Abstract
Diabetes mellitus, obesity, and dyslipidemia increase risk for cardiovascular disease, and expose the heart to high plasma fatty acid (FA) levels. Recent studies suggestthat distinct FA species are cardiotoxic (e.g., palmitate), while others are cardioprotective (e.g., oleate), although the molecular mechanisms mediating these observations are unclear. The purpose of the present study was to investigate the differential effects of distinct FA species (varying carbon length and degree of saturation) on adult rat cardiomyocyte (ARC) gene expression.ARCs were initialy challenged with 0.4 mM octanoate (8:0), palmitate (16:0), stearate (18:0), oleate (18:1), or linoleate (18:2) for 24 h. Microarray analysis revealed differential regulation of gene expression by the distinct FAs; the order regarding the number of genes whose expression was influenced by a specific FA was octanoate (1,188) > stearate (740) > palmitate (590) > oleate (83) > linoleate (65). In general, cardioprotective FAs (e.g., oleate) increased expression of genes promoting FA oxidation to a greater extent than cardiotoxic FAs (e.g., palmitate), whereas the latter induced markers of endoplasmic reticulum and oxidative stress. Subsequent RT-PCR analysis revealed distinct time- and concentration-dependent effects of these FA species, in a gene-specific manner. For example, stearate- and palmitate-mediated ucp3 induction tended to be transient (i.e., initial high induction, followed by subsequent repression), whereas oleate-mediated induction was sustained. These findings may provide insight into why diets high in unsaturated FAs (e.g., oleate) are cardioprotective, whereas diets rich in saturated FAs (e.g., palmitate) are not.
Keywords: β-oxidation, endoplasmic reticulum stress, gene expression
Fatty acids (FAs) are more than just a substrate for oxidative metabolism. Fatty acids (and their derivatives) have been shown to influence transcription, translation, enzymatic activities, cellular signaling, ion homeostasis, membrane fluidity, and cell survival (1–7). It is therefore not surprising that cells have evolved mechanisms facilitating a balance between FA availability and rates of FA utilization. For example, when exposed to elevated circulating FAs, organs such as the heart, skeletal muscle, and liver respond by inducing genes that promote FA utilization, a response involving a number of distinct molecular mechanisms (8). One such mechanism involves a family of nuclear receptors known as peroxisome proliferator-activated receptors (PPARs) (9). Three PPAR isoforms are known, of which PPARα and PPARβ/δ are highly expressed in tissues exhibiting relatively high rates of FA oxidative metabolism (e.g., liver, skeletal muscle, and heart), whereas PPARγ is highly expressed in lipogenic tissues (e.g., adipose tissue and liver) (10). Upon activation through direct binding of FAs, PPARs, in combination with their heterodimerization partners, retinoid X receptors (RXRs), induce genes promoting FA utilization (9, 11). For example, prolonged increases in circulating FA levels (e.g., fasting, high-fat feeding, diabetes mellitus) are associated with increased expression of β-oxidation enzymes [and increased rates of FA oxidation (FAO)] in peripheral tissues such as liver, skeletal muscle, and heart, which is attenuated in PPARα- and PPARβ/δ-null mice (12–15). It should be noted that PPARs are not the only mechanism by which FAs influence gene expression. Several additional mechanisms exist, which include alterations in LXR, CHREBP, HNF-4α, and TLR4 activity (8). However, to date, studies on these non-PPAR-mediated mechanisms have focused primarily on extra-cardiac tissues (e.g., liver).
Important previously published studies investigating the direct effects of FAs on myocardial gene expression have utilized neonatal cardiomyocytes, wherein the effects of a single FA species on a limited number of candidate genes are typically investigated (16, 17). Limitations to this approach include the observation that neonatal cardiomyocytes differentially express many of the known nuclear receptors (e.g., PPARα) and coactivators (e.g., PGC1) involved in responsiveness of the myocardium to FAs, compared with adult hearts (18). In addition, not all FA species function in a biologically identical manner. Variations in carbon chain length and degree of saturation dictate not only the metabolic fate of the FA but also its biological influence on the cell. For example, long-chain saturated FAs exhibit increased lipotoxic properties (e.g., palmitate-induced apoptosis), an effect that can be attenuated by the long-chain monounsaturated FA oleate (19, 20). To date, determination of the direct effects of distinct FA species on adult cardiomyocytes has not been approached in a systematic, unbiased manner. The purpose of the present study was therefore to investigate the global transcriptional response of adult rat cardiomyocytes (ARCs) to five distinct FA species, which varied in carbon chain length and degree of saturation. We hypothesized that novel insight into the mechanisms of cardioprotective versus cardiotoxic effects might be revealed. Here we report that long-chain unsaturated FAs (e.g., oleate and linoleate)induce genes known to promote β-oxidation, whereas long-chain saturated FAs (e.g., palmitate and stearate) induce markers of endoplasmic reticulum (ER) and oxidative stress.
MATERIALS AND METHODS
Animals
Male Wistar rats (Charles River, Fredrick, NY; 200 g initial weight) were housed at the Animal Care Center of the University of Texas Health Science Center at Houston under controlled conditions (23 ± 1°C; 12 h light/12 h dark cycle), and received standard laboratory chow and water ad libitum. All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of Texas Health Science Center at Houston.
Isolated ARCs
Isolated ARCs were prepared using protocols previously described (21, 22). Freshly isolated cardiomyocytes were cultured overnight in serum-free DMEM-containing laminin-ciated plates. The cells were next (i.e., at time 0) challenged with one of five distinct FAs [octanoate (8:0), palmitate (16:0), stearate (18:0), oleate (18:1), and linoleate (18:2)] at a final concentration of 0.1 mM, 0.2 mM, or 0.4 mM. All FAs were conjugated to defatted BSA, which was present in the culture medium at a final concentration of 1%. Control cells were cultured in the presence of 1% BSA alone. BSA and BSA-FA conjugates were dialyzed overnight, sterile filtered, and the FA concentration confirmed by spectrophotometric assay (Wako). After 0, 3, 6, 12, 24, or 48 h of challenge, cardiomyocytes were harvested in TriReagent, and stored at −80°C prior to RNA isolation.
Microarray analysis
Microarray gene expression studies were performed using Sentrix BeadChips and the BeadStation system from Illumina, Inc. (San Diego, CA). Microarrays were performed according to the manufacturer's guidelines. Following extraction through standard procedures (23), total RNA was converted to cDNA by reverse transcription using ArrayScript reverse transcriptase and T7-(dT)24 primers, followed by second-strand synthesis to generate double-stranded cDNA (Ambion, Inc.; Austin, TX). After purification, the cDNA was converted to biotin-labeled cRNA, hybridized to a rat Ref-12 BeadChip array (Illumina, Inc.) and stained with strepavidin-Cy3 for visualization. The rat Ref-12 BeadChips contain sequences representing approximately 22,000 curated genes and putative expressed sequence tags, with an average of 30 replicates for each gene/transcript on each BeadChip. Quality standards for hybridization, labeling, staining, background signal, and basal level of housekeeping gene expression were verified for all BeadChips.
After the probe arrays were scanned, the resulting images were first analyzed using the BeadStudio software (Illumina, Inc.), which calculates the mean fluorescence signal across all replicates of each gene/transcript, along with the probability that the mean signal for each gene/transcript on the chip is greater than background (i.e., detection score). Genes/transcripts were defined as being significantly expressed above background when the average detection score across all replicates was greater than 0.75.
Quantitative RT-PCR
Quantitative RT-PCR of samples was performed using previously described methods (24, 25). Specific quantitative assays were designed from rat sequences available in GenBank. Taqman assays utilized for cytosolic thioesterase 1 (cte1), cyclophilin, malonyl-CoA decarboxylase (mcd), mitochondrial thioesterase 1 (mte1), pyruvate dehydrogenase kinase 4 (pdk4), and uncoupling protein 3 (ucp3) have been published previously (13, 22, 26, 27), and those for adipose differentiation-related protein (adrp) and pleckstrin homology-like domain a1 (phlda1) are reported in supplementary Table I. Standard RNA was made for all assays by the T7 polymerase method (Ambion) using total RNA isolated from rat hearts. The correlation between the Ct (the number of PCR cycles required for the fluorescent signal to reach a detection threshold) and the amount of standard was linear over at least a 5-log range of RNA for all assays (data not shown). Quantitative RT-PCR data were normalized to cyclophilin expression, and expressed relative to the control group.
