Abstract
Manα6(Manα3)Manβ4GlcNAcβ4GlcNAc-R is the core structure of the major processed protein N-glycans produced by insect cells. Ultimately, this paucimannose type structure is produced by an unusual β-N-acetylglucosaminidase, which removes the terminal N-acetylglucosamine residue from the upstream intermediate, Manα6(GlcNAcβ2Manα3)Manβ4GlcNAcβ4GlcNAc-R. Because the N-glycan processing pathways leading to the production of this intermediate are probably identical in insects and higher eukaryotes, the presence or absence of this specific, processing β-N-acetylglucosaminidase is a key factor distinguishing the processing pathways in these two different types of organisms. Recent studies have shown that the fused lobes (fdl) gene encodes the specific, processing β-N-acetylglucosaminidase of Drosophila melanogaster. However, there are conflicting reports on the identity of the gene encoding this enzyme in the lepidopteran insect, Spodoptera frugiperda. One has suggested that a gene alternatively designated SfGlcNAcase-3 or SfHex encodes this function, whereas another has suggested that this gene encodes a broad-spectrum β-N-acetylglucosaminidase that functions in glycan and chitin degradation. In this study we resolved this conflict by molecularly cloning an S. frugiperda fdl ortholog (Sf-fdl) and demonstrating that it encodes a product with the substrate specificity expected of the processing β-N-acetylglucosaminidase. Moreover, we showed that the endogenous levels of specific, processing β-N-acetylglucosaminidase activity were significantly reduced in S. frugiperda cells engineered to express a double-stranded RNA derived from the Sf-fdl gene. These results indicate that Sf-fdl encodes the specific, processing β-N-acetylglucosaminidase of S. frugiperda and validate our previous suggestion that the broad-spectrum β-N-acetylglucosaminidase encoded by the SfGlcNAcase-3/SfHex gene is more likely to be involved in N-glycan and/or chitin degradation.
Insects and other lower eukaryotes such as nematodes and plants occupy an interesting evolutionary niche in glycobiology because they produce N-glycoproteins but typically process their N-linked glycans less extensively than mammals (1, 2). This difference between lower and higher eukaryotic protein N-glycosylation pathways is biotechnologically significant because insects and plants are used to produce recombinant mammalian glycoproteins for many different biomedical research applications (3–7). Our research focuses on insects, and we believe the evolutionary status and widespread use of insect-based systems for recombinant glycoprotein production demands a much deeper understanding of their protein N-glycosylation pathways.
Insect and mammalian protein N-glycosylation pathways each begin with the co-translational transfer of N-glycan precursors to nascent proteins (1, 8). These precursors are subsequently trimmed and elongated by enzymes localized in the endoplasmic reticulum and Golgi apparatus of insect and mammalian cells to produce a common intermediate with the structure Manα6(GlcNAcβ2Manα3)Manβ4GlcNAcβ4GlcNAc-R. In mammalian cells, this intermediate is elongated by various glycosyltransferases to produce complex N-glycans, which often have terminal sialic acid residues. In contrast, insect cells usually fail to elongate this same intermediate and convert it instead to paucimannose N-glycans with the core structure Manα6(Manα3)Manβ4GlcNAcβ4GlcNAc-R. An unusual β-N-acetylglucosaminidase is responsible for the production of these structures (9). This enzyme specifically removes the terminal N-acetylglucosamine residue from the α3 branch of Manα6(GlcNAcβ2Manα3)Manβ4GlcNAcβ4GlcNAc-R, simultaneously eliminating the intermediate required for N-glycan elongation and producing the core paucimannose glycan typically found on insect cell-derived N-glycoproteins. This same enzyme is also responsible for the production of core paucimannose N-glycans in nematodes (10, 11). Thus, the presence of a processing β-N-acetylglucosaminidase is a key difference between the protein N-glycosylation pathways of lower and higher eukaryotes.
In a seminal study on this topic, Altmann et al. (9) demonstrated that IPLB-Sf21AE, a cell line derived from the lepidopteran insect Spodoptera frugiperda (12), has a membrane-associated β-N-acetylglucosaminidase activity that can specifically cleave the terminal N-acetylglucosamine residue from the α3 branch of a biantennary N-glycan in vitro. Subsequently, it was shown that cell lines derived from Estigmene acrea, another lepidopteran insect, produce hybrid and complex N-glycans containing terminal N-acetylglucosamine or galactose residues because they lack this intracellular β-N-acetylglucosaminidase activity (13). Together, these studies strongly supported the idea that the N-glycosylation pathway of at least some insect cells includes a processing β-N-acetylglucosaminidase, as described above. However, unequivocal proof of this concept awaited the isolation of an insect gene encoding this enzyme together with evidence that the gene product has the exquisite substrate specificity of the N-glycan processing enzyme.
The first proof of this kind was provided by a more recent study from the Altmann group (14) in which they demonstrated that the Drosophila melanogaster fused lobes (Dm-fdl)2 gene encodes the specific, processing β-N-acetylglucosaminidase in this organism. Importantly, this study demonstrated that the Dm-fdl gene product has several features distinguishing it from degradative hexosaminidases and chitinases, which also have β-N-acetylglucosaminidase activities. These features include its specificity for the terminal N-acetylglucosamine residue linked to the α3 branch of N-glycan substrates and its inability to degrade chito-oligosaccharides. Furthermore, it was shown that flies with no functional fdl gene produced a higher proportion of N-glycans with terminal N-acetylglucosamine residues linked to the α3 branch as compared with wild type. These findings together with the finding that the D. melanogaster hexosaminidase genes (hexo1 and hexo-2) encode enzymes that can cleave chito-oligosaccharides, but not N-glycans, strongly suggested that Dm-FDL is the β-N-acetylglucosaminidase responsible for N-glycan processing in this fly. The properties of Dm-FDL also were consistent with the idea that it is the fly ortholog of the lepidopteran insect N-glycan processing enzyme first detected by Altmann et al. (9) in microsomal membranes from IPLB-Sf21AE cells.
