Abstract
Chromatin endowed by histone modifications governs chromatin structure, which in turn represents a means to regulate cellular processes, including transcription and heterochromatin formation. Recent evidence revealed a plethora of enzymes that catalyze specific histone modifications for epigenetic maintenance, and dysregulation of which contributes to tumorigenesis and developmental defects. The histone methyltransferase SET8 (also known as Pr-Set7) was previously reported to monomethylate Lys20 of histone H4. However, the temporal and spatial control of SET8 activity remains elusive. Here, we provide evidence to support that SET8 monomethylates Lys20 of histone H4 during S phase by tethering to proliferating cell nuclear antigen via a putative proliferating cell nuclear antigen-interacting protein box. In addition, we show that SET8 function is required for S phase progression. Finally, deletion of SET8 in mice causes embryonic lethality, suggesting that SET8 plays an important role in mammalian embryogenesis.
Covalent modifications of histone tails play important roles in regulating chromatin dynamics. Recent genetic and immunochemical analyses suggest that these histone “marks” are of fundamental importance in controlling the recruitment of regulatory factors that ensure cell proliferation and survival (1–3). In particular, histone methylation, which occurs predominantly on histones H3 and H4 at arginine and lysine residues, has been implicated in DNA repair, maintenance of telomere length homeostasis, heterochromatin formation, and mitotic regulation (4–9). Accordingly, a number of modules have been demonstrated to recognize these specific histone modifications such that effector complexes are brought to close proximity of the chromatin to elicit their specific cellular functions (10–12).
Histone methyltransferases, including SET8, Suv4-20h1/2, NSD1, and Ash1, have been shown previously to methylate histone H4 at Lys20 (H4-K20) (8, 13–19). Specifically, H4-K20 trimethylation have been associated with constitutive pericentromeric heterochromatin formation, whereas mono- and dimethylated H4-K20 histones have been suggested to recruit the malignant brain tumor protein L3MBTL1, which is important for chromatin condensation (20–22). In addition, we proposed previously that dimethylated H4-K20 might play a role in the recruitment of the DNA damage-response protein 53BP1 to chromatin (4). Despite mounting evidence for the biological functions of H4-K20 methylation, exactly how these epigenetic marks are perpetuated during DNA replication remains largely unexplored.
Given that SET8 expression is cell cycle-regulated, we decided to further probe how SET8 might be required for cell cycle progression. Using a TAP5 scheme, we identified PCNA as a SET8-associated protein. Our studies suggest that the function of SET8 in H4-K20 monomethylation is coupled to S phase by its physical tethering to PCNA, failure of which results in improper S phase progression, S/G2 checkpoint arrest, and embryonic lethality in mice.
MATERIALS AND METHODS
Antibodies—Anti-histone H4 and anti-monomethylated H4-K20 antibodies were purchased from Upstate Cell Signaling. Anti-SET8 antibodies were from Abcam and Upstate Cell Signaling. Anti-actin and anti-FLAG (M2) antibodies were obtained from Sigma. Horseradish peroxidase-conjugated GST was from Santa Cruz Biotechnology Inc. Anti-PCNA (PC10) and anti-glyceraldehyde-3-phosphate dehydrogenase antibodies were from Chemicon.
Cell Culture and Transfection—HeLa and 293T cells were cultured in RPMI 1640 medium supplemented with 5% fetal calf serum, 5% bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin and maintained in 5% CO2 at 37 °C. Cell transfection was performed using FuGENE 6 (Roche Applied Science) and Lipofectamine 2000 (Invitrogen) following the manufacturers' protocols.
Construction of set8 and Its Mutants—The set8 cDNA was obtained from American Type Culture Collection and subcloned into the entry vector pDONR201 (Gateway Technology). Site-directed mutagenesis was performed according to standard procedures to obtain the YMAA mutant using primers 5′-ACC GGG CAG TCA AAG ATC TAT TCC GCC GCG AGC CCG AAC AAA TGC TCT GGA ATG-3′ and 5′-CAT TCC AGA GCA TTT GTT CGG GCT CGC GGC GGA ATA GAT CTT TGA CTG CCC GGT-3′. For mammalian expression of wild-type and set8 mutants, their respective cDNAs in the entry vector was transferred into a Gateway-compatible destination vector harboring an N-terminal TAP tag containing SFB. For GST fusion protein production, SET8 was subcloned into pDEST15, and GST-SET8 was purified according to standard procedures. His-tagged PCNA was purified from bacteria using a nickel column (Roche Applied Science). The siRNA-resistant SET8 construct was generated by site-directed mutagenesis using primers 5′-AAA ACC TAC TGC GTG GAT GCA ACT AGG GAA ACA AAA TCG CCT AGG AAG ACT G-3′ and 5′-AGT CTT CCT AGG CGA TTT GTT TCC CTA GTT GCA TCC ACG CAG TAG GTT TTG CTC AG-3′.
