Abstract
The anti-tumor synthetic triterpenoid 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO)-imidazolide (CDDO-Im) ectopically activates the transforming growth factor β (TGFβ)-Smad pathway and extends the duration of signaling by an undefined mechanism. Here we show that CDDO-Imdependent persistence of Smad2 phosphorylation is independent of Smad2 phosphatase activity and correlates with delayed TGFβ receptor degradation and trafficking. Altered TGFβ trafficking parallels the dispersal of EEA1-positive endosomes from the perinuclear region of CDDO-Im-treated cells. The effect of CDDO-Im on the EEA1 compartment led to an analysis of the cytoskeleton, and we observed that CDDO-Im alters microtubule dynamics by disrupting the microtubule-capping protein, Clip-170. Interestingly, biotinylated triterpenoid was found to localize to the polarity complex at the leading edge of migrating cells. Furthermore, CDDO-Im disrupted the localization of IQGAP1, PKCζ, Par6, and TGFβ receptors from the leading edge of migrating cells and inhibited TGFβ-dependent cell migration. Thus, the synthetic triterpenoid CDDO-Im interferes with TGFβ receptor trafficking and turnover and disrupts cell migration by severing the link between members of the polarity complex and the microtubule network.
Transforming growth factor β (TGFβ)4 family members regulate many cellular functions, including proliferation and differentiation, and TGFβ is a potent apoptotic agent in many cells, including early stage epithelial tumors (1). However, in late stage epithelial tumors, TGFβ becomes a metastatic agent and stimulates epithelial to mesenchymal transition (EMT) and cell migration (2-5). Signaling by TGFβ growth factor is initiated via ligand-induced heteromeric complex formation of the Ser/Thr kinase type I (TβRI) and type II (TβRII) transmembrane receptors (6). The phosphorylation of receptor-regulated Smad proteins R-Smad2 and R-Smad3 is facilitated by Smad anchor for receptor activation protein, which binds the receptors and recruits R-Smad to the membrane of EEA1-positive early endosomes (7). Early endosomes also contain other modulators of TGFβ receptor signaling, such as hepatocyte growth factor-regulated tyrosine kinase substrate (8) and cytoplasmic promyelocytic leukemia protein (9).
Inactivation of the TGFβ signaling pathway is carried out by several mechanisms. Phosphorylated Smad2 is targeted by the nuclear phosphatase PPM1A (10), and the receptors are the target of inhibitory Smad7, which interacts with TβRI in lipid rafts/caveolae and recruits the E3 ligases, Smurf1 and Smurf2, which direct ubiquitin-dependent degradation of the TGFβ receptors (11-13). Perturbation of lipid rafts increases signaling and reduces the rate of receptor degradation (14-18). Thus, the signal transduction pathway initiated by cell surface TGFβ receptor complex is dependent on receptor internalization and trafficking via distinct endocytic pathways (19).
Endocytosis of cell surface proteins is dependent on the microtubule cytoskeleton (20-22). Microtubules are dynamic protein filaments that span the cell interior and provide a mechanical framework for chromosome sorting, cell polarity, and organelle localization, among other functions (23-28). Microtubules grow and shrink from their plus-ends, and their minus-ends are usually located at the microtubule-organizing center but are also found at the apical domain in epithelial cells (29).
Polarized migration of cells as well as the movement of vesicles along the microtubules is dependent on molecular motors and microtubule-binding proteins, such as the capping protein, Clip-170 (30, 31). Furthermore, the association of microtubule-bound Clip-170 at the cell membrane with the Rac1/Cdc42-binding protein, IQGAP1, is an essential mediator between microtubules and the leading edge of migrating cells (32, 33). Cdc42 also binds to the Par6-PKCζ polarity complex, which in turn links Cdc42 to APC and the microtubule network to form a focal point for directional cellular movement (26). These molecular links, in combination with the observation that Par6 associates with TGFβ receptors (34), have introduced a new area of study to the field of TGFβ-dependent signaling in cell migration and metastasis (2).
Chemotherapeutic agents that block TGFβ-dependent signaling and TGFβ-dependent metastasis have been a major focus in cancer chemotherapy (35). Recently, 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid (CDDO), has been shown to be a promising cancer therapeutic agent that is currently in Phase I clinical trials (36). Interestingly, both CDDO and its imidazolide derivative, CDDO-Im, have been shown to synergistically increase cellular responses to factors such as TGFβ in cell culture studies (37-39). The mechanism whereby CDDO-Im does this has yet to be elucidated. In this study, we demonstrate that CDDO-Im alters TGFβ cell signaling and receptor trafficking and inhibits cell migration by disrupting cytoskeletal attachments to the polarity complex.
EXPERIMENTAL PROCEDURES
Cell Lines and Antibodies
Mv1Lu cells were cultured in minimal essential medium supplemented with 1% nonessential amino acids. Mv1Lu cells stably transfected with the HA epitope-tagged TGFβ type II receptor (HAT cells) were cultured in minimal essential medium plus 1% nonessential amino acids and 0.3 mg/ml hygromycin. COS-7, C2C12, HEK293, and Rat2 cells were cultured in Dulbecco's modified Eagle's medium. All media were supplemented with 10% fetal bovine serum unless otherwise stated. Texas Red-conjugated phalloidin, anti-caveolin-1 (610059), anti-EEA1 (610456), anti-Rac1 (610650), anti-GM130 (610822), anti-Smad2 (610842), and anti-LAMP1 (555798) antibodies were purchased from BD Transduction Laboratories (Mississauga, Canada). Monoclonal anti-tubulin (Tub2.1), anti-FLAG (M2), and polyclonal anti-actin (A2668) antibodies were purchased from Sigma. Polyclonal anti-PKCζ (C-20), anti-TGFβ type II receptor (C-16), anti-Par6 (H-90), anti-IQGAP1 (H-109), anti-HA (Y-11), and goat anti-lamin A/C antibodies (sc-6215) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti-phospho-Smad2 (AB3849) antibodies were purchased from Chemicon (Temecula, CA). The polyclonal phospho-Par6 (Ser-345) antibody was used as previously described (34). The polyclonal anti-Clip-170 (N-terminal) antibody was a generous gift from Dr. K. Kaibuchi (Nagoya University, Nagoya, Japan), and the polyclonal anti-calnexin was kindly provided by Dr. J. J. M. Bergeron and P. H. Cameron (McGill University, Montreal, Canada). The GFP-tagged Clip-170 construct was a generous gift from Dr. F. Perez (CNRS, Paris, France).
Affinity Labeling
Cells were preincubated in control media or media containing 1 μm CDDO-Im for 1 h at 37 °C, placed on ice, and incubated with 250 pm 125I-TGFβ in KRH plus 0.5% bovine serum albumin at 4 °C for 2 h. Following cross-linking with disuccinimidyl suberate, cells were lysed (time 0) or incubated at 37 °C for 2, 4, or 8 h prior to lysis. Receptors were visualized by SDS-PAGE and quantified using PhosphorImager analysis (Amersham Biosciences).
Subcellular Fractionation
Cytoskeleton—Fractions containing the cytoskeleton were separated from those containing detergent-solubilized membranes and cytosol by the method described by Contin et al. (40). Briefly, COS-7 cells were incubated for 3 h in control medium or media containing 10 μm nocodazole or 1 μm CDDO-Im and then rinsed with microtubule stabilization buffer (90 mm Mes (pH 6.7), 1 mm EGTA, 1 mm MgCl2, 10% (v/v) glycerol) that had been preheated to 37 °C. Cells were then lysed with microtubule stabilization buffer containing 10 μm placitaxel (Sigma), 0.5% Triton X-100, and protease inhibitors for 4 min at 37 °C. The solubilized fractions were then collected. To collect the remaining cellular structures containing the cytoskeleton, SDS-PAGE sample buffer was added to the culture dishes. Following scraping and passaging through a syringe, the fractions containing the cytoskeleton were collected.
To analyze the partitioning of cytoskeletal proteins, fractions containing the soluble proteins or the cytoskeleton were subjected to SDS-PAGE followed by immunoblotting with anti-Rac1, anti-tubulin, anti-actin, anti-IQGAP1, anti-EEA1, anti-calnexin, anti-LAMP1, anti-GM130, or anti-Clip-170 antibodies.
Nuclear Fractionation—To quantify the amount of Smad2 present in the nucleus in the presence of CDDO-Im, subcellular fractionation was carried out as described by Cong and Varmus (41). Briefly, cells were incubated with 0.5 mm TGFβ for 30 min in the absence or presence of 1 μm CDDO-Im, washed, and incubated in media containing Me2SO (control), 1 μm CDDO-Im, 10 μm SB431542, or both compounds (CDDO-Im + SB431542) at 37 °C for an additional 1 or 4 h. The cells were then scraped into ice-cold PBS and collected by centrifugation at 1000 × gav for 5 min. The cells were then washed once again with phosphate-buffered saline and resuspended in hypotonic buffer (10 mm Hepes-KOH, 10 mm NaCl, 1 mm KH2PO4, 5 mm NaHCO3, 1 mm CaCl2, 0.5 mm MgCl2, pH 7.4). After a 5-min incubation on ice, cells were homogenized with 50 strokes using a Dounce homogenizer. Cells were then centrifuged at 1000 × gav for 5 min. The pellets were washed twice with hypotonic buffer and resuspended in nuclear isolation buffer (10 mm Tris (pH 7.5), 300 mm sucrose, 0.1% Nonidet P-40). The pellets in the nuclear isolation buffer were then further homogenized 50 times using a Dounce homogenizer and centrifuged at 1000 × gav for 5 min. The nuclear pellet was resuspended with nuclear isolation buffer with 1% Triton X-100 to generate the nuclear fraction. The nuclear fractions were subjected to SDS-PAGE and immunoblotted with mouse anti-Smad2 or goat anti-lamin A/C antibodies. The levels of Smad2 were quantitated using QuantityOne software (Bio-Rad) and normalized to the amount of lamin A/C in each fraction.