Statistical analysis
For the microarry study, differential expression of genes/transcripts, relative to control values, was calculated by one-way ANOVA, with correction for type I error using false-discovery rate (FDR) adjustment (SAS Corp.). Scheffe post hoc analysis was used to perform passive comparisons between treatment groups, for genes exhibiting significant main effects after FDR adjustment. Ontology analyses were performed in order to group genes from the final lists using the Onto-Tools package of ontology from Wayne State University (http://vortex.cs.wayne.edu/projects.htm). Normalized data have been submitted to the GEO archive, and are available at http://www.ncbi.nlm.nih.gov/geo/. For RT-PCR data, ANOVA was conducted to investigate the main effects of concentration separately for each FA and time, followed by post hoc pair-wise comparisons. The significance level for post hoc pair-wise comparisons was adjusted using a conservative Bonferroni approach.Stata version 8.0 (Stata Corp.; San Antonio, TX) was used to perform this analysis. The null hypothesis of no treatment effects was rejected at P < 0.05.
RESULTS
Genome-wide effects of distinct FAs on ARC gene expression
In an attempt to improve our understanding of the transcriptional response of ARCs to distinct FAs, gene expression profiling was performed through the use of microarrays. ARCs were therefore challenged with 0.4 mM octanoate, palmitate, stearate, oleate, or linoleate for 24 h, after which RNA was isolated and utilized for gene expression analysis; exposure of ARCs to 0.4 mM oleate for 24 h has previously been shown to elicit maximal effects on metabolic gene expression (13, 22). Control cells were treated in a manner identical to that used for the experimental groups, with the exception that no FA was added to the medium. Of the approximate 22,000 genes/transcripts interrogated through microarray analysis, approximately 11,500 were expressed in control ARCs. Supplementary Table II reports the number of genes differentially expressed in ARCs following challenge with distinct FA species. Somewhat surprisingly, of the five FA species investigated, challenging ARCs with the medium-chain FA octanoate resulted in the largest number (1,188) of differences in gene expression, relative to control cells. The saturated long-chain FAs palmitate and stearate influenced the expression of a similar number of genes compared with one another (590 and 740, respectively, relative to control), whereas the unsaturated long-chain FAs oleate and linoleate influenced expression of the smallest number of genes (83 and 65, respectively, relative to control). This sub-stratification based on the number of differentially expressed genes for medium-chain, saturated long-chain, and unsaturated long-chain FAs was also mirrored at the levels of induced versus repressed genes (see supplementary Table III). For all FAs investigated, approximately two-thirds of the differentially expressed genes were induced, and approximately one-third were repressed, compared with control cells. In addition, a high level of similarity was observed between the genes influenced by palmitate and stearate, and between oleate and linoleate (see supplementary Fig. I).
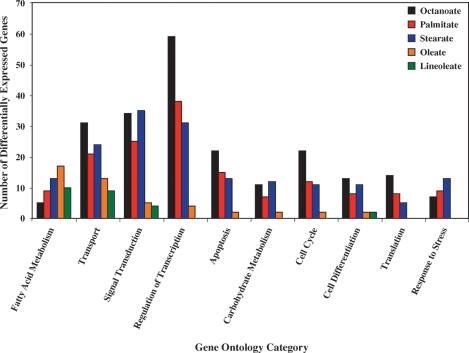
Gene ontology analysis was next performed for those genes identified as being differentially expressed in the five distinct FA treatment groups. Figure 1 shows an enrichment of genes in multiple gene ontology categories that would be expected to influence myocardial biology. These include genes regulating transcription, signaling, cell survival,and metabolism. In the case of palmitate and stearate, genes influencing apoptosis were highly represented; in particular, these saturated FAs induced a number of genes involved in ER and oxidative stress (Table 1). In contrast,FA/lipid metabolism was particularly enriched for both oleate- and linoleate-challenged ARCs. Table 2 shows that not only does oleate induce a larger number of these genes (relative to other FA species), it generally induces these genes to a greater extent. For example, ucp3 is induced 8.8-fold by oleate, but is not differentially expressed following challenge with palmitate. However, examples in which metabolic genes are differentially expressed following palmitate but not oleate challenge also exist, including adrp (induced 9.5-fold by palmitate, with no effect by oleate).
Fig. 1.
Gene ontology analysis of differentially expressed genes 24 h after challenging adult rat cardiomyocytes (ARCs) with octanoate, palmitate, stearate, oleate, or linoleate. Data are presented as number of differentially expressed genes.
TABLE 1.
Differential effects of distinct fatty acids on ARC expression of genes influencing apoptosis, cell survival, and stress response
| Process | Gene | Protein | Octanoate | Palmitate | Stearate | Oleate | Linoleate |
|---|---|---|---|---|---|---|---|
| ER stress-induced apoptosis | atf3 | Activating transcription factor 3 (ATF-3) | 7.3↑ | 6.5↑ | 6.1↑ | NS | NS |
| ddit3 | DNA damage-inducible transcript 3 | NS | 2.7↑ | 2.6↑ | NS | NS | |
| herpud1 | Homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain member 1 protein | 1.8↑ | 2.5↑ | 1.9↑ | NS | NS | |
| hspa5 | Heat-shock 70 kDa protein-5 | 4.1↑ | 3.0↑ | NS | NS | NS | |
| phlda1 | Pleckstrin homology-like domain, family A, member 1 | NS | 5.9↑ | 5.8↑ | NS | NS | |
| xbp1 | X-box binding protein 1 | 2.4↑ | 2.2↑ | NS | NS | NS | |
| Induction of apoptosis | bcl10 | B-cell CLL/lymphoma 10 | 2.8↑ | 2.1↑ | NS | NS | NS |
| cidea_predicted | Cell death-inducing DFFA-like effector a | NS | NS | NS | 2.1↑ | 2.2↑ | |
| nme3 | Non-metastatic cells 3, protein expressed in | 2.5↓ | 1.6↓ | NS | NS | NS | |
| pycard | PYD and CARD domain containing | 5.8↓ | 5.7↓ | 3.2↓ | 2.2↓ | NS | |
| siah2 | Seven in absentia homolog 2 | 2.6↑ | 2.8↑ | 2.3↑ | NS | NS | |
| stk17b | Serine/threonine kinase 17b | 4.4↑ | 5.1↑ | 5.3↑ | NS | NS | |
| tde1 | Tumor differentially expressed 1 | NS | 1.3↑ | 1.3↑ | NS | NS | |
| tieg | TGFB inducible early growth response | NS | 2.7↑ | 2.7↑ | 2.6↑ | NS | |
| trp53inp1 | Tumor protein p53-inducible nuclear protein 1 | 2.2↑ | NS | 1.7↑ | NS | NS | |
| Anti-apoptosis | angptl4 | Angiopoietin-like 4 | NS | 12.8↑ | NS | 18.9↑ | 15.6↑ |
| atf5 | Activating transcription factor 5 | NS | 2.8↑ | NS | NS | NS | |
| bdnf | Brain-derived neurotrophic factor | 2.7↑ | 3.2↑ | NS | NS | NS | |
| cfl1 | Cofilin 1 (non-muscle) | 1.8↑ | 1.8↑ | NS | NS | NS | |
| dnajb9 | DnaJ (Hsp40) homolog, subfamily B, member 9 | 7.8↑ | 7.7↑ | NS | NS | NS | |
| gclc | Glutamate-cysteine ligase, catalytic subunit | NS | 3.8↑ | NS | NS | NS | |
| gnrh1 | Gonadotrophin-releasing hormone 1 | 3.1↑ | 2.6↑ | NS | NS | NS | |
| msh2 | MutS homolog 2 | 1.8↓ | 1.8↓ | NS | NS | NS | |
| prkdc_predicted | Protein kinase, DNA-activated, catalytic polypeptide | 3.0↓ | 2.5↓ | 2.2↓ | NS | NS | |
| prnp | Prion protein (p27-30) | 3.1↑ | 2.2↑ | 2.1↑ | NS | NS | |
| rtkn | Rhotekin | 1.8↓ | 2.0↓ | 2.8↓ | NS | NS | |
| sgk | Serum/glucocorticoid regulated kinase | 6.0↑ | 5.1↑ | 6.9↑ | NS | NS | |
| tra1_predicted | Transformation/transcription domain-associated protein | 3.2↑ | 2.7↑ | NS | NS | NS | |
| tpt1 | Tumor protein, translationally-controlled 1 | NS | NS | 1.8↑ | NS | NS | |
| txndc5_predicted | Thioredoxin domain containing 5 | NS | 2.1↑ | NS | NS | NS | |
| Response to oxidative stress and DNA damage | gadd45a | Growth arrest and DNA-damage-inducible, α | 2.6↑ | 4.0↑ | 4.2↑ | NS | NS |
| gadd45b | Growth arrest and DNA-damage-inducible, β | NS | 2.1↑ | 3.0↑ | NS | NS | |
| gss | Glutathione synthetase | NS | 1.8↑ | NS | NS | NS | |
| hao1 | Hydroxyacid oxidase | NS | NS | 3.0↑ | NS | NS | |
| hmox2 | Heme oxygenase (decycling) 2 | NS | NS | 1.5↑ | NS | NS | |
| txnrd1 | Thioredoxin reductase 1 | NS | 2.8↑ | NS | NS | NS |
ARC, adult rat cardiomyocyte; ER, endoplasmic reticulum; NS, nonsignificant.