siRNAs—The SET8 siRNAs used were 5′-GCA ACT AGA GAG ACA AAT C-3′ and 5′-GAT TGA AAG TGG GAA GGA A-3′. The non-targeting control siRNA sequence was described previously (23). To deplete endogenous SET8, U2OS cells were transfected twice using Oligofectamine (Invitrogen) with the indicated siRNAs at 24-h intervals. For reconstitution experiments, siRNA-resistant FLAG-tagged SET8 or YMAA was electroporated into siRNA-treated U2OS cells 24 h after the second transfection. Cells were subsequently harvested for cell cycle analysis or lysed for immunoblot experiments.
Binding Assays—For co-immunoprecipitation studies, constructs encoding SFB-tagged SET8 and its mutants were transiently transfected into 293T cells using Lipofectamine 2000. Cells were lysed in NETN buffer (100 mm NaCl, 1 mm EDTA, 20 mm Tris-HCl (pH 8), and 0.5% Nonidet P-40), cleared by centrifugation, and incubated with streptavidin beads for 2 h at 4 °C. Beads were washed, boiled in sample buffer, separated by SDS-PAGE, and immunoblotted with the indicated antibodies. To detect endogenous interaction between SET8 and PCNA, 293T cell lysate was incubated with protein A-conjugated anti-SET8 polyclonal antibody.
Immunofluorescence Staining—Cells cultured on coverslips were washed with phosphate-buffered saline and incubated in methanol/acetone (1:1) for 20 min at –20 °C. For detection of endogenous SET8, HeLa cells were preincubated in 10 μm MG132 prior to fixation. Samples were subsequently blocked with 5% goat serum, incubated with primary antibody for 30 min, washed three times, and then incubated with secondary antibody for 30 min. Cells were subsequently stained with 4′,6-diamidino-2-phenylindole to visualize nuclear DNA. Coverslips were mounted on glass slides with antifade solution and visualized using a Nikon ECLIPSE E800 fluorescence microscope.
Cell Cycle Expression Profile of SET8 and H4-K20 Monomethylation—HeLa cells arrested with nocodazole were released into fresh medium without nocodazole to monitor the expression profile of SET8 and H4-K20 monomethylation status during different cell cycle phases. At the indicated time points, cells were collected and lysed, and proteins were quantified by Western blot analysis. Cell cycle progression was monitored by FACS analysis.
Cell Cycle Analysis—HeLa cells were transiently transfected with the N-terminal mutant of SET8 or mock-transfected. 16 h post-transfection, cells were incubated in 0.5 mm mimosine for 20 h to arrest cells at G1/S. Cells were subsequently released into the cell cycle, and samples were fixed in 70% ethanol for FACS analysis at the indicated time points.
Generation of set8-deficient Mice—Embryonic stem cell clone RRB075 was obtained from BayGenomics and injected into C57BL/6 blastocysts to generate chimeric mice. The chimeras were then crossed with C57BL/6 females, and heterozygous mice with successful germ line transmission of the targeted allele were used for mating experiments.