Subsequently, two laboratory groups independently reported molecular cloning of genes encoding β-N-acetylglucosaminidases from Sf9 cells, which are a clonal derivative of the IPLB-Sf21AE cell line (15, 16). Our group described the isolation of three β-N-acetylglucosaminidase genes from Sf9 cells, which were designated SfGlcNAcase-1,-2,-3 (16). SfGlcNAcase-1 was clearly distinct from the other two, which were nearly identical to each other and likely allelic variants of the same gene. Further analysis of the SfGlcNAcase-1 and SfGlcNAcase-3 gene products showed that they had high sequence homology to known hexosaminidases and that each also had β-N-acetylglucosaminidase activity when assayed against relevant substrates. However, neither had the tight α3 branch specificity of the processing enzyme activity originally described by Altmann et al. (9). In fact, each could remove the terminal N-acetylglucosamine residues from either the α3 or the α6 branch of various N-glycan substrates, and each also could release N-acetylglucosamine monomers from a chito-oligosaccharide substrate. Accordingly, we concluded that none of these S. frugiperda genes encoded the N-glycan processing enzyme but more likely encoded broad-spectrum β-N-acetylglucosaminidases involved in N-glycan and chitin degradation. In a similar study, Tomiya et al. (15) also molecularly cloned two allelic variants of an Sf9 cell β-N-acetylglucosaminidase gene, which they termed Sfhex. Further analysis of the Sfhex gene product, which is identical to the gene product we designated SfGlcNAcase-3, confirmed that the SfGlcNAcase-3/SfHex gene product lacks the α3 branch specificity of the processing enzyme activity originally described by Altmann et al. (9). However, because this enzyme had a 2–5-fold higher preference for the terminal N-acetylglucosamine residue on the α3 branch of an N-glycan substrate, Tomiya et al. (15) concluded that the SfGlcNAcase-3/SfHex gene encodes the processing β-N-acetylglucosaminidase of Sf9 cells.
In the present report we provide data that resolve the discrepancy in the conclusions drawn from these two previous reports. Briefly, we molecularly cloned a new β-N-acetylglucosaminidase cDNA from Sf9 cells, which turned out to be the S. frugiperda ortholog of the Dm-fdl gene. This gene, which we have designated Sf-fdl, encodes a membrane-associated product that specifically cleaves the terminal N-acetylglucosamine residue from the α3 branch of N-glycan substrates, which has virtually no activity against chito-oligosaccharide substrates and which has precisely the same pH profile as the activity originally identified by Altmann et al. (9) in IPLB-Sf21AE cell microsomes. Furthermore, Sf9 cells engineered to express an Sf-fdl-specific double-stranded RNA had significantly lower levels of specific, processing β-N-acetylglucosaminidase activity. These results indicate that the specific, processing β-N-acetylglucosaminidase activity originally detected by Altmann et al. (9) is encoded by the Sf-fdl gene. The definitive identification of this new gene and the demonstration that it can be used to suppress a key, endogenous N-glycan processing activity sets the stage for an effort to create a transformed Sf9 cell line lacking this activity, which would be an improved host for recombinant glycoprotein production by baculovirus expression vectors.
EXPERIMENTAL PROCEDURES
Cells and Cell Culture—Sf9 cells, which are a subclone of the IPLB-Sf21-AE cell line derived from S. frugiperda ovaries (12), were routinely maintained as shake flask cultures in either TNM-FH medium containing 10% fetal bovine serum (HyClone, Logan, UT) or ESF 921 serum-free medium (Expression Systems, Woodland, CA) as described previously (16).
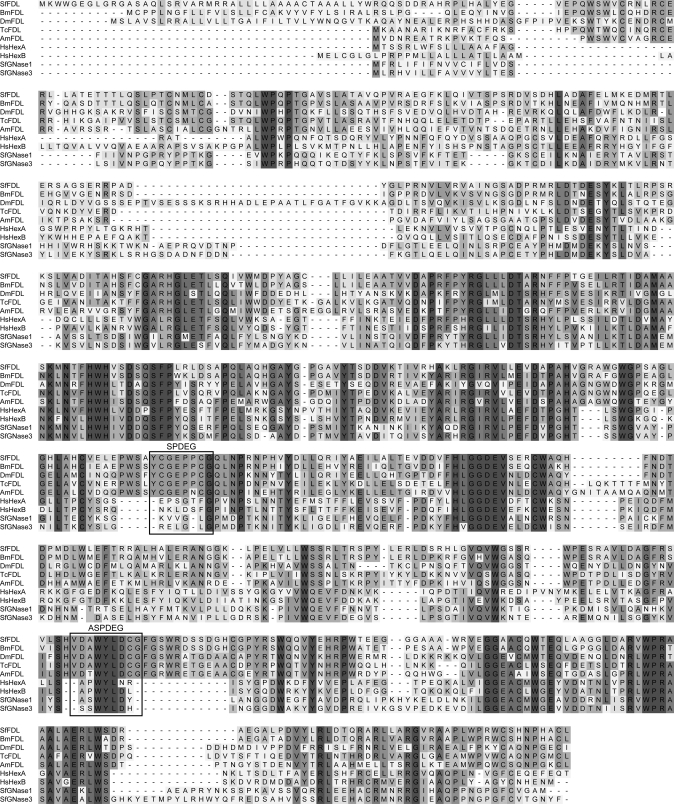
Molecular Cloning of an fdl Gene Homolog from Sf9 Cells—The Apis mellifera, Bombyx mori, and Tribolium castaneum genomic databases were searched using tBLASTn (17) with the derived amino acid sequence of Dm-FDL isoform C (accession number NM_165909) as the query. Exons encoding fragments of putative processing β-N-acetylglucosaminidases were joined in silico using an online splice site prediction algorithm available through the NetGene2 Server (18), and the predicted amino acid sequences were aligned using ClustalX version 1.83 (19) with the default settings. Highly conserved amino acid sequences were used to design degenerate oligonucleotide primers (SPDEG and ASPDEG; Fig. 1 and Table S1), which were then used for PCRs (20) with Sf9 genomic DNA or cDNA as the templates. The methods used to isolate genomic DNA, total RNA, and cDNA from Sf9 cells and to degenerate PCR conditions are given in the supplemental data. The degenerate PCRs yielded specific amplification products of about the expected size (420 bp), which were recovered, purified, and directly sequenced using the degenerate PCR primers specified above. The resulting nucleotide sequences were assembled using ContigExpress (Vector NTI Advance 10.3.0; Invitrogen). These data were used to design gene-specific primers for primary and nested 5′- and 3′-RACE reactions, which are described in the supplemental data. These reactions yielded the full-length, putative Sf-fdl gene sequence shown in supplemental Fig. S1. Subsequently, the Sf-fdl open reading frame was amplified from Sf9 cell genomic DNA and cDNA, and accuracy of the amplification products was checked by sequencing, as described in the supplemental data.
FIGURE 1.
