Abstract
To identify the genes controlling plasma concentrations of triglycerides (TGs), FFAs, and glucose, we carried out a quantitative trait loci (QTL) analysis of the closely related mouse strains New Zealand Black (NZB/B1NJ) and New Zealand White (NZW/LacJ), which share 63% of their genomes. The NZB × NZW F2 progeny were genotyped and phenotyped to detect QTL, and then comparative genomics, bioinformatics, and sequencing were used to narrow the QTL and reduce the number of candidate genes. Triglyceride concentrations were linked to loci on chromosomes (Chr) 4, 7, 8, 10, and 18. FFA concentrations were affected by a significant locus on Chr 4, a suggestive locus on Chr 16, and two interacting loci on Chr 2 and 15. Plasma glucose concentrations were affected by QTL on Chr 2, 4, 7, 8, 10, 15, 17, and 18. Comparative genomics narrowed the QTL by 31% to 86%; haplotype analysis was usually able to further narrow it by 80%. We suggest several candidate genes: Gba2 on Chr 4, Irs2 on Chr 8, and Ppargc1b on Chr 18 for TG; A2bp1 on Chr 16 for FFA; and G6pc2 on Chr 2 and Timp3 on Chr 10 for glucose.
Keywords: quantitative trait loci gene, comparative genomics, haplotype
During lipolysis, triglycerides (TGs) are broken down into FFAs and glycerol by the lipolytic enzymes (1). Several lines of evidence have shown the association of plasma FFA with the development of insulin resistance and type 2 diabetes (2). First, elevated FFA could alter glucose metabolism by affecting access to insulin-sensitive cells (adipose and muscle), as well as by reducing glucose transport into muscle (3). Second, chronically elevated FFA may also impair insulin secretory function through toxic effects on pancreatic β-cells, as predicted by the “lipotoxicity hypothesis” (4). Finally, increased flux of FFA into the liver, particularly from lipolysis of visceral adipose depots, may lead to excessive endogenous glucose production (5).
The levels of plasma lipids and glucose are partially genetically determined, and these determinants involve multiple genes (6, 7). Because of inherent difficulties in carrying out linkage analyses for complex traits in humans, inbred strains of mice have been used as a powerful tool for identifying quantitative trait loci (QTL) that contribute to variations in circulating levels of lipids and glucose (7–9). Studies in mice not only have revealed a large number of QTL that regulate lipid levels but also have shown that there is a high degree of concordance with human QTL that regulate the same traits (7). However, only a few QTL genes that regulate lipids or glucose have so far been identified.
Usually one chooses to intercross mouse strains that maximize the difference in both phenotype and genotype. However, the advent of thousands of single-nucleotide polymorphisms (SNPs) has enhanced the ability to genotypically distinguish even closely related strains. By intercrossing closely related strains that nevertheless differ in phenotype, one might enhance the probability of finding QTL and even causative genes, because the complexity of the system is reduced. For this reason, we crossed the closely related strains NZB/B1NJ (NZB) and NZW/LacJ (NZW), which differ in plasma TG, FFA, and glucose. The similar genome of these two strains facilitates the finding of QTL genes because such genes are likely to be located only in the regions polymorphic between the two strains. We report the detection of several QTL and suggest several candidate genes after narrowing the QTL by comparative genomics, haplotype analysis, and sequencing.
MATERIALS AND METHODS
Animals and diets
NZB × NZW F1 mice were obtained from The Jackson Laboratory (Bar Harbor, ME) and mated to produce 264 F2 progeny. Mice were maintained in a temperature- and humidity-controlled environment with a 14 h light/10 h dark cycle and given unrestricted access to food and acidified water. Weanling mice were fed standard chow containing 19% protein and 6% fat (LabDiets, 5K52, St. Louis, MO) until they were 8 weeks old, and then they were fed an atherogenic diet containing 50 kcal% glucose and 32 kcal% fat (30 kcal% butter fat and 2 kcal% corn oil), 1% (w/w) cholesterol, and 0.5% cholic acid as described previously (10) until they were euthanized at 16 weeks of age. This study was conducted in conformity with the Public Health Service Policy on Humane Care and Use of Laboratory Animals. Experiments were reviewed and approved by the Institutional Animal Care and Use Committee of The Jackson Laboratory.
Measuring plasma TG, FFA, and glucose
Blood samples were obtained from chow-fed mice at 8 weeks of age and from the atherogenic diet-fed mice at 16 weeks of age. Animals were fasted for 4 h in the morning. Blood from the retro-orbital sinus was collected in tubes containing EDTA and was centrifuged at 9,000 rpm for 5 min. Plasma was frozen at −20°C until analyzed. We measured plasma FFA, glucose, and TG concentrations within a week of collection using an enzymatic reagent kit (Beckman Coulter, Fullerton, CA) according to the manufacturer's recommendations on the Synchron CX Delta System (Beckman Coulter).
Genotyping
We digested mouse tail tips using proteinase K and isolated DNA using phenol-chloroform. The DNAs were resuspended in 10 mM Tris HCl (pH 8.0). To genotype F2 progeny, we used 147 SNPs spaced approximately 20 Mb apart that discriminated between NZB and NZW. SNPs were genotyped by the Allele-Typing Service at the Jackson Laboratory in conjunction with KBiosciences (Hoddesdon, UK). Reported genetic map positions were retrieved from the Mouse Genome Database (http://informaticsjaxorg).
Determining the similarity of strains NZB and NZW
To determine the percentage of genome that was shared by strains NZB and NZW, we used a total of 7,870,134 SNPs derived from Perlegen (http://mouse.perlegen.com/mouse) and Broad (http://www.broad.mit.edu/snp/mouse/). We inferred the haplotype and imputed the missing data using software written in C (11). We implemented a hidden Markov model to identify the regions where the mouse genome displays a haplotype block structure, to assign individual strains to local dominant haplotypes, and to infer the genotypes of missing SNP alleles. Within each inferred block, strains were grouped by the inferred local haplotype states. This was done for a large set of strains, and the number of distinct haplotypes at any one genomic location ranged from two to five. The percentage of SNPs located in identical haplotype blocks was used to calculate the similarity of the NZB and NZW genomes.
QTL analysis
TG and FFA data were log-transformed to obtain a normal distribution, but glucose exhibited a normal distribution and was not transformed. QTL analysis was performed by using R/qtl (version 1.07-12, available at http://www.rqtl.org) and Pseudomarker (version 2.03, available at http://www.jax.org/staff/churchill/labsite) software packages. A three-step QTL analysis was conducted to search for main effects and pair-wise gene interactions and then to integrate all of the main and interacting QTL phenotype associations into a multiple regression. In single-locus scans, the sex was first included as an additive covariate in the genome scan to account for overall differences in phenotypes between the sexes. A second set of scans included an interaction between sex and the putative QTL at each locus to identify sex-specific QTL. The difference in LOD scores (ΔLOD) between these two scans constitutes a test for sex-specific effects. We applied a significance threshold of LOD > 2.0 corresponding to P < 0.01, based on the 2 degree of freedom χ2 distribution of the log likelihood ratio, for QTL-by-sex interactions (12). QTL were deemed significant if they either met or exceeded the 95% genome-wide adjusted threshold, which was assessed by 1,000-permutation analysis for each trait; they were deemed suggestive if they either met or exceeded the 37% genome-wide adjusted threshold but were not significant. QTL confidence intervals were calculated according to posterior probability (13). Simultaneous pair-wise genome scans were performed to search for pairs of interacting loci (13). This method examines all pairs of marker loci for association with trait levels in a two-dimensional genome scan. In the regression analysis, we combined all significant and suggestive QTL and interactions in a multiple regression model. Terms that did not meet the nominal 0.01 level in the regression were eliminated in a backward stepwise manner with the exception that main-effect terms involved in a significant interaction were retained. Final models were reported for each trait.
