Abstract
Lipid transfer inhibitor protein (LTIP) is a physiologic regulator of cholesteryl ester transfer protein (CETP) function. We previously reported that LTIP activity is localized to LDL, consistent with its greater inhibitory activity on this lipoprotein. With a recently described immunoassay for LTIP, we investigated whether LTIP mass is similarly distributed. Plasma fractionated by gel filtration chromatography revealed two LTIP protein peaks, one coeluting with LDL, and another of ∼470 kDa. The 470 kDa LTIP complex had a density of 1.134 g/ml, indicating ∼50% lipid content, and contained apolipoprotein A-I. By mass spectrometry, partially purified 470 kDa LTIP also contains apolipoproteins C-II, D, E, J, and paraoxonase 1. Unlike LDL-associated LTIP, the 470 kDa LTIP complex does not inhibit CETP activity. In normolipidemic subjects, ∼25% of LTIP is in the LDL-associated, active form. In hypercholesterolemia,this increases to 50%, suggesting that lipoprotein composition may influence the status of LTIP activity. Incubation (37°C) of normolipidemic plasma increased active, LDL-associated LTIP up to 3-fold at the expense of the inactive pool. Paraoxon inhibited this shift by 50%. Overall, these studies show that LTIP activity is controlled by its reversible incorporation into an inactive complex. This may provide for short-term fine-tuning of lipoprotein remodeling mediated by CETP.
Keywords: apolipoprotein F, fast-protein liquid chromatography, gel filtration, apolipoprotein A-I, mass spectrometry, hypercholesterolemia
In plasma, cholesteryl ester transfer protein (CETP) mediates the net transfer of cholesteryl ester (CE) from LDL and HDL to VLDL in return for triglyceride (TG) (1, 2). This remodeling of lipoprotein composition alters the metabolism of lipoproteins and ultimately influences both the quality and quantity of lipoproteins in plasma (3–5). Physiologically, CETP may be regulated by at least two proteins. Apolipoprotein C-I, which resides primarily on HDL, has been reported to inhibit CETP in vitro, and studies with transgenic animals have demonstrated its ability to suppress CETP activity in vivo (6, 7). Its mode of action is via modification of the surface charge of HDL, resulting in weakened CETP-HDL interactions and thus suppression of lipid transfer events with HDL (8). A second regulatory protein is lipid transfer inhibitor protein (LTIP), also known as apolipoprotein F (9). Although the mechanism of inhibition of CETP by LTIP has not been firmly established, it also appears to function by disrupting the interaction of CETP with its substrate (10). However, the effects of LTIP on lipid transfer events are quite distinct from that of apolipoprotein C-I. Unlike apolipoprotein C-I, LTIP activity is associated with LDL (11). The addition of exogenous LTIP to native plasma reduces the participation of LDL in lipid transfer events but leads to a dose-dependent increase in the efflux of CE from HDL to VLDL. This enhanced CETP activity on HDL results in HDL particles that are markedly better substrates for lecithin:cholesterol acyltransferase (11). We have proposed that this increased CETP activity on HDL happens because plasma VLDL TG, not CETP, concentration is rate limiting for net lipid transfer in normal plasma (12, 13). So, because CETP-mediated lipid transfers from LDL and HDL to VLDL in essence compete for a limited supply of TG, in the presence of LTIP less VLDL TG is expended by transfer to LDL, allowing CE-TG exchange between VLDL and HDL to be increased.
Under steady-state binding conditions with isolated lipoproteins, CETP has similar affinity for all lipoproteins (10), and in vitro measurements of CETP activity show that CETP displays no significant preference for lipoprotein classes when lipoproteins are compared on an equivalentsurface basis (14). Yet it appears in whole plasma that HDL is the preferred CETP substrate. Notably though, in assays with isolated lipoproteins and CETP, the addition of plasma levels of LTIP completely reconstitute the 2-fold preference of CETP for HDL that exists in native plasma (14). This strongly suggests that the enhanced lipid transfer from HDL in control plasma, which has been generally attributed to a preferential interaction between CETP and HDL, is the consequence of LTIP activity. In aggregate, these data show that LTIP does not simply reduce CETP activity, but alters the overall pattern of CETP-mediated lipidflux between lipoproteins.