TABLE 2.
Differential effects of distinct fatty acids on ARC expression of genes influencing fatty acid metabolism
| Process | Gene | Protein | Octanoate | Palmitate | Stearate | Oleate | Linoleate |
|---|---|---|---|---|---|---|---|
| FA uptake | slc27a1 | Fatty acid transport protein 1 (FATP1) | NS | NS | NS | 3.7↑ | 3.4↑ |
| cd36 | Fatty acid translocase (FAT) | NS | NS | NS | 2.3↑ | NS | |
| LOC499984 | Similar to FAT | NS | 1.9↑ | NS | 2.1↑ | NS | |
| LOC499985 | Similar to FAT | NS | NS | NS | 2.5↑ | 2.3↑ | |
| acsl1 | Long-chain acyl-CoA synthetase 1 (ACSL1) | NS | 2.9↑ | NS | 2.6↑ | NS | |
| Mitochondrial β-oxidation | cpt1a | Carnitine palmitoyltransferase 1A (CPT1A) | NS | NS | NS | 4.3↑ | NS |
| cpt1b | Carnitine palmitoyltransferase 1B (CPT1B) | NS | 1.8↑ | NS | 2.0↑ | 2.1↑ | |
| cpt2 | Carnitine palmitoyltransferase 2 (CPT2) | 1.8↓ | NS | NS | 1.6↑ | NS | |
| mlycd | Malonyl-CoA decarboxylase (MCD) | NS | 1.9↑ | 2.0↑ | 2.2↑ | 2.1↑ | |
| slc25a20 | Carnitine/acylcarnitine translocase (CACT) | NS | 1.9↑ | 1.6↑ | 2.5↑ | 2.1↑ | |
| slc25a29 | Carnitine/acylcarnitine translocase-like (CACL) | NS | 2.0↓ | NS | 1.8↓ | 1.9↓ | |
| acad9 | Very long chain acyl-CoA dehydrogenase (VLCAD) | 1.9↓ | 1.5↓ | NS | NS | NS | |
| crat | Carnitine acetyltransferase (CAT) | NS | 2.1↑ | 1.8↑ | 2.3↑ | 2.2↑ | |
| etfdh | Electron-transferring flavoprotein dehydrogenase (ETFDH) | NS | 2.2↑ | 2.1↑ | 2.3↑ | 2.2↑ | |
| hadha | Hydroxyacyl-CoA dehydrogenase, α subunit (HADHα) | NS | NS | 1.4↑ | 1.5↑ | NS | |
| hadhb | Hydroxyacyl-CoA dehydrogenase, β subunit (HADHβ) | NS | 1.6↑ | 1.8↑ | 1.6↑ | 1.6↑ | |
| decr1 | 2,4-Dienoyl-CoA reductase 1 (DECR1) | NS | 2.2↑ | 2.0↑ | 2.6↑ | 2.3↑ | |
| Triglyceride metabolism | gpam | Glycerol-3-phosphate acyltransferase, mitochondrial (GPAM) | NS | NS | NS | 3.2↑ | NS |
| lipe | Hormone-sensitive lipase (HSL) | NS | 8.7↑ | 6.2↑ | 8.3↑ | 6.6↑ | |
| aqp7 | Aquaporin 7 (AQP7) | NS | 7.1↑ | 6.0↑ | 9.6↑ | 7.2↑ | |
| LOC361676 | Adipose triglyceride lipase (ATGL) | NS | 3.3↑ | 3.0↑ | 4.1↑ | 3.2↑ | |
| LOC501283 | Lipid storage droplet protein 5 (LSDP5) | NS | NS | NS | 12.0↑ | 7.3↑ | |
| adrp | Adipose differentiation-related protein (ADRP) | NS | 9.5↑ | 10.7↑ | NS | NS | |
| Phospholipid metabolism | chkb | Choline kinase β (CHKβ) | NS | 2.3↑ | 2.0↑ | 2.5↑ | 2.0↑ |
| impa2 | Inositol(myo)-1(or 4)-monophosphatase 2 (IMPA2) | NS | 1.8↑ | NS | 2.2↑ | 1.8↑ | |
| pik3c2g | Phosphoinositide-3-kinase, class 2, γ polypeptide (PIK3C2γ) | NS | 18.6↑ | 8.2↑ | 19.7↑ | 12.3↑ | |
| pik4cb | Phosphoinositol 4-kinase, catalytic, β polypeptide (PIK4Cβ) | NS | 2.3↑ | NS | 2.0↑ | 1.9↑ | |
| ppap2b | Phosphatidic acid phosphatase type 2B (PPAP2B) | NS | 2.2↑ | NS | 2.4↑ | NS | |
| Peroxisomal metabolism | acox1 | Acyl-CoA oxidase 1 (ACOX1) | NS | NS | NS | 1.9↑ | 1.8↑ |
| ech1 | Enoyl-CoA hydratase 1, peroxisomal (ECH1) | NS | 2.3↑ | 1.8↑ | 2.8↑ | 2.3↑ | |
| hsd17b4 | Hydroxysteroid (17β) dehydrogenase 4 (HSD17β4) | 1.7↓ | NS | NS | 1.4↑ | 1.4↑ | |
| LOC315973 | Acyl-CoA dehydrogenase family member 11 (ACAD11) | NS | NS | NS | 2.0↑ | NS | |
| nudt7_predicted | Peroxisomal CoA diphosphatase (NUDT7) | NS | 6.0↑ | 4.3↑ | 5.7↑ | 4.3↑ | |
| peci | Peroxisomal D3,D2-enoyl-CoA isomerase (PECI) | NS | 2.5↑ | 2.5↑ | 2.6↑ | 2.2↑ | |
| FA-mediated futile cycling | bach | Acyl-CoA thioesterase 7 (ACOT7) | NS | 3.5↑ | 3.4↑ | 3.8↑ | 3.2↑ |
| cte1 | Cytosolic thioesterase 1 (CTE1) | NS | 83.4↑ | 47.8↑ | 56.3↑ | NS | |
| mte1 | Mitochondrial thioesterase 1 (MTE1) | NS | 20.4↑ | NS | 14.7↑ | NS | |
| ucp2 | Uncoupling protein 2 (UCP2) | NS | 2.8↑ | 2.9↑ | 3.6↑ | 2.8↑ | |
| ucp3 | Uncoupling protein 3 (UCP3) | NS | NS | 4.9↑ | 8.8↑ | NS | |
| Other | grcc3f_predicted | Membrane-bound O-acyltransferase domain containing 5 (MBOAT5) | NS | 2.7↑ | 2.6↑ | 2.3↑ | 2.2↑ |
| pdk4 | Pyruvate dehydrogenase kinase 4 (PDK4) | NS | 23.1↑ | 34.4↑ | 28.1↑ | NS |
FA, fatty acid.