Phospho-Smad2 Time Course—Cells were incubated with 0.5 mm TGFβ for 30 min, in the absence or presence of 1 μm CDDO-Im, washed, and incubated in media containing Me2SO (control), 1 μm CDDO-Im, 10 μm SB431542 (Sigma), or both compounds (CDDO-Im + SB431542) at 37 °C for an additional 1 or 4 h prior to lysis. Cell lysates were then analyzed by SDS-PAGE and immunoblotted with mouse anti-Smad2 or rabbit anti-phospho-Smad2 antibodies. Quantitation of the amount of phosphorylated Smad2 was carried out using QuantityOne software and normalized to Smad2 levels.
Immunofluorescence Microscopy
Smad2 Nuclear Accumulation—Cells were incubated with 0.5 mm TGFβ for 30 min in the absence or presence of 1 μm CDDO-Im, washed, and incubated in media containing Me2SO (control), 1 μm CDDO-Im, 10 μm SB431542, or both compounds (CDDO-Im + SB431542) at 37 °C for an additional 1, 4, or 8 h prior to fixation and permeabilization. Cells were then immunostained with anti-Smad2/3 antibody followed by Cy2-conjugated secondary antibody. DAPI staining was used to visualize cell nuclei. To quantify the amount of Smad2 nuclear localization, nuclear versus cytoplasmic intensity profiles were generated from individual cells using ImagePro software. The quantitation of ≥100 cells/condition from three experiments was graphed as the nuclear signal minus the cytoplasmic signal versus time ± S.D.
Receptor Traffic—Receptor internalization studies were carried out as previously described (15). Briefly, HAT cells expressing extracellularly HA-tagged TβRII receptors were incubated with biotinylated-TGFβ for 2 h at 4 °C, washed, and incubated with Cy3-streptavidin (Jackson Immunoresearch, West Grove, PA). Cells were then washed and incubated at 37 °C for 1 h with or without CDDO-Im. After the 37 °C incubation, cells were fixed and incubated with mouse monoclonal anti-EEA1 antibodies and rabbit polyclonal anti-caveolin-1 antibodies. Anti-EEA1 and anti-caveolin-1 were detected using anti-mouse Cy5-conjugated antibodies and anti-rabbit Cy2-conjugated antibodies (Jackson Laboratories Inc.), respectively. Acid washing was carried out as previously described (15). Images were captured using an Olympus 1X81 inverted microscope equipped with fluorescence optics and deconvolved using ImagePro software.
EEA1 Distribution—To visualize the effect of CDDO-Im on EEA1-positive endosomes, we assessed the staining pattern of the EEA1 compartment via immunofluorescence microscopy as described above with the exception that Cy2-labeled secondary antibodies were used. DAPI staining was used to visualize cell nuclei.
Staining observed to be localized to an area no greater than one-half of the cell area and located more intensely on one side of the nucleus was scored as “perinuclear-positive.” If the intensity and distribution of the stain was equal throughout the cell body, it was scored as “dispersed.” The quantitation of ≥100 cells/condition from three experiments was graphed ± S.D.
Cytoskeleton Studies—To visualize the effect of CDDO-Im on the microtubule cytoskeleton, cells were incubated for 1 h with or without 1 μm CDDO-Im or 10 μm nocodazole (Sigma) at 37 °C. Following fixation and permeabilization, cells were incubated with monoclonal anti-tubulin antibody followed by fluorescein isothiocyanate-conjugated secondary antibodies. To assess the effect of CDDO-Im on the actin cytoskeleton, cells were incubated with 1 μm CDDO-Im or 10 μm cytochalasin B for 1 h prior to fixation and permeabilization. The cells were then incubated for 30 min with Texas Red-conjugated phalloidin. Images were collected from a Leica DMIRE inverted microscope.
Scratch Assay—Rat2 fibroblasts were grown to confluence, and the cell monolayer was scratched with a sterile pipette tip to create a “wound.” For bright field or DIC microscopy, images were collected using an Olympus IX81 inverted microscope. For immunofluorescence studies, cells were fixed, permeabilized, and incubated with anti-Clip-170, anti-tubulin, anti-Rac1, anti-TβRII, anti-IQGAP1, anti-Par6, or anti-PKCζ antibodies. Following fluorescently tagged secondary antibody incubation, the nuclei of cells were stained with DAPI, and cells were visualized using an Olympus IX81 microscope controlled by QED in vivo software (Olympus).
The quantitation of the number of cells containing proteins at the leading edge of the cell was carried out using ≥100 cells/condition from three experiments ± S.D. Briefly, a cell containing a positive immunofluorescence signal only along the edge of the plasma membrane that was directly adjacent to the scratch was scored as positive for leading edge localization. If a cell contained no signal along the edge adjacent to the scratch or contained dispersed signal along the complete cell periphery, it was scored as negative for leading edge staining.
Clip-170 Movies—Rat2 fibroblasts were microinjected with 0.25 nm cDNA encoding GFP-tagged Clip-170 protein and incubated at 37 °C for 4 h. The cells were then incubated in control medium or media containing 0.1 or 1 μm CDDO-Im. Expressed GFP-tagged Clip-170 was visualized by immunofluorescence, and time lapse images were collected using a Leica DMIRE inverted microscope utilizing Openlab 3.0 software.
Cell Transfection
Cells were transfected with the constructs described in the figure legends using the calcium phosphate method as previously described (15).
Immunoprecipitation
Cells were lysed (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 0.5% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, and mixture of protease inhibitors) and centrifuged at 15,000 × gav at 4 °C for 5 min. Aliquots of supernatants were collected for analysis of total protein concentration. The remaining cell lysates were incubated with primary antibody, followed by incubation with protein G-Sepharose. The precipitates were washed three times with lysis buffer, eluted with Laemmli sample buffer, and subjected to SDS-PAGE and immunoblot analysis.
CDDO-biotin Subcellular Localization
Rat2 fibroblasts were grown to 100% confluence and scratched to create a wound. The cells were then incubated at 37 °C for 6 h to allow for the establishment of polarity. The cells were then fixed, permeabilized, and incubated with Rac1 antibodies followed by Cy2-labeled secondary antibodies. The cells were then incubated with 10 μm biotin, CDDO, or CDDO-biotin for 2 h, followed by incubation with Cy3-labeled streptavidin and DAPI. Cells were visualized using an Olympus IX81 inverted microscope.
Protein Concentration
Protein concentrations of lysates were measured using the Lowry method (Fisher).
RESULTS
The triterpenoid, CDDO-imidazolide (CDDO-Im) extends TGFβ signal transduction in several cell lines (37, 39); however, the mechanism(s) and functional consequences have yet to be fully elucidated.
In order to study the mechanism of how CDDO-Im extends TGFβ-dependent signaling, we first attempted to identify cell lines in which TGFβ-dependent Smad2 phosphorylation is transient. We treated various cell lines with a 30-min pulse of TGFβ and investigated the state of Smad2 phosphorylation by immunoblot analysis using a phospho-Smad2-specific antibody. We observed that the phosphorylation of Smad2 was transient in Rat2, COS-7, and C2C12 cells (≤4.5 h) whereas it was prolonged (>8 h) in HepG2 and HeLa cells (Fig. 1) (data not shown). In Rat2 and COS-7 cells, Smad2 phosphorylation was observed 30 min after TGFβ stimulation and attenuated over the remainder of the time course (Fig. 1, A and B, lanes 1-4). In C2C12 cells, Smad2 phosphorylation returned to background levels within 1 h of ligand removal from the culture medium (Fig. 1C, lanes 1-4). Having observed transient Smad2 phosphorylation in Rat2, C2C12, and COS-7 cells, we next assessed the effect of co-incubating cells with TGFβ and CDDO-Im. We observed that CDDO-Im increased both the extent and duration of TGFβ-dependent Smad2 phosphorylation in all of the three cell types tested (Fig. 1, lanes 5-8).
FIGURE 1.
CDDO-Im extends TGFβ-dependent Smad2 phosphorylation independently of Smad2 phosphatase activity. Rat2 (A), COS-7 (B), or C2C12 (C) cells were incubated with 0.5 mm TGFβ for 30 min in the absence or presence of 1 μm CDDO-Im, washed, and incubated in media containing Me2SO (DMSO) (Control), 1 μm CDDO-Im, 10 μm SB431542, or both compounds (CDDO-Im + SB431542) at 37 °C for an additional 1 or 4 h prior to lysis. One hundred micrograms of cell lysates were then processed for SDS-PAGE and immunoblotted with anti-phosphospecific Smad2 (α-P-S2) or Smad2/3 (α-S2/3) antibodies. The bands corresponding to phosphoserine-modified Smad2 (P-S2) and Smad2 (S2) are indicated. Representative blots from four experiments are shown.