In preliminary studies with a recently developed antisera against LTIP (15), we observed that significant LTIP mass is found in both LDL and HDL. The observation of the latter, although consistent with recent shotgun proteomic analysis of HDL (16), is contrary to our previous studies showing LTIP to be associated only with LDL (9, 11). To resolve this discrepancy, we have characterized the plasma forms of LTIP. We report that LTIP exists in an active andan inactive form, and that LTIP activity is regulated by its reversible sequestration into the inactive complex. This provides a mechanism for fine-tuning the control of CETP activity by LTIP.
METHODS
Reagent preparation
Human plasma, obtained from the Cleveland Clinic Blood Bank, was separated into VLDL, LDL, HDL, and d> 1.21 g/ml (lipoprotein-free) fractions by sequential ultracentrifugation (17). HDL3 was isolated as the 1.125–1.21 g/ml density fraction and further purified by a second, identical centrifugation step. LDL was radiolabeled with [3H]CE by a dispersion transfer method (18). CETP and LTIP were partially purified from frozen plasma as previously described (19). CETP and LTIP activities were measured by their ability to stimulate CE transfer from [3H]CE-LDL to HDL, or to inhibit CETP-mediated CE transfer from [3H]CE-LDL to HDL, respectively, as previously published (18, 20, 21). LTIP mass in plasma and plasma fractions was determined by an ELISA (15).
Gel filtration studies
Plasma and other plasma-derived samples were fractionated by fast-protein liquid chromatography (FPLC) on tandem Superose 6 HR columns (GE Healthcare; Piscataway, NJ). Samples (≤0.5 ml) were applied to columns equilibrated in 0.9% NaCl, 0.02% EDTA, 0.02% NaN3, pH 7.4, and eluted at a rate of 0.3 ml/min. Starting 60 min after sample application, 1 ml fractions were collected. Fractions were analyzed for cholesterol content by enzymatic assay (Wako Diagnostics; Richmond, VA) and for LTIP content by ELISA (15). Alternatively, plasma (0.75 ml) was fractionated by chromatography on a 1 × 50 cm Bio-Gel A 5m (Bio-Rad; Hercules, CA) column equilibrated in 50 mM Tris-HCl, 150 mM NaCl, 0.02% EDTA, 0.02% NaN3, 0.5% BSA, pH 7.4. Fractions were assayed as above. Both gel filtration methods were performed at room temperature.
Density gradient ultracentrifugation
Plasma (1.5 ml) was adjusted to a density of 1.21 g/ml with solid NaBr and layered under 3.5 ml of density 1.063 g/ml NaBr salt solution. Samples were centrifuged (80,000 rpm) in a VTi80 rotor for 90 min at 4°C. Sequential fractions (0.5 ml) were collected from the top of the tube by pumping 1.47 g/ml NaBr saltsolution into the bottom of the tube. Density was determined by refractometry. Samples were dialyzed against 0.9% NaCl, 0.02% EDTA, 0.02% NaN3, pH 7.4 before measuring LTIP content by ELISA (15).
1-D gel electrophoresis
Samples were resolved on premade 1% agarose gels (Corning; Palo Alto, CA) following the manufacturer's instructions. Gels were dried, and lipid was detected by 0.025% Fat Red 7B in 60% methanol. Alternatively, after electrophoresis, resolved bands were transferred to polyvinylidene fluoride membranes by passive adsorption. Membranes were blocked (2 h) with 5% nonfat powdered milk, 2% calf serum, 1% BSA in 0.9% NaCl, 0.1% EDTA, pH 7.6, then reacted with rabbit anti-human LTIP (9, 15) or preimmune rabbit sera. Immune complexes were detected by reaction of blots with goat anti-rabbit IgG conjugated with HRP followed by treatment with ECL (Perkin Elmer; Boston, MA) (9).