RESULTS AND DISCUSSION
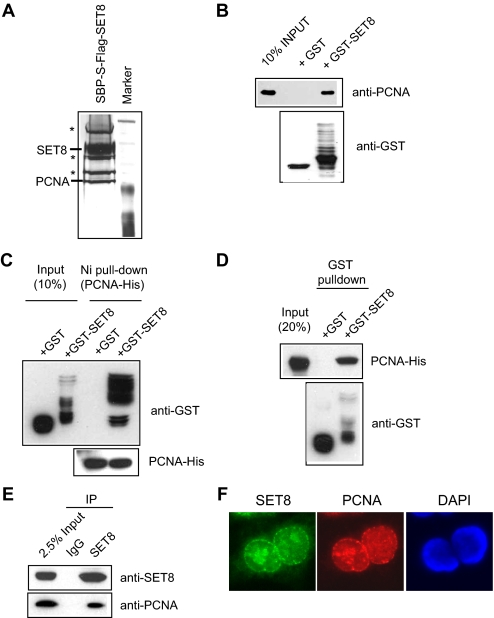
Identification of PCNA as a SET8-interacting Protein—To explore how the histone methyltransferase activity of SET8 might be regulated in vivo, we generated a 293T cell line overexpressing triple tagged (SFB) SET8 for the identification of potential SET8-interacting proteins. Following a TAP scheme, the SET8 complex was eluted and separated by SDS-PAGE, and protein bands were excised and subjected to mass spectrometry analysis. Several of these associated proteins were identified as various heat shock proteins and ribosomal proteins (as indicated by asterisks in Fig. 1A), which are common contaminants that normally interact with overexpressed proteins. Interestingly, one of these associated proteins was found to be PCNA (Fig. 1A).
FIGURE 1.
SET8 interacts with PCNA in vitro and in vivo. A, identification of PCNA as a SET8-interacting partner. Shown is a silver stain of SET8-associated proteins separated by SDS-PAGE. B, GST-SET8 but not GST alone interacts with PCNA. C and D, SET8 directly interacts with PCNA. E, SET8 interacts with PCNA in vivo. F, SET8 localizes with PCNA in replication foci. IP, immunoprecipitation; DAPI, 4′,6-diamidino-2-phenylindole.
To confirm that SET8 can specifically interact with PCNA, we performed a pulldown experiment using bacterially purified GST-SET8 fusion protein or GST alone. Consistent with TAP results, PCNA interacted with GST-SET8 but not GST (Fig. 1B), suggesting that SET8 interacts directly with PCNA in vitro. Consistent with a direct interaction, bacterially purified histidine-tagged PCNA precipitated with GST-SET8 but not GST alone (Fig. 1, C and D). More important, this interaction was also detected in vivo between endogenous proteins (Fig. 1E). Because PCNA is a key component of replication forks and forms discrete replication foci during S phase, we next tested whether SET8 colocalizes with PCNA in indirect immunofluorescence experiments. Cytological observation of SET8 revealed extensive overlap of SET8 with PCNA in S phase cells (Fig. 1F). Taken together, these results suggest that PCNA is a SET8-associated protein.
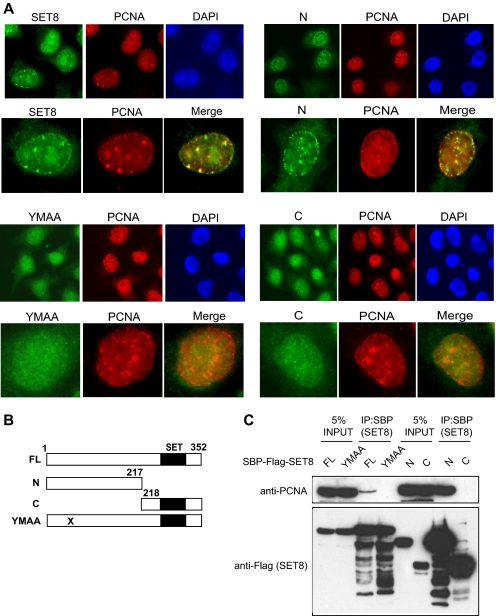
SET8 Interacts with PCNA via a PIP Box at Its N Terminus—To investigate how SET8 might interact with PCNA, we generated deletion mutants that encode either the N or C terminus of the protein. The histone methyltransferase activity of SET8 is located in the C terminus, where the conserved SET domain resides (Fig. 2B). Visual inspection also identified a potentially divergent PIP box in the N terminus of SET8 (QSKIYSYM). Many PCNA-associated proteins have been shown to interact with PCNA via their corresponding PIP box, which has the consensus motif QXX(M/L/I)XX(F/Y)(F/Y) (24). To test whether SET8 might similarly interact with PCNA via its putative PIP box, we generated a double point mutation that resulted in amino acid changes from YM to AA at resides 57 and 58 (YMAA). We first transfected these constructs into HeLa cells to examine which region of SET8 is required for its localization to PCNA foci. Resembling endogenous SET8, full-length SET8 formed discrete foci that overlapped with PCNA foci. Likewise, the N-terminal truncation mutant also localized to PCNA foci, suggesting that SET8 might interact with PCNA via its N terminus. Consistent with this hypothesis, we did not observe discernible foci in cells expressing FLAG-tagged YMAA or the C-terminal mutant (Fig. 2A), suggesting that SET8 is recruited to replication foci by binding to PCNA via its putative PIP box.