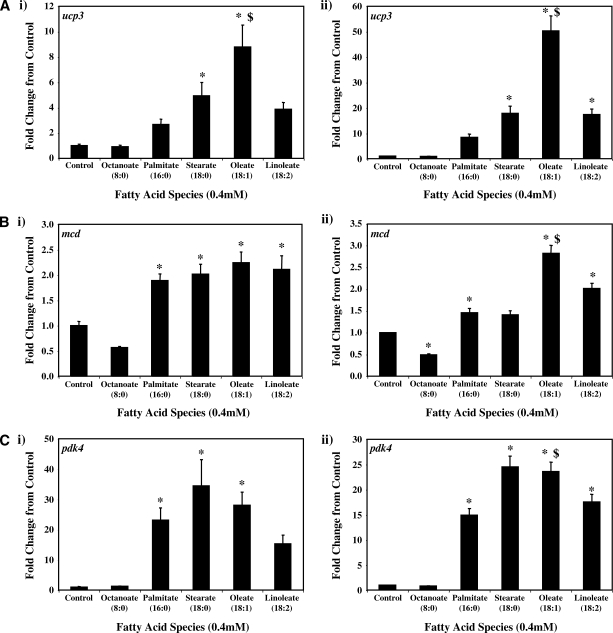
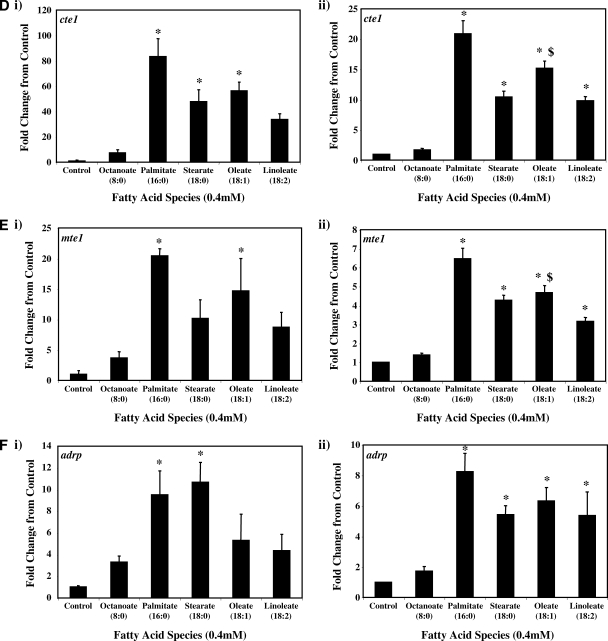
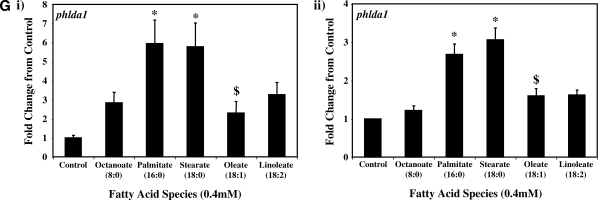
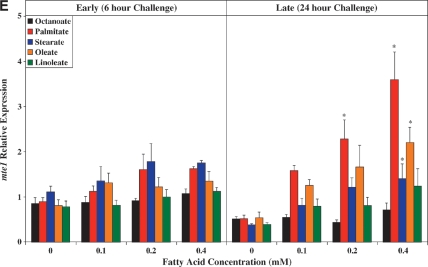
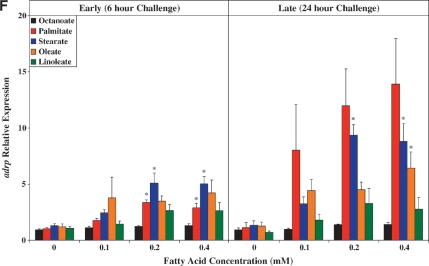
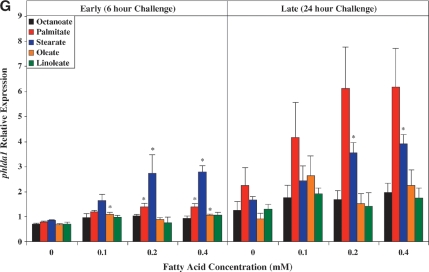
Real-time RT-PCR was next employed to validate a number of genes identified as being differentially expressed in ARCs following challenge with distinct FA species. Seven genes were chosen: ucp3 (promotes mitochondrial FA export),mcd (promotes mitochondrial FA import), pdk4 (represses carbohydrate oxidation), cte1 (promotes fatty acyl-CoA hydrolysis), mte1 (promotes fatty acyl-CoA hydrolysis), adrp (lipid droplet binding protein), and phlda1 (ER stress marker). Figure 2 compares the differential effects of distinct FAs on these genes, as assessed by microarray versus quantitative RT-PCR analysis. Similar to our microarray data, we find that two of the five metabolic genes (i.e., ucp3 and mcd) were induced to the greatest extent by oleate, compared with the other FAs (Fig. 2A, B, respectively). Stearate and oleate induced pdk4 to a similar extent (Fig. 2C), whereas cte1 and mte1 were induced to the greatest extent by palmitate (Fig. 2D, E, respectively). Finally, palmitate and stearate induced adrp and phlda1 to the greatest extent (Fig. 2F, G, respectively).
Fig. 2.
RT-PCR validation of microarray analysis. Effects of distinct fatty acid (FA) species on ARC expression of ucp3 (A), mcd (B), pdk4 (C), cte1 (D), mte1 (E), adrp (F), and phlda1 (G), as assessed by microarray analysis (i) and real-time RT-PCR (ii). All FA species were conjugated to 1% BSA, and were present at a concentration of 0.4 mM. Control cells were cultured in the absence of FA, but in the presence of 1% BSA. ARCs were challenged for 24 h. Individual gene expression data were initially normalized to the housekeeping gene cyclophilin and are presented as values relative to the control group. Data are shown as mean ± SEM for between 7 and 8 observations (microarray analysis) or 21 observations (RT-PCR) per group. * P < 0.05, control versus specific FA; $ P < 0.05, oleate versus palmitate.
Time- and concentration-dependent effects of distinct FAs on metabolic gene expression
Given the large number of metabolic genes identified by our microarray studies as being differentially regulated by distinct FA species, we decided to investigate the dynamics by which specific FA species induce genes. Exposure of cardiomyocytes to a FA at a single concentration, for a singleduration, may provide only limited information regarding the response of the cardiomyocyte to that FA. For example, it is possible that palmitate and stearate rapidly induce expression of FAO-promoting genes (e.g., ucp3) at lower concentrations, whereas higher concentrations (such as 0.4 mM) activate counterregulatory mechanisms.We therefore investigated the time- and concentration-dependent effects of distinct FAs on the selected genes previously utilized for microarray validation purposes. Here, ARCs were challenged with various concentrations of the investigated FAs (i.e., 0 mM, 0.1 mM, 0.2 mM, 0.4 mM) for multiple durations (i.e., 0, 3, 6, 12, 24, 48 h). For the purposes of clarity, Fig. 3 reports the acute (6 h) and chronic (24 h) concentration-dependent effects of the distinct FAs. Data for the entire time course are presented in supplementary Figs. II–VIII.