Heparin-agarose chromatography
Plasma (0.5 ml) was applied to a 1 ml column of heparin-agarose (MP Biomedicals; Solon, OH) equilibrated in 50 mM Tris-HCl, 150 mM NaCl, 0.02% EDTA, 0.02% NaN3, pH 7.4. The column was eluted with the same buffer, and the three 0.5 ml fractions of highest protein content (OD280) were pooled. VLDL and LDL were quantitatively removed under these conditions. Pooled samples were assayed for LTIP mass by ELISA. To measure LTIP activity, fractions were first rendered lipoprotein free by dextran sulfate-manganese precipitation (19, 22), dialyzed extensively against 50 mM Tris-HCl, 150 mM NaCl, 0.02% EDTA, 0.02% NaN3, pH 7.4; then inhibitor activity was measured in a [3H]CE-LDL to HDL lipid transfer assay as previously described (18).
Western blot analysis
Samples were reduced and denatured as previously described (15) then applied to 7.5% or 4–20% polyacrylamide gels (Cambrex BioScience; Rockland, ME). Subsequently, proteins were electrotransferred to polyvinylidene fluoride membranes (23). Membranes were blocked with 5% dry milk and 1% calf serum in PBS, and then reacted with the indicated antibody in the same buffer. Bound primary antibodies were reacted with the appropriate HRP-conjugated secondary antibody (1/1000 dilution) (Calbiochem; San Diego, CA) and complexes were detected by chemiluminesence (ECL; Perkin Elmer) -mediated autoradiography. Rabbit anti-human LTIP was prepared as previously described (15). Other antibodies to human antigens included: goat anti-apolipoprotein A-I and goat anti-apolipoprotein C-I (Academy Biomedical Co. Inc.; Houston, TX), goat anti-apolipoprotein E (Calbiochem, San Diego, CA), rabbit anti-LCAT and rabbit anti-phospholipid transfer protein (PLTP) (Novus Biologicals; Littleton, CO), and goat anti-α2 macroglobulin (MP Biomedicals; Solon,OH).
Mass spectrometry
An FPLC fraction enriched in LTIP was precipitated with acetone and run on a Criterion 10–20% gradient SDS-PAGE gel (Bio-Rad). Bands were visualized by Coomassie blue staining, excised,destained, reduced/alkylated, and digested with trypsin (20 ng/ml in 50 mM ammonium bicarbonate) overnight at room temperature. Peptides were extracted with 50% acetonitrile, 5% formic acid for liquid chromatography-tandem mass spectrometry (LC-tandem MS) analysis.
Two microliter volumes of the extract were injected into a self-packed 10 cm × 75 μm id Phenomenex (Torrance, CA) Jupiter C18 reversed-phase capillary chromatography column interfaced with a ThermoFisher (Waltham, MA) LCQ-Deca ion trap mass spectrometer system with a Protana (Toronto, Canada) microelectrospray ion source. Peptides were eluted from the column by a linear acetonitrile/0.05 M acetic acid gradient at a flow rate of 0.2 μl/min. Peptide molecular weights and product ion spectra to determine amino acid sequence in successive instrument scans were obtained. The data were analyzed by comparison with the National Center for Biotechnology Information nonredundant database using the search program TurboSequest. All matching spectra were verified by manual interpretation. The interpretation process was also aided by additional searches using the programs Mascot and Fasta as needed.
Human plasma
Blood was drawn from study volunteers following written informed consent. The Institutional Review Board of the Cleveland Clinic approved the study protocol. Plasma was isolated from fasting whole blood collected in EDTA tubes. Samples were maintained at 4°C until use. Lipoprotein/lipid measurements on whole plasma were performed as previously described (24). Unless indicated, donors were normolipidemic.