Statistical analysis
One-way ANOVA was used for determining whether the mean phenotype values of progeny with different genotypes at a specific marker were significantly different. Linear regression analysis was performed to assess the association of various traits. Differences were considered statistically significant at P < 0.05. Data were analyzed using Graphpad Prism (Windows v5.00; GraphPad Software, San Diego, CA).
Comparative genomics and haplotype analysis
Homologous chromosomal regions between mouse and human were found at http://www.informatics.jax.org/reports/homologymap/mouse_human.shtml. When human and mouse QTL were located in the homologous locations, we assumed they were caused by the same gene and reduced the mouse QTL region to that homologous to the human QTL. If a confidence interval was not provided in the original report, we used a 1-logarithm of the odds ratio (1-LOD) score drop from the QTL peak; if no chromosome LOD score plot was provided, we used a confidence interval of 20 Mb on either side of the reported peak.
The reduced region from comparative genomics was further restricted by haplotype analysis (14). In the first step, we examined a dense SNP map and excluded those genomic regions within the QTL where NZB and NZW had an identical SNP pattern. Such regions are likely to be identical by descent (IBD) and do not contain ancestral polymorphisms. This step assumes that the mutation causing the QTL is ancestral rather than recent. Such an assumption is correct most of the time with two strains, and when a different pair of strains have found the same QTL, it is almost certain that the mutation is ancestral. The second step of haplotype analysis compares haplotypes of additional strains if the QTL has been found previously in a cross with different strains. This step assumes that QTL found in the same location in different crosses are nevertheless caused by the same QTL gene. The haplotype analysis was performed by using SNPs of all the strains that were parents of the QTL crosses in current and previous studies. We found those regions where the strains carrying the allele that increased the trait were identical but differed from the strains carrying the allele that decreased the trait. For example, for glucose QTL on chromosome (Chr) 2, we searched the SNPs in regions where the high-allele strains BKS and NZB had identical SNPs, low-allele strains C3H and NZW had identical SNPs, and these SNPs differed between the high and low alleles. SNPs were downloaded from the Mouse Phenome Database (www.phenome.org).
Candidate gene analysis
Gene lists for each QTL were extracted from Ensembl (www.ensembl.org). Those genes that mapped to the regions where the haplotypes were shared among the strains carrying the allele for increased phenotype but different from the strains carrying the allele for decreased phenotype were added to the list of positional candidate genes.
Sequencing
The genomic sequences of Gba2 (ENSMUST00000030189), Ppargc1b (ENSMUSG00000033871), and Timp3 (ENSMUSG00000020044) were obtained from the Ensembl (http://www.ensembl.org) mouse genome assembly, and primers were designed to amplify each exon plus at least 50 nucleotides of the adjacent introns. (For primer pairs used for standard PCR, see supplementary Table I.) Purified PCR products were subjected to thermocycle sequencing on capillary-based machines by the Jackson Laboratory DNA Sequence Laboratory. The sequence was analyzed using Sequencher software (version 4.1.4, GeneCodes Technology, Ann Arbor, MI).
Timp3 expression examined by real-time PCR
Total RNA was extracted from five NZB and five NZW male mice by using TRIzol® (Gibco BRL; Burlington, ON) following the manufacturer's instructions. The RNA was quantified with NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies; Wilmington, DE). The RNA quality was assessed with a 2100 Bioanalyzer instrument using RNA Nano Chips (Agilent Technologies; Santa Clara, CA). Two micrograms of total RNA was primed with random hexamers to synthesize cDNA in a total volume of 20 μl using the Omniscript RT Kit (QIAGEN; Valencia, CA). cDNA samples were mixed with SYBR Green Master Mix (Applied Biosystems; Foster City, CA) and gene-specific primers in a total volume of 25 μl. The primer pairs are: Timp3 forward 5′- TTGAAGAAAAGAGCGGCAGT and reverse 5′- GCTTCTTTCCCACCACTTTG; and β-actin forward 5′- CTTCTTGGGTATGGAATCC and reverse 5′- GCTCAGGAGGAGCGGTGAT. PCR was performed in 96-well optical reaction plates with an ABI PRISM 7500 Sequence Detection System (Applied Biosystems). Cycling parameters were 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 60°C. After PCR, a dissociation curve was constructed by increasing temperature from 65°C to 95°C for detection of PCR product specificity. A cycle threshold (Ct) value was recorded for each sample. PCR reactions were set up in triplicates and the mean of the three Cts was calculated. For each strain, the expression of the Timp3 gene was normalized to the expression of β-actin. A comparative Ct (ΔΔCt) was applied to the raw Ct values to find a relative gene expression between strains.
RESULTS
Similarity of the NZB and NZW genomes
Strains NZB and NZW belong to the New Zealand family of mice, and their genomes were expected to have regions that are IBD as defined by the haplotype block structure inferred from the SNPs. Using a total of 7,870,134 SNPs between strains NZB and NZW, we found that 4,976,141 SNPs were in the same haplotype. Thus, 63.2% of the genome was IBD between strains NZB and NZW. The degree of IBD by chromosome ranged from 51.4% for Chr 11 to 76.6% for Chr 19 (Table 1).
TABLE 1.
Genome similarity between strains NZB and NZW
| Chr | Total SNPa | Same Haplotype | IBD Proportion | Chr | Total SNPa | Same Haplotype | IBD Proportion |
|---|---|---|---|---|---|---|---|
| 1 | 694,809 | 431,308 | 62.1 | 11 | 259,028 | 133,086 | 51.4 |
| 2 | 524,667 | 318,659 | 60.7 | 12 | 396,114 | 237,299 | 59.9 |
| 3 | 509,892 | 305,120 | 59.8 | 13 | 399,930 | 262,492 | 65.6 |
| 4 | 476,425 | 290,790 | 61.0 | 14 | 345,783 | 189,866 | 54.9 |
| 5 | 496,888 | 340,661 | 68.6 | 15 | 337,461 | 228,232 | 67.6 |
| 6 | 509,545 | 366,856 | 72 | 16 | 305,078 | 222,442 | 72.9 |
| 7 | 405,733 | 219,105 | 54 | 17 | 266,421 | 141,367 | 53.1 |
| 8 | 444,910 | 276,603 | 62.2 | 18 | 291,266 | 202,005 | 69.4 |
| 9 | 361,571 | 207,465 | 57.4 | 19 | 222,031 | 170,114 | 76.6 |
| 10 | 399,126 | 281,196 | 70.5 | X | 223,456 | 151,475 | 67.7 |
Chr, chromosome; IBD, identical by descent; SNP, single-nucleotide polymorphism.
The SNPs are from Perlegen (www.perlegen.com/mouse/) and Broad (http://www.broad.mit.edu/snp/mouse/).
Plasma TG, FFA, and glucose profiles of NZB, NZW, and their F1 progeny
Plasma TG, FFA, and glucose concentrations for the parental and NZB × NZW F1 mice are summarized in Table 2. TG concentrations were 28.7% and 22.2% (P < 0.01) higher in female NZW animals as compared with female NZB mice fed chow and atherogenic diets, respectively, but similar in males of both strains on both diets. FFA concentrations were 19.1% and 25.7% (P < 0.05) higher in male NZW than in NZB animals on chow and atherogenic diet, respectively, but were similar in female NZW and NZB mice on both diets. Glucose levels were significantly higher (P < 0.01) in NZW as compared with sex- and diet-matched NZB mice. TG levels were significantly higher in F1 mice compared with both parental strains (P < 0.01). FFA levels in F1 female mice were comparable to the higher levels observed in NZW mice, but the difference from NZB was significant only in males. Glucose levels in F1 mice were somewhat lower than either parent; the difference was significant only when F1 mice were compared with NZW mice. In summary, NZW mice had significantly higher levels of TG (females), FFA (males), and glucose (both sexes) compared with NZB mice.