Hexosaminidase multiple sequence alignment. This figure shows an alignment of the Sf-FDL amino acid sequence with known or putative hexosaminidase amino acid sequences. The shading is directly proportional to the degree of amino acid conservation. The regions used to design the SPDEG and ASPDEG primers (Table S1) for degenerate PCR are boxed. The sources of the known or putative hexosaminidase sequences and their GenBank™ accession numbers are: SfFDL, S. frugiperda FDL (EU334747); BmFDL, putative B. mori FDL (derived from BAAB01153187 and BAAB01083831); DmFDL, D. melanogaster FDL (NP_725179); TcFDL, putative T. castaneum FDL (XM_962583.1); AmFDL, putative A. mellifera FDL (derived from NW_001253565); HsHexA, Homo sapiens hexosaminidase A (NP_000511); HsHexB, H sapiens hexosaminidase B (NP_000512); SfGNase1, S. frugiperda β-N-acetylglucosaminidase-1 (ABB76924); SfGNase3, S. frugiperda β-N-acetylglucosaminidase 3/Sfhex (ABB76926). Amino acid sequences were aligned using ClustalX Version 1.83 with the default settings, and the image was generated using DSGene. The overall amino acid sequence identities between Sf-FDL and Dm-FDL or SfGlcNAcase-3/SfHex are 44 and 29%, respectively.
Isolation of Recombinant Baculoviruses Encoding Native or GST-tagged Soluble Domains of β-N-Acetylglucosaminidases—Baculovirus transfer plasmids encoding the relevant full-length, untagged N-acetylglucosaminidases or their GST-tagged soluble domains were produced by using PCR to amplify the appropriate nucleotide sequences, as described in the supplemental data, and error-free clones were identified by sequencing. The transfer plasmids encoding the native proteins were used to produce recombinant baculoviruses by the BaculoDirect™ method (Invitrogen) according to the manufacturer's protocol, whereas those encoding the GST-tagged soluble domains were used to produce viruses by a standard allelic transplacement method (3, 4) with Bsu36I-digested BacPAK6 viral DNA (21) as the target for homologous recombination. Each recombinant baculovirus vector was plaque-purified, amplified in Sf9 cells, and titered by plaque assay on Sf9 cells, as described previously (4). The recombinant viruses encoding the native β-N-acetylglucosaminidases were designated AcSfGlcNAcase-3 (16), AcDm-FDL, and AcSf-FDL, whereas those encoding the N-terminal GST-tagged ectodomains of these enzymes were designated AcGSTSfGlcNAcase-3, AcGSTDm-FDL, and AcGSTSf-FDL, respectively. The parental virus used to produce these viruses, which also served as a negative control for some of the experiments included in this study, was Autographa californica nucleopolyhedrovirus (AcMNPV).
Expression of Recombinant Proteins in Insect Cells—Sf9 cells were seeded into 100 ml of ESF 921 medium in 250-ml DeLong flasks (Corning Glass Works, Corning, NY) and allowed to grow to a density of about 1.5–2.0 × 106 cells/ml at 28 °C and 125 rpm in a model 4580 rotary platform shaker-incubator (Forma Scientific, Inc., Marietta, OH). The cells were then infected with the appropriate baculovirus at a multiplicity of infection of about 1 plaque-forming unit/cell and incubated for another 72 h under the same conditions.
Isolation of Purified Microsomal Fractions—The isolation of microsomal fractions from baculovirus-infected Sf9 cells has been described previously (16). Microsomes were solubilized in β-N-acetylglucosaminidase assay buffer (100 mm citrate phosphate buffer, pH 6.0) containing 0.5% (v/v) Triton-X-100, total protein concentrations were determined using a commercial bicinchoninic acid assay (Pierce), and samples containing equal amounts of total protein were assayed for β-N-acetylglucosaminidase activity as described below.
Glutathione Affinity Chromatography—The GST-tagged ectodomains of the various β-N-acetylglucosaminidases examined in this study were purified from the extracellular fraction of Sf9 cells infected with AcGSTSfGlcNAcase-3, AcGSTDm-FDL, or AcGSTSf-FDL as described in the supplemental data. The purity of the final affinity-purified protein preparations was assessed by SDS-PAGE with Coomassie Blue staining (supplemental Fig. S3A), and GST-tagged protein content was measured by SDS-PAGE and immunoblotting with a GST-specific antiserum (supplemental Fig. S3B).
β-N-Acetylglucosaminidase Activity Assays—Enzyme activity assays were performed using either solubilized microsomal fractions or affinity-purified recombinant proteins isolated from baculovirus-infected Sf9 cells. For the microsomal membrane assays, microsomes were prepared and extracted as described above, and samples containing equal amounts of total protein were assayed in a total volume of 100 μl containing 20 pmol of various pyridylamine (PA)-tagged glycan substrates, which were generously provided by Drs. Friedrich Altmann and Iain Wilson of the Glycobiology group in the Department of Chemistry, University of Natural Resources and Applied Life Sciences, Vienna. The enzymatic activity of the affinity-purified recombinant proteins was assayed under identical conditions, except the amounts of purified protein used for these assays were equalized by immunoblotting rather than by total protein assays. The substrates used in this study included GlcNAcβ2Manα6 (GlcNAcβ2Manα3)Manβ4GlcNAcβ4GlcNAc-PA (GnGn), GlcNAcβ2Manα6(Manα3)Manβ4GlcNAcβ4GlcNAc-PA (GnM), and Manα6(GlcNAcβ2Manα3)Manβ4GlcNAcβ4GlcNAc-PA (MGn). After being incubated for various times at 37 °C, each reaction was diluted to 150 μl with water, and the products were analyzed by reverse phase high performance liquid chromatography as described previously (22). GnGn, GnM, MGn, and also Manα6(Manα3)Manβ4GlcNAcβ4GlcNAc-PA (MM) were used as standards for the chromatographic analyses.
RNA Interference—In general, the RNA interference approach used in this study involved transforming Sf9 cells with an immediate early expression plasmid encoding an inverted repeat derived from a portion of the Sf-fdl coding sequence, with the inverted repeat separated by a D. melanogaster white gene intron, as originally described by Lee and Carthew (23). A detailed description of the construction of this expression plasmid is given in the supplemental data. Ultimately, a sequence-verified Sf-fdl inverted repeat and white gene intron cassette was inserted into pIE1HR3 (24) to produce a plasmid designated pIE1HR3SfFdlRNAi. This plasmid was then used along with pIE1Neo to co-transfect Sf9 cells using a modified calcium phosphate method, as described previously (4), and neomycin-resistant clones were isolated by limiting dilution, as described previously (25). The levels of specific, processing β-N-acetylglucosaminidase activity in the parental and transformed cells were compared by HPLC analysis of the products obtained by reacting microsomal membrane preparations with GnGn, as described above.