RESULTS
Distribution of LTIP in normolipidemic plasma
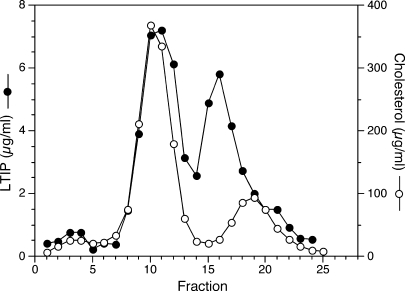
We previously observed that LTIP, as assessed by its activity, was exclusively associated with LDL in plasma fractionated by gel filtration (11). Similar results were obtained when antibodies prepared against synthetic peptide fragments of LTIP were used to detect LTIP in plasma samples fractionated by agarose electrophoresis (9). Because we recently obtained a polyclonal antibody against intact LTIP, we reexamined the plasma distribution to validate the previous results. To this end, plasma was fractionated by FPLC gel filtration chromatography using tandem Superose 6 columns and fractions were assayed for LTIP mass. Although we validated the association of LTIP with LDL, we were surprised to find that most LTIP is associated with a smaller-molecular-weight fraction (Fig. 1). This lower-molecular-weight form of LTIP elutes just before HDL, although these two peaks are not completely resolved by this method. This fraction is subsequently referred to as the 470 kDa fraction, because it has an apparent molecular weight of 468 ± 67 (n = 11). We have verified this bimodal distribution using traditional gel filtration media (Bio-Gel A 5m) (data not shown), as we used in the published studies cited above, thus indicating that these findings are not unique to the FPLC gel filtration matrix. The distribution of LTIP was not sensitive to changes in the ionic strength of the chromatography buffer in that the same profiles were obtained with buffers containing 0, 150, or 300 mM NaCl. Additionally, the presence of 0.5% BSA in the chromatography buffer was without effect on the LTIP elution pattern. Collectively, these results indicate that this novel bimodal elution pattern of LTIP is consistently observed under a variety of chromatographic conditions. Among normolipidemic subjects, the distribution of LTIP between the LDL and 470 kDa peaks was 24 ± 4% and 76 ± 4% (n= 4), respectively.
Fig. 1.
Distribution of lipid transfer inhibitor protein (LTIP) in plasma. Plasma (0.3 ml) was applied to tandem Superose 6 columns, and eluted fractions were assayed for LTIP mass and cholesterol. Fraction collection began 60 min after sample application. Inset: Western blot analysis of select column fractions and partially purified LTIP separated by 4–20% SDS-PAGE and reacted with anti-LTIP. Molecular mass standards are shown in kDa units. Data are representative of multiple experiments.
Properties of the 470 kDa LTIP complex
To characterize the 470 kDa LTIP complex, plasma was fractionated by ultracentrifugation. Either by single or sequential ultracentrifugation spins, the bulk of LTIP was recovered in the HDL-density range (1.063 < d < 1.21 g/ml) (Table 1). As assessed by FPLC gel filtration analysis, the 470 kDa component of LTIP was recovered in the HDL fraction, showing that it remained intact even after treatment with high concentrations of NaBr (>2 M) to adjust sample density and exposure to high centrifugal force. LTIP association with LDL was less stable under these conditions.Ultracentrifuge-derived LDL contained only 33.2 ± 12% (n = 6) of the LTIP found on FPLC-isolated LDL (data not shown). LTIP was not found in VLDL or in the lipoprotein-free (d > 1.21 g/ml) fraction under these conditions (Table 1).
TABLE 1.
Distribution of LTIP immunoreactivity in ultracentrifugal fractions of plasma
| Sample | Density Fraction | LTIP |
|---|---|---|
| g/ml | μg/fraction | |
| A | <1.019 | ND |
| >1.019 | 120.6 ± 36.9 | |
| B | <1.063 | 19.3 ± 6.4 |
| >1.063 | 93.9 ± 13.4 | |
| C | <1.21 | 92.3 ± 3.3 |
| >1.21 | ND | |
| D | <1.019 | ND |
| 1.019–1.063 | 19.9 ± 3.3 | |
| 1.063–1.21 | 97.3 ± 13.4 | |
| >1.21 | ND |
LTIP, lipid transfer inhibitor protein; ND, none detected. Plasma was fractionated by single-step or sequential ultracentrifugation at the densities indicated. Following dialysis, the LTIP content of each fraction was determined by ELISA. Mean ± SD.
