hyperactivity disorder or personality traits, such as sexual behavior, have been suggested, but so far no real functional effects of the repeats have been documented (5–8). Despite intensive research during the last decade, many characteristics and signaling pathways still remain to be elucidated for the D4 receptor and dopamine receptors in general.
Ubiquitin is a 76-amino acid polypeptide that can be covalently attached to lysine residues of target proteins. Recently, it is becoming more and more clear that several signaling pathways are influenced by ubiquitination (9). Ubiquitination of membrane proteins has been shown to mark these for degradation by the proteasome in the ER-associated degradation pathway or for lysosomal degradation after endocytosis, but it can also have much broader functions, varying from internalization, trafficking, and signaling to even providing a framework for many ubiquitin-dependent interactions. Nevertheless, of thousands of known GPCRs, only few of them, namely the β2-adrenergic receptor (10), the chemokine receptor CXCR4 (11, 12), the V2 vasopressin receptor (13), the follitropin receptor (14), δ- and μ-opioid receptors (15, 16), PAR-1 (17), PAR-2 (18), and the thyrotropin-releasing hormone receptor (19), have been described to undergo ubiquitination. Generally, the actual ubiquitination process is performed by the coordinated action of three different classes of enzymes; ubiquitin is first activated by the ubiquitin-activating enzyme E1, followed by transfer to the ubiquitin-conjugating enzyme E2. Ubiquitin ligases (E3) are multiprotein complexes that catalyze the final reaction by ligating the ubiquitin to the substrate protein and also provide specificity toward the substrate. Two major classes of E3 ligases have been characterized: HECT (homologous to E6-associated protein carboxyl terminus)-type E3s that display catalytic activity and RING (really interesting new gene)-type E3s that bring the E2 and the substrate in close proximity to each other (20, 21). For many RING-type E3s, selectivity can be achieved by Cullin proteins, which recruit substrates to a core ubiquitination machinery via different adaptor proteins that show substrate specificity (22). Typical models are the SCF (Skp1-Cullin1-F-box) complexes consisting of a small RING finger factor Roc1 (RING of Cullins; also known as Rbx1 or Hrt1) that brings the associated E2 to the complex by binding with the Cul1 protein and whereby F-box proteins serve as adaptors. Another well defined E3 ligase is the ECS (elongin C-Cullin2-SOCS) complex, based on a Cul2 type of protein that binds Roc1 and the Skp1-related protein elongin C, using BC-box-containing proteins as adaptors. Recently, it was discovered that members of the large family of BTB (broad complex, Tramtrack, and Bric à Brac) domain-containing proteins define a new class of adaptors in E3 ligases that are based on the Cul3 type of Cullin proteins (21, 23–26). These BTB proteins provide substrate specificity through other protein-protein interaction domains, such as MATH domains (Meprin and TRAF Homology) or Kelch domains (originally identified in the Drosophila melanogaster Kelch protein). However, these BTB protein adaptors bind directly to Cul3, whereas F-box and BC-box proteins interact with their Cullin partners via Skp1 and elongin C, respectively. Although more than 200 BTB proteins have been identified in humans, only a few BCR3 (BTB-Cul3-Roc1) E3 complexes have been identified so far (21–23). The first described example of a mammalian Cul3-based E3 ligase system with known substrate is the Keap1-Cul3 complex. Keap1 is a BTB-Kelch protein that binds to Cul3 and Nrf2 via its BTB and Kelch domains, respectively. Recent work from several groups has demonstrated a role for Keap1 as an adaptor that targets the Nrf2 protein to the Cul3-based E3 ligase for subsequent ubiquitination of Nrf2 (27–30).
We performed a yeast two-hybrid screening and identified a BTB-Kelch protein, KLHL12, as a novel interaction partner of the D4 receptor. KLHL12, also known as C3IP1 or hDKIR, has an N-terminal BTB domain and a C-terminal Kelch domain, linked by a central BACK domain (for “BTB and C-terminal Kelch”) (31). KLHL12 was originally identified to be important in forming ringlike structures in mammalian cells, comparable with the function of the Kelch protein in D. melanogaster (32). We provide evidence that KLHL12 specifically interacts with the polymorphic repeats present in the IC3 of the D4 receptor. We further demonstrate that KLHL12 promotes ubiquitination of the D4 receptor by targeting this receptor to the Cul3-based E3 ligase complex. Moreover, we also provide evidence for the first time that ubiquitination seems to be a common modification for all types of dopamine receptors.
EXPERIMENTAL PROCEDURES
Plasmids and Antibodies—The coding sequence of KLHL12 was amplified from the pOTB7-KLHL12 Vector (I.M.A.G.E. Clone ID: IRAL p962K062) by PCR using primers anchored with NotI and XhoI recognition sequences. The NotI-XhoI fragment was cloned into the NotI-XhoI sites of the Etag pCAGGS/A20 vector (33) to generate Etag KLHL12. A similar PCR-based approach was used to generate the deletion mutants Etag KLHL12ΔKelch (aa 1–280) and Etag KLHL12ΔBTB (aa 118–568). Details concerning primers used and cloning strategy are available upon request. The integrity of all constructed KLHL12 vectors was confirmed by sequence analysis. The plasmids HA D4.2, HA D4.2Δ(249–280), corresponding to a hypothetical D4.0 receptor, HA D4.2Δ(233–253)Δ(283–307), HA D4.4, HA D4.7, and FLAG D4.4, were kindly provided by Dr. Van Tol (University of Toronto, Canada). Expression constructs for the HA D1, HA D2L, and HA D5 were purchased from UMR cDNA Resource Center (available on the World Wide Web). The c-Myc Cul3 vector was a kind gift from Dr. Rivero Crespo (Universität zu Köln, Cologne, Germany). The HA-tagged Cul2, Cul3, and dominant-negative Cul3 (C-terminal deletion containing the Roc1-binding domain) constructs were kindly provided by Dr. Hannink (University of Missouri, Colombia, MO). The vector for expression of HA-tagged Roc1 was provided by Dr. Furukawa (University of North Carolina, Chapel Hill, NC). The FLAG-tagged ubiquitin expression construct was a kind gift from Dr. Dikic (Goethe Universität, Frankfurt, Germany). Yeast two-hybrid constructs encoding the IC3 domains of D4.2, D4.4, and D4.7 and of D1, D5, M1-muscarinic, and β2-adrenergic receptor were kindly provided by Dr. Van Tol (University of Toronto, Canada) and Dr. Levenson (Pennsylvania State College of Medicine, Hershey, PA), respectively. Antibodies used were mouse monoclonal anti-HA (clone 16B12; Covance Research Products, Berkeley, CA), mouse monoclonal anti-c-Myc (clone 4A6; Upstate Biotechnology, Inc., Lake Placid, NY), mouse monoclonal anti-Etag (Amersham Biosciences), horseradish peroxidase-conjugated mouse monoclonal anti-Etag (Amersham Biosciences), mouse monoclonal anti-ubiquitin clone P4D1 (Cell Signaling, Danvers, MA), rabbit polyclonal anti-D4 receptor (Chemicon, Temecula, CA), rabbit polyclonal anti-D4 receptor (kind gift from Dr. Rivera, University of Malaga (Malaga, Spain) (34)), goat polyclonal anti-Cul3 clone C18 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), rabbit polyclonal anti-Cul3 clone H293 (Santa Cruz Biotechnology), and mouse monoclonal anti-FLAG M2 (Sigma). Horseradish peroxidase-conjugated sheep anti-mouse antibodies were purchased from Amersham Biosciences. For detection of the ubiquitination signal, horseradish peroxidase-conjugated anti-FLAG M2 mouse monoclonal antibody (Sigma) was used.
Yeast Two-hybrid Screening—For the yeast two-hybrid screening, we used the Matchmaker GAL4 Two-hybrid System 3 (BD Biosciences) according to the manufacturer's instructions. The pAS-1 vector, encoding the fusion between the DNA-binding domain of GAL4 and the third intracellular loop, corresponding to amino acids 219–336 of the D4.4 receptor, was used as bait (35). The bait plasmid, transformed into the yeast strain YRG2, was screened against a human fetal brain MATCHMAKER cDNA Library (BD Biosciences). Positive clones were selected on medium lacking leucine, tryptophan, and histidine and further tested for expression of the reporter gene β-galactosidase by the nitrocellulose filter lift assay (36).
Cell Culture and Transfection—HEK293T cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen), supplemented with 10% fetal calf serum, penicillin (100 units/ml), and streptomycin (100 μg/ml) in a controlled environment (37 °C, 98% humidity, 5% CO2). HEK293T cells were transfected in 10-cm dishes, using the calcium phosphate method. The amount of plasmid DNA used for transfection is indicated for each experiment. 48 h post-transfection, cells were washed twice with ice-cold phosphate-buffered saline and harvested, and the cell pellet was frozen at –70 °C for at least 1 h before lysis.
Preparation of Mouse Brain Lysates—Mice were killed, and brains were immediately frozen at –70 °C upon dissection. Brain tissue was homogenized by mixing in a buffer containing 150 mm NaCl, 50 mm Tris-HCl, pH 7.5, and freshly added protease inhibitors (2.5 μg/ml aprotinin, 1 mm PEFA-block, 10 μg/ml leupeptin, 10 mm β-glycerolphosphate). Subsequently, the homogenized tissue was lysed by the addition of a lysis buffer containing 150 mm NaCl, 10 mg/ml sodium deoxycholate, 50 mm Tris-HCl, pH 7.5, 2% Nonidet P-40, 0.2% SDS, supplemented with freshly added protease inhibitors (2.5 μg/ml aprotinin, 1 mm PEFA-block, 10 μg/ml leupeptin, 10 mm β-glycerolphosphate) to a final volume ratio of homogenization buffer/lysis buffer of 1:1. Upon rotation at 4 °C for 1 h, the lysate was cleared by centrifugation at 8000 × g for 10 min at 4 °C and treated further as described below.