RESULTS AND DISCUSSION
Isolation and Characterization of an fdl Gene Homolog from Sf9 Cells—Our effort to isolate an fdl gene homolog from Sf9 cells was informed and facilitated by the availability of genome sequence data from several insect species and also by our previous efforts to isolate the gene encoding the processing β-N-acetylglucosaminidase activity from this cell line. tBLASTn analyses of the A. mellifera, B. mori, and T. castaneum genomes with the predicted amino acid sequence of Dm-FDL as the query yielded putative exons from each species encoding peptides related to Dm-fdl (data not shown). We subsequently used a splice site prediction algorithm to join the relevant exons and identify open reading frames encoding at least partial, putative β-N-acetylglucosaminidases from each insect species. Importantly, a ClustalW alignment revealed that these open reading frames encoded amino acid sequences that were conserved in the Dm-fdl gene product but not in the SfGlcNAcase-1 or SfGlcNAcase-3 gene products identified in our previous study (Fig. 1; Ref. 16). These conserved sequences (boxed in Fig. 1) were used to design degenerate oligonucleotides for high fidelity PCRs with Sf9 cDNA or genomic DNA as the templates. These PCRs yielded an amplification product of about the expected size (420 bp) that appeared to be specific because it was not observed in control reactions in which either one of the degenerate oligonucleotides was excluded (data not shown). This product was directly sequenced, and the derived translation product was found to be highly similar to the corresponding fragments of the known D. melanogaster and putative A. mellifera, B. mori, and T. castaneum FDL proteins. Accordingly, we used the nucleotide sequence of this amplification product to design gene-specific primers for 5′- and 3′-RACE reactions.
The 5′-RACE reactions yielded a specific 1.4-kilobase amplification product that overlapped with the sequence of the original degenerate PCR product, extended it by 1180 bp in the 5′ direction, and included a potential translational initiation site (data not shown). The 3′-RACE reactions yielded a specific 1.0-kilobase amplification product that also overlapped with the sequence of the original degenerate PCR product, extended it by 734 bp in the 3′ direction, and encoded a translational termination site. A contiguous nucleotide sequence of 2319 bp was assembled by joining the sequences of the degenerate amplimer, the 5′-RACE product, and the 3′-RACE product. The accuracy of this sequence was confirmed by PCR with gene-specific primers using both Sf9 cDNA and genomic DNA as the templates followed by direct sequencing of the products as described under “Experimental Procedures.”
In Silico Analysis of the Sf9 Cell fdl Gene Homolog—The full-length Sf-fdl nucleotide sequence and derived amino acid sequence of the Sf-FDL polypeptide are shown in supplemental Fig. S1. The nucleotide sequence includes a single long open reading frame of 1896 bp, which has a GC content of 69%. The theoretical product of this open reading frame is a polypeptide consisting of 631 amino acids, which has a calculated molecular mass of 70,530 Da and a calculated isoelectric point of 7.18. The theoretical protein also has an N-terminal transmembrane domain (underlined in supplemental Fig. S1) which extends from amino acids 25 to 42 with in/out topology according to the TMHMM and TopPred2 algorithms (26). Thus, the putative S. frugiperda FDL polypeptide appears to be a type II transmembrane protein with a short cytoplasmic tail. This is consistent with the idea that the Sf-fdl gene encodes an N-glycan processing enzyme because all N-glycan processing enzymes characterized to date have been predicted or shown to be transmembrane proteins with type II topology (27–29). The putative Sf9 cell enzyme also includes two potential N-glycosylation sites, which are boxed in the amino acid sequence shown in supplemental Fig. S1.
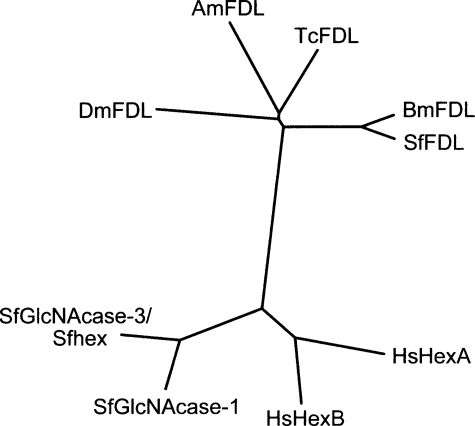
A phylogenetic analysis (Fig. 2) showed that the predicted Sf-fdl gene product is related to known hexosaminidases, including the human α (30)- and β (31)-hexosaminidases as well as to SfGlcNAcase-1 (16) and SfGlcNAcase-3/SfHex (15, 16) as expected (Fig. 2). Strikingly, however, this analysis also revealed that the predicted Sf-fdl gene product is much more closely related to the Dm-fdl gene product (14) than to either of the S. frugiperda hexosaminidase gene products despite the fact that Spodoptera and Drosophila belong to distinct insect orders, which diverged well over 300 million years ago (32). The predicted Sf-fdl gene product was most closely related to the putative B. mori fdl gene product and was also closely related to the putative T. castaneum and A. mellifera fdl gene products. Together, these results indicated that we had successfully isolated a Dm-fdl gene homolog from Sf9 cells. In addition, the much closer relationship between the Dm-fdl and Sf-fdl gene products coupled with the more distant relationship between the Sf-fdl and SfGlcNAcase-3/SfHex gene products supported the idea that the newly isolated Sf-fdl gene was more likely to encode the specific, processing β-N-acetylglucosaminidase in Sf9 cells. Additional experiments were undertaken to test this hypothesis.
FIGURE 2.
Hexosaminidase phylogenetic tree. This figure shows an unrooted phylogenetic tree depicting the phylogenetic relationships among the known or putative hexosaminidases included in Fig. 1. Protdist (PHYLIP Version 3.66) was used to generate a distance matrix from the multiple sequence alignment shown in Fig. 1 with the Jones-Taylor-Thornton model. An unrooted tree was then produced from the distance matrix using the neighbor-joining method (Neighbor; PHYLIP package) and drawn using the PHYLIP drawtree postscript generator.