The fractions isolated by sequential ultracentrifugation steps (sample D, Table 1) were further analyzed by agarose electrophoresis. As shown previously (9), LTIP isolated in the LDL-density range comigrated with LDL (Fig. 2). Likewise, although differing in size from HDL, the 470 kDa LTIP fraction isolated in the HDL-density range had the same electrophoretic mobility as bulk HDL.
Fig. 2.
Agarose gel electrophoresis. Plasma was fractionated by sequential density ultracentrifugation to yield the three major lipoprotein fractions and the d > 1.21 g/ml lipoprotein-free fraction. Samples were subsequently electrophoresed on 1% agarose gels. Lipoproteins were visualized by staining with Fat Red 7B. The samples on other gels were transferred to polyvinylidene fluoride and reacted with anti-LTIP or preimmune antisera as indicated.
To further characterize the 470 kDa LTIP fraction, plasma was subjected to density gradient ultracentrifugation. LTIP associated with LDL was recovered in the top of the gradient, and accounted for 23% of the applied LTIP. The 470 kDa LTIP complex floated at a mean density of 1.134 (n = 2), indicating that it has a density that lies at the low end of the HDL3 density range. Based on this density,the 470 kDa LTIP complex contains approximately 49% lipid and 51% protein.
Two approaches were taken to identifying possible protein components of the 470 kDa complex. In the first, plasma was fractionated by FPLC gel filtration, and then eluted fractions were analyzed for their protein content by Western blot (Fig. 3). Peak LTIP mass was measured in fraction 20. As expected, this peak preceded that of HDL (fraction 23), as identified by apolipoprotein A-I distribution, and by separate cholesterol analyses. Interestingly, LTIP-containing fractions overlapping the main apolipoprotein A-I peak contained both native LTIP (Mr = ∼26 kDa) and also an ∼19 kDa immunoreactive band. This smaller form of LTIP is consistent with the carbohydrate-free form of this protein (15). LCAT and PLTP, which have been previously reported to be at least partially bound to HDL (25, 26), eluted slightly later than LTIP. Apolipoprotein C-I, an inhibitor of CETP, had anelution profile that largely mirrored that of apolipoprotein A-I.
Fig. 3.
Apolipoprotein distribution in LTIP-containing fractions. Plasma (500 μl) was applied to tandem Superose 6 columns, and fractions were collected beginning 55 min after sample application. Apolipoprotein content was determined by Western blot. For apolipoprotein A-I, apolipoprotein C-I, and LTIP, samples were separated on 4–20% PAGE gels. For LCAT and phospholipid transfer protein (PLTP), 10% gels were used. Molecular mass standards are shown on the left-hand side of each blot, and are in kDa units. The HDL cholesterol peak was in fraction 23. HC, IgG heavy chain.
To further characterize the proteins contained in the 470 kDa LTIP fraction, this complex was purified from plasma (approximately 120-fold) by density ultracentrifugation under conditions that isolated HDL3 (1.125 < d < 1.21 g/ml). This fraction was then further purified by FPLC gel filtration (Fig. 4A). Fractions enriched in LTIPbut poor in HDL were pooled, and proteins were identified by LC-tandem MS. The LTIP/protein ratio of the pool was increased an additional ∼3-fold by the FPLC step. Multiple proteins were identified (Fig. 4B). Except for apolipoprotein A-I, most other candidate proteins were present in similar amounts, ranging 2–3-fold based on the intensity of Coomassie staining. Quantitative immunoabsorption of apolipoprotein A-I resulted in the complete removal of LTIP, showing the direct association of these two proteins (data not shown). All identified proteins, except for albumin, have been previously identified as components of HDL. Collectively, the compositional and physical characteristics of the 470 kDa complex identify it as a unique HDL subclass.
Fig. 4.