We next investigated if the effect of CDDO-Im on the duration of Smad2 phosphorylation could be due to an inhibition of Smad2 phosphatase activity. In order to test this, we employed one of the strategies used by Lin et al. (10) to identify PPM1A as the nuclear Smad2 phosphatase. Briefly, the TGFβ type I receptor inhibitor, SB431542, was added to the cell culture media after a preincubation of the cells with TGFβ. The addition of the inhibitor would allow assessment of whether TGFβ receptor activity would influence the effect of CDDO-Im on TGFβ-dependent Smad2 phosphorylation. In Rat2, COS-7, and C2C12 cells, the SB431542 inhibitor greatly reduced the phosphorylation of Smad2 within 1 h of incubation, indicating that these cell types have functional phosphatase activity (Fig. 1, lanes 9-12). We next assessed if CDDO-Im would influence Smad2 phosphatase by co-incubating cells with SB431542 and CDDO-Im. We observed that CDDO-Im did not extend TGFβ-dependent Smad2 phosphorylation in Rat2, COS-7, or C2C12 cells when SB431542 was present in the culture media (Fig. 1, lanes 13-16). These results suggest that the Smad2 phosphatase was not a CDDO-Im target, since CDDO-Im was unable to extend the duration of Smad2 phosphorylation after TGFβ receptor inactivation.
As a second line of investigation, we assessed TGFβ-dependent Smad2 nuclear translocation using immunofluorescence microscopy (Fig. 2). We found that in the absence of TGFβ, Smad2 staining was observed throughout the cytoplasm of C2C12 cells (Fig. 2A). After 1 h of TGFβ stimulation, Smad2 staining was only observed in the nuclei of cells regardless of treatment (Fig. 2, B-E, top panels). Eight hours after TGFβ was removed from the cell culture medium, Smad2 cytoplasmic staining reappeared in both control and CDDO-Im-treated cells (Fig. 2, B and C, bottom panels).
FIGURE 2.
CDDO-Im extends TGFβ-dependent Smad2 nuclear accumulation independently of Smad2 phosphatase activity. Unstimulated C2C12 cells (A) or C2C12 cells pulsed with 0.5 mm TGFβ for 30 min in the absence or presence of 1 μm CDDO-Im were incubated in media containing Me2SO (DMSO) (B), 1 μm CDDO-Im (C), 10 μm SB431542 (D), or both compounds (E) at 37 °C for 1 or 8 h. The cells were then processed for immunofluorescence microscopy and probed with DAPI (blue) to indicate nuclei and anti-Smad2/3 (Smad2) antibodies (green). Nuclear and cytoplasmic Smad2 from three experiments ± S.D. were quantitated using ImagePro software and graphed as nuclear-cytoplasmic Smad2 versus time (F). Bar, 10 μm.
Consistent with the phospho-Smad2 time course studies, the TGFβ type I receptor inhibitor, SB431542, increased cytoplasmic Smad2 staining after 8 h of incubation (Fig. 2D), and this was also observed if the cells were co-incubated with SB431542 and CDDO-Im (Fig. 2E). These results were supported by the quantitation of three experiments (Fig. 2F), and similar results were obtained with the use of Rat2 or COS-7 cells (data not shown).
Finally, to further evaluate TGFβ-dependent nuclear accumulation of Smad2 in the presence or absence of CDDO-Im and/or SB431542, we carried out subcellular fractionation studies. We assessed TGFβ-dependent Smad2 nuclear accumulation by immunoblotting isolated nuclear fractions with anti-Smad2 antibodies (supplemental Fig. 1). In control cells, we observed Smad2 in the nuclear fractions after 0.5 h of TGFβ stimulation and the signal attenuated after 4.5 h of stimulation (supplemental Fig. 1, lanes 1-3). This was accentuated if the cells were incubated with CDDO-Im (supplemental Fig. 1, lanes 4-6). We observed that the TGFβ type I receptor inhibitor, SB431542, decreased the amount of Smad2 in the nuclear fractions after 4 h of incubation (supplemental Fig. 1, lanes 7-9), and this was also observed if the cells were co-incubated with SB431542 and CDDO-Im (supplemental Fig. 1, lanes 10-12). Taken together, these results support the conclusion that CDDO-Im increases TGFβ-dependent Smad2 phosphorylation and nuclear accumulation in a process that is independent of Smad2 phosphatase activity.
CDDO-Im Interferes with TGFβ Receptor Degradation and Trafficking—Efficient phosphorylation and nuclear translocation of Smad2 is dependent on the proper targeting of TGFβ receptors to the early endosomal compartment (8, 9, 15, 42-45). Inhibition of the signal transduction pathway is dependent on Smad2 phosphatase activity (10) and on the association of TGFβ receptors with the Smad7-Smurf2 complex, which inhibits TGFβ receptor kinase activity and promotes receptor complex degradation (11, 12). Since we concluded that Smad2 phosphatase activity was not targeted by CDDO-Im, we attempted to determine if CDDO-Im regulates the kinetics of Smad activation by altering TGFβ receptor degradation. We therefore assessed TGFβ receptor turnover in C2C12 and COS-7 cells by affinity-labeling receptors with 125I-TGFβ at 4 °C followed by incubation at 37 °C, SDS-PAGE, and PhosphorImager analysis. We observed that both C2C12 and COS-7 cells exhibited a receptor half-life of ∼3 h and that treating cells with CDDO-Im greatly reduced the rate of TGFβ receptor degradation (Fig. 3A). We also carried out TGFβ receptor degradation studies in mink lung epithelial cells, Mv1Lu cells, a cell line that has been used to study TGFβ signaling and degradation (12, 13). In Mv1Lu cells, the receptor half-life was observed to be ∼3 h (Fig. 3A). Consistent with the results observed with COS-7 and C2C12 cells, the rate of TGFβ receptor degradation was delayed to at least 8 h in the CDDO-Im-treated Mv1Lu cells.
FIGURE 3.
CDDO-Im delays TGFβ receptor degradation and trafficking. A, C2C12, COS-7, or Mv1Lu cells were affinity-labeled with 125I-TGFβ at 4 °C, cross-linked, and lysed (zero time control) or incubated at 37 °C for 2, 4, or 8 h prior to lysis. One hundred micrograms of cell lysates were then subjected to SDS-PAGE followed by autoradiography and PhosphorImager analysis. The bands representing TβRI or TβRII are indicated on the left side of each panel. Total receptor levels were quantified and graphed (right panel) as receptor levels (percentage of control) versus time (n = 3 ± S.D.). B, Mv1Lu cells stably expressing TβRII were incubated at 4 °C with biotinylated TGFβ (bTGFβ), followed by streptavidin Cy3 (red). The cells were then incubated at 37 °C in the absence (left; Control) or presence (right) of 1 μm CDDO-Im for 1 h. Cells were then fixed, permeabilized, and immunostained with anti-caveolin-1 (Cav-1; green) and anti-EEA1 (EEA1; blue) antibodies. Areas of interest (white boxes) are magnified for both the control and CDDO-Im-treated cells and are shown below each panel (insets). Representative cells shown indicate the co-localization of biotinylated TGFβ with either caveolin-1 or EEA1 indicated by yellow or magenta arrowheads, respectively. The contour (dotted line) of each cell was established using bright field images and overlaid on the DAPI nuclear stain. Representative micrographs from three experiments are shown. Bar, 10 μm.
A delay in receptor degradation might be due to receptor exclusion from the compartment where degradation occurs or to a reduction in receptor trafficking to that compartment. To clarify this, we investigated receptor internalization and co-localization with EEA1- or caveolin-1-positive vesicles in the same cell. As we have previously reported, Mv1Lu cells expressing HA-tagged TβRII internalize biotinylated TGFβ into both EEA1- and caveolin-1-positive vesicles, as assessed by deconvolution immunofluorescence microscopy (15) (Fig. 3B, left). In the presence of CDDO-Im, the amount of TGFβ receptor trafficking was reduced, since the majority of receptors after 1 h of incubation at 37 °C did not accumulate in the perinuclear regions of cells (Fig. 3B, right). However, co-localization with EEA1 or caveolin-1-positive compartments, even in regions close to the plasma membranes of cells, was still readily apparent (Fig. 3B, inset). Although this suggested that TGFβ receptors had indeed internalized from the plasma membrane, we wanted to test this further by carrying out acid washing experiments (supplemental Fig. 2). Briefly, Mv1Lu cells stably expressing TβRII were incubated at 4 °C with biotinylated TGFβ followed by streptavidin-Cy3. The cells were then either acid-washed to remove any cell surface labeling or incubated at 37 °C in the absence or presence of 1 μm CDDO-Im for 1 h prior to incubation in acidic conditions. We observed that fluorescent probes bound to cell surface receptors at 4 °C were susceptible to acid washing and were removed. However, receptor-bound probes were not removed by acidic treatment after the cells had been incubated at 37 °C for 1 h either in the presence or absence of CDDO-Im (supplemental Fig. 2). This demonstrated that although CDDO-Im altered TGFβ receptor trafficking, it did not inhibit receptor internalization from the plasma membrane.