Co-immunoprecipitation Assays and Immunoblot Analysis—For detection of protein expression or co-immunoprecipitation assays, cells were lysed in 500 μl of radioimmune precipitation buffer without EDTA or EGTA (150 mm NaCl, 5 mg/ml sodium deoxycholate, 50 mm Tris-HCl, pH 7.5, 1% Nonidet P-40, 0.1% SDS) supplemented with freshly added protease inhibitors (2.5 μg/ml aprotinin, 1 mm PEFA-block, 10 μg/ml leupeptin, 10 mm β-glycerolphosphate, 10 mm NaF). After rotating for 1 h at 4 °C, cell lysates were cleared by centrifugation at 8000 × g for 10 min at 4 °C. To detect protein expression, 5× Laemmli buffer (5% SDS, 50% glycerol, 65 mm Tris-HCl, pH 6.8, 0.2% bromophenol blue) supplemented with freshly added dithiothreitol (final concentration 25 mm) was added. The samples were denatured at 37 °C for 10 min, loaded onto a 10% acrylamide gel and subjected to SDS-PAGE. Resolved proteins were subsequently blotted onto a nitrocellulose membrane (Protran, Whatman; Schleicher & Schuell). Finally, the membranes were subjected to immunoblot analysis to detect proteins.
For immunoprecipitation (IP), 2 μg of antibody was added to the cleared lysate, followed by incubation at 4 °C for 4 h under continuous rotation. 20 μl of washed immobilized protein A (A-beads; Pierce) was added to the samples, followed by overnight incubation at 4 °C under continuous rotation. After washing the immunoprecipitates three times with lysis buffer (supplemented with freshly added protease inhibitors), bound proteins were eluted from the beads by heating the samples for 10 min at 37 °C under continuous shaking in elution buffer (radioimmune precipitation buffer, 2× Laemmli buffer (4% SDS, 20% glycerol, 26 mm Tris-HCl, pH 6.8, 0.08% bromophenol blue) and 1 m dithiothreitol in the respective ratio of 8:10:1). The eluates were then subjected to SDS-PAGE, followed by immunoblotting as described above. For immunoprecipitation of the FLAG-tagged D4.4 receptor, 25 μl of pre-equilibrated EZview Red anti-FLAG M2 affinity gel (FLAG-beads; Sigma) was added to the cleared lysate. Upon rotation at 4 °C for at least 1 h, the immunoprecipitates were washed three times with lysis buffer. Finally, bound proteins were eluted from the beads and subjected to SDS-PAGE and immunoblotting, as described above.
Ubiquitination Assays in Eukaryotic Cells—Immunoprecipitation was performed as described above, but N-ethylmaleimide (final concentration of 10 mm) was additionally added to the radioimmune precipitation lysis buffer. To perform double (sequential) immunoprecipitation assays, the washed beads were heated three times in 50 μl of elution buffer. A quarter of the first eluate was subjected to SDS-PAGE and immunoblotting to confirm IP and to check for ubiquitination. The combined three eluate fractions were further diluted with radioimmune precipitation lysis buffer (850 μl; again supplemented with protease inhibitors and N-ethylmaleimide), and a second immunoprecipitation round was performed. The immunoprecipitates were washed three times with lysis buffer, heated in elution buffer for 10 min at 37 °C, and analyzed as described above.
siRNA—For knockdown of Cul3 or KLHL12 protein expression levels, siGENOME SMARTpools (Dharmacon) were purchased, each consisting of a mixture of four different RNA duplexes. In each experiment, siCONTROL nontargeting siRNA pool 1 (Dharmacon) was used in parallel as a negative control. Transfection of siRNA in HEK293T cells was performed with the calcium phosphate method to a final concentration of 100 nm for each siRNA.
RESULTS
Yeast Two-hybrid Screening with the IC3 of the Human D4 Receptor—To identify proteins that interact with the D4 receptor, the IC3 domain of the D4.4 receptor (aa 219–336) was used as a bait to screen a (fetal) human brain cDNA library. Of the 1.2 × 106 clones screened, several positive clones were isolated. The library plasmids were isolated from these clones and analyzed by restriction digestion. Sequencing of a 2.7-kb cDNA insert and data base searches at NCBI revealed a perfect match to the protein sequence and 3′-untranslated region of KLHL12. Sequence analysis of the 2.7-kb cDNA revealed that it contains the BACK domain and the six Kelch repeats of KLHL12. The entire KLHL12 ORF encodes a protein of 568 amino acids with a calculated molecular mass of 62.5 kDa.
The specificity of the interaction between the IC3 of the D4.2, D4.4, and D4.7 receptor and KLHL12 (aa 118–568) was confirmed in yeast. Therefore, YRG2 was retransformed with pAS-D4.2 (aa 219–304), pAS-D4.4 (aa 219–336), or pAS-D4.7 (aa 219–384) vector as bait and pACT-KLHL12 (aa 118–568) as prey, including the respective empty vectors as negative controls. Detection of histidine- and β-galactosidase-expressing colonies confirmed the specific interaction of the IC3s of D4.2, D4.4, and D4.7 receptor with KLHL12 (aa 118–568) (data not shown).
To test whether the interaction was restricted to the D4 receptor, several other GPCRs were used as bait. No interaction was found between KLHL12 and the IC3 of D1, D5, M1-muscarinic, or β2-adrenergic receptors (data not shown). These data at least suggest that KLHL12 is not a common GPCR-interacting protein.
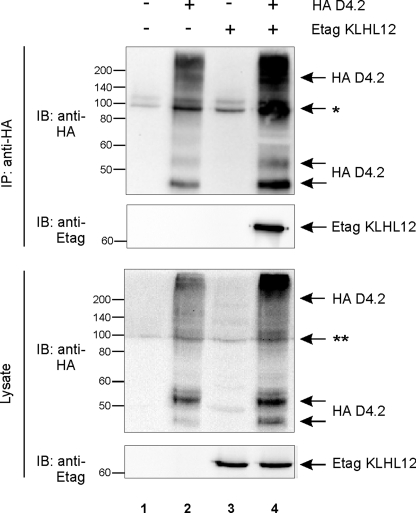
KLHL12 Interacts Specifically with the D4 Receptor in Mammalian Cells—To confirm the interaction between the D4 receptor and KLHL12 in mammalian cells, we performed co-immunoprecipitation studies. HEK293T cells were transiently transfected with expression constructs for both the D4 receptor and KLHL12 (Fig. 1). In the lysates, several bands for the HA D4.2 receptor were detected, of which the band with a molecular mass of ∼52–54 kDa represents mature, fully processed receptor, whereas the lower band with a molecular mass of ∼46–48 kDa represents immature, ER-retained receptor (37). The D4-specific but smeary pattern visible at the top of the gel could represent oligomeric receptor forms and receptor forms with different post-translational modification patterns (see also further). Immunoprecipitation of HA D4.2 receptor resulted in clear co-purification of Etag KLHL12 (Fig. 1, lane 4), indicating that KLHL12 also specifically interacts with the D4 receptor in mammalian cells. This interaction was also confirmed for the HA D4.4 and HA D4.7 receptor variants (data not shown).
FIGURE 1.
KLHL12 interacts with the D4 receptor in eukaryotic cells. HEK293T cells were transiently transfected with plasmids expressing HA D4.2 receptor (5.5 μg) and/or Etag KLHL12 (5.5 μg), as indicated. 48 h post-transfection, cells were lysed. 5% of the lysates were subjected to SDS-PAGE and subsequent immunoblotting (IB) with anti-HA and anti-Etag to detect expression of HA D4.2 receptor and Etag KLHL12, respectively (bottom). The rest of the lysates were subjected to immunoprecipitation (IP) with anti-HA. The D4-KLHL12 association was examined through subsequent immunoblotting with anti-Etag. Immunoprecipitation of the HA D4.2 was confirmed by subsequent immunoblotting with anti-HA. *, this signal represents a combination of immunoglobulin G, a nonspecific binding signal from the anti-HA antibody, and a D4-specific signal in lanes 2 and 4. **, a combined nonspecific signal from anti-HA and a D4-specific signal in lanes 2 and 4.
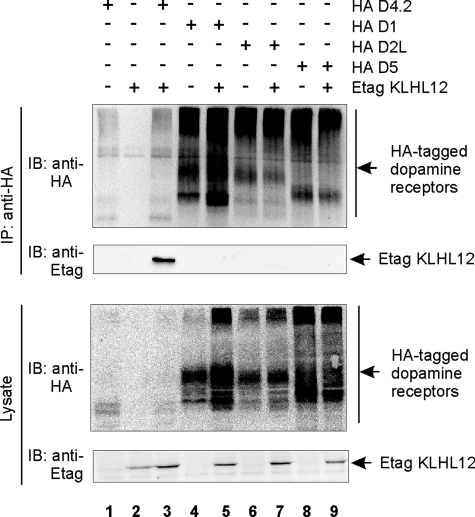
To test the specificity of this interaction in human cells, we performed co-immunoprecipitation studies in HEK293T cells, coexpressing Etag KLHL12 together with HA D1, D2L, and D5 receptors. We could again specifically detect co-immunoprecipitation of Etag KLHL12 with HA D4.2 receptor (Fig. 2, lane 3), whereas no interaction was found with the HA D1 (lane 5), HA D2L (lane 7), or HA D5 (lane 9) receptor. These results suggest that KLHL12 specifically interacts with the D4 type of dopamine receptors.