FIGURE 2.
SET8 interacts with PCNA and localizes to replication foci via a putative PIP box. A, immunofluorescence studies of full-length SET8, its N- and C-terminal truncation mutants, and the PIP mutant (YMAA) and their colocalization with PCNA foci. B, schematic illustration of full-length (FL) SET8 and its mutants. C, PCNA coprecipitates with full-length SET8 and its N-terminal mutant, but not the YMAA and C-terminal mutants. DAPI, 4′,6-diamidino-2-phenylindole; IP, immunoprecipitation.
To confirm these findings, we also performed co-immunoprecipitation experiments in 293T cells. The results show that although full-length SET8 and its N-terminal truncation mutant coprecipitated with PCNA, the YMAA and C-terminal mutants failed to do so (Fig. 2C). From these results, we concluded that SET8 interacts with PCNA via a PIP box, which is important for its localization to replication foci.
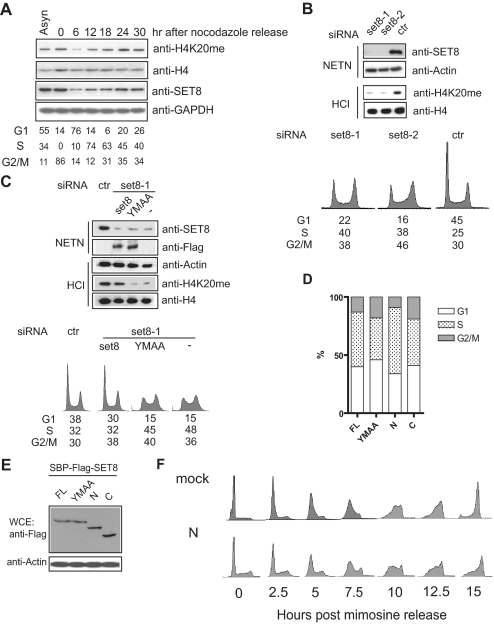
SET8-dependent H4-K20 Monomethylation Is Important for S Phase Progression—Because SET8 specifically interacts with PCNA and colocalizes with PCNA at replication forks, we speculated that SET8 may function in S phase. Consistent with a possible S phase role of SET8, we observed that H4-K20 monomethylation is coupled to expression of SET8, which starts to accumulate during S phase and peaks at mitosis (Fig. 3A) (7). Moreover, SET8 depletion resulted in decreased H4-K20 monomethylation, with a concomitant accumulation of S/G2 phase cell population (Fig. 3B). To examine whether the function of SET8 in cell cycle progression requires its targeting to the replication fork via PCNA, siRNA-resistant SET8 or YMAA was reintroduced into SET8-depleted cells. Indeed, upon depletion of endogenous SET8, only cells expressing siRNA-resistant SET8, but not its YMAA mutant, restored monomethylated H4-K20 and allowed progression through S/G2 phase. (Fig. 3C).
FIGURE 3.
SET8 is involved in S phase progression. A, expression profile of SET8 and H4-K20 monomethylation status during cell cycle progression. Histone H4 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as loading controls. Asyn, asynchronized. B, depletion of SET8 accumulates cells in S/G2 phase, which coincides with down-regulated H4-K20 monomethylation. C, function of SET8 in S/G2 progression requires its interaction with PCNA. Reintroduction of siRNA-resistant SET8 but not YMAA restored normal cell cycle progression. D, FACS analysis of HeLa cells transiently transfected with set8 or its mutants. E, expression of SET8 and its mutants in transiently transfected HeLa cells. F, expression of a dominant-negative SET8 mutant results in improper S phase progression. ctr, control; FL, full-length; WCE, whole cell extract.