Interestingly, we noted that caveolin-1-positive vesicles were closer to the plasma membrane in CDDO-Im-treated cells (Fig. 3B). Moreover, co-localization of TGFβ receptors with the caveolin-1-positive compartment became quite prominent. The EEA1-positive vesicles also appeared to have an altered subcellular distribution in cells treated with CDDO-Im.
To further assess the effect of CDDO-Im on the EEA1-positive endosomal compartment, we stained untreated or CDDO-Im-treated cells with anti-EEA1 antibodies and scored the number of cells that demonstrated a perinuclear staining pattern versus cells with a dispersed EEA1 staining pattern (Fig. 4). In cells that were not incubated with CDDO-Im, 72 ± 5% of the cells counted displayed EEA1 in a perinuclear distribution, whereas CDDO-Im-treated cells only had 27 ± 4% perinuclear staining (Fig. 4B). These data indicate that although CDDO-Im does not affect TGFβ entrance into either the EEA1 or caveolin-1-positive compartments, it interferes with the dynamics of their trafficking. Furthermore, CDDO-Im leads to the dispersal of the EEA1 compartment and an accumulation of caveolin-1-positive structures in close proximity to the plasma membranes of cells.
FIGURE 4.
CDDO-Im disperses the EEA1 early endosomal compartment. A, vehicle-treated Mv1Lu cells (Control) or Mv1lu cells treated with 1 μm CDDO-Im for 1 h at 37 °C were fixed, permeabilized, and immunostained with anti-EEA1 antibodies (EEA1; green). The nuclei were visualized using DAPI staining (blue). A representative cell (inset) was magnified and is shown at the bottom. Bar, 10 μm. B, three experiments were carried out as described in A. One hundred cells from each experimental condition were analyzed, scored on the basis of perinuclear or dispersed staining of EEA1 protein, and graphed (n = 3 ± S.D.).
CDDO-Im Interferes with TGFβ-dependent Cell Migration—In order to investigate a functional consequence of extending TGFβ-dependent Smad signaling by CDDO-Im, we assessed TGFβ-dependent cell migration, because CDDO-Im as well as the parental CDDO compound exhibit potent antimetastatic activity in animal models (36). To assess polarized cell movement, we carried out “wound healing” assays using Rat2 fibroblasts. In this assay, regions of confluent Rat2 cells were removed, and cell migration into the cell-free space was assessed (Fig. 5A). When cells were incubated in medium containing low serum, the “wound” was observed even after 12 h of incubation at 37 °C (Fig. 5A, top). The addition of TGFβ to the media containing low serum induced the cells at the edge of the scratch to migrate perpendicularly into the cell-free space (Fig. 5A, middle). Interestingly, the co-incubation of TGFβ and CDDO-Im inhibited TGFβ-dependent cell migration (Fig. 5A, bottom).
FIGURE 5.
CDDO-Im delays TGFβ-dependent cell migration but does not disrupt the association of TGFβ receptors with Par6 protein. A, confluent monolayers of Rat2 fibroblasts were scratched and incubated at 37 °C for 12 h in media supplemented with 0.2% fetal bovine serum (Control; top), media supplemented with 0.2% fetal bovine serum plus 0.5 nm TGFβ (middle), or media supplemented with 0.2% fetal bovine serum plus 0.5 nm TGFβ and 0.5 μm CDDO-Im (bottom). Cells were then fixed and imaged using bright field microscopy. The dotted lines indicate the starting point of cell migration. Representative micrographs from three experiments are shown. Bar, 0.1 mm. B, HEK293 cells were transiently transfected with cDNA encoding the proteins indicated and incubated in the absence or presence of 1 μm CDDO-Im for 2 h prior to lysis and immunoprecipitation (IP) with anti-Par6 antibodies. The immunoprecipitates were then subjected to SDS-PAGE and immunoblotting with anti-HA or anti-FLAG antibodies to reveal protein complexes. One hundred micrograms of total protein lysates were immunoblotted (bottom) to assess protein expression. The bands corresponding to TβRI, TβRII, wild type Par6 (Par6 wt.), Par6 lacking the PB1 domain (Par6-ΔPB1), and IgG heavy chain (IgG HC) are indicated. Representative immunoblots from four experiments are shown.
One possibility for this inhibition could be due to a dissociation of TGFβ receptors from the polarity complex protein Par6. This is based on the observation that the association between TGFβ receptors and Par6 is essential for EMT and cell migration (2). We therefore carried out co-immunoprecipitation studies and observed that TGFβ type I and II receptors associate with Par6 in untreated or CDDO-Im-treated cells (Fig. 5B). To assess the specificity of association, we used a Par6 mutant that lacks the PB1 domain (Par6-ΔPB1). This domain was previously shown to be essential for Par6-TGFβ receptor association (34). As expected, we observed little association between the Par6-ΔPB1 mutant and TGFβ receptors (Fig. 5B). We next assessed the phosphorylation of Par6 by the TGFβ type II receptor, using phosphoserine 345-specific antibodies. The phosphorylation of Ser-345 by TGFβ type II receptors has been shown to be necessary for TGFβ-dependent EMT (34). As seen in the co-immunoprecipitation studies, we did not observe a change in TGFβ receptor-dependent phosphorylation of Par6 in the presence of CDDO-Im (data not shown).
These results suggested that the inhibition of TGFβ-dependent migration by CDDO-Im was not dependent on the association of Par6 with TGFβ receptors or its phosphorylation and that other cellular targets could be part of the underlying mechanism(s).
CDDO-Im Alters Cytoskeletal Dynamics—The cellular cytoskeleton regulates subcellular position of organelles, vesicular traffic, and cell polarity and cell migration (46, 47). Since we observed that CDDO-Im affects all of these processes in various cell lines, we examined if the cytoskeletal network is a potential target for this compound. First, we evaluated the actin cytoskeleton in control and CDDO-Im-treated cells by staining for filamentous actin (F-actin) using Texas Red-labeled phalloidin (Fig. 6A, top and middle). In untreated cells, F-actin was observed as both a membrane-bound network, as well as stress fibers. In CDDO-Im-treated cells, the pattern of F-actin staining was indistinguishable from control cells, suggesting that the actin cytoskeleton is not a target of CDDO-Im. As a positive control, we treated cells with cytochalasin B, which depolymerizes the actin cytoskeleton, and observed marked structural differences compared with control or CDDO-Im-treated cells (Fig. 6A, bottom).
FIGURE 6.
CDDO-Im does not affect the actin cytoskeleton but alters the microtubule cytoskeleton. A, control Mv1Lu cells (top) or cells treated with 1 μm CDDO-Im (middle) or 10 μm cytochalasin B (bottom) for 1 h were fixed and permeabilized. To visualize the actin cytoskeleton and the nuclei, cells were incubated with Texas Red (TR)-phalloidin (red) and DAPI stain (blue), respectively. Representative micrographs from three experiments are shown. Bar, 10 μm. B, Mv1Lu cells stably expressing the HA-tagged TβRII were incubated at 4 °C with biotinylated TGFβ followed by strepavidin-Cy3 (red; bTGFβ) and transferred to 37 °C in the absence (Control) or presence of 1 μm CDDO-Im, 10 μm nocodazole, or 10 μm cytochalasin B. The cells were fixed, permeabilized, and probed with anti-β-tubulin antibodies (Tubulin; green). The nuclei were visualized using DAPI staining (blue). The micrographs of the microtubule staining for the control (i) or the CDDO-Im-treated cells (ii) were magnified to demonstrate microtubule structural differences (right). Representative micrographs from five experiments are shown. Bar, 10 μm.
We next assessed how CDDO-Im affected the microtubule network (Fig. 6B). Although CDDO-Im did not cause depolymerization of the microtubule network, the compound had a marked effect on the organization and orientation of microtubules. In control cells, the microtubule network radiated outward from the microtubule organizing center toward the cell periphery (Fig. 6B, i). However, in CDDO-Imtreated cells, microtubules emanating from the microtubule organizing center clearly lacked the characteristic pattern of the microtubule network, appearing to snake through the cytoplasm in a less organized fashion (Fig. 6B, ii). As positive and negative controls, we treated cells with nocodazole (which causes complete microtubule disruption) and cytochalasin B (which does not alter microtubule networking), respectively. As expected, nocodazole but not cytochalasin B depolymerized microtubules and interfered with the trafficking of TGFβ receptors. We also examined TGFβ receptor localization after CDDO-Im treatment and observed that the drug interfered with the distribution of TGFβ receptors. Thus, trafficking of TGFβ receptors to the perinuclear region is dependent on proper microtubule organization and polarity. Together, these data indicate that CDDO-Im alters the microtubule network in a fashion that is distinct from the microtubule-depolymerizing drug, nocodazole.