FIGURE 2.
KLHL12 specifically interacts with the D4 type of dopamine receptors. HEK293T cells were transiently transfected with plasmids expressing Etag KLHL12 (4 μg), HA D4.2 receptor (4 μg), HA D1 receptor (4 μg), HA D2L receptor (4 μg), and/or HA D5 receptor (4 μg), as indicated. 48 h post-transfection, cells were lysed, and 5% of the lysates were subjected to SDS-PAGE followed by immunoblotting (IB) with anti-HA and anti-Etag to detect HA-tagged dopamine receptors or Etag KLHL12, respectively. The rest of the lysates were subjected to immunoprecipitation (IP) with anti-HA. Receptor-KLHL12 associations were examined through subsequent immunoblotting (IB) with anti-Etag. Immunoprecipitation of the receptors was confirmed by immunoblotting with anti-HA.
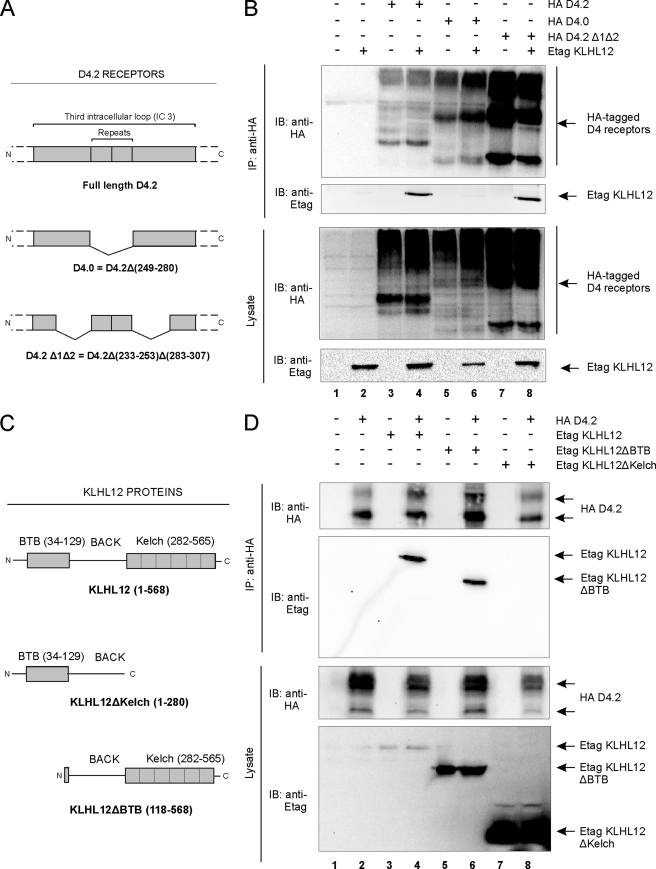
Domain Mapping of the Interaction Reveals a Role for the Polymorphism in the Third Intracellular Loop of the D4 Receptor and the Kelch Domain of KLHL12—To further characterize this novel interaction between the D4 receptor and KLHL12, we investigated which regions of the D4 receptor are involved. As described above, the yeast two-hybrid screening was performed using the IC3 of the D4 receptor as a bait. Therefore, we used mutants of the D4.2 receptor with deletions in this IC3 region (Fig. 3A); these mutants are still functional, as confirmed by GTPγS binding assays, and show plasma membrane localization, as visible through immunofluorescence microscopy.6 Co-immunoprecipitation studies in HEK293T cells showed that KLHL12 could still interact with the mutant D4.2 Δ1Δ2 receptor (Fig. 3B, lane 8). These data suggest that the regions flanking both sides of the polymorphism in the IC3 are not required for interaction of the D4 receptor with KLHL12. In contrast, we could not co-immunoprecipitate KLHL12 with the mutant D4.0 receptor, in which amino acids 249–280 are deleted (Fig. 3B, lane 6). Together, these results suggest that the repeats in the IC3 are important for interaction of the D4 receptor with KLHL12.
FIGURE 3.
Domain mapping of the D4 receptor-KLHL12 interaction. A, schematic illustration of the HA D4.2 receptor protein and deletion mutants used in this study. B, HEK293T cells were transiently transfected with the plasmids expressing Etag KLHL12 (5.5 μg) together with the HA D4.2 receptors (5.5 μg) depicted in A, in the indicated combinations. 48 h post-transfection, cells were lysed, and 5% of the lysates were subjected to SDS-PAGE. The presence of all three HA D4 receptors and Etag KLHL12 was revealed by immunoblotting (IB) with anti-HA and anti-Etag, respectively. The rest of the lysates was subjected to immunoprecipitation (IP) with anti-HA. The KLHL12-D4 receptor association was examined by immunoblotting with anti-Etag, whereas the immunoprecipitation was confirmed by subsequent immunoblotting with anti-HA. C, schematic illustration of the wild type KLHL12 protein and deletion mutants used in this study. The depicted sequences were fused to an N-terminal Etag. D, HEK293T cells were transiently transfected with plasmids expressing the HA D4.2 receptor (5.5 μg), Etag KLHL12 (5.5 μg), and the deletion mutants Etag KLHL12ΔBTB (5.5 μg) and Etag KLHL12ΔKelch (5.5 μg). Co-immunoprecipitation was examined by subsequent immunoprecipitation-immunoblot, as described in B. The expression of the HA D4.2 receptor and the Etag KLHL12 full-length and deletion mutant proteins in lysates was confirmed by immunoblotting with anti-HA and anti-Etag, respectively (bottom).
Similarly, we wanted to pinpoint the region in KLHL12 important for interaction with the D4 receptor. Co-immunoprecipitation studies in HEK293T cells using deletion mutants of KLHL12 (Fig. 3C) showed that deletion of the BTB region does not disrupt the interaction with the receptor, since co-immunoprecipitation of the KLHL12ΔBTB mutant with the D4.2 receptor could be clearly detected (Fig. 3D, lane 6). We note although that the interaction of the receptor with KLHL12ΔBTB might be weaker than with wild type KLHL12, given the higher expression level but lower co-purification of the ΔBTB mutant (Fig. 3D, compare lane 6 with lane 4). In contrast, by deleting the Kelch domain from KLHL12, interaction with the receptor is completely abolished (Fig. 3D, lane 8). These results suggest that the Kelch domain of KLHL12, but not the BTB domain, is required for interaction with the D4 receptor.
To summarize, these data show that the polymorphism in the IC3 of the D4 receptor and the Kelch domain of KLHL12 are important for interaction of these proteins with each other.
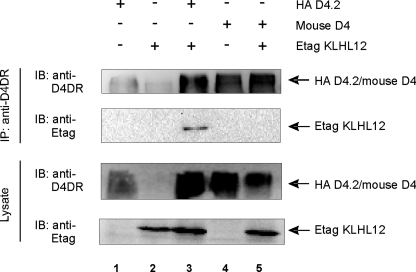
KLHL12 Does Not Interact with the Mouse D4 Receptor, Suggesting That the D4-KLHL12 Interaction Is Human (Primate)-specific—We have demonstrated that the polymorphism in the IC3 of the human D4 receptor is essential for interaction with KLHL12. Interestingly, this polymorphism is primate-specific (3) and thus not present in rodent (mouse or rat) D4 receptors. Therefore, we investigated whether KLHL12 would interact with the mouse D4 receptor via co-immunoprecipitation studies in HEK293T cells. We could not detect co-immunoprecipitation of KLHL12 with the mouse D4 receptor (Fig. 4, lane 5), in contrast to a clear co-purification with the human D4 receptor (lane 3; also see Fig. 1). These results are in accordance with our previous data, showing that the polymorphic repeats in the IC3 of the D4 receptor are important for interaction with KLHL12 (Fig. 3B). Although KLHL12 of human origin was used in these interaction studies, we note that the human and mouse variants of KLHL12 are very similar (amino acid similarity of 99.39%), suggesting that, most probably, no interaction occurs between the mouse D4 receptor and mouse KLHL12.
FIGURE 4.
KLHL12 does not interact with the mouse D4 receptor. HEK293T cells were transiently transfected with plasmids expressing human HA D4.2 receptor (5.5 μg), mouse D4 receptor (not tagged; 5.5 μg), or Etag KLHL12 (5.5 μg), as indicated. 48 h post-transfection, cells were lysed, and 5% of the lysate was subjected to SDS-PAGE followed by immunoblotting (IB) with anti-D4DR (Chemicon) and anti-Etag to detect dopamine receptors or Etag KLHL12, respectively. The rest of the lysates were subjected to immunoprecipitation (IP) with anti-D4DR (Chemicon). Receptor-KLHL12 associations were examined through subsequent immunoblotting with anti-Etag. Immunoprecipitation of the receptors was confirmed by immunoblotting with anti-D4DR.