To further test whether endogenous SET8 might associate with PCNA to elicit its role in cell cycle regulation, we transiently transfected cells with full-length SET8 and its mutants and analyzed the cell cycle profile 36 h post-transfection (Fig. 3, D and E). We reasoned that the N terminus of SET8 might function as a dominant negative to inhibit endogenous SET8 function because it interacts with PCNA but does not have methyltransferase activity. In agreement with this hypothesis, we observed an increase in S phase cell population in cells transfected with the N-terminal mutant compared with those transfected with the YMAA or C-terminal mutant, which do not interact with PCNA.
To further examine how SET8 might be required for cell cycle progression, we again took advantage of the N-terminal mutant, which binds to PCNA but does not encode the SET domain. Mimosine arrest and release experiments revealed that expression of this dominant-negative SET8 mutant inhibited S phase progression (Fig. 3F). Together, these results substantiate that via its interaction with PCNA, SET8 is important for S phase progression.
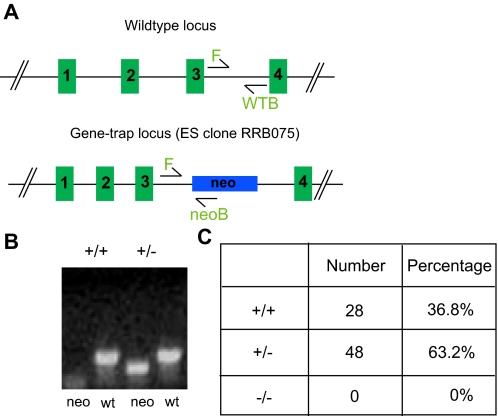
SET8 Deficiency Results in Embryonic Lethality in Mice—To explore the biological significance of SET8 function in vivo, we injected blastocytes and generated chimeras using an embryonic stem cell clone in which the set8 genomic locus was disrupted by insertion of a neomycin cassette (Fig. 4A). Genotyping of viable offspring from heterozygous matings revealed only wild-type mice or mice heterozygous for set8, suggesting that SET8 is required for embryogenesis (Fig. 4, B and C).
FIGURE 4.
SET8 is required for mouse development. A, schematic illustration of the wild-type (wt) set8 genomic locus and the gene-trapped locus. WTB, wild-type reverse primer; neoB, neo reverse primer. B, PCR analysis of F1 offspring. C, genotyping results obtained with offspring from the breeding of heterozygous set8 mice. EC, embryonic stem cell.
Epigenetic modifications are perpetuated during cell division and determine chromatin architecture and transcriptional control. One would envision that, like DNA methylation, histone modifications are also inherited from parental cells by daughter cells. Indeed, it was demonstrated recently that one of these histone marks, viz. histone H3 Lys9, is methylated by G9a during DNA replication by virtue of its interaction with the loading factor PCNA. Interestingly, our finding that SET8 also interacts with PCNA and localizes to replication foci suggests that additional histone-modifying enzymes might follow similar strategies to maintain chromatin states.
The cell cycle profile of SET8 expression correlates with H4-K20 monomethylation, which is supportive of the proposed coupling of SET8 at DNA replication sites. Our results are complementary to a recent study reporting a role for SET8 in genome replication and stability (25). Using RNA interference, the authors showed that SET8 depletion results in replicative stress and activation of the DNA damage response (25, 26). More important, the authors showed that cells accumulate in S/G2 phase upon reintroduction of methyltransferase mutants, indicating the importance of SET8-mediated H4-K20 monomethylation during DNA replication (25). These results are entirely consistent with our findings (Fig. 3). Moreover, our study provides a mechanistic basis as to how SET8 function is coupled to DNA replication: via a direct physical interaction between SET8 and PCNA.
A recent study suggested a role for mono- and dimethylated H4-K20 histones in chromatin condensation processes (20). Given that SET8 localizes to mitotic chromosomes and is required for mitosis, it would be interesting to examine whether SET8-mediated H4-K20 methylation is essential for the recruitment of L3MBTL1. Our findings that SET8 mediates H4-K20 during DNA replication and is required for S phase progression suggest that SET8 might maintain H4-K20 methylation status, which is important for subsequent chromatin condensation and explains the observed defects in mitosis in SET8-deficient larvae (6).