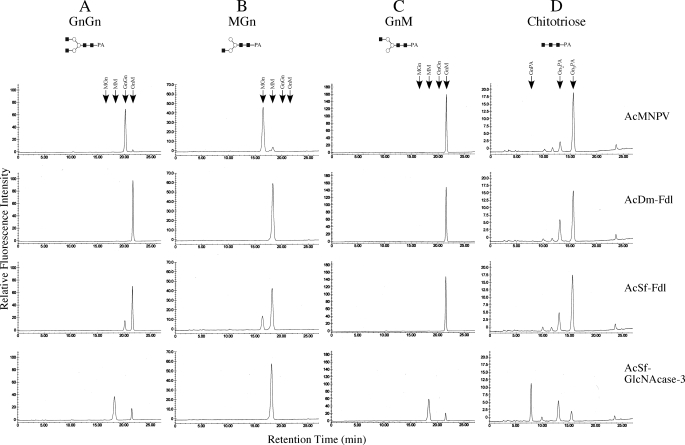
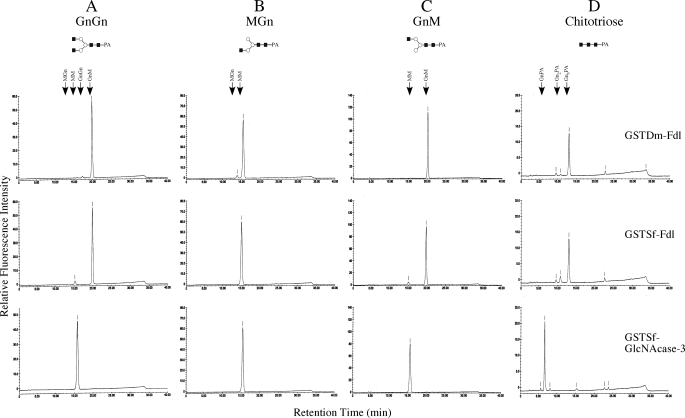
Expression and Biochemical Analysis of the Native Sf-fdl Gene Product—The full-length Sf-FDL coding sequence was subcloned into a baculovirus transfer plasmid, and the resulting construct was used to isolate a recombinant baculovirus, AcSf-FDL, that was used in turn for high level expression of the native cDNA product in insect cells. The parental baculovirus (AcMNPV) was used as a negative control, and recombinant baculoviruses encoding Dm-FDL (AcDm-FDL) or Sf-GlcNAcase-3/SfHex (AcSfGlcNAcase-3) were used to directly compare the enzymatic activities of the Sf-fdl, Dm-fdl, and Sf-GlcNAcase-3/SfHex gene products. Individual Sf9 cell cultures were infected with the appropriate baculoviruses, and then crude microsomal membrane fractions were prepared and assayed for enzymatic activity with various PA-tagged glycans as substrates, as described under “Experimental Procedures.” The results showed that negative control microsomes from AcMNPV-infected cells had very little effect on GnGn, whereas microsomes isolated from either AcDm-FDL- or AcSf-FDL-infected cells converted this substrate to GnM (Fig. 3A, top three panels). In contrast, parallel assays showed that microsomes from AcGlcNAcase-3-infected cells converted GnGn to both GnM and MM (Fig. 3A, bottom panel). These results indicated that Sf-FDL and Dm-FDL specifically removed only the terminal N-acetylglucosamine residue from the α3-branch of GnGn, whereas SfGlcNAcase-3/SfHex had a broader spectrum of activity, which allowed it to remove the terminal N-acetylglucosamine residues from both branches of GnGn.
FIGURE 3.
Substrate specificity of native Sf-FDL, Dm-FDL, and SfGlcNAcase-3/SfHex. Various glycan substrates, including GnGn (A), MGn (B), GnM (C), and chitotriose (D) were incubated for 16 h with microsomal fractions containing 10 μg of total protein from Sf9 cells infected with AcMNPV, AcDm-FDL, AcSf-FDL, or AcGlcNAcase-3. The reaction products were then recovered and analyzed by reverse phase HPLC, as described under “Experimental Procedures.” The arrows show the elution times for each of the relevant glycans.
This tentative conclusion was supported by the results of additional assays in which other glycans were used as substrates. Microsomes from cells infected with AcDm-FDL, AcSf-FDL, or AcSfGlcNAcase-3 all removed the terminal N-acetylglucosamine residue from the α3-branch of MGn to produce MM, as expected (Fig. 3B, lower three panels). Significantly, however, microsomes from AcDm-FDL and AcSf-FDL (Fig. 3C, middle two panels)-infected cells failed to remove the terminal N-acetylglucosamine from the α6-branch of GnM, whereas those from AcSfGlcNAcase-3-infected cells (Fig. 3C, bottom panel) clearly converted GnM to MM. These results confirmed that Sf-FDL and Dm-FDL are more highly specific enzymes that remove only the terminal N-acetylglucosamine residue from the α3-branch of glycan substrates in these assays. This substrate specificity distinguishes these enzymes from the SfGlcNAcase-3/SfHex gene product, as this latter enzyme readily removed the terminal N-acetylglucosamine residues from both branches of biantennary glycan substrates.
The relatively broader spectrum of activity observed with the SfGlcNAcase-3/SfHex gene product was underscored by the ability of microsomes from AcSfGlcNAcase-3-infected cells to efficiently convert chitotriose to chitobiose and chitobiose to a PA-tagged N-acetylglucosamine residue (Fig. 3D, bottom panel). In contrast, microsomes from AcDm-FDL- or AcSf-FDL-infected cells (Fig. 3D, middle two panels) converted only small amounts of chitotriose to chitobiose and produced no PA-tagged N-acetylglucosamine. In fact, microsomes from AcMNPV-infected cells (Fig. 3D, top panel) produced nearly as much chitobiose as the microsomes from AcDm-FDL- or AcSf-FDL-infected cells, suggesting that the apparent ability of these latter two enzymes to hydrolyze chitotriose was actually an artifact resulting from the presence of a contaminating chitinase activity in the crude microsomal preparations.
The results of the experiments described in this section show that the Sf-fdl gene product is orthologous to the Dm-fdl gene product, which is responsible for N-glycan processing in D. melanogaster (14), and paralogous to the SfGlcNAcase-3/SfHex gene product, which we previously concluded was more likely to be responsible for N-glycan and/or chitin degradation in S. frugiperda (16). Furthermore, the isolation of a Dm-fdl ortholog from Sf9 cells, its clear substrate specificity, and the relatively nonspecific nature of the SfGlcNAcase-3/SfHex gene product strongly suggest that the former, not the latter, is the N-glycan processing enzyme in Sf9 cells.