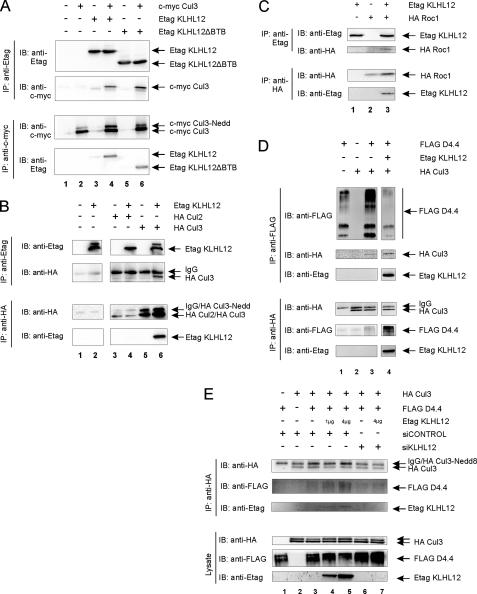
The D4 Receptor Is Recruited to a Cul3-based E3 Ubiquitin Ligase Complex via KLHL12 as an Adaptor—It is now well documented that BTB proteins can function as adaptors in E3 ubiquitin ligase complexes (21, 23–26). Furthermore, it has recently been shown that KLHL12 interacts with Cul3 (38). Therefore, we investigated the possibility that KLHL12 could function as an adaptor to target the D4 receptor to a Cul3-based E3 ubiquitin ligase complex. We could clearly demonstrate co-immunoprecipitation of c-Myc Cul3 and Etag KLHL12 in HEK293T cells (Fig. 5A), thereby confirming the results of Angers and co-workers (38). Although many BTB domain-containing proteins interact with Cul3 through their BTB domain, we were still able to find functional interaction of Cul3 with the KLHL12ΔBTB mutant protein (Fig. 5A, lane 6). These results suggest that the BTB domain of KLHL12 is not absolutely necessary for interaction with Cul3.
FIGURE 5.
Complex forming of D4 receptor-KLHL12-Cul3 in a E3 ubiquitin ligase complex. A, HEK293T cells were transiently transfected with plasmids expressing c-Myc Cul3 (5.5 μg), Etag KLHL12 (5.5 μg), or the deletion mutant Etag KLHL12ΔBTB (5.5 μg), as indicated. 48 h post-transfection, cells were lysed. Half of the lysate was used for immunoprecipitation (IP) of KLHL12 with anti-Etag (top), whereas the other half was used for immunoprecipitation of Cul3 with anti-c-Myc (bottom). Immunoprecipitation in both directions was confirmed by immunoblotting (IB) with anti-Etag or anti-c-Myc, respectively. Co-immunoprecipitation of c-Myc Cul3 was detected by immunoblotting with anti-c-Myc (top), whereas co-immunoprecipitation of Etag KLHL12 or Etag KLHL12ΔBTB was detected by subsequent immunoblotting with anti-Etag (bottom). Cul3-Nedd is the neddylated form of the Cul3 protein. A representative blot of different independent experiments is shown. B, HEK293T cells were transiently transfected with plasmids expressing Etag KLHL12 (5.5 μg), HA Cul2 (5.5 μg), and HA Cul3 (5.5 μg), as indicated. 48 h after transfection, cells were lysed. Half of the lysate was used for IP with anti-HA (bottom), whereas the other half was used for immunoprecipitation with anti-Etag (top). Confirmation of the respective immunoprecipitations and detection of KLHL12-Cullin associations were visualized by subsequent immunoblotting with anti-Etag to detect Etag KLHL12 and anti-HA to detect HA Cul2 or HA Cul3. All depicted lanes originate from the same gel. C, HEK293T cells were transiently transfected with plasmids expressing Etag KLHL12 (4 μg) and HA Roc1 (4 μg). Cell lysis and subsequent immunoprecipitation in both directions were performed as described in B. Detection of HA Roc1 or Etag KLHL12 was performed via immunoblotting with anti-HA or anti-Etag, respectively. D, HEK293T cells were transiently transfected with plasmids expressing FLAG D4.4 receptor (4 μg), Etag KLHL12 (4 μg), and HA Cul3 (4 μg), as indicated. 48 h post-transfection, cells were lysed, and the lysate was divided into two halves. One half was subjected to immunoprecipitation of the FLAG D4.4 receptor via FLAG-beads, whereas the other half was subjected to immunoprecipitation of HA Cul3 with anti-HA. Co-immunoprecipitation of Cul3 was detected by subsequent immunoblotting with anti-HA, whereas co-immunoprecipitation of the D4.4 receptor was detected by immunoblotting with anti-FLAG. Association of KLHL12 in the immunoprecipitates was confirmed by immunoblotting with anti-Etag. E, HEK293T cells were transiently transfected with plasmids expressing FLAG D4.4 receptor (4 μg), Etag KLHL12 (1 or 4 μg), and HA Cul3 (4 μg), in combination with siCONTROL or siKLHL12 (final concentration 100 nm), as indicated. 48 h post-transfection, cells were lysed, and 5% of the lysates were subjected to SDS-PAGE, followed by immunoblotting with anti-HA, anti-FLAG, and anti-Etag to detect HA Cul3, FLAG D4.4, and Etag KLHL12, respectively. The rest of the lysates were subjected to immunoprecipitation with anti-HA. Co-immunoprecipitation of the D4.4 receptor was detected by immunoblotting with anti-FLAG. Association of KLHL12 in the immunoprecipitates was confirmed by immunoblotting with anti-Etag.
Although it is commonly accepted that BTB proteins, functioning as adaptors in E3 ubiquitin ligase complexes, typically interact with the Cul3 type of Cullin proteins, there are different types of E3 ubiquitin ligase complexes, in which different types of Cullin proteins are used. Therefore, we tested the specificity of KLHL12 for interaction with Cul3. Co-immunoprecipitation studies in HEK293T cells did not reveal association of Cul2 and KLHL12 with each other (Fig. 5B, lane 4). Although in this experiment, the lack of a KLHL12-Cul2 interaction could be due to a lower level of Cul2 expression compared with Cul3 expression, we were not able to detect KLHL12-Cul2 interaction in three independent experiments (data not shown). These results confirm that KLHL12 indeed specifically interacts with the Cul3 type of Cullin proteins.
To further characterize the E3 ubiquitin ligase complex, we studied the involvement of Roc1 (equivalent to Rbx1 or Hrt1) (Fig. 5C). Roc1 interacts with Cul3 and is also carrier of the E2 ubiquitin-conjugating enzyme, in this way bringing this enzyme to the E3 ligase complex. After co-expressing Etag KLHL12 in HEK293T cells together with an HA-tagged Roc1 protein, we were able to show co-immunoprecipitation of Roc1 and KLHL12 with each other (Fig. 5C, lane 3). These results suggest that KLHL12 and Roc1 could be part of the same complex, using Cul3 as an intermediate interaction partner. As our results show, there was no need for overexpression of exogenous Cul3 to detect interaction between KLHL12 and Roc1. Moreover, the observed interaction did not increase upon co-expression of c-Myc-tagged or HA-tagged wild type Cul3 (see supplemental Fig. 1, lanes 4 and 8, respectively). Possibly, endogenous Cul3 levels are sufficiently high in HEK293T cells to support the interaction. However, the interaction could not be blocked upon co-expression of an HA-tagged dominant negative form of Cul3, in which the Roc1-binding domain is deleted (see supplemental Fig. 1, lane 6). Together, our current data do not fully exclude the possibility that Roc1 interacts directly with KLHL12.
Since we have shown that KLHL12 can interact both with the D4 receptor and with Cul3, we suggest that KLHL12 functions as an adaptor between the D4 receptor and Cul3. In this way, the D4 receptor could be targeted to a Cul3-based E3 ubiquitin ligase complex and become a substrate for subsequent ubiquitination (Fig. 6A). Upon co-expression of the D4 receptor and Cul3 in HEK293T cells, we could clearly demonstrate co-immunoprecipitation of the D4 receptor and Cul3 with each other under endogenous KLHL12 levels (Fig. 5D, lane 3). Both our reverse transcription-PCR data (results not shown) and the data of Angers et al. (38) suggest that endogenous KLHL12 levels are sufficiently high to explain the observed interaction. However, the interaction becomes significantly stronger upon co-expression of exogenous Etag KLHL12 (Fig. 5D, lane 4). To verify the role of KLHL12 as an adaptor and to exclude a possible direct interaction between the D4 receptor and Cul3, we knocked down KLHL12 protein expression levels via siRNA against KLHL12. Although we could not detect silencing of endogenous KLHL12, due to the lack of a high affinity antibody, we observed complete silencing of overexpressed Etag KLHL12 (Fig. 5E, compare lane 7 with lane 5). The increased interaction of the D4 receptor and Cul3 upon KLHL12 overexpression (lanes 4 and 5 compared with lane 3) is nearly completely abolished upon knockdown of KLHL12 (compare lane 6 with lane 3 and lane 7 with lane 5). Altogether, these data strongly indicate that the D4 receptor can form a complex with Cul3, whereby KLHL12 serves as an adaptor protein.
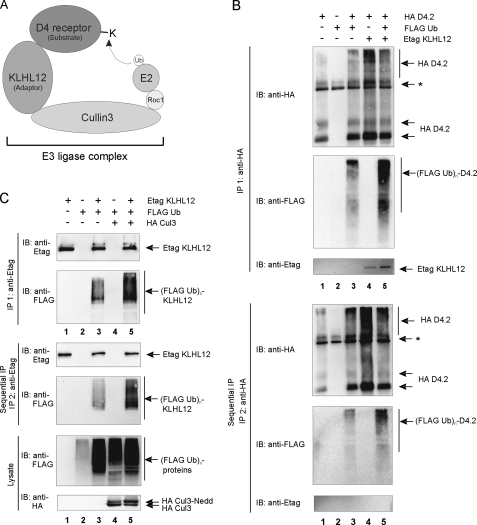
FIGURE 6.