TABLE 2.
Plasma concentrations of TG, FFA, and glucose in NZB, NZW, and F1 (NZB × NZW) progeny at 8 weeks of age on chow diet and after consumption of the atherogenic diet for 8 weeks
| Chow Diet
|
Atherogenic Diet
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mice | TG | FFA | Glucose | TG | FFA | Glucose | ||||||
| mg/dl | mEq/l | mg/dl | mg/dl | mEq/l | mg/dl | |||||||
| Female | ||||||||||||
| NZB (n = 10) | 66 ± 10 | 1.49 ± 0.2 | 155 ± 11 | 72 ± 14 | 1.72 ± 0.3 | 161 ± 10 | ||||||
| NZW (n = 10) | 85 ± 9b | 1.73 ± 0.3 | 174 ± 12b | 88 ± 12b | 1.90 ± 0.2 | 186 ± 11b | ||||||
| F1 (n = 7) | 112 ± 14a | 1.81 ± 0.3 | 152 ± 13d | 121 ± 24a | 2.04 ± 0.2 | 154 ± 15d | ||||||
| Male | ||||||||||||
| NZB (n = 10) | 87 ± 14 | 1.52 ± 0.2 | 168 ± 12 | 98 ± 15 | 1.67 ± 0.1 | 171 ± 16 | ||||||
| NZW (n = 10) | 89 ± 13 | 1.81 ± 0.2c | 189 ± 13b | 91 ± 9 | 2.10 ± 0.3c | 193 ± 16b | ||||||
| F1 (n = 6) | 115 ± 16a | 2.13 ± 0.2b | 158 ± 16d | 134 ± 13a | 2.27 ± 0.3b | 145 ± 23d | ||||||
FFA, free fatty acid; TG, triglyceride. Data are presented as the means ± SD.
P < 0.01 versus NZB and NZW.
P < 0.01 versus NZB.
P < 0.05 versus NZB.
P < 0.01 versus NZW.
Identification of main-effect QTL in the F2 population
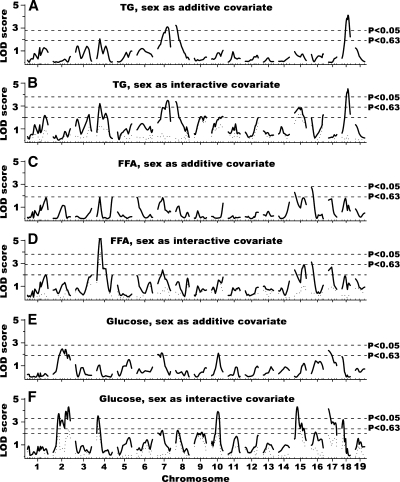
To identify QTL influencing plasma TG, FFA, and glucose, we performed single-locus genome scans on 264 F2 progeny from the NZB × NZW intercross. These scans included sex as an additive covariate to account for overall average differences between the sexes (Fig. 1A, C, E), and sex as interactive covariate to identify sex-specific QTL (Fig. 1B, D, F). The difference in LOD (ΔLOD) between the scans with sex as an additive or interactive covariate constitutes a test for QTL-by-sex interaction, and a ΔLOD of 2 or greater, as shown by the dotted line in Fig. 1B, D, F, represents a significant difference in QTL between the sexes. Details of the QTL including peak marker locus, LOD score, 95% confidence interval, allele conferring the higher value, and significance as determined by 1,000 permutation tests are summarized in Table 3.
Fig. 1.
Genome-wide scans for triglyceride (TG), FFA, and glucose. Sex as additive covariate (A, C, E); sex as interactive covariate (B, D, F). The horizontal dashed lines represent suggestive (P = 0.63) and significant (P = 0.05) levels as determined by 1,000-permutation test. The dotted lines at the bottom of B, D, and F depict the ΔLOD between scans A and B, C and D, and E and F, respectively. ΔLOD > 2.0 (denoted by the lower horizontal dashed line in B, D, and F) indicates quantitative trait loci (QTL) that differ significantly between sexes (P < 0.05).
TABLE 3.
Significant and suggestive QTL for plasma TG, FFA, and glucose
| LOD Scorec
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Locus Namea | Chr | Peak Mb | 95% CI | Sex as Additive Covariate | Sex as Interactive Covariate | Nearest Marker | High Allele (Sex)e | |||||||
| Mb | ||||||||||||||
| TG | ||||||||||||||
| None | 4 | 43 | 21–65 | 2.1 | 3.2 | rs3725792 | NZW | |||||||
| Tgq4 | 7 | 101 | 58–116 | 3.1 | 3.5 | rs3670069 | NZW | |||||||
| Tgq5 | 8 | 21 | 0–50 | 3.2 | 3.4 | rs3691954 | NZW | |||||||
| Tgq6 | 18 | 68 | 52–78 | 4.1 | 4.5 | rs4231907 | NZB | |||||||
| Tgq7b | 10 | 48 | — | 1.0 | 1.7 | rs3722617 | — | |||||||
| Tgq8b | 10 | 107 | — | 1.2 | 2.1 | rs13480757 | — | |||||||
| FFA | ||||||||||||||
| Ffaq2 | 4 | 43 | 30–60 | 2.0 | 5.4d | rs3725792 | NZW (M) | |||||||
| None | 16 | 6 | 0–25 | 2.7 | 3.1 | rs3090912 | NZW | |||||||
| Ffaq3b | 2 | 69 | — | 1.2 | 1.3 | rs3726142 | — | |||||||
| Ffaq4b | 15 | 10 | 0–32 | 2.3 | 2.0 | rs3676680 | NZB | |||||||
| Glucose | ||||||||||||||
| Bglu4 | 2 | 72 | 60–88 | 2.2 | 3.6 | rs3687515 | NZB | |||||||
| Bglu5 | 2 | 145 | 120–175 | 2.2 | 4.3d | rs3022911 | NZW (M) | |||||||
| Bglu6 | 4 | 32 | 21–43 | 1.1 | 3.5d | rs3725792 | NZB (M) | |||||||
| None | 7 | 56 | 26–78 | 2.2 | 2.2 | rs3708035 | NZB | |||||||
| Bglu7 | 10 | 87 | 74–100 | 2.1 | 3.9d | rs3089366 | NZW (M) | |||||||
| Bglu8 | 15 | 29 | 15–70 | 1.2 | 4.3d | rs4230721 | NZW (F), NZB (M) | |||||||
| Bglu9 | 17 | 22 | 0–45 | 2.3 | 4.1 | rs4231344 | NZB | |||||||
| Bglu10 | 18 | 33 | 0–50 | 1.8 | 3.3 | rs3705122 | NZB (M) | |||||||
| Bglu11b | 8 | 24 | — | 0.4 | 2.3 | rs3691954 | — | |||||||
CI, confidence interval; LOD, logarithm of the odds ratio; QTL, quantitative trait loci.
The QTL were named if they were significant or if they were suggestive but confirmed QTL that were found previously. The nomenclature of Bglu and Tgq for blood glucose QTL and triglyceride QTL is adopted from previous QTL for these traits (8, 9). Ffaq is used for FFA QTL.
Interacting QTL are selected from pairwise gene interaction search.
LOD score values were obtained from single QTL analysis. The suggestive and significant LOD score thresholds were determined by 1,000 permutation tests for each trait, the significant LOD scores were bolded. With sex as additive covariate, the suggestive/significant LODs are 1.9/2.8 for TG, 1.9/2.8 for FFA, and 1.9/2.9 for glucose. With sex as interactive covariate, the suggestive and significant LODs are 2.9/3.7 for TG, 2.9/3.8 for FFA, and 2.3/3.3 for glucose.