Dysregulation of chromatin dynamics has profound effects in cell proliferation and development. This is exemplified by clinical mutations in genes that encode proteins involved in the maintenance of histone modifications (27–33). Similarly, our finding that abrogation of set8 in mice leads to embryonic lethality strongly indicates a critical role for histone modifications in early embryogenesis. In summary, we propose that SET8 mediates H4-K20 methylation during S phase by coupling to PCNA, which is required for S phase progression and mouse embryogenesis.
Acknowledgments
We thank Irene Ward for initiating the SET8 knock-out project and Dr. Hübscher for the PCNA-His construct.
This work was in part supported by National Institutes of Health Grant CA100109 (to J. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TAP, tandem affinity purification; PCNA, proliferating cell nuclear antigen; GST, glutathione S-transferase; SFB, S tag/FLAG tag/streptavidin-binding peptide; siRNA, small interfering RNA; FACS, fluorescence-activated cell sorter; PIP, PCNA-interacting protein.
References
- 1.Berger, S. L. (2007) Nature 447 407–412 [DOI] [PubMed] [Google Scholar]
- 2.Mellor, J. (2006) Cell 126 22–24 [DOI] [PubMed] [Google Scholar]
- 3.Schreiber, S. L., and Bernstein, B. E. (2002) Cell 111 771–778 [DOI] [PubMed] [Google Scholar]
- 4.Botuyan, M. V., Lee, J., Ward, I. M., Kim, J. E., Thompson, J. R., Chen, J., and Mer, G. (2006) Cell 127 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benetti, R., Gonzalo, S., Jaco, I., Schotta, G., Klatt, P., Jenuwein, T., and Blasco, M. A. (2007) J. Cell Biol. 178 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karachentsev, D., Sarma, K., Reinberg, D., and Steward, R. (2005) Genes Dev. 19 431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rice, J. C., Nishioka, K., Sarma, K., Steward, R., Reinberg, D., and Allis, C. D. (2002) Genes Dev. 16 2225–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishioka, K., Rice, J. C., Sarma, K., Erdjument-Bromage, H., Werner, J., Wang, Y., Chuikov, S., Valenzuela, P., Tempst, P., Steward, R., Lis, J. T., Allis, C. D., and Reinberg, D. (2002) Mol. Cell 9 1201–1213 [DOI] [PubMed] [Google Scholar]
- 9.Kourmouli, N., Jeppesen, P., Mahadevhaiah, S., Burgoyne, P., Wu, R., Gilbert, D. M., Bongiorni, S., Prantera, G., Fanti, L., Pimpinelli, S., Shi, W., Fundele, R., and Singh, P. B. (2004) J. Cell Sci. 117 2491–2501 [DOI] [PubMed] [Google Scholar]
- 10.Lee, M. G., Villa, R., Trojer, P., Norman, J., Yan, K. P., Reinberg, D., Di Croce, L., and Shiekhattar, R. (2007) Sciences (N. Y.) 318 447–450 [DOI] [PubMed] [Google Scholar]
- 11.Wynder, C., Hakimi, M. A., Epstein, J. A., Shilatifard, A., and Shiekhattar, R. (2005) Nat. Cell Biol. 7 1113–1117 [DOI] [PubMed] [Google Scholar]
- 12.Schneider, J., Wood, A., Lee, J. S., Schuster, R., Dueker, J., Maguire, C., Swanson, S. K., Florens, L., Washburn, M. P., and Shilatifard, A. (2005) Mol. Cell 19 849–856 [DOI] [PubMed] [Google Scholar]
- 13.Yin, Y., Liu, C., Tsai, S. N., Zhou, B., Ngai, S. M., and Zhu, G. (2005) J. Biol. Chem. 280 30025–30031 [DOI] [PubMed] [Google Scholar]
- 14.Xiao, B., Jing, C., Kelly, G., Walker, P. A., Muskett, F. W., Frenkiel, T. A., Martin, S. R., Sarma, K., Reinberg, D., Gamblin, S. J., and Wilson, J. R. (2005) Genes Dev. 19 1444–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Couture, J. F., Collazo, E., Brunzelle, J. S., and Trievel, R. C. (2005) Genes Dev. 19 1455–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang, J., Feng, Q., Ketel, C. S., Wang, H., Cao, R., Xia, L., Erdjument-Bromage, H., Tempst, P., Simon, J. A., and Zhang, Y. (2002) Curr. Biol. 12 1086–1099 [DOI] [PubMed] [Google Scholar]