Ubiquitination of the D4 receptor and KLHL12 in eukaryotic cells. A, schematic illustration of the D4 receptor-KLHL12-Cul3 E3 ubiquitin ligase complex. E2 represents the ubiquitin-conjugating enzyme; Roc1 (Rbx1 and Hrt1) is the RING finger factor; Cul3 serves as the scaffold protein for both Roc1 and KLHL12. The D4 receptor is targeted to the E3 ligase by the adaptor protein KLHL12 and functions as a substrate for subsequent ubiquitination by E2. B, HEK293T cells were transiently transfected with plasmids encoding the HA D4.2 receptor (4 μg), FLAG-tagged ubiquitin (FLAG Ub; 4 μg) and Etag KLHL12 (4 μg) as indicated. 48 h post-transfection, cells were lysed. The lysates were subjected to a first immunoprecipitation (IP 1) with anti-HA. Bound proteins were eluted from the washed immunoprecipitates; a quarter of the eluate was subjected to SDS-PAGE and subsequent immunoblotting (IB) with anti-HA to confirm immunoprecipitation of the HA D4.2 receptor and anti-FLAG antibody to detect ubiquitination (top; (FLAG Ub)n-D4.2). Detection of Etag KLHL12 in the immunoprecipitates was confirmed by immunoblotting with anti-Etag. The rest of the eluate was diluted with lysis buffer and subjected to a second immunoprecipitation round (IP 2) with anti-HA. Bound receptor was eluted from the washed immunoprecipitates. The eluate was again subjected to SDS-PAGE and subsequent immunoblotting with anti-HA to confirm the second immunoprecipitation of D4.2 receptor and anti-FLAG to detect specific ubiquitination of the D4.2 receptor (bottom). The exclusion of KLHL12 from the second immunoprecipitates was confirmed by immunoblotting with anti-Etag. *, the signal is a combination of IgG, a nonspecific signal from the anti-HA antibody, and a D4-specific signal in lanes 1, 3, 4, and 5. C, HEK293T cells were transiently transfected with plasmids expressing Etag KLHL12 (4 μg), FLAG Ub (4 μg), and HA Cul3 (4 μg). 48 h post-transfection, cells were lysed. A small fraction of each lysate was subjected to SDS-PAGE to confirm the expression of FLAG Ub and HA Cul3 by immunodetection with anti-FLAG and anti-HA, respectively. The rest of the lysates were subjected to a first immunoprecipitation using anti-Etag. Bound proteins were eluted from the beads; 20% of the eluate was subjected to SDS-PAGE and subsequent immunoblotting with anti-Etag to confirm immunoprecipitation of Etag KLHL12 and with anti-FLAG to detect ubiquitinated Etag KLHL12 (top; (FLAG Ub)n-KLHL12). The rest of the eluate was diluted with lysis buffer and subjected to a second immunoprecipitation round with anti-Etag. Bound Etag KLHL12 was eluted from the washed immunoprecipitates and again subjected to SDS-PAGE. The second immunoprecipitation of Etag KLHL12 was confirmed by subsequent immunoblotting with anti-Etag, whereas specific ubiquitination of Etag KLHL12 was detected by immunoblotting with anti-FLAG.
KLHL12 Promotes Ubiquitination of the D4 Receptor—Since our results clearly indicate that the D4 receptor can form a complex with KLHL12 and Cul3, we propose a model in which KLHL12 targets the D4 receptor to a Cul3-based E3 ubiquitin ligase complex (Fig. 6A). According to this model, the D4 receptor and the E2 ubiquitin-conjugating enzyme are brought in close proximity to each other through KLHL12 and Cul3 so that the D4 receptor can be targeted for ubiquitination. To test whether the D4 receptor gets ubiquitinated in living cells, we performed a ubiquitination assay in HEK293T cells, transiently transfected with HA D4.2 receptor and FLAG ubiquitin. After cell lysis, the HA D4.2 receptor was immunoprecipitated with anti-HA. The smeary pattern, observed after immunoblotting with anti-FLAG, is typical for ubiquitinated proteins (Fig. 6B, lane 3). This result suggests that the D4 receptor can undergo ubiquitination. To exclude the possibility that the smeary pattern represents ubiquitinated proteins that are associated with the D4.2 receptor, rather than the receptor itself, a sequential immunoprecipitation was performed. After a first immunoprecipitation, the diluted samples were subjected to a second immunoprecipitation round with anti-HA. At this stage, all protein-protein interactions are destroyed due to the denaturing conditions during elution from the beads after the first IP. Since ubiquitin is covalently attached to target proteins, this modification remains unaffected under these conditions. Using this procedure, we could still clearly detect FLAG ubiquitin signal (Fig. 6B, bottom, lane 3), thereby confirming that the D4 receptor can be ubiquitinated in eukaryotic cells.
For some BTB-Kelch proteins, such as Keap1, GAN1, and ENC1, it has been demonstrated that they can undergo Cul3-dependent ubiquitination themselves (39). Ubiquitination of KLHL12 was tested in HEK293T cells overexpressing Etag KLHL12 and FLAG ubiquitin. The IP of KLHL12 was confirmed by immunoblotting with anti-Etag (Fig. 6C, top, lanes 1 and 3), and a strong ubiquitin signal was detected by subsequent immunoblotting with anti-FLAG (lane 3). Using the sequential immunoprecipitation assay described above, we could demonstrate that the ubiquitin signal is specific for KLHL12 (Fig. 6C, bottom, lane 3), indicating that KLHL12 can indeed undergo ubiquitination. To demonstrate the role of Cul3 in this process, we investigated the effect of Cul3 overexpression on the ubiquitination level of KLHL12. Upon co-expression of Cul3, the ubiquitination level of KLHL12 increased significantly, both after the first (Fig. 6C, top, compare lane 5 with lane 3) and the second immunoprecipitation round (bottom, compare lane 5 with lane 3). These results suggest that KLHL12 ubiquitination is indeed Cul3-mediated.
Next, we examined the effect of KLHL12 overexpression on the ubiquitination level of the D4 receptor. We could clearly detect an increased ubiquitin signal upon co-expression of Etag KLHL12, both after the first (Fig. 6B, top, compare lane 5 with lane 3) and after the second immunoprecipitation round of the D4 receptor (Fig. 6B, bottom, compare lane 5 with lane 3). The significant positive effect of KLHL12 overexpression on the ubiquitination level of the D4 receptor is in agreement with our hypothesis that KLHL12 serves as an adaptor to target the D4 receptor for ubiquitination.
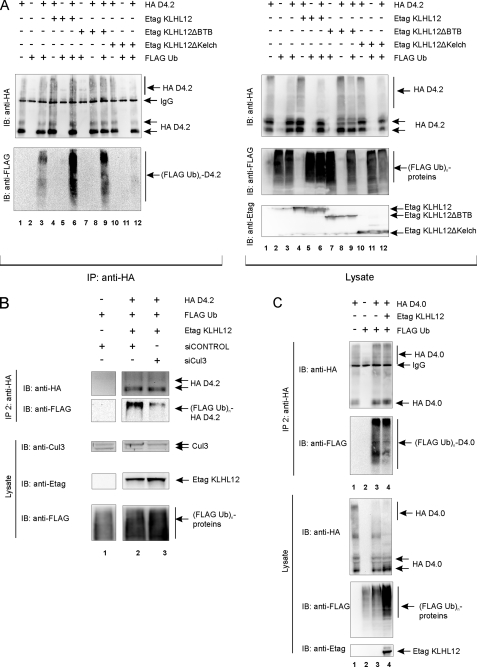
To substantiate this, we next tested the effect of the different KLHL12 mutants on the ubiquitination level of the D4 receptor. We expect that both interaction with the D4 receptor and Cul3 is required for KLHL12 to target the receptor for ubiquitination. Both full-length KLHL12 and KLHL12ΔBTB caused a significant increase of D4.2 receptor ubiquitination (Fig. 7A, left, compare lanes 6 and 9 with lane 3). This indicates that this ΔBTB mutant, which still interacts with the receptor and Cul3, remains capable of serving as an adaptor to target the D4.2 receptor to the E3 ligase complex for ubiquitination. Co-expression of KLHL12ΔKelch, which does not interact with the D4 receptor, did not result in increased ubiquitination of the D4.2 receptor (Fig. 7A, left, compare lane 12 with lane 3), thereby confirming that interaction with the receptor is essential for the observed effect. The same results were obtained upon sequential double immunoprecipitation of the D4.2 receptor (data not shown). Altogether, these data confirm our hypothesis that KLHL12 exerts its positive effect on the ubiquitination level of the D4 receptor by direct interaction with the D4 receptor.
FIGURE 7.