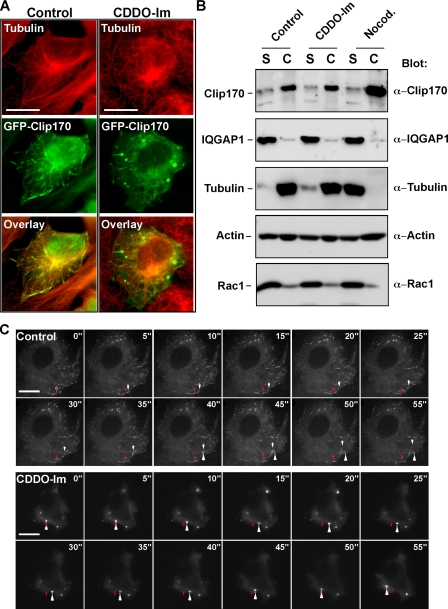
To further investigate the mechanism of CDDO-Im-dependent microtubule network interference, we first investigated microtubule-capping proteins, which modulate membrane association of the microtubule both at the plasma membrane and with vesicles (48). Clip-170 is a major capping protein that associates with the plus-end of growing microtubules and links the microtubule to vesicles and to the plasma membrane via interactions with IQGAP1 (32). Given the meandering nature of the microtubules in CDDO-Im-treated cells, we postulated that membrane and vesicular attachment might also be affected by CDDO-Im.
To investigate the effect of CDDO-Im on Clip-170 association with microtubules, we transiently transfected Rat2 fibroblasts with GFP-tagged Clip-170 and assessed its localization by immunofluorescence microscopy. In control cells, Clip-170 was observed to be associated with the positive end of growing microtubules (Fig. 7A, left), but in CDDO-Im-treated cells, Clip-170 was redistributed to large puncta in the cytoplasm and was no longer associated with microtubules (Fig. 7A, right). To further address this, we examined Clip-170 association with the cytoskeleton by biochemical fractionation. For this, we solubilized membranes and separated the soluble fraction from the insoluble cytoskeleton (Fig. 7B), a method that effectively removes the majority of cellular organelles from the cytoskeleton fraction (supplemental Fig. 3). As expected, tubulin and most of actin protein was in the cytoskeletal fraction in control cells (Fig. 7B), whereas in nocodazole-treated cells, tubulin, but not actin, partitioned with the soluble fraction. Consistent with the immunofluorescence studies, CDDO-Im of cells did not induce a solubilization and loss of tubulin from the cytoskeletal fraction. Interestingly, Clip-170 was consistently present in the cytoskeletal fraction, regardless of the treatment. This was surprising, because we assumed that the dissociation of Clip-170 from microtubules by either nocodazole or CDDO-Im treatment would cause the majority of the Clip-170 to be concentrated in the soluble fraction. However, Clip-170 remained in the cytoskeletal fraction, suggesting that either CDDO-Im did not induce complete Clip-170 dissociation from the cytoskeleton or that Clip-170 aggregated in punctate masses in response to CDDO-Im. In order to distinguish between the two possibilities, we visualized the dynamics of Clip-170 mobility in real time by microinjecting cDNA encoding GFP-Clip-170 into Rat2 fibroblasts and carrying out time lapse immunofluorescence microscopy. In control cells, the GFP-Clip-170 followed the growing microtubules (Fig. 7C and supplemental Movie 1). In the CDDO-Im treated cells, the punctate aggregates that contained the GFP-Clip-170 remained mobile at lower triterpenoid concentrations (0.1 μm; Fig. 7C and supplemental Movie 2) but were immobilized at higher doses (1 μm; supplemental Movie 3), thereby stalling the formation of a normal microtubule network. These data indicate that CDDO-Im affects the organization of the microtubular network by interfering with the function of the capping protein Clip-170.
FIGURE 7.
CDDO-Im induces Clip-170 to dissociate from microtubules. A, Rat2 fibroblasts transiently expressing GFP-Clip-170 were incubated in the absence (left) or presence of 1 μm CDDO-Im (right) for 1 h and were then fixed, permeabilized, and immunostained with anti-β-tubulin antibodies (red). The co-localization of GFP-Clip-170 (green) and microtubules (red) results in yellow staining. Bar, 10 μm. B, COS-7 cells were incubated in the absence (Control) or presence of 1 μm CDDO-Im or 10 μm nocodazole (Nocod.) for 1 h and then subjected to lysis at 37 °C to separate soluble proteins (S) from the cytoskeleton (C). Following processing for SDS-PAGE, cell lysates were immunoblotted with antibodies raised against Clip-170, IQGAP1, tubulin, actin, or Rac1. The relative position of each resolved protein is indicated. Representative blots from four experiments are shown. C, Rat2 fibroblasts were microinjected with a GFP-Clip-170 cDNA and incubated for 4 h at 37 °C. The movement of GFP-tagged Clip-170 in control cells (top time course) or cells incubated with 0.1 μm CDDO-Im (bottom time course) were imaged using time lapse immunofluorescence microscopy. The starting point of movement of a representative Clip-170 cap is indicated by a red bar, and the white arrow follows the movement of the Clip-170 signal along microtubules in the control cell and in large aggregates in the CDDO-Im-treated cell. Representative images from three experiments are shown. Bar, 10 μm.
CDDO-Im Targets the Polarity Complex—In order to evaluate the cellular target of triterpenoids, we attempted to identify the subcellular localization of triterpenoids by utilizing a biotinylated version of CDDO. The biotinylated compound elicits identical cellular responses as the CDDO-Im, albeit at higher concentrations (49), and allows for the assessment of triterpenoid subcellular localization by immunofluorescence microscopy (Fig. 8).
FIGURE 8.
Subcellular targeting of biotinylated CDDO. Rat2 fibroblasts were grown to 100% confluence and scratched to create a wound. After incubation for 6 h to allow cell polarization and migration, cells were fixed, permeabilized, and incubated with monoclonal anti-Rac1 antibodies (Rac1; green) and biotin (A), CDDO (B), or CDDO-biotin (C), followed by Cy2-labeled anti-mouse antibody and Cy3-labeled streptavidin. The co-localization of Rac1 (green) with CDDO-biotin (red) at the leading edge of migrating cells is demonstrated in the inset (yellow arrowheads). The blue arrow indicates the direction of cell movement, and DIC microscopy was included to visualize the leading edge of migrating cells. Representative images from four experiments are shown. Bar, 10 μm.
As controls, biotin or CDDO did not exhibit any fluorescence signal (Fig. 8, A and B). However, we observed that CDDO-biotin localized in the nuclei and at the cell membrane in patches that were consistent with the leading edge of migrating cells. To confirm this possibility, we immunostained scratched Rat2 fibroblasts using CDDO-biotin and Rac1 antibodies and observed that Rac1 co-localized with biotinylated CDDO-biotin at the leading edge of the migrating cells (Fig. 8C).
We next examined the subcellular localization of molecules known to be involved in the leading edge of polarized cells (Fig. 9). To carry this out, we first assessed the morphology of the leading edge of migrating Rat2 cells by DIC microscopy and observed that untreated cells were elongated and had distinct lamellipodia, whereas CDDO-Im-treated cells were round. In order to assess this in a dynamic fashion, we carried out a real time study where Rat2 fibroblasts were wounded and incubated in control or CDDO-Im-containing media over time. Bright field images were collected over 13 h and arranged in a movie (supplemental Movie 4).
FIGURE 9.
CDDO-Im interferes with cell morphology and the localization of Clip-170 at the leading edge of migrating cells. A, Rat2 fibroblasts were grown to 100% confluence and scratched to create a wound. The cells were then incubated for 4 h at 37 °C and treated with control medium or medium containing 1 μm CDDO-Im for an additional 2 h before being fixed and processed for DIC microscopy. The dotted lines indicate the starting point of cell migration. Representative micrographs from three experiments are shown. The blue arrows indicate the direction of cellular movement. Bar, 10 μm. B, model of microtubule association with a subset of polarity complex proteins at the leading edge of migrating cells. The microtubule capping protein, Clip-170, associates with IQGAP1. PKCζ is shown in this model as a member of the polarity complex, and the blue arrow indicates the direction of cellular movement. C, Rat2 fibroblasts were grown to 100% confluence, scratched, and then incubated for 6 h at 37 °C. Cells were then incubated an additional 2 h in control medium (Control; top) or in media containing 1 μm CDDO-Im (middle) or 10 μm nocodazole (bottom) prior to fixation, permeabilization, and immunostaining with anti-Clip-170 (Clip-170) and anti-tubulin (Tubulin) antibodies. The scratches were made in the horizontal plane above the cells shown, and the leading edges of migrating cells containing microtubule ends and Clip-170 are indicated with green and red arrowheads, respectively. The yellow arrowheads show the co-localization of microtubule ends and Clip-170. The blue arrows indicate the direction of cellular movement. Bar, 10 μm.