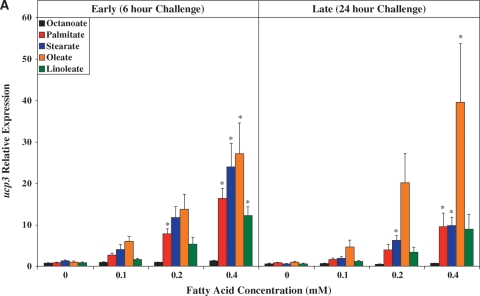
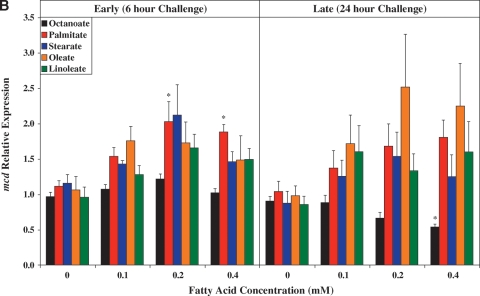
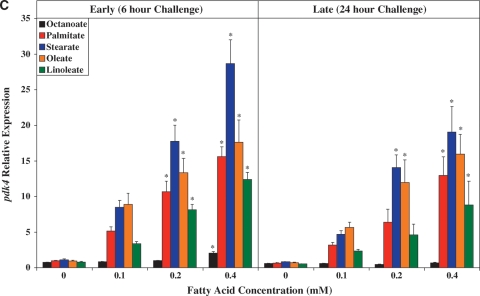
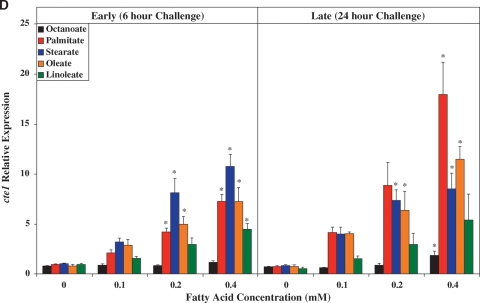
Fig. 3.
Time- and concentration-dependent effects of distinct FA species on ARC expression of ucp3 (A), mcd (B), pdk4 (C), cte1 (D), mte1 (E), adrp (F), and phlda1 (G). All FA species were conjugated to 1% BSA, and were present at a concentration of 0.1 mM, 0.2 mM, or 0.4 mM. Control cells were cultured in the absence of FA, but in the presence of 1% BSA. ARCs were challenged with FAs either acutely (6 h) or chronically (24 h). Individual gene expression data were initially normalized to the housekeeping gene cyclophilin and are presented as values relative to the control group (i.e., time zero). Data are shown as mean ± SEM for between 4 and 5 observations. * P < 0.05, control (i.e., zero concentration) versus respective FA at distinct concentration.
Figure 3A shows that with the exception of octanoate, the FAs investigated induced ucp3 expression in a concentration-dependent manner. Acutely (i.e., 6 h), palmitate, stearate, and oleate induced ucp3 to a similar extent. However, induction of ucp3 was transient in nature for palmitate and stearate, whereas the effects of oleate persisted chronically (i.e., 24 h). A similar pattern was observed for mcd (Fig.3B), although the level of induction of this gene was markedly lower, compared with that of ucp3.
As for ucp3, pdk4 was rapidly induced by all long-chain FAs investigated, in a concentration-dependent manner (Fig. 3C). However, stearate acutely induced pdk4 to the greatest extent, whereas chronically, stearate and oleate had similar effects. Figure 3D, E shows that expression of cte1 and mte1 is influenced by distinct FAs in a similar manner to one another. Acutely, these thioesterases are induced by stearate to the greatest extent, whereas chronically, induction is greatest with palmitate. Virtually identical profiles were observed with adrp (Fig. 3F) and phlda1 (Fig. 3G).
DISCUSSION
The primary goal of the present study was to investigate whether FAs of distinct carbon chain length and degree of saturation elicit differential influences on ARC gene expression.We report that: 1) microarray profiling reveals that long-chain unsaturated FAs (e.g., oleate) induce a larger number of genes promoting FA oxidation compared with long-chain saturated FAs (such as palmitate); 2) palmitate and stearate induced markers of ER stress, as well as oxidative and DNA damage; 3) palmitate and stearate tended to induce FA-responsive metabolic genes only transiently; and 4) the short-chain FA octanoate influenced expression of a greater number of genes, relative to long-chain FAs. These observations reveal differential effects of distinct FAs on the transcriptional response of ARCs, and may ultimately aid in elucidation of the mechanisms contributing to the cardioprotective versus the cardiotoxic effects of FAs on the heart.
The cardioprotective FA oleate induces genes promoting FA oxidation
Distinct FA species have divergent functions in the heart. For example, long-chain saturated FAs (e.g., palmitate and stearate) have been shown to induce apoptosis in cardiomyocytes, whereas oleate (a monounsaturated long-chain FA) rescues cardiomyocytes from palmitate-induced lipotoxicity (19, 20). Fatty acid composition is altered by diet, resulting in altered expression of proteins that regulate FA metabolism, which, in turn, may influence the ability of the heart to avoid lipotoxic damage. We recently observed that feeding rats a diet rich in unsaturated FAs reduced cardiac ceramide content and the frequency of cardiomyocyte apoptosis compared with a high-saturated-fat diet (28). In addition, high-fat diets composed of either medium- or long-chain saturated FAs, or long-chain unsaturated FAs, differentially induced the expression of known PPARα-regulated genes (28, 29). However, one must use caution in drawing conclusions about the effects of chain length and saturation from in vivo feeding studies, because such studies do not allow for assessment of the direct effects of individual FAs. At present, the direct effects that various saturated and unsaturated FAs have on myocardial gene expression are poorly understood.
Through a comprehensive, unbiased transcriptional profiling (i.e., microarray analysis) we report that oleate (as well as linoleate) influenced the expression of a relatively small number of genes (83 for oleate), compared with octanoate, palmitate, and stearate, of which 75% were induced (see supplementary Table III). Furthermore,of these genes, a large proportion are known to promote FAO (Fig. 1). When comparisons are made between the cardioprotective FA oleate and the cardiotoxic FA palmitate, for genes promoting FA metabolism, several interesting observations are made. First, a larger number of these metabolic genes are induced by oleate compared with palmitate (Table 2). Furthermore, when both of these FAs induce the same gene, the effect is generally larger for oleate. These generalizations are particularly noticeable for genes involved in long-chain FA uptake and mitochondrial β-oxidation. In the case of mcd and ucp3 (both believed to promote FA oxidation), the greater induction by oleate was confirmed by RT-PCR (Fig. 2A, B).
A number of specific differentially expressed genes are worthy of additional discussion. MTE1 and UCP3 have been proposed to work in tandem, in which MTE1 hydrolyzes FA-CoAs to free FAs, which are subsequently exported out of the mitochondrial matrix by UCP3 (30). In doing so, MTE1/UCP3 probably promotes FA oxidation by preventing the sequestration of free CoA required for continued β-oxidation. However, a relatively high induction of mte1 by palmitate (Fig. 2C), in the absence of induction of ucp3 (Fig. 2E), would promote accumulation of free fatty acyl groups within the mitochondrial matrix (as a long-chain acyl-CoA synthetase has not been identified within the mitochondrial matrix). Accumulation of palmitate within the mitochondrial matrix would probably promote apoptosis. Another notable difference in the transcriptional effects of oleate versus palmitate is in regard to lipid storage droplet proteins (LSDPs). LSDPs bind to lipid droplets, thereby influencing the latters' stability and metabolic fate (31). Five LSDPs have been identified to date, of which ADRP and LSDP5 are highly expressed in the heart (32–34). Microarray analysis revealed that oleate induces lsdp5 gene expression to a greater extent than palmitate, whereas palmitate induces adrp gene expression to a greater extent than oleate (Table 2). Interestingly, ER-derived ADRP-coated lipid droplets have been shown to specifically activate xanthine oxidase in mammary gland epithelial cells, resulting in reactive oxygen species (ROS) production (31, 35). Given the strong supportive evidence that palmitate-induced lipotoxicity involves ROS production (36), and that xanthine oxidase inhibition has been shown to be cardioprotective (37), an attractive hypothesis regarding palmitate-induced apoptosis is that ADRP-rich lipid droplets induce ROS production through xanthine oxidase activation following palmitate exposure. Given that ADRP-rich lipid droplets are synthesized in the ER, and that ADRP-rich lipid droplets are enriched with saturated (palmitate and stearate) versus unsaturated(oleate) long-chain FAs (35), the possibility arises that this mechanism may contribute to palmitate-induced ER stress. A greater induction of LSDP5, in response to oleate, may attenuate ADRP-rich lipid droplet formation, thereby protecting the cardiomyocyte.