The LOD difference between sex as additive and interactive models is >2, indicating that the QTL differs between the sexes.
The sex is given if it is sex-specific QTL, M for male, F for female. For Bglu8, NZW is high allele in females while NZB is high in males.
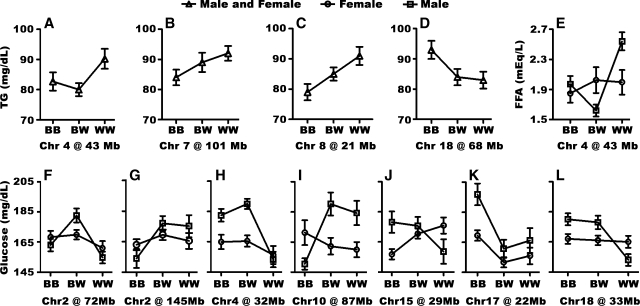
For TG, we identified three significant loci on Chr 7 at 101 Mb, Chr 8 at 21 Mb, and Chr 18 at 68 Mb, as well as one suggestive QTL on Chr 4; the chromosomal LOD plots are shown in Fig. 2A–D, and the allele effects are shown in Fig. 3A–D. All QTL for TG levels affect both sexes. For FFA, we identified one male-specific significant locus on Chr 4 at Mb 43 (Figs. 2A, 3E) and one suggestive QTL on Chr 16 at 6 Mb (Table 3). For glucose levels, we found significant loci on Chr 2, 4, 10, 15, 17, and 18 (Fig. 2A, E–I), as well as one suggestive locus on Chr 7. The QTL on Chr 2 was complex. We found evidence for two loci at Mb 72 and 145, but there might be others as well. Several QTL for glucose had sex-specific effects; the loci on Chr 2 at 145 Mb (Fig. 3G), Chr 4 at 32 Mb (Fig. 3H), Chr 10 at 87 Mb (Fig. 3I), and Chr 18 at 33 Mb (Fig. 3L) affected only males. The loci on the top of Chr 2 and Chr 17 affected glucose concentrations in both sexes, but the effect was stronger in males than in females (Fig. 3F, K), and the locus on Chr 15 at 29 Mb was higher in males in homozygous NZB mice, but higher in females in homozygous NZW mice (Fig. 3J).
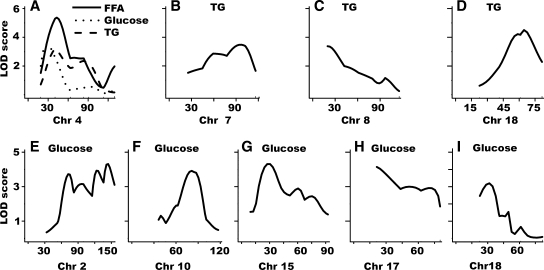
Fig. 2.
Chromosomal logarithm of the odds ratio (LOD) score plots for the significant QTL. The X-axis depicts the marker positions in Mb for each chromosome; the Y-axis depicts the LOD score. A: QTL on Chr 4 for TG, FFA, and glucose. B–D: TG. E–I: glucose.
Fig. 3.
The allele effects at significant QTL. The impact of the different alleles in F2 offspring at the peak of the QTL for TG (A–D), FFA (E), and glucose (F–L). Chromosome number and QTL position in Mb are given for each QTL. BB, homozygosity for NZB alleles; WW, homozygosity for NZW alleles; BW, heterozygosity. Error bars represent SEM.
QTL for TG, FFA, and glucose on proximal Chr 4
The LOD score plot for Chr 4 showed that the QTL for TG and FFA coincided well with double peaks at 43 Mb and 92 Mb, suggesting that these traits were determined by the same QTL genes. The QTL for glucose had only a single peak, which was at 32 Mb, near but not coincident with the peaks for the other two traits (Fig. 2A). For FFA and glucose, the QTL had significant LOD scores of 5.4 and 3.6, respectively, but for TG, this locus had only a suggestive LOD score of 3.2 (Table 3). The NZW allele at this locus behaved in a recessive manner to cause increased TG and FFA (Fig. 3A, E) and decreased glucose (Fig. 3G). Among individual F2 male mice, FFA levels were strongly correlated with higher plasma TG concentrations (R = 0.67; P = 1 × 10−15) but weakly correlated with lower glucose concentrations (R = 0.19; P = 0.04).
Epistasis and interacting QTL
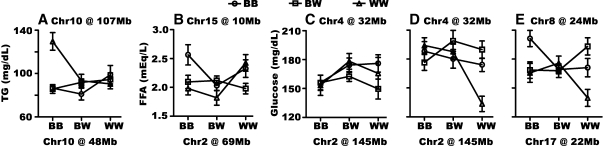
We also carried out pairwise genome scans to find interacting QTL using sex as an additive covariate. For TG, we found a significant interaction between two Chr 10 loci at 48 Mb and 107 Mb. The locus at 107 Mb did not affect TG concentrations by itself, but when combined with homozygous NZB allele at the locus at 48 Mb, TG increased by 45 mg/dl (Fig. 4A). For plasma FFA concentrations, a Chr 2 locus at 69 Mb interacted with a Chr 15 locus at 10 Mb. When both loci were homozygous NZB, FFA increased (Fig. 4B). For plasma glucose concentration, the pairwise genome scan revealed two significant interactions. The main scan QTL on Chr 2 (145 Mb) interacted with Chr 4 in a sex-specific fashion. When the Chr 2 locus was homozygous NZW, NZW alleles at Chr 4 had little effect on glucose levels in female mice (Fig. 4C), but decreased glucose in male animals (Fig. 4D). A second significant interaction was found between loci on Chr 8 and 17. The locus on Chr 8 did not affect glucose concentrations by itself, but its combined effect with the main scan QTL on Chr 17 on glucose concentrations was dramatic (Fig. 4E). When mice were homozygous NZB at both loci, glucose was 200 mg/dl; when mice were homozygous NZW allele at both loci, glucose decreased to 140 mg/dl.
Fig. 4.
The effects of gene interactions detected by the pairwise genome scan. BB, homozygosity for NZB alleles; WW, homozygosity for NZW alleles; BW, heterozygosity. Y-axes show mean values of TG (A), FFA (B), and glucose (C–E). Error bars represent SEM.
Regression analysis
To evaluate the relative contributions of each QTL when considered together, we combined all significant, suggestive, and interacting QTL in a multiple regression model, eliminating terms that did not meet the 0.01 significance level based on the multiple regression F test (Table 4). The results showed that we could account for 31% of the variation for TG, 21% for FFA, and 31% for glucose.
TABLE 4.