The positioning of CDDO-biotin at the leading edge of migrating cells (Fig. 8C) and the CDDO-Im-dependent loss of lamellipodia (Fig. 9A, bottom) prompted us to investigate the link between microtubules and the polarity complex, found at the leading edge of migrating cells (Fig. 9B). For this line of investigation, we carried wound-healing assays by first scratching Rat2 cells, allowing them to polarize and migrate for 6 h, and then incubated them in media containing CDDO-Im or nocodazole for an additional 2 h. This experimental approach would allow us to study the effects of the drugs on the fate of the polarity complex after it had been pre-established for 6 h. We therefore first assessed tubulin and Clip-170 in scratched Rat2 fibroblasts (Fig. 9C). In addition to the cell body, control cells displayed endogenous Clip-170 decorating the leading edge of migrating cells, where tubulin extended to the plasma membrane. In contrast, CDDO-Im caused endogenous Clip-170 to disperse from the leading edge into punctate structures reminiscent of those observed after GFP-Clip-170 expression (see Fig. 7A). We also examined the localization of tubulin and Clip-170 proteins in nocodazole-treated cells. As expected, microtubules were disrupted, but despite the accompanying alteration in cellular morphology, tubulin and Clip-170 staining remained co-localized to the leading edge in these cells (Fig. 9C, bottom). This was notably different from the CDDO-Im treated cells, where there was an absence of Clip-170 staining at the leading edge.
The association of microtubules to the leading edge of cells is facilitated by Clip-170, which links microtubules to IQGAP1 (32). Since Clip-170 failed to localize to the leading edge of CDDO-Imtreated cells, we examined if CDDO-Im also affects IQGAP1 targeting to the leading edge of polarized cells. In untreated cells, IQGAP1 localized to the leading edge of migrating cells, and CDDO-Im treatment abrogated this localization (Fig. 10A). Moreover, general microtubule destabilization with nocodazole did not affect IQGAP1 localization, consistent with our observation that this drug does not affect the polarity complex once it has already been established (Fig. 10, A and B). Finally, in order to determine if CDDO-Im causes a complete dissociation of proteins present in the polarity complex, we assessed the association of IQGAP1 with Rac1 (Fig. 10C). We did not find any appreciable dissociation of Rac1 that was co-immunoprecipitated with IQGAP1 antibodies either in CDDO-Im- or nocodazole-treated cells. These data, in conjunction with the observation that CDDO-Im induced the redistribution of IQGAP1 at the plasma membrane, suggested that the localization of Rac1 and IQGAP1 to the leading edge might be affected by triterpenoid treatment.
FIGURE 10.
Reduction of IQGAP1 localization at the leading edge of migrating cells in response to CDDO-Im treatment. A, cells were scratched and allowed to migrate into the wound for 6 h in order to establish cell polarity prior to incubation with control medium (top) or media containing 1 μm CDDO-Im (middle) or 10 μm nocodazole (bottom) for an additional 2 h. Cells were then fixed, permeabilized, and immunostained for endogenous Rac1 (green) and IQGAP1 (red) protein. A representative area of interest (white box) from each condition was enlarged and shown (inset). The green and red arrows indicate Rac1 and IQGAP1 at the leading edge of migrating cells, respectively. The co-localization of Rac1 with IQGAP1 is indicated by yellow arrowheads. Bar, 10 μm. B, quantitation of cells containing Rac1 or IQGAP1 at the leading edge of migrating cells was carried out as described under “Experimental Procedures” and graphed (n = 3 ± S.D.). C, untreated cells (Control) or cells incubated with either CDDO-Im or nocodazole (Nocod.) were lysed and immunoprecipitated with nonimmune IgG (Non-IM IgG) or anti-IQGAP1 antibodies (α-IQGAP1) and immunoblotted (Blot) with anti-IQGAP1 or anti-Rac1 antibodies. The relative mobilities of Rac1 or IQGAP1 are indicated on the left of each panel. One hundred micrograms of total protein lysates were immunoblotted and shown on the left. Representative blots from three experiments are shown.
To expand our analysis to other members of the polarity complex, we assessed the localization of Rac1 and PKCζ, both of which normally localize to the leading edge of cells undergoing polarized cell movement (Fig. 11). In untreated cells, both Rac1 and PKCζ were observed to co-localize at the leading edge of migrating cells (Fig. 11A). However, in response to CDDO-Im, we observed that although Rac1 localized to the membrane, it was less organized and dispersed along the cell membrane. Furthermore, PKCζ staining was altogether absent from the cell membrane of CDDO-Im-treated cells (Fig. 11B). Nocodazole treatment altered the morphology of leading edge cells, and Rac1 was now found in numerous protrusive structures all around the cell (Fig. 11C). In these cells, PKCζ remained co-localized with Rac1, consistent with the lack of change in IQGAP1 or Clip-170 localization observed after nocodazole treatment. These results indicate that the function and assembly of the polarity complex is disrupted in response to CDDO-Im treatment and that this effect is not dependent on general microtubule disruption, since polarity complex constituents remained associated in nocodazole-treated cells.
FIGURE 11.
CDDO-Im disrupts PKCζ localization at the leading edge of polarized cells. Rat2 fibroblast monolayers were scratched and allowed to grow into the wound for 6 h in order to polarize before being incubated in the absence (Control; A) or presence of 1 μm CDDO-Im (B) or 10 μm nocodazole (C) for an additional 2 h. Cells were then fixed, permeabilized, and immunostained for endogenous Rac1 (green) or PKCζ (red), using anti-Rac1 and anti-PKCζ antibodies, respectively. A representative area of interest (white box) from each condition was enlarged and shown (inset). The green and red arrows indicate Rac1 and PKCζ at the leading edge of migrating cells, respectively. The co-localization of Rac1 with PKCζ is indicated by yellow arrowheads. The blue arrows indicate the direction of movement. Representative images from four experiments are shown. Bar, 10 μm.
Based on our observations that CDDO-Im does not disrupt the association between Par6 and TGFβ receptors (Fig. 5B) but does disrupt the localization of members of the polarity complex (Figs. 9, 10 and 11), we predicted that the localization of TGFβ receptors and Par6 at the leading edge might be reduced and/or abrogated in the presence of CDDO-Im. To test this, we assessed the localization of Par6 and TβRII by immunofluorescence microscopy (Fig. 12). In untreated cells, we observed that both Par6 and TGFβ receptors were indeed present at the leading edge of migrating cells and that they both co-localized with Rac1 (Fig. 12). However, in response to CDDO-Im treatment, the amounts of Par6 and TβRII at the leading edge of cells were markedly reduced (Fig. 12). These results further support our observations that CDDO-Im disrupts TGFβ-dependent cell migration by disrupting the polarity complex.
FIGURE 12.
CDDO-Im disrupts Par6 and TGFβ receptor localization at the leading edge of migrating cells. Rat2 fibroblast monolayers were scratched and allowed to grow into the wound for 6 h before being incubated in the absence (Control), or presence of 1 μm CDDO-Im for an additional 2 h. Cells were then fixed, permeabilized, and immunostained for endogenous Rac1 (green) and Par6 (red) using anti-Rac1 and anti-Par6 antibodies (A) or for endogenous Rac1 (green) and TβRII (red) using anti-Rac1 and anti-TβRII antibodies, respectively (B). A representative area of interest (white box) from each condition was enlarged and shown (inset). The green arrowheads indicate Rac1, and red arrowheads indicate Par6 or TβRII at the leading edge of migrating cells. The co-localization of Rac1 with Par6 or TβRII is indicated by yellow arrowheads. The blue arrows indicate the direction of movement. Bar, 10 μm. Cells containing Rac1, Par6, or TβRII at the leading edge were quantitated from three experiments carried out as described in A and B ± S.D. and graphed (C).
Taken together, our results demonstrate that the triterpenoid CDDO-Im alters TGFβ receptor trafficking and signal transduction, microtubule-plasma membrane attachments, and vesicular transport. The mechanism of affecting cell polarity and migration is dependent on the disruption of Clip-170 capping of microtubules and disrupting microtubule attachments with the polarity complex. Furthermore, the co-localization of triterpenoid with Rac-1 at the leading edge of migrating cells positions it to interfere with cell polarity by disrupting the localization of IQGAP1, PKCζ, Par6, and TGFβ receptors at the leading edge of migrating cells.
DISCUSSION
TGFβ is a growth factor that acts as a tumor suppressor or promoter, depending on cellular context (4, 50). TGFβ receptors propagate several signaling pathways, two of which are essential for EMT: the Smad pathway (51, 52) and the Par6 pathway (34, 53). In this study, we found that CDDO-Im inhibits TGFβ-dependent cell migration. In order to identify the mechanism, we first assessed its effect on TGFβ signaling. As previously described in studies using U937 and HL60 cells (37, 39), we found that CDDO-Im extended the phosphorylation profile of Smad2; however, further investigation suggested that this was not due a decrease in Smad2 phosphatase activity (Figs. 1 and 2 and supplemental Fig. 1). We next assessed receptor trafficking and degradation. Previous studies indicated that by perturbing the lipid raft compartment, the distribution of internalized cell surface TGFβ receptors could be shifted from the caveolin-1-positive compartment to the EEA1 early endosomal compartment, leading to enhanced signaling and delayed receptor degradation (15). We observed that CDDO-Im did delay receptor degradation; however, unlike lipid raft-destabilizing agents, such as nystatin, CDDO-Im did not shift receptor equilibrium toward the caveolin-1-positive compartment. Instead, CDDO-Im delayed overall receptor traffic and induced both caveolin-1- and EEA1-positive vesicles to be localized to the peri-plasma membrane regions of cells (Figs. 3 and 4). This led us to conclude that the compound was affecting a cellular structural component and quite possibly the cytoskeleton, since both the actin and microtubule cytoskeleton have been shown to be important for the trafficking of vesicles from the plasma membrane to the cell interior (21, 46, 54, 55).