KLHL12 promotes D4 receptor ubiquitination via direct interaction with the receptor. A, HEK293T cells were transiently transfected with plasmids encoding the HA D4.2 receptor (4 μg), FLAG-tagged ubiquitin (FLAG Ub; 4 μg), full-length Etag KLHL12 (Etag KLHL12; 4 μg), or the deletion mutants Etag KLHL12ΔBTB (4 μg) and Etag KLHL12ΔKelch (4 μg) in the indicated combinations. 48 h post-transfection, cells were lysed. Small fractions of the lysates were subjected to SDS-PAGE and subsequent immunoblotting (IB) with anti-HA, anti-FLAG, and anti-Etag to confirm expression of HA D4.2 receptor, FLAG Ub, and Etag KLHL12, respectively (right). The rest of the lysates was subjected to immunoprecipitation (IP) of the receptor with anti-HA. The washed immunoprecipitates were boiled, and the eluate was subjected to SDS-PAGE. Subsequent immunoblotting with anti-HA confirmed immunoprecipitation of the HA D4.2 receptor, whereas ubiquitinated HA D4.2 receptor was detected by immunoblotting with anti-FLAG (left; (FLAG Ub)n-D4.2). B, HEK293T cells were transiently transfected with plasmids expressing the HA D4.2 receptor (HA D4.2; 4 μg), FLAG Ub (4 μg), and Etag KLHL12 (4 μg) in combination with siCONTROL or siCul3 as indicated. 48 h post-transfection, cells were lysed. A small fraction of each lysate was subjected to SDS-PAGE to confirm the expression of FLAG Ub and Etag KLHL12 by immunodetection with anti-FLAG and anti-Etag, respectively. Endogenous levels and silencing of Cul3 expression were detected via immunoblotting with anti-Cul3 (clone H293). The rest of the lysates were subjected to two sequential immunoprecipitation rounds, as described under “Experimental Procedures” and in Fig. 6B. After the second immunoprecipitation round, the eluate was subjected to SDS-PAGE and subsequent immunoblotting with anti-HA to confirm sequential immunoprecipitation of the D4.2 receptor and anti-FLAG to detect specific ubiquitination of the D4.2 receptor. C, HEK293T cells were transiently transfected with plasmids expressing the deletion mutant HA D4.0 receptor (HA D4.0; 4 μg), FLAG Ub (4 μg), and Etag KLHL12 (4 μg) in the indicated combinations. 48 h post-transfection, cells were lysed. Small fractions of the lysates were subjected to SDS-PAGE and subsequent immunoblotting with anti-HA, anti-FLAG, and anti-Etag to confirm expression of HA D4.0 receptor, FLAG Ub, and Etag KLHL12, respectively. The rest of the lysates were subjected to two sequential immunoprecipitation rounds, as described under “Experimental Procedures” and in Fig. 6B. After the second immunoprecipitation round, the eluate was subjected to SDS-PAGE and subsequent immunoblotting with anti-HA to confirm sequential immunoprecipitation of the D4.0 receptor and anti-FLAG to detect specific ubiquitination of the D4.0 receptor.
As demonstrated, KLHL12 functions as an adaptor to form a complex with the D4 receptor and Cul3 (Fig. 5, D and E). However, we were not able to rule out a possible direct interaction between KLHL12 and Roc1 (Fig. 5C and supplemental Fig. 1). To validate our model, as proposed in Fig. 6A, we therefore investigated the role of Cul3 in the ubiquitination of the D4 receptor. Overexpression of Cul3 did not promote the ubiquitination level of the D4 receptor (data not shown), probably due to sufficient levels of Cul3 in HEK293T cells (see also Fig. 5C). Therefore, we investigated the effect of siRNA against Cul3 on the KLHL12-mediated receptor ubiquitination (Fig. 7B). Upon cotransfection of the D4 receptor, KLHL12, and FLAG ubiquitin in HEK293T cells, the D4 receptor was purified via sequential immunoprecipitation as described before. Upon cotransfection of siCul3, a significant decrease in the ubiquitination level of the D4 receptor was observed (Fig. 7B, lane 3), compared with the control level (lane 2). Immunodetection of Cul3 in the cell lysates revealed a visible reduction of endogenous Cul3 expression levels upon siRNA treatment (compare lanes 2 and 3), whereas receptor levels remained unaffected. A slight increase in KLHL12 expression was observed, which is in accordance with the fact that KLHL12 ubiquitination and subsequent degradation is Cul3-dependent (38) (see also Fig. 6C). These results clearly demonstrate a role for Cul3 in KLHL12-mediated D4 receptor ubiquitination and support our model as depicted in Fig. 6A.
Since we have shown that the repeats in the IC3 of the D4 receptor are essential for interaction with KLHL12, we expect that the D4.0 receptor should no longer be susceptible to increased ubiquitination upon overexpression of KLHL12. The results in Fig. 7C clearly indicate that, although the D4.0 receptor shows a basal level of ubiquitination (lane 3), this signal is not longer enhanced upon co-expression of KLHL12 (lane 4). Again, these data were obtained upon performing a sequential double immunoprecipitation of the HA D4.0.
To summarize, we showed that KLHL12 targets the D4 receptor to the E3 ligase complex by a direct and simultaneous interaction of KLHL12 with the D4 receptor and Cul3, which gives rise to ubiquitination of the D4 receptor.
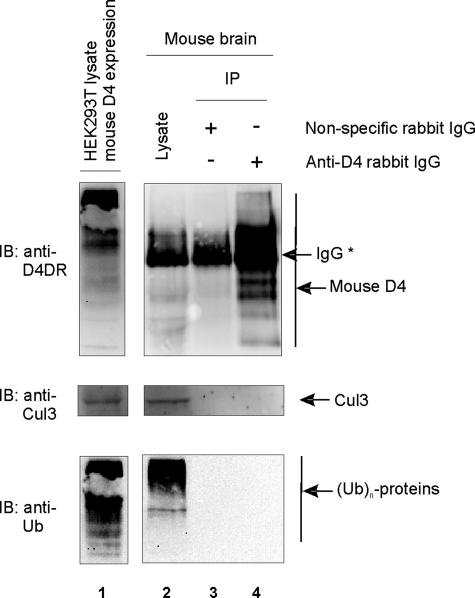
Interaction Studies in Mouse Brain Do Not Show Basal Recruitment of the D4 Receptor to the E3 Ligase or Basal Ubiquitination of the Receptor—To investigate whether we could isolate the D4-KLHL12-Cul3 complex from mouse brain lysates, we performed co-immunoprecipitation studies under endogenous protein expression levels (Fig. 8). We were able to detect endogenous levels of the D4 receptor and Cul3 (lane 2) in the mouse brain lysates, but we could not detect a specific signal for KLHL12, probably due to the lack of a high affinity antibody (data not shown). Although the D4 receptor was specifically immunoprecipitated from these mouse brains (Fig. 8, lane 4), we could not detect co-purification of Cul3 under these conditions (middle, lane 4).
FIGURE 8.
The mouse D4 receptor is not constitutively recruited to the E3 ligase complex and does not undergo basal ubiquitination. Mouse brain lysates were prepared as described under “Experimental Procedures.” Then nonspecific rabbit IgG antibody (2 μg) was added to half of the lysate, whereas a mix of anti-D4 antibody from Chemicon (1 μg) and anti-D4 antibody from Rivera and co-workers (34) (1 μg) was added to the other half. Before immunoprecipitation with A-beads, small fractions of the mixtures were subjected to SDS-PAGE. Endogenously expressed mouse D4 receptor and Cul3 were detected via immunoblotting with anti-D4 (Chemicon) and anti-Cul3 (clone C18), respectively (lane 2). The presence of ubiquitinated proteins in the brain lysates was confirmed via immunoblotting with anti-ubiquitin (clone P4D1). As a positive control, lysate was prepared from HEK293T cells, transiently transfected with mouse D4 receptor. Immunoblotting with anti-D4 (Chemicon), anti-Cul3 (clone C18), and anti-ubiquitin (clone P4D1) revealed overexpression of mouse D4 receptor and endogenously expressed Cul3 and ubiquitinated proteins, respectively (lane 1). After immunoprecipitation, immunoblotting with anti-Cul3 (clone C18) did not reveal co-purification of Cul3. Similarly, immunoblotting with anti-ubiquitin (clone P4D1) did not detect ubiquitination of the purified mouse D4 receptor. *, rabbit IgG (of nonspecific and anti-D4 antibodies).
Next, we investigated whether the mouse D4 receptor undergoes basal ubiquitination in mouse brains. Although a clear signal from ubiquitinated proteins is visible in the brain lysates (Fig. 8, bottom, lane 2), we were not able to detect ubiquitination signal in immunoprecipitates of the D4 receptor. Together, these data suggest that the D4 receptor is not constitutively recruited to an E3 ligase complex and does not undergo basal ubiquitination in mouse brain.
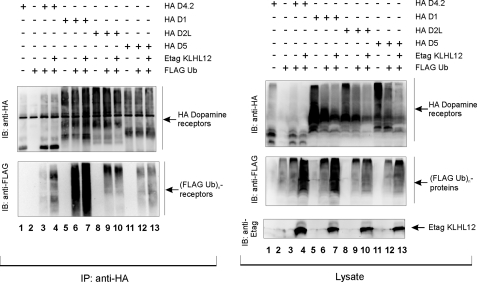
Ubiquitination of Dopamine Receptors Other than the D4 Is Not Mediated by KLHL12—We have not only shown that the D4 type of dopamine receptors undergoes ubiquitination, but we also found KLHL12 to be a corresponding adaptor. Since ubiquitination of the D4 receptor is a totally new finding and has not been described so far, we wanted to investigate whether other members of the dopamine receptor family also undergo ubiquitination. We therefore performed ubiquitination assays in HEK293T cells using HA-tagged constructs for D1, D5 (D1-type receptors), and D2L together with the HA D4.2 receptor. Besides ubiquitination of the D4.2 receptor (Fig. 9, left, lane 3), we were also able to detect ubiquitination signal for the D1 (lane 6), the D2L (lane 9), and the D5 receptor (lane 12). We ensured that we detected ubiquitination of the receptors themselves by validating these results in a sequential immunoprecipitation assay (data not shown). These data show that ubiquitination of dopamine receptors is not restricted to the D4 type but seems to be a common modification for all types of dopamine receptors. However, upon co-expression of Etag KLHL12 in these ubiquitination experiments in eukaryotic cells, we did not detect a significant effect on the ubiquitination level of the D1 (Fig. 9, left, lane 7), the D2L (lane 10), or D5 receptor (lane 13), whereas the effect was again clearly visible for the D4.2 receptor (lane 4). We note that, although a slight increase in ubiquitination might be detected for the D1 receptor (compare lane 7 with lane 6), also more D1 receptor was immunoprecipitated in this sample, so that the ratio of ubiquitin signal versus receptor signal remains virtually unaffected. Together, these results show that different types of dopamine receptors undergo ubiquitination, but only for the D4 receptor, KLHL12 serves as an adaptor, thereby giving specificity for the E3 ligase to the substrate.
FIGURE 9.