pH Optimum of the Sf-fdl Gene Product—In their seminal study on the endogenous processing β-N-acetylglucosaminidase activity in microsomal membranes isolated from Sf21 cells, Altmann et al. (9) found that it had a pH optimum for GnGn hydrolysis of 6.0. This was consistent with the idea that the activity measured in these assays was involved in N-glycan processing rather than degradation because a processing enzyme would be expected to reside in the Golgi apparatus and have a pH optimum around 6.0–6.5, whereas a degradative enzyme would be expected to reside in the lysosomal compartment and have a more acidic optimal pH. We found that the SfGlcNAcase-3/SfHex gene product had a pH optimum of 5.5 and took this as one line of evidence that this enzyme could not account for the processing activity identified by Altmann et al. (9) and was more likely to be involved in N-glycan or chitin degradation (16). Tomiya et al. (15) also found that the SfGlcNAcase-3/SfHex gene product had a pH optimum of 5.5, but concluded that it is involved in N-glycan processing in Sf9 cells because it has the same pH optimum as Dm-FDL (14), and it is at least partially active at the higher pH of secretory compartments, such as the trans-Golgi network (15). Hence, it was of interest to examine the optimal pH of the Sf-fdl gene product. Microsomal membranes were isolated from AcSf-FDL-infected Sf9 cells and assayed for GnGn hydrolysis at various pH values. The results showed that the pH optimum of the Sf-fdl gene product is 6.0 and that it has nearly optimal activity at pH 6.5 as well (supplemental Fig. S2). Thus, the pH optimum of the Sf-fdl gene product is identical to that of the processing activity originally identified in microsomal fractions from Sf21 cells by Altmann et al. (9). Furthermore, the range of optimal or near-optimal pH values for this enzyme more clearly encompasses the range of pH values found within late secretory pathway compartments, such as the trans-Golgi network, than the SfGlcNAcase-3/SfHex gene product. Thus, these results also support the idea that the Sf-fdl gene product, not the SfGlcNAcase-3/SfHex gene product, is the specific, processing β-N-acetylglucosaminidase in Sf9 cells.
Biochemical Analysis of the Purified Ectodomain of Sf-FDL—Each of the biochemical assays performed to this point in our study had involved the use of crude microsomes isolated from Sf9 cells infected with recombinant baculoviruses encoding the relevant β-N-acetylglucosaminidases. These assays were relevant because they mimicked the original assays of the endogenous processing β-N-acetylglucosaminidase activity in Sf21 cells and provided data on the substrate specificities of the full-length, native forms of each of the enzymes of interest. However, one criticism of these assays is that they did not involve the use of purified enzymes. To address this issue, we isolated recombinant baculoviruses encoding the N-terminal GST-tagged ectodomains of Sf-FDL, Dm-FDL, and SfGlcNAcase-3/SfHex, as described under “Experimental Procedures.” Each tagged enzyme was expressed in Sf9 cells and purified from the extracellular fraction using glutathione affinity chromatography, as described under “Experimental Procedures.” Analysis of the purified products by SDS-PAGE with Coomassie Blue staining (supplemental Fig. S3A) or immunoblotting with anti-GST (supplemental Fig. S3B) established that each had been effectively purified, and their concentrations were normalized. Subsequently, equivalent amounts of each purified protein preparation were assayed for β-N-acetylglucosaminidase activity using various glycan substrates.
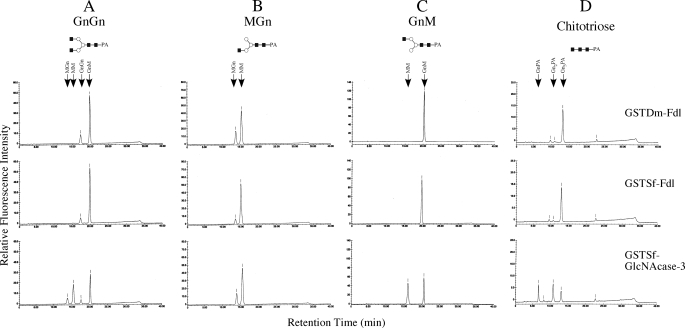
The results of these assays showed that the GST-tagged ectodomains of both Dm-FDL and Sf-FDL converted GnGn to GnM (Fig. 4A, top two panels), whereas the GST-tagged ectodomain of the GlcNAcase-3/Sfhex protein converted this substrate to GnM, MGn, and MM (Fig. 4A, bottom panel). All three enzymes removed the terminal N-acetylglucosamine residue from the α3-branch of MGn to produce MM (Fig. 4B) as expected, but only the SfGlcNAcase-3/SfHex protein removed the terminal N-acetylglucosamine from the α6-branch of GnM to produce MM (Fig. 4C, bottom panel). Neither Dm-FDL nor Sf-FDL had any detectable effect on this glycan (Fig. 4C, top two panels). Similarly, only the SfGlcNAcase-3/SfHex protein hydrolyzed chitotriose to produce chitobiose and PA-tagged N-acetylglucosamine monomers (Fig. 4D, bottom panel), whereas Dm-FDL and Sf-FDL had virtually no effect on this glycan (Fig. 4D, top two panels). An additional assay showed that Sf-FDL had very little effect on chitotriose when the substrate concentration was increased by 5-fold (data not shown).
FIGURE 4.
Substrate specificity of the GST-tagged ectodomains of Sf-FDL, Dm-FDL, and SfGlcNAcase-3/SfHex. Equal amounts of each enzyme were incubated for 2 h with GnGn (A), MGn (B), GnM (C), or chitotriose (D), and the reaction products were recovered and analyzed by reverse phase HPLC as described under “Experimental Procedures.” The arrows show the elution times for each of the relevant glycans.
These data supported the major conclusion drawn from the experiments performed with the full-length, untagged forms of these enzymes, which was that Dm-FDL and Sf-FDL are specific for the terminal N-acetylglucosamine on the α3-branch of biantennary N-glycan substrates, whereas SfGlcNAcase-3/SfHex has a much broader spectrum of β-N-acetylglucosaminidase activity. Again, the specificities of Sf-FDL and Dm-FDL are consistent with their proposed function in N-glycan processing and with the conclusion that the Sf-fdl gene encodes the membrane bound, processing β-N-acetylglucosaminidase activity originally identified in Sf21 cells by Altmann et al. (9).
To examine their substrate specificities more stringently, we incubated the purified, GST-tagged ectodomains of Dm-FDL, Sf-FDL, and SfGlcNAcase-3/SfHex with the various synthetic glycan substrates for 20 h to achieve a 10-fold increase in the enzyme assay times (Fig. 5). The results of these assays verified that Dm-FDL and Sf-FDL are highly specific even under this extreme condition. It can be seen that Sf-FDL produced tiny amounts of MGn from GnGn (Fig. 5A, middle panel) and tiny amounts of MM from GnM (Fig. 5C, middle panel). In addition, both Sf-FDL and Dm-FDL produced tiny amounts of chitobiose from chitotriose, and none of the aforementioned products was observed when the relevant glycan substrates were mock-digested with elution buffer alone (data not shown). Nevertheless, Sf-FDL is clearly much more specific than SfGlcNAcase-3/SfHex, and one might reasonably question the physiological relevance of the small amounts of conversion observed under these extreme in vitro reaction conditions. Finally, the results of two additional experiments demonstrated clearly that the purified, GST-tagged soluble domain of Sf-FDL used for these experiments retains full activity relative to fresh enzyme throughout the 20-h incubation period (data not shown).