Characterization of 470 kDa LTIP isolated from HDL3. A:HDL3 (8.8 mg protein) was isolated from plasma by sequential ultracentrifugation and applied to tandem Superose 6 columns. Eluted fractions were assayed for LTIP mass and cholesterol. B:Fractions containing high LTIP and low cholesterol, equivalent to fractions 14–15 in Panel A, were pooled, and the protein constituents were fractionated by SDS-PAGE. Excised bands were digested with trypsin, and the resultant peptides were identified by liquid chromatography-tandem mass spectrometry as described in the Methods. Molecular mass standards are shown in kDa units. The 33 kDa band also contained apolipoprotein E, PON1, and apolipoprotein A-I. PON1, serum paraoxonase 1; SAA4, serum amyloid A4. Lane A = molecular mass standards; lane B = 470kDa LTIP fraction.
Functional activity of the 470 kDa LTIP complex
We previously observed that LTIP inhibitory activity could only be detected in gel filtration fractions coeluting with LDL (11). However, we now show that most LTIP mass resides in a 470 kDa complex. This implies that LTIP in the 470 kDa complex may be inactive. To evaluate this, plasma was applied to heparin-agarose under conditions in which LDL was quantitatively retained and the 470 kDa LTIP fraction was recovered in the unbound fraction containing HDL. LTIP mass and LTIP activity were measured for the starting and unbound fractions, and calculated for the bound fraction. As expected, about 75% of the LTIP mass was recovered in the unbound fraction; however, a much smaller portion of LTIP activity was recovered in this fraction. Among six experiments, the specific activity of LTIP in the 470 kDa fraction averaged 21-fold less than that for LTIP in LDL. However, for these experiments, values ranged from 2.6- to 85-fold. This large variability was almost entirely due to differences in the activity of the 470 kDa fraction. This suggests that the delipidation step, which is necessary before samples can be assayed for LTIP activity, may partially and variably activate LTIP in the 470 kDa fraction. To avoid this potential complication, an alternate method was taken.
As illustrated above, the 470 kDa LTIP complex is partially separated from HDL3 by FPLC (Fig. 4A). To further examine the activity of the 470 kDa complex, we pooled samples from the FPLC fractionation to yield HDL-containing fractions with low (pool A) and high (pool B) 470 kDa LTIP content. CETP inhibitory activity was then measured directly by quantifying CETP activity in assays where the pooled samples served as the acceptor in a lipid transfer assay. Despite containing 75 times more LTIP, pool B displayed negligible inhibitory activity compared with pool A (Table 2). This low CETP inhibitory activity was not due to the inability of these assays to respond to LTIP, because exogenous LTIP elicited readily measurable inhibition, which was essentially the same regardless of the HDL pool assayed. Thus, the modest inhibitor activity found for the 470 kDa LTIP complex in the heparin-agarose studies above is not seen when this fraction is assayed directly. These studies show that LTIP contained in the 470 kDa complex is inactive.
TABLE 2.
Activity in the 470 kDa LTIP Fraction
| Sample | LTIP | LTIP Activity | Specific Activity | Inhibition by Exogenous LTIP |
|---|---|---|---|---|
| μg/assay | % inhibition | % inhibition/μg LTIP | % inhibition | |
| Pool A | 0.08 | 0 | — | 48.3 ± 0.6 |
| Pool B | 6.1 | 7.7 ± 8.6 | 1.2 | 40.2 ± 2.6 |
| LTIP | 0.91 | 45.4 ± 0.8 | 49.9 | — |
HDL3 was separated by Superose 6 chromatography, and HDL fractions essentially free of LTIP (Pool A) or those containing the 470 kDa component of LTIP (Pool B) were combined. Pools were assayed for LTIP activity in a [3H]CE-LDL (5 μg cholesterol) to HDL (10 μg cholesterol) transfer assay. The HDL contained in Pools A and B served as the acceptor in the lipid transfer assay. Additional purified LTIP was added to these assays to verify the capacity of these HDL fractions to support LTIP activity. The inhibitor activity of LTIP itself was determined in similar assays but with total, unfractionated HDL as the acceptor. Results are the mean ± SD (n = 2) and are representative of five experiments using fractions isolated from separate HDL3 preparations. Specific activity = % inhibition/μg LTIP.