- 17.Rayasam, G. V., Wendling, O., Angrand, P. O., Mark, M., Niederreither, K., Song, L., Lerouge, T., Hager, G. L., Chambon, P., and Losson, R. (2003) EMBO J. 22 3153–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beisel, C., Imhof, A., Greene, J., Kremmer, E., and Sauer, F. (2002) Nature 419 857–862 [DOI] [PubMed] [Google Scholar]
- 19.Schotta, G., Lachner, M., Sarma, K., Ebert, A., Sengupta, R., Reuter, G., Reinberg, D., and Jenuwein, T. (2004) Genes Dev. 18 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trojer, P., Li, G., Sims, R. J., III, Vaquero, A., Kalakonda, N., Boccuni, P., Lee, D., Erdjument-Bromage, H., Tempst, P., Nimer, S. D., Wang, Y. H., and Reinberg, D. (2007) Cell 129 915–928 [DOI] [PubMed] [Google Scholar]
- 21.Min, J., Allali-Hassani, A., Nady, N., Qi, C., Ouyang, H., Liu, Y., MacKenzie, F., Vedadi, M., and Arrowsmith, C. H. (2007) Nat. Struct. Mol. Biol. 14 1229–1230 [DOI] [PubMed] [Google Scholar]
- 22.Li, H., Fischle, W., Wang, W., Duncan, E. M., Liang, L., Murakami-Ishibe, S., Allis, C. D., and Patel, D. J. (2007) Mol. Cell 28 677–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huen, M. S., Grant, R., Manke, I., Minn, K., Yu, X., Yaffe, M. B., and Chen, J. (2007) Cell 131 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moldovan, G. L., Pfander, B., and Jentsch, S. (2007) Cell 129 665–679 [DOI] [PubMed] [Google Scholar]
- 25.Tardat, M., Murr, R., Herceg, Z., Sardet, C., and Julien, E. (2007) J. Cell Biol. 179 1413–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jorgensen, S., Elvers, I., Trelle, M. B., Menzel, T., Eskildsen, M., Jensen, O. N., Helleday, T., Helin, K., and Sorensen, C. S. (2007) J. Cell Biol. 179 1337–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwase, S., Lan, F., Bayliss, P., de la Torre-Ubieta, L., Huarte, M., Qi, H. H., Whetstine, J. R., Bonni, A., Roberts, T. M., and Shi, Y. (2007) Cell 128 1077–1088 [DOI] [PubMed] [Google Scholar]
- 28.Shi, Y. (2007) Nat. Rev. Genet. 8 829–833 [DOI] [PubMed] [Google Scholar]
- 29.Lan, F., Bayliss, P. E., Rinn, J. L., Whetstine, J. R., Wang, J. K., Chen, S., Iwase, S., Alpatov, R., Issaeva, I., Canaani, E., Roberts, T. M., Chang, H. Y., and Shi, Y. (2007) Nature 449 689–694 [DOI] [PubMed] [Google Scholar]
- 30.Tahiliani, M., Mei, P., Fang, R., Leonor, T., Rutenberg, M., Shimizu, F., Li, J., Rao, A., and Shi, Y. (2007) Nature 447 601–605 [DOI] [PubMed] [Google Scholar]
- 31.Krivtsov, A. V., and Armstrong, S. A. (2007) Nat. Rev. Cancer 7 823–833 [DOI] [PubMed] [Google Scholar]
- 32.Schlesinger, Y., Straussman, R., Keshet, I., Farkash, S., Hecht, M., Zimmerman, J., Eden, E., Yakhini, Z., Ben-Shushan, E., Reubinoff, B. E., Bergman, Y., Simon, I., and Cedar, H. (2007) Nat. Genet. 39 232–236 [DOI] [PubMed] [Google Scholar]
- 33.Fraga, M. F., Ballestar, E., Villar-Garea, A., Boix-Chornet, M., Espada, J., Schotta, G., Bonaldi, T., Haydon, C., Ropero, S., Petrie, K., Iyer, N. G., Perez-Rosado, A., Calvo, E., Lopez, J. A., Cano, A., Calasanz, M. J., Colomer, D., Piris, M. A., Ahn, N., Imhof, A., Caldas, C., Jenuwein, T., and Esteller, M. (2005) Nat. Genet. 37 391–400 [DOI] [PubMed] [Google Scholar]