FIGURE 5.
Over-digestion of glycan substrates with the GST-tagged ectodomains of Sf-FDL, Dm-FDL, and SfGlcNAcase-3/SfHex. Equal amounts of each enzyme were incubated for 20 h with GnGn (A), GnM (B), or chitotriose (C), and the reaction products were recovered and analyzed by reverse phase HPLC as described under “Experimental Procedures.” The arrows show the elution times for each of the relevant glycans.
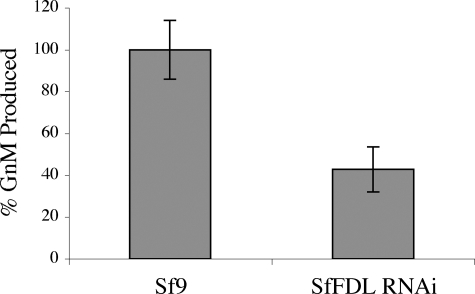
Inhibition of Specific, Processing β-N-Acetylglucosaminidase Activity by Sf-fdl-specific Double-stranded RNA—If the Sf-fdl gene encodes the specific, processing β-N-acetylglucosaminidase activity in Sf9 cells, it should be possible to reduce this activity by RNA interference with Sf-fdl-specific double-stranded RNA. To test this hypothesis, we constructed an immediate early expression plasmid encoding an inverted repeat sequence derived from a portion of the Sf-fdl coding sequence and used it to isolate a transformed Sf9 cell subclone designated SfFDL-RNAi, as described under “Experimental Procedures.” Microsomal membranes were then isolated from Sf9 or SfFDL-RNAi cells and used for β-N-acetylglucosaminidase activity assays with GnGn as the substrate. HPLC analysis of the reaction products showed that microsomal membranes from both cell types converted GnGn to GnM but produced no detectable MGn or MM (data not shown). Thus, the only type of endogenous β-N-acetylglucosaminidase activity that could be detected against GnGn in microsomal membranes from these cells was the specific, processing enzyme activity. This allowed us to demonstrate that the levels of specific, processing β-N-acetylglucosaminidase activity were reduced by >50% in microsomes from SfFDL-RNAi cells, as compared with the Sf9 cell control (Fig. 6). Similarly, we used chitotriose to demonstrate that microsomes from Sf9 cells have high levels of endogenous chitinolytic activity and that this activity is not significantly reduced in microsomes from Sf9FDL-RNAi cells (data not shown). The significant (p < 0.01) suppression of specific, processing β-N-acetylglucosaminidase activity in SfFDL-RNAi cells, which constitutively express Sf-fdl-specific double-stranded RNA, provided in vivo evidence that extended and supported our in vitro evidence, indicating that this activity is encoded by the Sf-fdl gene.
FIGURE 6.
Endogenous levels of specific, processing β-N-acetylglucosaminidase activity in parental Sf9 cells or a subclone encoding Sf-fdl-specific double-stranded RNA. Microsomal membrane preparations from Sf9 or SfFDL-RNAi cells were incubated for 16 h with GnGn, and the reaction products were analyzed by HPLC to compare the relative amounts of GnM produced. The plot shows the average results obtained in five replicate assays, with the average percentage of GnM produced by microsomes from the Sf9 controls set to 100%. The error bars show the S.D. and a one-way analysis of variance analysis showed that the two datasets are significantly different (p < 0.01).
Conclusion—The presence or absence of a specific, processing β-N-acetylglucosaminidase is a key difference in the protein N-glycosylation pathways of insects and higher eukaryotes. In insect systems, this function was first identified as an enzyme activity in crude microsomal membranes isolated from a cell line derived from the lepidopteran insect S. frugiperda (9). Efforts to molecularly clone the gene encoding this enzyme in these cells yielded two recent reports describing a single gene alternatively termed SfGlcNAcase-3 (16) and SfHex (15). Biochemical assays revealed that the SfGlcNAcase-3/SfHex gene product lacked the strict substrate specificity of the enzyme activity originally described by Altmann et al. in 1995 (9). Based upon this and other findings, we concluded that the SfGlcNAcase 3/SfHex gene product is more likely to be involved in glycan and chitin degradation than in N-glycan processing (16). In contrast, based upon a slight preference for the appropriate substrate, Tomiya et al. (15) concluded that this gene product is involved in N-glycan processing and hypothesized that it serves multifunctional roles in both N-glycan processing and glycan degradation in Sf9 cells. Independent efforts to molecularly clone the processing β-N-acetylglucosaminidase from D. melanogaster yielded a report describing the Dm-Fdl gene and the characteristics of the gene product (14). Based upon the substrate specificity of the gene product and the presence of a higher level of N-glycans containing terminal N-acetylglucosamine residues in mutant flies lacking a functional Fdl gene, Leonard et al. (14) concluded that Dm-Fdl encoded the β-N-acetylglucosaminidase involved in N-glycan processing in this fruit fly.
In the present study we isolated an Fdl gene from Sf9 cells and demonstrated that it encodes a membrane-associated β-N-acetylglucosaminidase with the same, strict substrate specificity of Dm-FDL and of the enzyme activity originally detected in S. frugiperda microsomal membranes (9). The fact that the Sf9 genome encodes a gene with a close phylogenetic relationship to Dm-fdl (and other, putative insect fdl genes), the fact that the Sf-fdl gene product is membrane-associated and has the strict substrate specificity and pH optimum profile of the original activity detected in S. frugiperda microsomes, and the fact that Sf9 cells engineered to express Sf-Fdl-specific DS RNA have lower levels of specific, processing β-N-acetylglucosaminidase activity all support the idea that the Sf-fdl gene encodes the β-N-acetylglucosaminidase involved in N-glycan processing in Sf9 cells. In addition, these findings support our previous conclusion that the broad-spectrum β-N-acetylglucosaminidase encoded by the SfGlcNAcase 3/SfHex gene is more likely involved in glycan and/or chitin degradation.
Acknowledgments
We thank Drs. Friedrich Altmann and Iain Wilson of the Glycobiology group in the Department of Chemistry, University of Natural Resources and Applied Life Sciences, Vienna for generously providing PA-labeled glycans and pIEBac-CG8824Myc for use in this project.