The cardiotoxic FA palmitate induces markers of ER and oxidative stress
When exposed to elevated levels of specific long-chain saturated FAs (e.g., palmitate, stearate) for prolonged periods of time (e.g., 24–48 h), certain cell types (e.g., pancreatic β-cells, cardiomyocytes) initiate apoptosis (19, 20, 36, 38, 39). Initial studies investigating the potential molecular mechanisms for this lipotoxicity have focused largely on mitochondrially based processes, such as inhibition of the electron transport chain (by ceramide, for example), ROS production, diminution of the inner-mitochondrial membrane proton motive force, and cytochrome c release (36, 40, 41). More recently, potential extra-mitochondrial mechanisms have been explored. Borradaile et al. (38) have recently provided evidence that palmitate-induced apoptosis in HeLa/COS cells is potentially mediated through the ER stress response. Furthermore, pharmacological stimulation of FAO reduced palmitate-induced ER stress and apoptosis, presumably because of increased oxidation of this lipotoxic FA (38).
Our microarray analysis revealed that the long-chain saturated FAs palmitate and stearate induced a markedly larger number of genes in ARCs, compared with oleate and linoleate (Table 1). We reasoned that genes known to impact apoptosis, whose expression is differentially influenced by palmitate/stearate versus oleate/linoleate, may provide vital clues to the mechanisms by which long-chain saturated FAs induce lipotoxicity. Following this logic,two genes known to promote ER stress-induced apoptosis (ddit3 and phlda1) were induced exclusively by palmitate and stearate; these observations were confirmed by RT-PCR for phlda1. Additional markers of ER stress (e.g., atf3 and herpud1) were also induced by both palmitate and stearate (although these genes were also induced by the medium-chain FA, octanoate). These observations are consistent with those of Borradaile et al. (38), which suggest that palmitate induces ER stress in HeLa/COS cells.
Previously published studies suggest that palmitate-mediated ROS generation in cardiomyocytes probably plays a role in palmitate-induced apoptosis (36). Consistent with this possibility, our microarray analysis reveals that exposure of ARCs to 0.4 mM palmitate for 24 h leads to an induction of multiple markers of oxidative stress and DNA damage (Table 1). These include gadd45a/b, gss,and txnrd1. Similar observations are seen with ARCs exposed to stearate, but not following exposure to oleate or linoleate (Table 1).
Acute versus chronic effects of FAs on ARC gene expression
An obvious question relates to whether alterations in FAO gene expression initiate apoptosis, whether initiation of apoptosis causes alterations in FAO gene expression, or whether these events are mutually exclusive. A time course study was therefore performed, to compare early (acute) changes versus late (chronic) changes in an attempt to provide potential insight regarding this issue. When comparisons are made between oleate and palmitate, we find that oleate induces ucp3 to a greater extent than does palmitate,both acutely and chronically. Consistent with palmitate-induced apoptosis being a later event, palmitate induces the ER stress marker phlda1 to a greater extent than does oleate only chronically. These data support the hypothesis that oleate induces cardioprotective genes, such as ucp3, which help protect against apoptosis. Consistent with this concept, McLeod et al. (42) have shown that ucp3 is cardioprotective during ischemia/reperfusion. Clearly, future studies are required to definitely determine whether promotion of FAO is indeed the mechanism by which oleate protects the myocardium from palmitate-induced apoptosis.
Identification of novel potential mechanisms for FA-induced cardiotoxicity/cardioprotection
Microarray analysis and subsequent gene ontology revealed differential effects of distinct FAs on cardiomyocyte processes (e.g., FA metabolism, apoptosis), as discussed above. However, several additional candidate genes for cardiotoxicity/cardioprotection were identified by our microarray analysis, several of which warrant further discussion. Given the opposing effects of palmitate/stearate versus oleate on cardiomyocyte survival, we reasoned that genes differentially expressed between palmitate- and oleate-challenged cells, as well as between stearate- and oleate-challenged cells, may reveal novel potential mechanisms involved in FA-induced cardiotoxicity/cardioprotection. One example is rgs7bp (regulator of G-protein signaling 7 binding protein; LOC499521). Expression of this gene is 553-fold higher in stearate-challenged cells, and 372-fold higher in palmitate-challenged cells, versus oleate-challenged cells; this is due to induction by both palmitate and stearate, as well as a slight repression by oleate (relative to control cells; see supplementary Table II). RGS7BP has been shown to modulate G protein signaling in the brain, although this function has not been investigated previously in the heart (43). Interestingly, RGS7BP requires reversible palmitoylation to anchor the protein at the cell surface, where it modulates G protein signaling; removal of palmitate from culture media causes relocalization of RGS7BP to the nucleus (43). Thus, challenging cardiomyocytes with palmitate would cause not only a dramatic induction of RGS7BP, but also anchoring of the protein to the cell surface. That RGS7BP-mediated alterations in cardiomyocyte signaling contribute to cardiotoxicity is a novel and testable hypothesis. This analytical approach also identified genes that were more highly expressed in oleate-challenged cardiomyocytes, relative to palmitate- and stearate-challenged cells (such as ucp3; see above discussion).These genes might be considered as candidates for cardioprotection. One example is mitocalcin (LOC501181). Expression of this gene is 14.8- and 6.9-fold higher in oleate-challenged cells relative to palmitate- and stearate-challenged cells, respectively (see supplementary Table II). MITOCALCIN is a novel mitochondrial Ca2+ binding protein with EF-hand and coiled-coil domains (44). Although no known function has been elucidated for this protein, knockdown of the gene in 2Y-3t cells results in cell death (44). One could therefore hypothesize that repression by palmitate and stearate contributes to apoptosis, perhaps through impairing mitochondrial Ca2+ handling.
Medium-chain FAs induced marked alterations in ARC gene expression
One surprising observation in the current study is that challenging ARCs with the medium-chain FA octanoate induced the largest number of changes in myocardial gene expression, as compared with the long-chain FAs investigated (see supplementary Table II). A total of 1,188 genes were differentially expressed in octanoate-exposed versus control ARCs. These observations are consistent with previously published studies showing that medium-chain FAs induce alterations in gene expression within other cell types, such hepatocytes and adipocytes (45–47). Clearly, the current study design is unable to delineate the precise mechanism(s) by which this occurs. However, one possible hypothesis arises upon inspection of the gene ontology analysis (Fig. 1). Although octanoate influences expression of the largest number of total genes, this FA has little effect on FA metabolism genes (presumably because of an inability to activate PPARα and/or PPARβ/δ). Palmitate influences expression of a larger number of genes involved in FA metabolism compared with octanoate, whereas the total number of differentially expressed genes decreases. In contrast, oleate influences expression of the greatest number of FA metabolism genes, whereas the total number of differentially expressed genes is very low. These observations led us to speculate that insufficient activation of pathways involved in FA utilization, when availability increases(such as the case with octanoate), is “sensed” by the cardiomyocyte, resulting in substantial alterations in gene expression. Given the large number of genes influenced, it could be hypothesized that a central gene expression mechanism might be involved. One such candidate is acetylation; increased carbon availability, in the absence of full oxidation/utilization, would provide acetyl groups for acetylation reactions. Indeed, studies in yeast have shown that acetylation-mediated alterations in gene expression are driven primarily through acetyl-CoA availability (48). If the hypothesis is correct, simple consumption of diets rich in medium-chain FAs (i.e., feeding studies) would probably not result in significant alterations in myocardial gene expression, in large part because of the energy demands of the beating heart (as compared with quiescent cells in culture).