Multiple regression ANOVA for single and interacting QTL
| Traits | Location: Chr@Mb | dfa | Variance (%)b | LOD Score | P |
|---|---|---|---|---|---|
| TG | Sex | 1 | 6.0 | 4.14 | 6 × 10−6 |
| Chr4@43 | 2 | 3.6 | 1.53 | 0.02 | |
| Chr7@101 | 2 | 6.0 | 3.87 | 3 × 10−5 | |
| Chr8@21 | 2 | 5.3 | 3.67 | 1 × 10−4 | |
| Chr18@68 | 2 | 2.9 | 3.22 | 0.006 | |
| Chr10@48 | 6 | 7.7 | 5.09 | 2 × 10−4 | |
| Chr10@107 | 6 | 8.0 | 5.29 | 1 × 10−4 | |
| Chr10@48:Chr10@107 | 4 | 6.9 | 4.68 | 1 × 10−4 | |
| Totals | 17 | 31.4 | 19.46 | ||
| FFA | Sex | 3 | 5.6 | 3.66 | 0.001 |
| Chr2@69 | 6 | 6.9 | 4.46 | 0.003 | |
| Chr4@43 | 4 | 7.6 | 4.89 | 3 × 10−4 | |
| Chr15@10 | 6 | 9.2 | 5.82 | 3 × 10−4 | |
| Chr2@69:Chr15@10 | 4 | 5.3 | 3.46 | 0.005 | |
| Sex : Chr4@43 | 2 | 4.9 | 3.21 | 9 × 10−4 | |
| Totals | 13 | 21.3 | 12.13 | ||
| Glucose | Sex | 9 | 9.6 | 6.86 | 9 × 10−4 |
| Chr2@145 | 12 | 13.6 | 9.51 | 9 × 10−5 | |
| Chr4@32 | 12 | 11.3 | 8.01 | 9 × 10−4 | |
| Chr7@56 | 2 | 3.0 | 2.23 | 0.008 | |
| Chr8@24 | 6 | 4.2 | 3.10 | 0.01 | |
| Chr17@22 | 6 | 5.2 | 3.85 | 0.009 | |
| Chr18@33 | 2 | 1.3 | 1.01 | 0.013 | |
| Sex:Chr2@145 | 6 | 7.0 | 5.07 | 0.002 | |
| Sex:Chr4@32 | 6 | 5.8 | 4.22 | 0.008 | |
| Chr2@145:Chr4@32 | 8 | 7.7 | 5.61 | 0.003 | |
| Sex:Chr2@145:Chr4@32 | 4 | 3.8 | 2.86 | 0.015 | |
| Chr8@24:Chr17@22 | 4 | 2.9 | 2.19 | 0.018 | |
| Totals | 29 | 31.1 | 19.62 |
df, degrees of freedom, includes main effect and any interactions.
Variance indicates the percentage of the total F2 phenotypic variance associated with each marker.
QTL were narrowed using bioinformatics tools
Four of the mouse QTL for TG (Chr 4, 7, 8, 18) were homologous to human QTL for TG (7); likewise, all eight QTL for glucose were homologous to human QTL for glucose (Table 5). Six of the glucose QTL, all except Chr 4 and 7, have been identified in other mouse crosses (Table 5). These factors of homologous human QTL and coincident QTL in other crosses, combined with the high similarity of the NZW and NZB genomes, facilitate the narrowing of the QTL regions using bioinformatics (14). Such narrowing is based on the assumption that the same gene causes colocalizing QTL in different crosses as well as homologous mouse and human QTL. We narrowed the Chr 4 QTL for TG and FFA, and all other significant mouse QTL for TG and glucose regions by first reducing the QTL to that portion homologous to the human QTL (Fig. 5B), a step that narrowed the QTL by a minimum of 31% for Chr 15 to a maximum of 86% for Chr 10. We then further reduced the QTL by using SNPs of all the strains that were parents of the QTL crosses and finding those regions where the strains carrying the allele that increased the trait were identical but differed from the strains carrying the allele that decreased the trait. Because of the high similarity of NZB and NZW, this step was particularly effective and caused further reduction of 85% for Chr 2, 84% for Chr 4, and greater than 90% for the remainder (Fig. 5C). The bioinformatics step reduced the positional candidate genes on Chr 10 locus for glucose to only 1, to only 2 for FFA on Chr 16, and to 8–40 genes for the remainder. (For the lists of candidate genes remaining, see supplementary Tables II, III.)
TABLE 5.
QTL for glucose identified in human homologous regions and other mouse crosses
| QTL | Chr | 95% CI | Human QTL Homologous Regiona (Ref) | Coincident QTL Crossb (Ref) |
|---|---|---|---|---|
| Mb | ||||
| Bglu4 | 2 | 60–90 | 2q241-311 (34) | BKS × C3H (35) |
| Bglu5 | 2 | 120–175 | 20p112-q131 (36) | B6 × C3H (37) |
| Bglu6 | 4 | 21–45 | 9p21-13 (38) | — |
| none | 7 | 26–78 | 19q12-19q134 (38, 39) | — |
| Bglu7 | 10 | 74–100 | 21q22 (40), 22q11-12 (41) | SM × A/J (42) |
| Bglu8 | 15 | 15–70 | 8q22-24 (40) | Balb/cJ × KK/Ta (F) (43), Akita × A/J (44) |
| Bglu9 | 17 | 0–45 | 6p213-212 (38), 6q253-qter (41) | B6 × NOD (45) |
| Bglu10 | 18 | 0–50 | 10p121-112, 5q31-32 (36),18q12-23(46) | NON × NZO (47), SMxA/J (42) |
Human homologous region of the mouse QTLs were retrieved from the Genome Orthology Map at http://www.informatics.jax.org/reports/homologymap/mouse_humans.html.
The strains that contribute the high allele are in boldface. For the NON × NZO cross, we obtained the high allele from Dr. Ed Leiter.
Fig. 5.
Narrowing QTL by bioinformatics. A: The 95% confidence interval and the number of genes in each QTL identified in cross NZB × NZW. B: Comparative genomics. The QTL narrowed by homology with human QTL. C: Haplotype analysis. Each QTL was reduced by haplotype analysis of the strains involved in the QTL crosses.
QTL candidate genes
Using the lists of candidate genes, we evaluated each by a literature search for gene function and SNPs that changed coding region polymorphisms; on the basis of these criteria, some strong candidate genes emerged.
For TG, the locus on Chr 4 was narrowed to 26 genes (Fig. 5; see supplementary Table II), and the strongest candidate gene was Gba2, encoding glucosidase β 2 (Table 6). Several lines of evidence supported the candidacy of this gene; it is located at 43.5 Mb and the QTL peak is 43 Mb, the haplotype of NZB and NZW differ, and Gba2 knock-out mice have decreased TG compared with the wild-type animals (15). We sequenced Gba2 in strains NZB and NZW and found a coding region polymorphism that changed the amino acid at position 385 from Ile (NZB) to Val (NZW). This amino acid variant is in a conserved region. Although there are 38 remaining genes in the Chr 8 interval, a potential candidate was Irs2, encoding insulin receptor substrate 2. Plasma TG levels in Irs2 knock-out mice are lower than those of wild-type animals (16). The locus on Chr 18 colocalized with a QTL previously identified in a (B6 × 129) F2 intercross (17), and haplotyping reduced the region to 12 genes (Fig. 5). We suggest the candidate gene Ppargc1b (61.4 Mb) encoding peroxisome proliferator-activated receptor γ coactivator 1 β. We sequenced this gene in strains NZB and NZW, and found three amino acid variants: Gly767Asp, Cys487Gly, and Ala146Ser (NZB,NZW) (Table 6). Ppargc1b is involved in the regulation of fatty acid oxidation and gluconeogenesis (18), and serum TG and FFA levels are significantly reduced in Ppargc1b knock-out mice (19).
TABLE 6.
QTL candidate genes
| Gene
|
Lines of Evidence
|
||||||
|---|---|---|---|---|---|---|---|
| Trait | Chr | QTL Peak | Name | Mb | Gene Numbera | AA Variantb | Knock-out Micec |
| Mb | |||||||
| TG, FFA | 4 | 43 | Gba2 | 43.5 | 26 | Ile 385 Val | Lower TG |
| TG | 8 | 20 | Irs2 | 11.0 | 38 | Lower TG | |
| TG | 18 | 68 | Ppargc1b | 61.4 | 12 | Ala146Ser Cys487GLy Gly767Asp | Lower TG |
| FFA | 16 | 6 | A2bp1 | 6 | 2 | Obesity | |
| Glucose | 2 | 72 | G6pc2 | 69.0 | 15 | Lower glucose | |
| Glucose | 10 | 87 | Timp3 | 85.7 | 1 | Higher glucose | |
Positional candidate genes reduced by using comparative genomics and haplotype analysis.