This work was supported by National Institutes of Health Grant RO1 GM49734. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Figs. S1–S3, and Table 1.
Footnotes
The abbreviations used are: Dm, D. melanogaster; AcMNPV, A. californica nucleopolyhedrovirus; FDL, fused lobes; GnGn, GlcNAcβ2Manα6 (GlcNAcβ2Manα3)Manβ4GlcNAcβ4GlcNAc-PA; GnM, GlcNAcβ2Manα6 (Manα3)Manβ4GlcNAcβ4GlcNAc-PA; MGn, Manα6(GlcNAcβ2Manα3) Manβ4GlcNAcβ4GlcNAc-PA; MM, Manα6(Manα3)Manβ4GlcNAcβ4GlcNAc-PA; PA, pyridylamine; Sf, S. frugiperda; RACE, rapid amplification of cDNA ends; GST, glutathione S-transferase; HPLC, high performance liquid chromatography; RNAi, RNA-mediated interference.
References
- 1.Marz, L., Altmann, F., Staudacher, E., and Kubelka, V. (1995) in Glycoproteins (Montreuil, J., Vliegenthart, J. F. G., and Schachter, H., eds) pp. 543–564, Elsevier Science Publishers B. V., Amsterdam
- 2.Marchal, I., Jarvis, D. L., Cacan, R., and Verbert, A. (2001) Biol. Chem. 382 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Reilly, D. R., Miller, L. K., and Luckow, V. A. (1992) Baculovirus Expression Vectors, W. H. Freeman and Co., New York
- 4.Summers, M. D., and Smith, G. E. (1987) Texas Agricultural Experimental Station Bulletin 1555, Texas Agricultural Experimental Station, College Station, TX
- 5.Jarvis, D. L. (1997) in Baculoviruses (Miller, L. K., ed) pp. 389–431, Plenum Press, New York
- 6.Fischer, R., Stoger, E., Schillberg, S., Christou, P., and Twyman, R. M. (2004) Curr. Opin. Plant Biol. 7 152–158 [DOI] [PubMed] [Google Scholar]
- 7.Ma, J. K., Drake, P. M., and Christou, P. (2003) Nat. Rev. Genet. 4 794–805 [DOI] [PubMed] [Google Scholar]
- 8.Kornfeld, R., and Kornfeld, S. (1985) Annu. Rev. Biochem. 54 631–664 [DOI] [PubMed] [Google Scholar]
- 9.Altmann, F., Schwihla, H., Staudacher, E., Glossl, J., and Marz, L. (1995) J. Biol. Chem. 270 17344–17349 [DOI] [PubMed] [Google Scholar]
- 10.Zhang, W., Cao, P., Chen, S., Spence, A. M., Zhu, S., Staudacher, E., and Schachter, H. (2003) Biochem. J. 372 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutternigg, M., Kretschmer-Lubich, D., Paschinger, K., Rendic, D., Hader, J., Geier, P., Ranftl, R., Jantsch, V., Lochnit, G., and Wilson, I. B. (2007) J. Biol. Chem. 282 27825–27840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaughn, J. L., Goodwin, R. H., Thompkins, G. J., and McCawley, P. (1977) In Vitro 13 213–217 [DOI] [PubMed] [Google Scholar]
- 13.Wagner, R., Geyer, H., Geyer, R., and Klenk, H. D. (1996) J. Virol. 70 4103–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leonard, R., Rendic, D., Rabouille, C., Wilson, I. B., Preat, T., and Altmann, F. (2006) J. Biol. Chem. 281 4867–4875 [DOI] [PubMed] [Google Scholar]
- 15.Tomiya, N., Narang, S., Park, J., Abdul-Rahman, B., Choi, O., Singh, S., Hiratake, J., Sakata, K., Betenbaugh, M. J., Palter, K. B., and Lee, Y. C. (2006) J. Biol. Chem. 281 19545–19560 [DOI] [PubMed] [Google Scholar]
- 16.Aumiller, J. J., Hollister, J., and Jarvis, D. L. (2006) Protein Expression Purif. 47 571–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997) Nucleic Acids Res. 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunak, S., Engelbrecht, J., and Knudsen, S. (1991) J. Mol. Biol. 220 49–65 [DOI] [PubMed] [Google Scholar]
- 19.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997) Nucleic Acids Res. 25 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Innis, M. A., and Gelfand, D. H. (1990) in PCR Protocols: A Guide to Methods and Applications (Innis, M. A., Gelfand, D. H., Sninsky, J. J., and White, T. J., eds) Academic Press, Inc., San Diego, CA
- 21.Kitts, P. A., and Possee, R. D. (1993) Biotechniques 14 810–817 [PubMed] [Google Scholar]
- 22.Altmann, F., Kornfeld, G., Dalik, T., Staudacher, E., and Glossl, J. (1993) Glycobiology 3 619–625 [DOI] [PubMed] [Google Scholar]
- 23.Lee, Y. S., and Carthew, R. W. (2003) Methods 30 322–329 [DOI] [PubMed] [Google Scholar]
- 24.Jarvis, D. L., Weinkauf, C., and Guarino, L. A. (1996) Protein Expression Purif. 8 191–203 [DOI] [PubMed] [Google Scholar]
- 25.Harrison, R. L., and Jarvis, D. L. (2007) Methods Mol. Biol. 388 299–316 [DOI] [PubMed] [Google Scholar]
- 26.von Heijne, G. (1992) J. Mol. Biol. 225 487–494 [DOI] [PubMed] [Google Scholar]
- 27.Paulson, J. C., and Colley, K. J. (1989) J. Biol. Chem. 264 17615–17618 [PubMed] [Google Scholar]
- 28.Breton, C., Mucha, J., and Jeanneau, C. (2001) Biochimie (Paris) 83 713–718 [DOI] [PubMed] [Google Scholar]
- 29.Field, M. C., and Wainwright, L. J. (1995) Glycobiology 5 463–472 [DOI] [PubMed] [Google Scholar]
- 30.Myerowitz, R., Piekarz, R., Neufeld, E. F., Shows, T. B., and Suzuki, K. (1985) Proc. Natl. Acad. Sci. U. S. A. 82 7830–7834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Dowd, B. F., Quan, F., Willard, H. F., Lamhonwah, A. M., Korneluk, R. G., Lowden, J. A., Gravel, R. A., and Mahuran, D. J. (1985) Proc. Natl. Acad. Sci. U. S. A. 82 1184–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaunt, M. W., and Miles, M. A. (2002) Mol. Biol. Evol. 19 748–761 [DOI] [PubMed] [Google Scholar]