Study limitations, perspectives, and future directions
The primary limitations of the current study include its descriptive nature and focus on alterations in gene expression.Conclusions regarding either the precise mechanisms by which distinct FAs differentially influence cardiomyocyte gene expression, or whether these alterations in gene expression manifest concomitant alterations in protein expression and activity cannot be made. As such, our observations are limited to generation of novel hypotheses that remain to be interrogated. Previous studies have shown that unsaturated long-chain FAs are more potent ligands for the PPARs, as compared with saturated FAs, consistent with the idea that PPARα and PPARβ/δ mediate the induction of genes promoting FA oxidation (49, 50). However, the mechanisms responsible for the greater number of genes influenced by palmitate, stearate, and octanoate remain unidentified. One cannot rule out the possibility that the transient nature in which palmitate and stearate influence ARC metabolic gene expression is secondary to initiation of apoptosis and/or acceleration of dedifferentiation. However, the current study is the first to investigate the effects of distinct FA species on adult cardiomyocytes. The majority of previously published studies have focused on the effects of a specific FA species on candidate gene expression in neonatal cardiomyocytes (16, 17). Given the prominent alterations in myocardial nuclear receptors and metabolic substrate switching during the first 3 weeks of life for the rodent, the present study provides an improved understanding of the direct effects of distinct FA species on the adult heart.
Summary
The present study profiles the transcriptional response of ARCs to distinct FAs. We report that long-chain saturated FAs influence ARC gene expression in a manner that is quite distinct from that of long-chain unsaturated FAs. Specifically,the long-chain unsaturated FAs oleate and linoleate influenced the expression of a relatively small subset of genes, within which many genes promoting FA oxidation were induced. In contrast, the long-chain saturated FAs palmitate and stearate influence expression of a larger number of genes, yet fewer are known to promote FA oxidation. Furthermore, a greater subset of genes involved in ER and oxidative stress were induced by palmitate. These observations may ultimately aid in the elucidation of the mechanisms contributing to the cardioprotective versus the cardiotoxic effects of FAs on the heart.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Philip Barger for constructive comments prior to submission of this manuscript.
Published, JLR Papers in Press, April 2, 2008.
Footnotes
This work was supported by USDA/ARS Grants 6250-51000-044 and 6250-51000-046 to M.E.Y. and M.S.B. and by National Institutes of Health National Heart, Lung, and Blood Institute Grant HL-074259.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the forms of eight figures and three tables.
References
- 1.Faergeman N., and J. Knudsen. 1997. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem. J. 323 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shimabukuro M., Y. Zhou, M. Levi, and R. Unger. 1998. Fatty acid-induced beta cell apoptosis: a link between obesity and diabetes. Proc. Natl. Acad. Sci. USA. 95 2498–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clarke S., R. Baillie, D. Jump, and M. Nakamura. 1997. Fatty acid regulation of gene expression. Its role in fuel partitioning and insulin resistance. Ann. N. Y. Acad. Sci. 827 178–187. [DOI] [PubMed] [Google Scholar]
- 4.Clark H., D. Carling, and D. Saggerson. 2004. Covalent activation of heart AMP-activated protein kinase in response to physiological concentrations of long-chain fatty acids. Eur. J. Biochem. 271 2215–2224. [DOI] [PubMed] [Google Scholar]
- 5.Thompson A. L., and G. J. Cooney. 2000. Acyl-CoA inhibition of hexokinase in rat and human skeletal muscle is a potential mechanism of lipid-induced insulin resistance. Diabetes. 49 1761–1765. [DOI] [PubMed] [Google Scholar]
- 6.Singer S., and G. Nicolson. 1972. The fluid mosaic model of the structure of cell membranes. Science. 175 720–731. [DOI] [PubMed] [Google Scholar]
- 7.Liu G. X., P. J. Hanley, J. Ray, and J. Daut. 2001. Long-chain acyl-coenzyme A esters and fatty acids directly link metabolism to K(ATP) channels in the heart. Circ. Res. 88 918–924. [DOI] [PubMed] [Google Scholar]
- 8.Pegorier J., C. Le May, and J. Girard. 2004. Control of gene expression by fatty acids. J. Nutr. 134 2444S–2449S. [DOI] [PubMed] [Google Scholar]
- 9.Barger P. M., and D. P. Kelly. 2000. PPAR signaling in the control of cardiac energy metabolism. Trends Cardiovasc. Med. 10 238–245. [DOI] [PubMed] [Google Scholar]
- 10.Lee C. H., P. Olson, and R. M. Evans. 2003. Minireview: lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology. 144 2201–2207. [DOI] [PubMed] [Google Scholar]
- 11.Kliewer S. A., K. Umesono, D. J. Noonan, R. A. Heyman, and R. M. Evans. 1992. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 358 771–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patsouris D., J. K. Reddy, M. Muller, and S. Kersten. 2006. Peroxisome proliferator-activated receptor alpha mediates the effects of high-fat diet on hepatic gene expression. Endocrinology. 147 1508–1516. [DOI] [PubMed] [Google Scholar]
- 13.Stavinoha M. A., J. W. RaySpellicy, M. F. Essop, C. Graveleau, E. D. Abel, M. L. Hart-Sailors, H. J. Mersmann, M. S. Bray, and M. E. Young. 2004. Evidence for mitochondrial thioesterase 1 as a peroxisome proliferator-activated receptor-alpha-regulated gene in cardiac and skeletal muscle. Am. J. Physiol. 287 E888–E895. [DOI] [PubMed] [Google Scholar]
- 14.Leone T., C. Weinheimer, and D. Kelly. 1999. A critical role for theperoxisome proliferator-activated receptor alpha (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc. Natl. Acad. Sci. USA. 96 7473–7478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng L., G. Ding, Q. Qin, Y. Huang, W. Lewis, N. He, R. M. Evans, M. D. Schneider, F. A. Brako, Y. Xiao, et al. 2004. Cardiomyocyte-restricted peroxisome proliferator-activated receptor-delta deletion perturbs myocardial fatty acid oxidation and leads to cardiomyopathy. Nat. Med. 10 1245–1250. [DOI] [PubMed] [Google Scholar]
- 16.van der Lee K. A., M. M. Vork, J. E. De Vries, P. H. Willemsen, J. F. Glatz, R. S. Reneman, G. J. Van der Vusse, and M. Van Bilsen. 2000. Long-chain fatty acid-induced changes in gene expression in neonatal cardiac myocytes. J. Lipid Res. 41 41–47. [PubMed] [Google Scholar]
- 17.Wang G. L., M. L. Moore, and J. B. McMillin. 2002. A region in the first exon/intron of rat carnitine palmitoyltransferase Ibeta is involved in enhancement of basal transcription. Biochem. J. 362 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehman J. J., P. M. Barger, A. Kovacs, J. E. Saffitz, D. M. Medeiros, and D. P. Kelly. 2000. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Invest. 106 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Vries J. E., M. M. Vork, T. H. Roemen, Y. F. de Jong, J. P. Cleutjens, G. J. van der Vusse, and M. van Bilsen. 1997. Saturated but not mono-unsaturated fatty acids induce apoptotic cell death in neonatal rat ventricular myocytes. J. Lipid Res. 38 1384–1394. [PubMed] [Google Scholar]