Amino acid change between strains NZB (left) and NZW (right).
Phenotype changed in mice deficient in the gene (KO or gene knocked out by homologous recombination).
For FFA, we reduced the locus on Chr 16 to 2 Mb using the haplotype between the strains NZB and NZW (Fig. 5). Only two genes, LOC665158 (5.68 Mb) and A2bp1 (5.69–7.32 Mb), are located within this region. The LOC665158 is uncharacterized, but A2bp1 encodes ataxin-2 binding protein-1. Ataxin-2 is a protein of unknown function, but its deficiency leads to adult-onset obesity (20).
For glucose, we reduced the Chr 2 proximal locus Bglu4 to 15 genes (see supplementary Table III). We suggest the candidate gene G6pc2 (69.0 Mb), encoding glucose-6-phosphatase catalytic 2, which is an islet-specific glucose-6-phosphatase (21). G6pc2 knock-out mice have significantly decreased blood glucose levels in both sexes, and female mice also exhibit decreased plasma triacylglycerol (22). The Bglu7 on Chr 10 was reduced to only gene Timp3, located at 85.7 Mb (Fig. 5, Table 6), which encodes tissue inhibitor of metalloproteinase 3 (TIMP3). We sequenced the entire coding region of Timp3 in strains NZB and NZW and found one synonymous (C in NZW to T in NZB, Cys12Cys) and one 3′ untranslated region (T in NZW to G in NZB) polymorphisms. We quantified the Timp3 expression using real-time PCR with mRNA extracted from male NZB and NZW livers and normalizing Timp3 expression levels to those of β-actin (Actb). The mRNA expression levels of Timp3 were significantly lower in NZW mice compared with those in NZB mice (P < 0.01) (Fig. 6). Insulin receptor-haploinsufficient mice that also are TIMP3 deficient have increased blood glucose levels and vascular inflammation due to increased tumor necrosis factor-α (23).
Fig. 6.
The expression of Timp3 in NZB and NZW mice. Total RNA was extracted from the livers of male mice. The mRNA expression levels of Timp3 were normalized to β-actin and expressed as mRNA copies per 1,000 copies of β-actin. Error bars indicate ± SEM.
DISCUSSION
The genome-wide scans of NZB × NZW F2 mice resulted in the localization of several novel QTL, particularly for TG and FFA levels. We also found two novel QTL for glucose and confirmed six previously identified QTL. Studies of multiple mouse strains (24) have shown that sex exerts a profound effect on the plasma lipids and glucose levels, and previous QTL analysis indicated that many QTL were sex specific, suggesting that it is important to differentiate between two sexes in QTL analysis. Analyzing the entire population with and without a sex-by-QTL interaction is a powerful approach, because analyzing males and females separately might result in failure to detect QTL effects due to decreased statistical power (12). In this study, most of the QTL for glucose had sex-specific effects. Sex also contributed significantly to the multi-locus model, accounting for 6% of the TG variance, 5.6% of the FFA variance, and 9.6% of the glucose variance.
The ultimate goal of QTL mapping is to identify the underlying genes. However, the confidence intervals of the QTL identified from intercrosses are often quite large. Until QTL are narrowed, identifying the underlying genes by positional cloning is challenging. Although several classic breeding strategies can be used to resolve QTL (25), such as congenic lines, advanced intercross lines, and recombinant inbred segregation test, their usefulness is limited by the available resources, such as time, animal space, and money. Bioinformatics tools are an efficient and economical approach for narrowing QTL and prioritizing candidate genes (14), which has been made possible by publicly accessible sequence, genotype, and expression databases. These tools, including comparative genomics and haplotype analysis, allow stepwise narrowing of a QTL interval, prioritizing candidate genes for further analysis with the potential of identifying the most probable candidate gene.
Most loci for TG and glucose identified in this study have been found in human homologous genomic locations. Because rodent and human QTL for the same trait often map to homologous genomic locations, comparison of these homology maps may reduce the QTL size in both species if one assumes that homologous QTL are caused by the same gene in both species. To date, there are many examples of correspondence between alleles in particular genes and phenotypes in mouse and human. For instance, Apoa5, a member of the apolipoprotein gene family, is newly identified as a gene that affects plasma TG levels in both human and mouse by comparative gene sequences (26, 27). However, the pitfall of comparative genomics would be homologous QTL caused by different genes in human and mouse. Because closely linked QTL are often found, there may be circumstance in which two genes controlling a trait are located on the same chromosome but one gene is polymorphic in mouse and the other gene is polymorphic in human. In such a case, we would fail to find the QTL genes by assuming the same gene caused the QTL in both species.
Because most genetic variation among inbred mouse strains is ancestral, it is possible to reduce the QTL regions by eliminating those regions that are IBD between the two strains as inferred by a shared SNP pattern. This was particularly effective for the related strains NZW and NZB because they shared 63.2% of their genomes. However, using the genomic similarity between NZB and NZW to reduce the QTL region also has a pitfall. If the mutation causing the QTL arose since these strains were separated, it could have occurred in a region that was not polymorphic in other genes, and those regions may have been eliminated. This pitfall is unlikely for QTL found in other crosses as well; for those, the mutation is ancestral.
In addition to the genes with direct evidence as QTL genes either from knock-out mice or reduction of the QTL region to only one or two genes by bioinformatics, we also suggest some other candidates based on known function. For the Chr 7 TG locus, the likely candidate gene is Thrsp, encoding the thyroid hormone-responsive SPOT14 homolog. This gene is located at 97.2 Mb, near the QTL peak at 101 Mb, and the SPOT 14 protein is involved in de novo lipogenesis (28). For glucose, the Chr 2 distal locus, Bglu5, was reduced to 40 genes (see supplementary Table III); the potential candidate gene is Ggtl3, encoding γ-glutamyltransferase (GGT)-like 3. Population-based epidemiological studies have prospectively demonstrated that baseline γ-GGT activity predicts development of type 2 diabetes (29). The Bglu6 on Chr 4 was reduced to 16 genes, two of which, Il11ra1 and Il11ra2, encode interleukin 11 receptor-α chains. Interleukin 11 inhibits NF-κB and AP-1 activation in islets and prevents type I diabetes in NOD mice (30, 31). The Bglu8 on Chr 15 was reduced to 12 genes; one of the promising candidates is Adcy8, encoding adenylate cyclase 8. This protein has been identified in insulin-secreting cells as one potential molecular target for synergism between glucose-dependent insulinotropic peptide receptor-mediated and glucose-mediated signaling (32). The Bglu10 on Chr 18 was reduced to 8 genes; the potential candidate gene is Ppp2r2b, encoding protein phosphatase 2 (PP2A)-regulatory subunit B β isoform. Emerging evidence implicates PP2A in insulin secretion and the dephosphorylation and inactivation of specific β cell phosphoprotein substrates (33).
In summary, we identified the QTL for TG, FFA, and glucose by intercrossing the related strains NZB and NZW. Furthermore, we narrowed the QTL by bioinformatics and suggest some strong candidate genes. These genes need to be tested further to determine whether they are indeed the QTL-causing genes; however, we present three lines of evidence to demonstrate that Timp3 is a QTL gene.
Supplementary Material
Acknowledgments
The authors are most grateful to Harry Whitmore and Fred Rumill for their invaluable help in mouse husbandry, and to Drs. E. H. Leiter and D. V. Serreze for helpful comments regarding the manuscript.
Abbreviations
Chr, chromosome
Ct, cycle threshold
FFA, free fatty acid
IBD, identical by descent
LOD, logarithm of the odds ratio
PPA, phosphatase A
SNP, single-nucleotide polymorphism
TG, triglyceride
QTL, quantitative trait loci
Published, JLR Papers in Press, March 24, 2008.