Further study revealed that although the actin cytoskeleton was not morphologically affected, the microtubule network became disorganized (Fig. 6). The disorganization of vesicular positioning in the cell is a hallmark of microtubule catastrophe brought on by destabilizing drugs, such as nocodazole (20). Indeed, the microtubule network will disperse if cells are treated with high triterpenoid concentrations (56). However, in the present study, we used lower concentrations of CDDO-Im, consistent with the concentrations used in animal studies (57-59), and found that CDDO-Im does not dissolve the microtubule cytoskeleton. Rather, we observed that microtubules in CDDO-Im-treated cells take on a looping morphology that is reminiscent of a lack of proper attachment to the cell membrane and loss of cell polarity. We therefore tested if microtubule-capping proteins might be affected, since they act as anchorage points both between microtubules and vesicles and between microtubules and the cell membrane (26, 48). The latter process involves a number of intermediate molecules, such as Clip-170, and members of the polarity complex, Cdc42-Rac1, via IQGAP1 (32, 33, 48).
Interestingly, Clip-170 and IQGAP1 were found to dissociate from the leading edge of cells in response to CDDO-Im. These observations explain both the loss of vesicular traffic to the cell interior and the loss of microtubule targeting and association with the leading edge of cells (Figs. 7, 9, and 10). Of note, nocodazole, a drug that disrupts microtubules, was unable to affect the polarity complex after the cells were allowed to polarize in the absence of drugs. These observations indicate that the effects of CDDO-Im on Clip-170 dissociation from microtubules and the loss of concentration of molecules in the polarity complex are distinct from compounds that dissociate Clip-170 from microtubules via microtubule catastrophe. Our results also suggest that the association of different members of the polarity complex have variable stability. Indeed, we found that Rac1 remains associated with IQGAP1 regardless of the pharmacological treatment (Fig. 10). This stable association is also reflected in the fractionation studies, since the majority of Rac1 and IQGAP1 both partitioned with the soluble fractions, whereas Clip-170 was concentrated in the cytoskeletal fractions (Fig. 7). However, the dissociation of Clip-170 from microtubules and the loss of PKCζ from the polarity complex were exquisitely sensitive to CDDO-Im treatment. This has implications not only for TGFβ-dependent cell migration but for migration dependent on other growth factors and receptors as well. Indeed, CDDO-Im inhibits serum-stimulated Rat2 fibroblast migration (supplemental Movie 4). However, in the case of TGFβ-dependent cell migration, our results indicate that the location of Par6 and TGFβ receptors may be important.
It was interesting that TGFβ receptors remained associated with Par6 even as TGFβ-dependent migration was abrogated by CDDO-Im. When we assessed the association of Par6 with TGFβ receptors, we detected no difference in the association or phosphorylation of Par6 in response to CDDO-Im treatment. Therefore, this aspect of TGFβ-dependent migration was not perturbed. The localization of the triterpenoid to the leading edge of migrating cells was intriguing (Fig. 8), since it may suggest that the mechanism of CDDO-Im block could be the modulation of proteins at this locus. We reasoned that perhaps the loss of IQGAP1, PKCζ, and Rac1 at the leading edge of migrating cells would be accompanied with a loss of Par6 and TGFβ receptors. This was confirmed when we assessed Par6 and TGFβ receptor localization by immunofluorescence microscopy and found both partners to be greatly reduced at this site (Fig. 12). Further investigation of how CDDO-Im modulates TGFβ receptor signaling will be interesting, since the concentration of TGFβ receptors in various subcellular locations has been shown to be essential to stimulate Smad2 phosphorylation on endosomal membranes (8, 9, 15, 42-45), EMT at tight junctions (34), and degradation of RhoA in lamellipodia and filopodia (60-62).
Finally, our results may also give some insight into the mechanism of the antimetastatic and antiproliferative effects of CDDO-Im in animal studies (57-59) and would be an interesting area of study particularly for understanding mechanisms of cell migration and metastasis in human cancer. A general class of antimetastatic agents, microtubule-destabilizing drugs, affect cell motility, migration, and metastasis (63). Therefore, combining anti-microtubule drugs with drugs such as CDDO-Im that target the attachment sites between microtubules and the cell membrane may be an effective therapeutic approach to metastasis.
Acknowledgments
We thank the members of the Wrana laboratory for advice and support, Kavitha Sengodan for technical assistance, and Boun Thai for technical assistance and critical evaluation of the manuscript.
This work was supported by National Cancer Institute of Canada (NCIC) Terry Fox Foundation Grant 017189 (to G. M. D. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1-3 and Movies 1-4.
Footnotes
The abbreviations used are: TGFβ, transforming growth factor β; CDDO, 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid; CDDO-Im, CDDO-imidazolide; CDDO-biotin, biotinylated CDDO; EMT, epithelial to mesenchymal transition; TβRI and TβRII, TGFβ Ser/Thr kinase type I and II membrane receptor, respectively; E3, ubiquitin-protein isopeptide ligase; HA, hemagglutinin; GFP, green fluorescent protein; Mes, 4-morpholineethanesulfonic acid; DAPI, 4′,6-diamidino-2-phenylindole; DIC, differential interference contrast.
References
- 1.Pardali, K., and Moustakas, A. (2007) Biochim. Biophys. Acta 1775 21-62 [DOI] [PubMed] [Google Scholar]
- 2.Bose, R., and Wrana, J. L. (2006) Curr. Opin. Cell Biol. 18 206-212 [DOI] [PubMed] [Google Scholar]
- 3.Muraoka, R. S., Dumont, N., Ritter, C. A., Dugger, T. C., Brantley, D. M., Chen, J., Easterly, E., Roebuck, L. R., Ryan, S., Gotwals, P. J., Koteliansky, V., and Arteaga, C. L. (2002) J. Clin. Invest. 109 1551-1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiery, J. P. (2002) Nat. Rev. Cancer 2 442-454 [DOI] [PubMed] [Google Scholar]
- 5.Xu, Z., Shen, M. X., Ma, D. Z., Wang, L. Y., and Zha, X. L. (2003) Cell Res. 13 343-350 [DOI] [PubMed] [Google Scholar]
- 6.Attisano, L., and Wrana, J. L. (2002) Science 296 1646-1647 [DOI] [PubMed] [Google Scholar]
- 7.Tsukazaki, T., Chiang, T. A., Davison, A. F., Attisano, L., and Wrana, J. L. (1998) Cell 95 779-791 [DOI] [PubMed] [Google Scholar]
- 8.Miura, S., Takeshita, T., Asao, H., Kimura, Y., Murata, K., Sasaki, Y., Hanai, J. I., Beppu, H., Tsukazaki, T., Wrana, J. L., Miyazono, K., and Sugamura, K. (2000) Mol. Cell. Biol. 20 9346-9355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin, H. K., Bergmann, S., and Pandolfi, P. P. (2004) Nature 431 205-211 [DOI] [PubMed] [Google Scholar]
- 10.Lin, X., Duan, X., Liang, Y. Y., Su, Y., Wrighton, K. H., Long, J., Hu, M., Davis, C. M., Wang, J., Brunicardi, F. C., Shi, Y., Chen, Y. G., Meng, A., and Feng, X. H. (2006) Cell 125 915-928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebisawa, T., Fukuchi, M., Murakami, G., Chiba, T., Tanaka, K., Imamura, T., and Miyazono, K. (2001) J. Biol. Chem. 276 12477-12480 [DOI] [PubMed] [Google Scholar]