- 20.Miller T. A., N. K. LeBrasseur, G. M. Cote, M. P. Trucillo, D. R. Pimentel, Y. Ido, N. B. Ruderman, and D. B. Sawyer. 2005. Oleate prevents palmitate-induced cytotoxic stress in cardiac myocytes. Biochem. Biophys. Res. Commun. 336 309–315. [DOI] [PubMed] [Google Scholar]
- 21.Durgan D. J., M. A. Hotze, T. M. Tomlin, O. Egbejimi, C. Graveleau, E. D. Abel, C. A. Shaw, M. S. Bray, P. E. Hardin, and M. E. Young. 2005. The intrinsic circadian clock within the cardiomyocyte. Am. J. Physiol. Heart Circ. Physiol. 289 H1530–H1541. [DOI] [PubMed] [Google Scholar]
- 22.Durgan D. J., J. K. Smith, M. A. Hotze, O. Egbejimi, K. D. Cuthbert, V. G. Zaha, J. R. Dyck, E. D. Abel, and M. E. Young. 2006. Distinct transcriptional regulation of long-chain acyl-CoA synthetase isoforms and cytosolic thioesterase 1 in the rodent heart by fatty acids and insulin. Am. J. Physiol. Heart Circ. Physiol. 290 H2480–H2497. [DOI] [PubMed] [Google Scholar]
- 23.Chomczynski P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162 159–169. [DOI] [PubMed] [Google Scholar]
- 24.Gibson U. E. M., C. A. Heid, and P. M. Williams. 1996. A novel method for real time quantitative RT-PCR. Genome Res. 6 995–1001. [DOI] [PubMed] [Google Scholar]
- 25.Heid C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6 986–994. [DOI] [PubMed] [Google Scholar]
- 26.Young M. E., G. W. Goodwin, J. Ying, P. Guthrie, C. R. Wilson, F. A. Laws, and H. Taegtmeyer. 2001. Regulation of cardiac and skeletal muscle malonyl-CoA decarboxylase by fatty acids. Am. J. Physiol. Endocrinol.Metab. 280 E471–E479. [DOI] [PubMed] [Google Scholar]
- 27.Young M. E., S. Patil, J. Ying, C. Depre, H. S. Ahuja, G. L. Shipley, S.M. Stepkowski, P. J. Davies, and H. Taegtmeyer. 2001. Uncoupling protein 3 transcription is regulated by peroxisome proliferator-activated receptor (alpha) in the adult rodent heart. FASEB J. 15 833–845. [DOI] [PubMed] [Google Scholar]
- 28.Okere I. C., M. P. Chandler, T. A. McElfresh, J. H. Rennison, V. Sharov, H. N. Sabbah, K. Y. Tserng, B. D. Hoit, P. Ernsberger, M.E. Young, et al. 2006. Differential effects of saturated and unsaturated fatty acid diets on cardiomyocyte apoptosis, adipose distribution, and serum leptin. Am. J. Physiol. Heart Circ. Physiol. 291 H38–H44. [DOI] [PubMed] [Google Scholar]
- 29.Okere I. C., M. P. Chandler, T. A. McElfresh, J. H. Rennison, T. A. Kung, B. D. Hoit, P. Ernsberger, M. E. Young, and W. C. Stanley. 2007. Carnitine palmitoyl transferase-I inhibition is not associated with cardiac hypertrophy in rats fed a high-fat diet. Clin. Exp. Pharmacol. Physiol. 34 113–119. [DOI] [PubMed] [Google Scholar]
- 30.Himms-Hagen J., and M. E. Harper. 2001. Physiological role of UCP3 may be export of fatty acids from mitochondria when fatty acid oxidation predominates: an hypothesis. Exp. Biol. Med. 226 78–84. [DOI] [PubMed] [Google Scholar]
- 31.Murphy D. J., and J. Vance. 1999. Mechanisms of lipid-body formation. Trends Biochem. Sci. 24 109–115. [DOI] [PubMed] [Google Scholar]
- 32.Brasaemle D. L., T. Barber, N. E. Wolins, G. Serrero, E. J. Blanchette-Mackie, and C. Londos. 1997. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J. Lipid Res. 38 2249–2263. [PubMed] [Google Scholar]
- 33.Dalen K. T., T. Dahl, E. Holter, B. Arnrtsen, C. Londos, C. Sztalryd, and H. I. Nebb. 2007. LSDP5 is a PAT protein specifically expressed in fatty acid oxidizing tissues. Biochim. Biophys. Acta. 1771 210–227. [DOI] [PubMed] [Google Scholar]
- 34.Yamaquchi T., S. Matsushita, K. Motojima, F. Hirose, and T. Osumi. 2006. MLDP, a novel PAT family protein localized to lipid droplets and enriched in the heart, is regulated by peroxisome proliferator-activated receptor alpha. J. Biol. Chem. 281 14232–14240. [DOI] [PubMed] [Google Scholar]
- 35.Heid H. W., M. Schnölzer, and T. W. Keenan. 1996. Adipocyte differentiation-related protein is secreted into milk as a constituent of milk lipid globule membrane. Biochem. J. 320 1025–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Listenberger L. L., D. S. Ory, and J. E. Schaffer. 2001. Palmitate-induced apoptosis can occur through a ceramide-independent pathway. J. Biol. Chem. 276 14890–14895. [DOI] [PubMed] [Google Scholar]
- 37.Pacher P., A. Nivorozhkin, and C. Szabo. 2006. Therapeutic effects of xanthine oxidase inhibitors: renaissance half a century after the discovery of allopurinol. Pharmacol. Rev. 58 87–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borradaile N. M., X. Han, J. D. Harp, S. E. Gale, D. S. Ory, and J. E. Schaffer. 2006. Disruption of endoplasmic reticulum structure and integrity in lipotoxic cell death. J. Lipid Res. 47 2726–2737. [DOI] [PubMed] [Google Scholar]
- 39.Hickson-Bick D. L., G. C. Sparagna, L. M. Buja, and J. B. McMillin. 2002. Palmitate-induced apoptosis in neonatal cardiomyocytes is not dependent on the generation of ROS. Am. J. Physiol. Heart Circ. Physiol. 282 H656–H664. [DOI] [PubMed] [Google Scholar]
- 40.Sparagna G. C., D. L. Hickson-Bick, L. M. Buja, and J. B. McMillin. 2000. A metabolic role for mitochondria in palmitate-induced cardiac myocyte apoptosis. Am. J. Physiol. Heart Circ. Physiol. 279 H2124–H2132. [DOI] [PubMed] [Google Scholar]
- 41.Hickson-Bick D. L., M. L. Buja, and J. B. McMillin. 2000. Palmitate-mediated alterations in the fatty acid metabolism of rat neonatal cardiac myocytes. J. Mol. Cell. Cardiol. 32 511–519. [DOI] [PubMed] [Google Scholar]
- 42.McLeod C. J., A. Aziz, R. F. Hoyt, J. P. J. McCoy, and M. N. Sack. 2005. Uncoupling proteins 2 and 3 function in concert to augment tolerance to cardiac ischemia. J. Biol. Chem. 280 33470–33476. [DOI] [PubMed] [Google Scholar]
- 43.Drenan R. M., C. A. Doupnik, M. Jayaraman, A. L. Buchwalter, K.M. Kaltenbronn, J. E. Huettner, M. E. Linder, and K. J. Blumer. 2006. R7BP augments the function of RGS7*Gbeta5 complexes by a plasma membrane-targeting mechanism. J. Biol. Chem. 281 28222–28231. [DOI] [PubMed] [Google Scholar]
- 44.Tominaga M., H. Kurihara, S. Honda, G. Amakawa, T. Sakai, and Y. Tomooka. 2006. Molecular characterization of mitocalcin, a novel mitochondrial Ca2+-binding protein with EF-hand and coiled-coil domains. J. Neurochem. 96 292–304. [DOI] [PubMed] [Google Scholar]
- 45.Massillon D., I. J. Arinze, C. Xu, and F. Bone. 2003. Regulation of glucose-6-phosphatase gene expression in cultured hepatocytes and H4IIE cells by short-chain fatty acids: role of hepatic nuclear factor-4alpha. J. Biol. Chem. 278 40694–40701. [DOI] [PubMed] [Google Scholar]
- 46.Guo W., W. Xie, and J. Han. 2006. Modulation of adipocyte lipogenesis by octanoate: involvement of reactive oxygen species. Nutr. Metab. (Lond). 3 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hillgartner F. B., and T. Charron. 1997. Arachidonate and medium-chain fatty acids inhibit transcription of the acetyl-CoA carboxylase gene in hepatocytes in culture. J. Lipid Res. 38 2548–2557. [PubMed] [Google Scholar]
- 48.Takahashi H., J. M. McCaffery, R. A. Irizarry, and J. D. Boeke. 2006. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol. Cell. 23 207–217. [DOI] [PubMed] [Google Scholar]
- 49.Forman B. M., J. Chen, and R. M. Evans. 1997. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc. Natl. Acad. Sci. USA. 94 4312–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu H. E., M. H. Lambert, V. G. Montana, D. J. Parks, S. G. Blanchard, P.J. Brown, D. D. Sternbach, J. M. Lehmann, G. B. Wisely, T. M. Willson, et al. 1999. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol. Cell. 3 397–403. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.