Footnotes
This work was funded by National Institutes of Health Grants HL-81162, HL-74086, HL-77796, and GM-764668), American Heart Association Grant 0725905T (to S.Z.), and National Cancer Institute Cancer Core Grant CA-034196 to The Jackson Laboratory.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three tables.
References
- 1.Boden G., and G. I. Shulman. 2002. Free fatty acids in obesity and type 2 diabetes: defining their role in the development of insulin resistance and beta-cell dysfunction. Eur. J. Clin. Invest. 32 (Suppl.): 14–23. [DOI] [PubMed] [Google Scholar]
- 2.Bergman R. N., and M. Ader. 2000. Free fatty acids and pathogenesis of type 2 diabetes mellitus. Trends Endocrinol. Metab. 11 351–356. [DOI] [PubMed] [Google Scholar]
- 3.Randle P. J. 1998. Regulatory interactions between lipids and carbohydrates: the glucose fatty acid cycle after 35 years. Diabetes Metab. Rev. 14 263–283. [DOI] [PubMed] [Google Scholar]
- 4.Shimabukuro M., Y. T. Zhou, M. Levi, and R. H. Unger. 1998. Fatty acid-induced beta cell apoptosis: a link between obesity and diabetes. Proc. Natl. Acad. Sci. USA. 95 2498–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rebrin K., G. M. Steil, L. Getty, and R. N. Bergman. 1995. Free fatty acid as a link in the regulation of hepatic glucose output by peripheral insulin. Diabetes. 44 1038–1045. [DOI] [PubMed] [Google Scholar]
- 6.Aitman T. J., T. Gotoda, A. L. Evans, H. Imrie, K. E. Heath, P. M. Trembling, H. Truman, C. A. Wallace, A. Rahman, C. Dore, et al. 1997. Quantitative trait loci for cellular defects in glucose and fatty acid metabolism in hypertensive rats. Nat. Genet. 16 197–201. [DOI] [PubMed] [Google Scholar]
- 7.Wang X., and B. Paigen. 2005. Genome-wide search for new genes controlling plasma lipid concentrations in mice and humans. Curr. Opin. Lipidol. 16 127–137. [DOI] [PubMed] [Google Scholar]
- 8.Su Z., Y. Li, J. C. James, M. McDuffie, A. H. Matsumoto, G. A. Helm, J. L. Weber, A. J. Lusis, and W. Shi. 2006. Quantitative trait locus analysis of atherosclerosis in an intercross between C57BL/6 and C3H mice carrying the mutant apolipoprotein E gene. Genetics. 172 1799–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su Z., Y. Li, J. C. James, A. H. Matsumoto, G. A. Helm, A. J. Lusis, and W. Shi. 2006. Genetic linkage of hyperglycemia, body weight and serum amyloid-P in an intercross between C57BL/6 and C3H apolipoprotein E-deficient mice. Hum. Mol. Genet. 15 1650–1658. [DOI] [PubMed] [Google Scholar]
- 10.Nishina P. M., J. Verstuyft, and B. Paigen. 1990. Synthetic low and high fat diets for the study of atherosclerosis in the mouse. J. Lipid Res. 31 859–869. [PubMed] [Google Scholar]
- 11.Szatkiewicz J. P., G. L. Beane, Y. Ding, L. Hutchins, F. Pardo-Manuel de Villena, and G. A. Churchill. 2008. An imputed genotype resource for the laboratory mouse. Mamm. Genome. 19 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solberg L. C., A. E. Baum, N. Ahmadiyeh, K. Shimomura, R. Li, F. W. Turek, G. A. Churchill, J. S. Takahashi, and E. E. Redei. 2004. Sex- and lineage-specific inheritance of depression-like behavior in the rat. Mamm. Genome. 15 648–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sen S., and G. A. Churchill. 2001. A statistical framework for quantitative trait mapping. Genetics. 159 371–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiPetrillo K., X. Wang, I. M. Stylianou, and B. Paigen. 2005. Bioinformatics toolbox for narrowing rodent quantitative trait loci. Trends Genet. 21 683–692. [DOI] [PubMed] [Google Scholar]
- 15.Yildiz Y., H. Matern, B. Thompson, J. C. Allegood, R. L. Warren, D. M. Ramirez, R. E. Hammer, F. K. Hamra, S. Matern, and D. W. Russell. 2006. Mutation of beta-glucosidase 2 causes glycolipid storage disease and impaired male fertility. J. Clin. Invest. 116 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashimoto H., T. Arai, A. Takeguchi, K. Hioki, Y. Ohnishi, K. Kawai, M. Ito, R. Suzuki, T. Yamauchi, M. Ohsugi, et al. 2006. Ontogenetic characteristics of enzyme activities and plasma metabolites in C57BL/6J:Jcl mice deficient in insulin receptor substrate 2. Comp. Med. 56 176–187. [PubMed] [Google Scholar]
- 17.Ishimori N., R. Li, P. M. Kelmenson, R. Korstanje, K. A. Walsh, G. A. Churchill, K. Forsman-Semb, and B. Paigen. 2004. Quantitative trait loci that determine plasma lipids and obesity in C57BL/6J and 129S1/SvImJ inbred mice. J. Lipid Res. 45 1624–1632. [DOI] [PubMed] [Google Scholar]
- 18.Yoon J. C., P. Puigserver, G. Chen, J. Donovan, Z. Wu, J. Rhee, G. Adelmant, J. Stafford, C. R. Kahn, D. K. Granner, et al. 2001. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 413 131–138. [DOI] [PubMed] [Google Scholar]
- 19.Sonoda J., I. R. Mehl, L. W. Chong, R. R. Nofsinger, and R. M. Evans. 2007. PGC-1beta controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proc. Natl. Acad. Sci. USA. 104 5223–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiehl T. R., A. Nechiporuk, K. P. Figueroa, M. T. Keating, D. P. Huynh, and S. M. Pulst. 2006. Generation and characterization of Sca2 (ataxin-2) knockout mice. Biochem. Biophys. Res. Commun. 339 17–24. [DOI] [PubMed] [Google Scholar]
- 21.Arden S. D., T. Zahn, S. Steegers, S. Webb, B. Bergman, R. M. O'Brien, and J. C. Hutton. 1999. Molecular cloning of a pancreatic islet-specific glucose-6-phosphatase catalytic subunit-related protein. Diabetes. 48 531–542. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y., C. C. Martin, J. K. Oeser, S. Sarkar, O. P. McGuinness, J. C. Hutton, and R. M. O'Brien. 2007. Deletion of the gene encoding the islet-specific glucose-6-phosphatase catalytic subunit-related protein autoantigen results in a mild metabolic phenotype. Diabetologia. 50 774–778. [DOI] [PubMed] [Google Scholar]
- 23.Federici M., M. L. Hribal, R. Menghini, H. Kanno, V. Marchetti, O. Porzio, S. W. Sunnarborg, S. Rizza, M. Serino, V. Cunsolo, et al. 2005. Timp3 deficiency in insulin receptor-haploinsufficient mice promotes diabetes and vascular inflammation via increased TNF-alpha. J. Clin. Invest. 115 3494–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Svenson K. L., R. Von Smith, P. A. Magnani, H. R. Suetin, B. Paigen, J. K. Naggert, R. Li, G. A. Churchill, and L. L. Peters. 2007. Multiple trait measurements in 43 inbred mouse strains capture the phenotypic diversity characteristic of human populations. J. Appl. Physiol. 102 2369–2378. [DOI] [PubMed] [Google Scholar]