- 12.Kavsak, P., Rasmussen, R. K., Causing, C. G., Bonni, S., Zhu, H., Thomsen, G. H., and Wrana, J. L. (2000) Mol. Cell 6 1365-1375 [DOI] [PubMed] [Google Scholar]
- 13.Ogunjimi, A. A., Briant, D. J., Pece-Barbara, N., Le Roy, C., Di Guglielmo, G. M., Kavsak, P., Rasmussen, R. K., Seet, B. T., Sicheri, F., and Wrana, J. L. (2005) Mol. Cell 19 297-308 [DOI] [PubMed] [Google Scholar]
- 14.Chen, C. L., Huang, S. S., and Huang, J. S. (2006) J. Biol. Chem. 281 11506-11514 [DOI] [PubMed] [Google Scholar]
- 15.Di Guglielmo, G. M., Le Roy, C., Goodfellow, A. F., and Wrana, J. L. (2003) Nat. Cell Biol. 5 410-421 [DOI] [PubMed] [Google Scholar]
- 16.Ito, T., Williams, J. D., Fraser, D. J., and Phillips, A. O. (2004) J. Biol. Chem. 279 25326-25332 [DOI] [PubMed] [Google Scholar]
- 17.Schwartz, E. A., Reaven, E., Topper, J. N., and Tsao, P. S. (2005) Biochem. J. 390 199-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, X. L., Topley, N., Ito, T., and Phillips, A. (2005) J. Biol. Chem. 280 12239-12245 [DOI] [PubMed] [Google Scholar]
- 19.Le Roy, C., and Wrana, J. L. (2005) Nat. Rev. Mol. Cell. Biol. 6 112-126 [DOI] [PubMed] [Google Scholar]
- 20.D'Arrigo, A., Bucci, C., Toh, B. H., and Stenmark, H. (1997) Eur. J. Cell Biol. 72 95-103 [PubMed] [Google Scholar]
- 21.Pelkmans, L., Kartenbeck, J., and Helenius, A. (2001) Nat. Cell Biol. 3 473-483 [DOI] [PubMed] [Google Scholar]
- 22.Pol, A., Martin, S., Fernandez, M. A., Ferguson, C., Carozzi, A., Luetterforst, R., Enrich, C., and Parton, R. G. (2004) Mol. Biol. Cell 15 99-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burkhardt, J. K. (1998) Biochim. Biophys. Acta 1404 113-126 [DOI] [PubMed] [Google Scholar]
- 24.Galjart, N., and Perez, F. (2003) Curr. Opin. Cell Biol. 15 48-53 [DOI] [PubMed] [Google Scholar]
- 25.Musch, A. (2004) Traffic 5 1-9 [DOI] [PubMed] [Google Scholar]
- 26.Raftopoulou, M., and Hall, A. (2004) Dev. Biol. 265 23-32 [DOI] [PubMed] [Google Scholar]
- 27.Siegrist, S. E., and Doe, C. Q. (2007) Genes Dev. 21 483-496 [DOI] [PubMed] [Google Scholar]
- 28.Tirnauer, J. S., and Bierer, B. E. (2000) J. Cell Biol. 149 761-766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearson, C. G., and Bloom, K. (2004) Nat. Rev. Mol. Cell. Biol. 5 481-492 [DOI] [PubMed] [Google Scholar]
- 30.Goode, B. L., Drubin, D. G., and Barnes, G. (2000) Curr. Opin. Cell Biol. 12 63-71 [DOI] [PubMed] [Google Scholar]
- 31.Perez, F., Diamantopoulos, G. S., Stalder, R., and Kreis, T. E. (1999) Cell 96 517-527 [DOI] [PubMed] [Google Scholar]
- 32.Fukata, M., Watanabe, T., Noritake, J., Nakagawa, M., Yamaga, M., Kuroda, S., Matsuura, Y., Iwamatsu, A., Perez, F., and Kaibuchi, K. (2002) Cell 109 873-885 [DOI] [PubMed] [Google Scholar]
- 33.Watanabe, T., Wang, S., Noritake, J., Sato, K., Fukata, M., Takefuji, M., Nakagawa, M., Izumi, N., Akiyama, T., and Kaibuchi, K. (2004) Dev. Cell 7 871-883 [DOI] [PubMed] [Google Scholar]
- 34.Ozdamar, B., Bose, R., Barrios-Rodiles, M., Wang, H. R., Zhang, Y., and Wrana, J. L. (2005) Science 307 1603-1609 [DOI] [PubMed] [Google Scholar]
- 35.Yingling, J. M., Blanchard, K. L., and Sawyer, J. S. (2004) Nat. Rev. Drug Discov. 3 1011-1022 [DOI] [PubMed] [Google Scholar]
- 36.Liby, K. T., Yore, M. M., and Sporn, M. B. (2007) Nat. Rev. Cancer 7 357-369 [DOI] [PubMed] [Google Scholar]
- 37.Ji, Y., Lee, H. J., Goodman, C., Uskokovic, M., Liby, K., Sporn, M., and Suh, N. (2006) Mol. Cancer Ther. 5 1452-1458 [DOI] [PubMed] [Google Scholar]
- 38.Mix, K. S., Coon, C. I., Rosen, E. D., Suh, N., Sporn, M. B., and Brinckerhoff, C. E. (2004) Mol. Pharmacol. 65 309-318 [DOI] [PubMed] [Google Scholar]
- 39.Suh, N., Roberts, A. B., Birkey Reffey, S., Miyazono, K., Itoh, S., ten Dijke, P., Heiss, E. H., Place, A. E., Risingsong, R., Williams, C. R., Honda, T., Gribble, G. W., and Sporn, M. B. (2003) Cancer Res. 63 1371-1376 [PubMed] [Google Scholar]
- 40.Contin, M. A., Sironi, J. J., Barra, H. S., and Arce, C. A. (1999) Biochem. J. 339 463-471 [PMC free article] [PubMed] [Google Scholar]
- 41.Cong, F., and Varmus, H. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 2882-2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hayes, S., Chawla, A., and Corvera, S. (2002) J. Cell Biol. 158 1239-1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Itoh, F., Divecha, N., Brocks, L., Oomen, L., Janssen, H., Calafat, J., Itoh, S., and ten Dijke, P. (2002) Genes Cells 7 321-331 [DOI] [PubMed] [Google Scholar]
- 44.Panopoulou, E., Gillooly, D. J., Wrana, J. L., Zerial, M., Stenmark, H., Murphy, C., and Fotsis, T. (2002) J. Biol. Chem. 277 18046-18052 [DOI] [PubMed] [Google Scholar]
- 45.Runyan, C. E., Schnaper, H. W., and Poncelet, A. C. (2005) J. Biol. Chem. 280 8300-8308 [DOI] [PubMed] [Google Scholar]
- 46.Apodaca, G. (2001) Traffic 2 149-159 [DOI] [PubMed] [Google Scholar]
- 47.Murray, J. W., and Wolkoff, A. W. (2003) Adv. Drug Deliv. Rev. 55 1385-1403 [DOI] [PubMed] [Google Scholar]
- 48.Galjart, N. (2005) Nat. Rev. Mol. Cell. Biol. 6 487-498 [DOI] [PubMed] [Google Scholar]
- 49.Yore, M. M., Liby, K. T., Honda, T., Gribble, G. W., and Sporn, M. B. (2006) Mol. Cancer Ther. 5 3232-3239 [DOI] [PubMed] [Google Scholar]
- 50.Sporn, M. B. (2006) Cytokine Growth Factor Rev. 17 3-7 [DOI] [PubMed] [Google Scholar]
- 51.Kang, Y., He, W., Tulley, S., Gupta, G. P., Serganova, I., Chen, C. R., Manova-Todorova, K., Blasberg, R., Gerald, W. L., and Massague, J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 13909-13914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Levy, L., and Hill, C. S. (2005) Mol. Cell. Biol. 25 8108-8125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barrios-Rodiles, M., Brown, K. R., Ozdamar, B., Bose, R., Liu, Z., Donovan, R. S., Shinjo, F., Liu, Y., Dembowy, J., Taylor, I. W., Luga, V., Przulj, N., Robinson, M., Suzuki, H., Hayashizaki, Y., Jurisica, I., and Wrana, J. L. (2005) Science 307 1621-1625 [DOI] [PubMed] [Google Scholar]
- 54.Mundy, D. I., Machleidt, T., Ying, Y. S., Anderson, R. G., and Bloom, G. S. (2002) J. Cell Sci. 115 4327-4339 [DOI] [PubMed] [Google Scholar]
- 55.Parton, R. G., Joggerst, B., and Simons, K. (1994) J. Cell Biol. 127 1199-1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Couch, R. D., Ganem, N. J., Zhou, M., Popov, V. M., Honda, T., Veenstra, T. D., Sporn, M. B., and Anderson, A. C. (2006) Mol. Pharmacol. 69 1158-1165 [DOI] [PubMed] [Google Scholar]
- 57.Hyer, M. L., Croxton, R., Krajewska, M., Krajewski, S., Kress, C. L., Lu, M., Suh, N., Sporn, M. B., Cryns, V. L., Zapata, J. M., and Reed, J. C. (2005) Cancer Res. 65 4799-4808 [DOI] [PubMed] [Google Scholar]
- 58.Lapillonne, H., Konopleva, M., Tsao, T., Gold, D., McQueen, T., Sutherland, R. L., Madden, T., and Andreeff, M. (2003) Cancer Res. 63 5926-5939 [PubMed] [Google Scholar]
- 59.Place, A. E., Suh, N., Williams, C. R., Risingsong, R., Honda, T., Honda, Y., Gribble, G. W., Leesnitzer, L. M., Stimmel, J. B., Willson, T. M., Rosen, E., and Sporn, M. B. (2003) Clin. Cancer Res. 9 2798-2806 [PubMed] [Google Scholar]
- 60.Wang, H. R., Ogunjimi, A. A., Zhang, Y., Ozdamar, B., Bose, R., and Wrana, J. L. (2006) Methods Enzymol. 406 437-447 [DOI] [PubMed] [Google Scholar]
- 61.Wang, H. R., Zhang, Y., Ozdamar, B., Ogunjimi, A. A., Alexandrova, E., Thomsen, G. H., and Wrana, J. L. (2003) Science 302 1775-1779 [DOI] [PubMed] [Google Scholar]
- 62.Zhang, Y., Wang, H. R., and Wrana, J. L. (2004) Cell Cycle 3 391-392 [DOI] [PubMed] [Google Scholar]
- 63.Verrills, N. M., and Kavallaris, M. (2005) Curr. Pharm. Des. 11 1719-1733 [DOI] [PubMed] [Google Scholar]