Ubiquitination of dopamine receptor types other than the D4 is not mediated by KLHL12. HEK293T cells were transiently transfected with plasmids expressing the HA D4.2 receptor (4 μg), HA D1 receptor (4 μg), HA D2L receptor (4 μg), HA D5 receptor (4 μg), FLAG-tagged ubiquitin (FLAG Ub; 4 μg) and Etag KLHL12 (4 μg), as indicated. 48 h post-transfection, cells were lysed. Small parts of the lysates were subjected to SDS-PAGE and subsequent immunoblotting with the indicated antibodies. The rest of the lysate was subjected to immunoprecipitation (IP) with anti-HA. The washed immunoprecipitates were denatured, and the eluate was subjected to SDS-PAGE. Immunoblotting (IB) with anti-HA confirmed the immunoprecipitation of the various types of HA-tagged dopamine receptors, whereas ubiquitination of these receptors was detected by immunoblotting with anti-FLAG.
DISCUSSION
In this study, we have identified the BTB-Kelch protein KLHL12 as a novel interaction partner for the third intracellular loop (IC3) of the human dopamine D4 receptor in a yeast two-hybrid screening. Further experiments in yeast indicate that KLHL12 does not interact with the IC3s of other dopamine receptors (D1 and D5) or with the IC3s of more distantly related GPCRs, such as the β2-adrenergic receptor or M1-muscarinic receptor. Interaction data in mammalian cells confirm that the interaction is restricted to the D4 type of dopamine receptors (Figs. 1 and 2). Together, these results suggest that KLHL12 is not a common GPCR-interacting protein. Domain mapping of KLHL12 indicated that the Kelch domain is necessary for interaction with the D4 receptor (Fig. 3, C and D). Furthermore, domain mapping of the D4 receptor clearly demonstrates an important role for the polymorphism in its third intracellular loop, since interaction with KLHL12 is completely abolished upon deletion of the repeats but not with a receptor mutant lacking both sides of these repeats (Fig. 3, A and B). These findings provide a first molecular link to unravel the functional relevance of this D4 receptor polymorphism, which has been investigated intensively and has been linked to major neurological disorders, such as attention deficit hyperactivity disorder (6–8). Interestingly, this polymorphism is primate-specific (3) and thus not present in rodent (mouse or rat) D4 receptor or other species. The fact that we were not able to detect an interaction between the mouse D4 receptor and KLHL12 (Fig. 4), is in accordance with our data indicating that this polymorphism is necessary for interaction with KLHL12. We note that the KLHL12 protein, used in these experiments, is of human origin. Therefore, we cannot rule out a possible interaction of the mouse KLHL12 variant with the mouse D4 receptor. However, mouse and human KLHL12 only differ by four amino acids (NCBI data base: human KLHL12 gi:11056006; mouse KLHL12 gi:81875867); the first two reside in the BTB and BACK domain, respectively (Ser82 and Asn217 in humans versus Ala82 and Asp217 in mice, respectively), and the other two are in the Kelch domain (Val427 and Thr551 in humans versus Ile427 and Ala551 in mice, respectively). Assuming that only the Kelch domain is important for interaction (see Fig. 3D) and considering the nature of the latter two amino acid differences, we expect no significant structural differences between the mouse and human KLHL12 variants, suggesting that there is no interaction between mouse KLHL12 and the mouse D4 receptor. Together, these results indicate that the D4-KLHL12 interaction could be human (primate)-specific.
Recent work has shown that several members of the large family of BTB proteins can function as adaptors in Cul3-based E3 ubiquitin ligases (21, 23–26). We therefore investigated the possibility that KLHL12 also functions as an adaptor in a Cul3-based E3 ligase complex. Several data confirm this hypothesis. First, we have demonstrated that KLHL12 can interact specifically with the Cul3 type of Cullin proteins in eukaryotic cells (Fig. 5, A and B). Second, we could demonstrate association of KLHL12 and Roc1, the carrier of the E2 ubiquitin-conjugating enzyme (Fig. 5C). The observed interaction did not increase upon overexpression of Cul3, suggesting that the endogenous Cul3 levels are sufficiently high in HEK293T cells. However, the interaction could also not be blocked upon overexpression of a Cul3 deletion mutant, deficient in Roc1 binding (see supplemental Fig. 1). Although at this moment we cannot formally exclude a direct interaction of KLHL12 with Roc1, we demonstrated that Cul3 is in fact involved in the ubiquitination of the D4 receptor (Fig. 7B). This observation supports the possible formation of the classic E3 ligase, in which the adaptor protein recruits the substrate protein to Roc1 via Cul3, in accordance with most cases described in the literature. Third, it has recently been shown that KLHL12 functions as an adaptor in a Cul3-based E3 ligase complex (38). Nevertheless, Angers et al. (38) were not able to show interaction of Cul3 with a mutant KLHL12 protein in which the BTB domain was deleted, whereas our own data show that deletion of this BTB domain does not disrupt the interaction with Cul3 (Fig. 5A). The discrepancy between both findings could be due to differences in experimental approach. Namely, we performed co-immunoprecipitation studies in cells (HEK293T) with a ΔBTB mutant containing amino acids 118–568 of KLHL12 (Fig. 3C) instead of co-immunoprecipitation studies after in vitro translation of the respective proteins in reticulocyte lysate using a ΔBTB mutant that only contained amino acids 131–568 (38). Therefore, it might be possible that the region between amino acids 118 and 131 of KLHL12 is essential for interaction with Cul3. Remarkably, however, interaction remains to be detected between Cul3 and the KLHL12ΔBTB mutant (aa 118–568), since it is generally accepted that BTB proteins bind to Cul3 via their BTB domain. Nevertheless, our subsequent ubiquitination experiments in HEK293T cells are in accordance with these data, since we demonstrate that the KLHL12ΔBTB mutant (aa 118–568) still functions as an adaptor in an E3 ligase complex (Fig. 7A). Similar discrepancies in Cul3 binding were described for the BTB-Kelch protein Keap1. Whereas Furukawa and Xiong (28) demonstrated that the BTB domain of Keap1 mediated Cul3-binding, Kobayashi et al. (29) showed that deletion of the BTB domain did not affect association with Cul3.
Following the observation that KLHL12 interacts both with the D4 receptor and Cul3, we have validated the hypothesis that the D4 receptor could be a potential target for a Cul3-based E3 ubiquitin ligase. Our data show the existence of complex formation of the D4 receptor and Cul3 in HEK293T cells, even under conditions of endogenous KLHL12 levels (Fig. 5D). The observed interaction is, however, significantly stronger upon overexpression of exogenous KLHL12. Moreover, we were able to show that the interaction was blocked upon down-regulation of KLHL12 via siRNA (Fig. 5E). Together, these data indicate that the D4 receptor does not directly interact with Cul3 but is recruited to the E3 ligase via KLHL12 as an adaptor. We further demonstrate that this formation specifically targets the D4 receptor for ubiquitination. Several data support this conclusion. First, ubiquitination assays in eukaryotic cells show that the D4 receptor undergoes ubiquitination, which is strongly increased upon overexpression of exogenous KLHL12 (Fig. 6B). By performing sequential immunoprecipitation assays, we convincingly demonstrate that this increase in ubiquitination signal is specific for the receptor itself and not merely the result of ubiquitination of other proteins. The latter had to be taken into account, since we also provide evidence for Cul3-dependent ubiquitination of KLHL12 itself (Fig. 6C). Second, KLHL12 promotes ubiquitination of the D4 receptor through direct interaction with the D4 receptor, since no increase in D4 ubiquitination is observed when a D4 receptor mutant (D4.0) is used that no longer interacts with KLHL12 (Fig. 7C). In accordance with these results is the observation that the KLHL12ΔBTB mutant, which is still capable of interacting with both the D4 receptor and Cul3, still promotes ubiquitination of the D4 receptor (Fig. 7A). Third, KLHL12 specifically promoted ubiquitination of the D4 receptor, but not of other dopamine receptor subtypes (Fig. 9). Again, these data are in agreement with our interaction studies indicating that KLHL12 interacts specifically with the D4 receptor.
Since we have demonstrated that the D4 receptor is recruited to the E3 ligase via the adaptor protein KLHL12 in the human cell system HEK293T, we investigated whether we could isolate in vivo D4-KLHL12-Cul3 complex from mouse brains. Although we were able to detect Cul3 expression and specifically detect and isolate the D4 receptor in these mouse brain lysates, we could not show complex forming of Cul3 with the receptor (Fig. 8). Although we cannot exclude interaction of mouse KLHL12 with mouse D4 receptor in vivo, our previous observations indicate that the D4-KLHL12 association could be human (primate)-specific. In this case, KLHL12 could not function as an adaptor to target the D4 receptor to the E3 ligase in mouse, thereby explaining why we could not detect the D4-Cul3 complex. However, it is possible that in mice, as in humans, proteins other than KLHL12 could function as adaptors to recruit the D4 receptor. In this case, it is very likely that the D4 receptor is not constitutively recruited to the E3 ligase, but only after certain stimuli or under certain physiological conditions, which remain to be elucidated. Accordingly, we could not detect basal ubiquitination of the mouse D4 receptor in brain tissue (Fig. 8). Altogether, these results suggest that in mouse brain, the D4 receptor is not constitutively recruited to the E3 ligase and thereby does not undergo basal ubiquitination.