- 25.Flint J., W. Valdar, S. Shifman, and R. Mott. 2005. Strategies for mapping and cloning quantitative trait genes in rodents. Nat. Rev. Genet. 6 271–286. [DOI] [PubMed] [Google Scholar]
- 26.Pennacchio L. A., M. Olivier, J. A. Hubacek, J. C. Cohen, D. R. Cox, J. C. Fruchart, R. M. Krauss, and E. M. Rubin. 2001. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science. 294 169–173. [DOI] [PubMed] [Google Scholar]
- 27.Pennacchio L. A., and E. M. Rubin. 2003. Apolipoprotein A5, a newly identified gene that affects plasma triglyceride levels in humans and mice. Arterioscler. Thromb. Vasc. Biol. 23 529–534. [DOI] [PubMed] [Google Scholar]
- 28.Freake H. C., and J. H. Oppenheimer. 1987. Stimulation of S14 mRNA and lipogenesis in brown fat by hypothyroidism, cold exposure, and cafeteria feeding: evidence supporting a general role for S14 in lipogenesis and lipogenesis in the maintenance of thermogenesis. Proc. Natl. Acad. Sci. USA. 84 3070–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee D. H., M. H. Ha, J. H. Kim, D. C. Christiani, M. D. Gross, M. Steffes, R. Blomhoff, and D. R. Jacobs, Jr. 2003. Gamma-glutamyltransferase and diabetes—a 4 year follow-up study. Diabetologia. 46 359–364. [DOI] [PubMed] [Google Scholar]
- 30.Lgssiar A., M. Hassan, P. Schott-Ohly, N. Friesen, F. Nicoletti, W. L. Trepicchio, and H. Gleichmann. 2004. Interleukin-11 inhibits NF-kappaB and AP-1 activation in islets and prevents diabetes induced with streptozotocin in mice. Exp. Biol. Med. (Maywood). 229 425–436. [DOI] [PubMed] [Google Scholar]
- 31.Nicoletti F., P. Zaccone, I. Conget, R. Gomis, C. Moller, P. L. Meroni, K. Bendtzen, W. Trepicchio, and S. Sandler. 1999. Early prophylaxis with recombinant human interleukin-11 prevents spontaneous diabetes in NOD mice. Diabetes. 48 2333–2339. [DOI] [PubMed] [Google Scholar]
- 32.Delmeire D., D. Flamez, S. A. Hinke, J. J. Cali, D. Pipeleers, and F. Schuit. 2003. Type VIII adenylyl cyclase in rat beta cells: coincidence signal detector/generator for glucose and GLP-1. Diabetologia. 46 1383–1393. [DOI] [PubMed] [Google Scholar]
- 33.Kowluru A. 2005. Novel regulatory roles for protein phosphatase-2A in the islet beta cell. Biochem. Pharmacol. 69 1681–1691. [DOI] [PubMed] [Google Scholar]
- 34.Busfield F., D. L. Duffy, J. B. Kesting, S. M. Walker, P. K. Lovelock, D. Good, H. Tate, D. Watego, M. Marczak, N. Hayman, et al. 2002. A genomewide search for type 2 diabetes-susceptibility genes in indigenous Australians. Am. J. Hum. Genet. 70 349–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moritani M., K. Togawa, H. Yaguchi, Y. Fujita, Y. Yamaguchi, H. Inoue, N. Kamatani, and M. Itakura. 2006. Identification of diabetes susceptibility loci in db mice by combined quantitative trait loci analysis and haplotype mapping. Genomics. 88 719–730. [DOI] [PubMed] [Google Scholar]
- 36.Chiu Y. F., L. M. Chuang, C. F. Hsiao, Y. J. Hung, M. W. Lin, Y. T. Chen, J. Grove, E. Jorgenson, T. Quertermous, N. Risch, et al. 2005. An autosomal genome-wide scan for loci linked to pre-diabetic phenotypes in nondiabetic Chinese subjects from the Stanford Asia-Pacific Program of Hypertension and Insulin Resistance Family Study. Diabetes. 54 1200–1206. [DOI] [PubMed] [Google Scholar]
- 37.Kayo T., H. Fujita, J. Nozaki, X. E., and A. Koizumi. 2000. Identification of two chromosomal loci determining glucose intolerance in a C57BL/6 mouse strain. Comp. Med. 50 296–302. [PubMed] [Google Scholar]
- 38.Pratley R. E., D. B. Thompson, M. Prochazka, L. Baier, D. Mott, E. Ravussin, H. Sakul, M. G. Ehm, D. K. Burns, T. Foroud, et al. 1998. An autosomal genomic scan for loci linked to prediabetic phenotypes in Pima Indians. J. Clin. Invest. 101 1757–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.An P., B. I. Freedman, C. L. Hanis, Y. D. Chen, A. B. Weder, N. J. Schork, E. Boerwinkle, M. A. Province, C. A. Hsiung, X. Wu, et al. 2005. Genome-wide linkage scans for fasting glucose, insulin, and insulin resistance in the National Heart, Lung, and Blood Institute Family Blood Pressure Program: evidence of linkages to chromosome 7q36 and 19q13 from meta-analysis. Diabetes. 54 909–914. [DOI] [PubMed] [Google Scholar]
- 40.Frayling T. M., C. M. Lindgren, J. C. Chevre, S. Menzel, M. Wishart, Y. Benmezroua, A. Brown, J. C. Evans, P. S. Rao, C. Dina, et al. 2003. A genome-wide scan in families with maturity-onset diabetes of the young: evidence for further genetic heterogeneity. Diabetes. 52 872–881. [DOI] [PubMed] [Google Scholar]
- 41.Sale M. M., B. I. Freedman, C. D. Langefeld, A. H. Williams, P. J. Hicks, C. J. Colicigno, S. R. Beck, W. M. Brown, S. S. Rich, and D. W. Bowden. 2004. A genome-wide scan for type 2 diabetes in African-American families reveals evidence for a locus on chromosome 6q. Diabetes. 53 830–837. [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi M., T. Ohno, A. Tsuji, M. Nishimura, and F. Horio. 2003. Combinations of nondiabetic parental genomes elicit impaired glucose tolerance in mouse SMXA recombinant inbred strains. Diabetes. 52 180–186. [DOI] [PubMed] [Google Scholar]
- 43.Shike T., S. Hirose, M. Kobayashi, K. Funabiki, T. Shirai, and Y. Tomino. 2001. Susceptibility and negative epistatic loci contributing to type 2 diabetes and related phenotypes in a KK/Ta mouse model. Diabetes. 50 1943–1948. [DOI] [PubMed] [Google Scholar]
- 44.Takeshita S., M. Moritani, K. Kunika, H. Inoue, and M. Itakura. 2006. Diabetic modifier QTLs identified in F2 intercrosses between Akita and A/J mice. Mamm. Genome. 17 927–940. [DOI] [PubMed] [Google Scholar]
- 45.Deruytter N., O. Boulard, and H. J. Garchon. 2004. Mapping non-class II H2-linked loci for type 1 diabetes in nonobese diabetic mice. Diabetes. 53 3323–3327. [DOI] [PubMed] [Google Scholar]
- 46.Merriman T. R., H. J. Cordell, I. A. Eaves, P. A. Danoy, F. Coraddu, R. Barber, F. Cucca, S. Broadley, S. Sawcer, A. Compston, et al. 2001. Suggestive evidence for association of human chromosome 18q12-q21 and its orthologue on rat and mouse chromosome 18 with several autoimmune diseases. Diabetes. 50 184–194. [DOI] [PubMed] [Google Scholar]
- 47.Leiter E. H., P. C. Reifsnyder, K. Flurkey, H. J. Partke, E. Junger, and L. Herberg. 1998. NIDDM genes in mice: deleterious synergism by both parental genomes contributes to diabetogenic thresholds. Diabetes. 47 1287–1295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.