It is of interest to note that we observed for the first time a basal level of ubiquitination of all dopamine receptors examined (Fig. 9). Although ubiquitination of GPCRs has been described to have numerous possible effects, from tagging a protein for degradation to internalization, trafficking, and signaling, the functional relevance toward dopamine receptors remains to be elucidated. Many questions remain unsolved, such as how this process is dynamically regulated, what triggers ubiquitination, and what is the fate of the ubiquitinated receptors. Although we have identified KLHL12 to be a specific adaptor for the D4 receptor, the corresponding adaptors for other types of dopamine receptors remain to be identified. Interestingly, a basal ubiquitination signal was also detected for the D4.0 receptor mutant, deficient in KLHL12 binding (Fig. 7C). This suggests that other adaptors besides KLHL12 could also exist for the D4 receptor or that ubiquitination of this receptor can be regulated through alternative or redundant systems.
In summary, our data show for the first time ubiquitination of dopamine receptors, and we identified KLHL12 as an adaptor in a Cul3-based E3 ligase complex for specific ubiquitination of the human D4 type receptor. We also identified for the first time a protein specifically interacting with the repeat region in the IC3 of the D4 receptor, which should help to further unravel the role of this remarkable polymorphism.
Acknowledgments
We are very grateful to Beatrice Coornaert for performing the initial yeast two-hybrid screening. Furthermore, we are thankful for the excellent technical support by Béatrice Lintermans and Anneleen Spooren and for help with mouse brain dissections by Dr. Anne Derore.
This work was supported by Fonds voor Wetenschappelijk Onderzoek (FWO) Vlaanderen and Geneeskundige Stichting Koningin Elisabeth. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GPCR, G-protein-coupled receptor; IC3, third intracellular loop; HEK293, human embryonic kidney 293; IP, immunoprecipitation; E1, ubiquitin-activating enzyme; E2, ubiquitin carrier protein; E3, ubiquitin-protein isopeptide ligase; aa, amino acids; HA, hemagglutinin; siRNA, small interfering RNA; GTPγS, guanosine 5′-3-O-(thio)triphosphate.
R. Bader and H. H. M. Van Tol, unpublished data.
References
- 1.Missale, C., Nash, S. R., Robinson, S. W., Jaber, M., and Caron, M. G. (1998) Physiol. Rev. 78 189–225 [DOI] [PubMed] [Google Scholar]
- 2.Vallone, D., Picetti, R., and Borrelli, E. (2000) Neurosci. Biobehav. Rev. 24 125–132 [DOI] [PubMed] [Google Scholar]
- 3.Oak, J. N., Oldenhof, J., and Van Tol, H. H. (2000) Eur. J. Pharmacol. 405 303–327 [DOI] [PubMed] [Google Scholar]
- 4.Van Tol, H. H., Wu, C. M., Guan, H. C., Ohara, K., Bunzow, J. R., Civelli, O., Kennedy, J., Seeman, P., Niznik, H. B., and Jovanovic, V. (1992) Nature 358 149–152 [DOI] [PubMed] [Google Scholar]
- 5.Ben Zion, I. Z., Tessler, R., Cohen, L., Lerer, E., Raz, Y., Bachner-Melman, R., Gritsenko, I., Nemanov, L., Zohar, A. H., Belmaker, R. H., Benjamin, J., and Ebstein, R. P. (2006) Mol. Psychiatry 11 782–786 [DOI] [PubMed] [Google Scholar]
- 6.Helmeste, D. M., and Tang, S. W. (2000) Jpn. J. Pharmacol. 82 1–14 [DOI] [PubMed] [Google Scholar]
- 7.Paterson, A. D., Sunohara, G. A., and Kennedy, J. L. (1999) Neuropsychopharmacology 21 3–16 [DOI] [PubMed] [Google Scholar]
- 8.Wong, A. H., Buckle, C. E., and Van Tol, H. H. (2000) Eur. J. Pharmacol. 410 183–203 [DOI] [PubMed] [Google Scholar]
- 9.DiAntonio, A., and Hicke, L. (2004) Annu. Rev. Neurosci. 27 223–246 [DOI] [PubMed] [Google Scholar]
- 10.Shenoy, S. K., McDonald, P. H., Kohout, T. A., and Lefkowitz, R. J. (2001) Science 294 1307–1313 [DOI] [PubMed] [Google Scholar]
- 11.Marchese, A., and Benovic, J. L. (2001) J. Biol. Chem. 276 45509–45512 [DOI] [PubMed] [Google Scholar]
- 12.Marchese, A., Raiborg, C., Santini, F., Keen, J. H., Stenmark, H., and Benovic, J. L. (2003) Dev. Cell 5 709–722 [DOI] [PubMed] [Google Scholar]
- 13.Martin, N. P., Lefkowitz, R. J., and Shenoy, S. K. (2003) J. Biol. Chem. 278 45954–45959 [DOI] [PubMed] [Google Scholar]
- 14.Cohen, B. D., Bariteau, J. T., Magenis, L. M., and Dias, J. A. (2003) Endocrinology 144 4393–4402 [DOI] [PubMed] [Google Scholar]
- 15.Chaturvedi, K., Bandari, P., Chinen, N., and Howells, R. D. (2001) J. Biol. Chem. 276 12345–12355 [DOI] [PubMed] [Google Scholar]
- 16.Petaja-Repo, U. E., Hogue, M., Laperriere, A., Bhalla, S., Walker, P., and Bouvier, M. (2001) J. Biol. Chem. 276 4416–4423 [DOI] [PubMed] [Google Scholar]
- 17.Wolfe, B. L., Marchese, A., and Trejo, J. (2007) J. Cell Biol. 177 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacob, C., Cottrell, G. S., Gehringer, D., Schmidlin, F., Grady, E. F., and Bunnett, N. W. (2005) J. Biol. Chem. 280 16076–16087 [DOI] [PubMed] [Google Scholar]
- 19.Cook, L. B., Zhu, C. C., and Hinkle, P. M. (2003) Mol. Endocrinol. 17 1777–1791 [DOI] [PubMed] [Google Scholar]
- 20.Glickman, M. H., and Ciechanover, A. (2002) Physiol. Rev. 82 373–428 [DOI] [PubMed] [Google Scholar]
- 21.Pintard, L., Willems, A., and Peter, M. (2004) EMBO J. 23 1681–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krek, W. (2003) Nat. Cell Biol. 5 950–951 [DOI] [PubMed] [Google Scholar]
- 23.Furukawa, M., He, Y. J., Borchers, C., and Xiong, Y. (2003) Nat. Cell Biol. 5 1001–1007 [DOI] [PubMed] [Google Scholar]
- 24.Geyer, R., Wee, S., Anderson, S., Yates, J., and Wolf, D. A. (2003) Mol. Cell 12 783–790 [DOI] [PubMed] [Google Scholar]
- 25.Pintard, L., Willis, J. H., Willems, A., Johnson, J. L., Srayko, M., Kurz, T., Glaser, S., Mains, P. E., Tyers, M., Bowerman, B., and Peter, M. (2003) Nature 425 311–316 [DOI] [PubMed] [Google Scholar]
- 26.Xu, L., Wei, Y., Reboul, J., Vaglio, P., Shin, T. H., Vidal, M., Elledge, S. J., and Harper, J. W. (2003) Nature 425 316–321 [DOI] [PubMed] [Google Scholar]
- 27.Cullinan, S. B., Gordan, J. D., Jin, J., Harper, J. W., and Diehl, J. A. (2004) Mol. Cell. Biol. 24 8477–8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furukawa, M., and Xiong, Y. (2005) Mol. Cell. Biol. 25 162–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kobayashi, A., Kang, M. I., Okawa, H., Ohtsuji, M., Zenke, Y., Chiba, T., Igarashi, K., and Yamamoto, M. (2004) Mol. Cell. Biol. 24 7130–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, D. D., Lo, S. C., Cross, J. V., Templeton, D. J., and Hannink, M. (2004) Mol. Cell. Biol. 24 10941–10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stogios, P. J., and Prive, G. G. (2004) Trends Biochem. Sci. 29 634–637 [DOI] [PubMed] [Google Scholar]
- 32.Mai, A., Jung, S. K., and Yonehara, S. (2004) Exp. Cell Res. 300 72–83 [DOI] [PubMed] [Google Scholar]
- 33.De Valck, D., Heyninck, K., Van Criekinge, W., Vandenabeele, P., Fiers, W., and Beyaert, R. (1997) Biochem. Biophys. Res. Commun. 238 590–594 [DOI] [PubMed] [Google Scholar]
- 34.Khan, Z. U., Gutierrez, A., Martin, R., Penafiel, A., Rivera, A., and De La Calle, A. (1998) J. Comp. Neurol. 402 353–371 [DOI] [PubMed] [Google Scholar]
- 35.Oldenhof, J., Vickery, R., Anafi, M., Oak, J., Ray, A., Schoots, O., Pawson, T., von Zastrow, M., and Van Tol, H. H. (1998) Biochemistry 37 15726–15736 [DOI] [PubMed] [Google Scholar]
- 36.De Martelaere, K., Lintermans, B., Haegeman, G., and Vanhoenacker, P. (2007) Cell. Signal. 19 278–288 [DOI] [PubMed] [Google Scholar]
- 37.Van Craenenbroeck, K., Clark, S. D., Cox, M. J., Oak, J. N., Liu, F., and Van Tol, H. H. (2005) J. Biol. Chem. 280 19350–19357 [DOI] [PubMed] [Google Scholar]
- 38.Angers, S., Thorpe, C. J., Biechele, T. L., Goldenberg, S. J., Zheng, N., MacCoss, M. J., and Moon, R. T. (2006) Nat. Cell Biol. 8 348–357 [DOI] [PubMed] [Google Scholar]
- 39.Zhang, D. D., Lo, S. C., Sun, Z., Habib, G. M., Lieberman, M. W., and Hannink, M. (2005) J. Biol. Chem. 280 30091–30099 [DOI] [PubMed] [Google Scholar]