Factors influencing the association of LTIP with the 470 kDa complex
In normolipidemic plasma, we observed that approximately one-quarter of LTIP is associated with LDL and the remaining is associated with the 470 kDa complex. However, we previously observed in hypercholesterolemic subjects that LTIP activity is increased 3-fold over control, but this increase was associated with a comparatively smaller LTIP mass change, as estimated by immunoblot (27). Also, among hypercholesterolemic subjects, elevated LTIP activity consistently exceeds the rise that would be predicted from LTIP mass changes (27). These data suggest that in hypercholesterolemic subjects, the distribution of LTIP between active and inactive pools may be altered, favoring the activated (LDL-associated) state. To investigate this, hypercholesterolemic plasma was fractionated by FPLC and the distribution of LTIP was determined. As Fig. 5 illustrates, in hypercholesterolemic plasma, approximately 50% of LTIP is associated with LDL. This distribution was typical of the three subjects studied. The total LTIP concentration in these subjects averaged only 12% above control, indicating that the percentage shift seen in the FPLC profiles reflects an absolute increase in LDL-associated LTIP and a net decrease in LTIP in the 470 kDa complex.
Fig. 5.
LTIP distribution in hypercholesterolemic plasma. Plasma (500 μl) was applied to tandem Superose 6 columns, and fractions were collected 60 min after sample application. LTIP mass, as determined by ELISA, and cholesterol were measured as described in the Methods. The hypercholesterolemic plasma sample contained 368 mg/dl cholesterol (LDL 271 mg/dl, HDL 78 mg/dl), and 97 mg/dl triglyceride. This LTIP profile was typical of that seen in three hypercholesterolemic subjects.
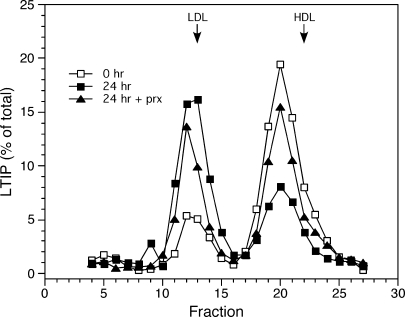
To examine whether this altered distribution of LTIP reflects the expanded LDL pool in hypercholesterolemic plasma, we supplemented normolipidemic plasma with purified LDL. In four of five experiments, doubling the plasma LDL content, which approximated the LDL content in hypercholesterolemic plasma, did not induce a shift of LTIP from the 470 kDa fraction to LDL. This suggests that the distribution of LTIP between the active and inactive pools is primarily controlled by factors other than LDL pool size. One possibility is lipoprotein chemical composition, because this is commonly aberrant in hyperlipidemic settings. To investigate this possibility, normolipidemic plasma was incubated overnight to permit LCAT, PLTP, CETP, and other activities in plasma to modify lipoprotein composition. Under these conditions, the LTIP distribution in plasma shifted dramatically toward LDL (Fig. 6). In this plasma, LDL-associated LTIP accounted for ∼20% of LTIP at time zero, but 61% after incubation at 37°C for 24 h. This shift in LTIP distribution was time dependent (data not shown). Blocking LCAT activity with paraoxon reduced the shift in LTIP by half, resulting in only 40% of LTIP being associated with LDL after incubation (Fig.6). This shift in LTIP association also required the presence of LDL. This was demonstrated in studies with VLDL-LDL-depleted plasma (heparin-agarose unbound fraction). Overnight 37°C incubation of this 470 kDa-containing fraction did not change the elution profile of LTIP.
Fig. 6.
Incubation induced dissociation of LTIP from the inactive complex. Aliquots of normolipidemic plasma were incubated at 37°C for 0 or 24 h in the absence or presence of 1 mM paraoxon (prx), as indicated, then applied (500 μl) to tandem Superose 6 columns. LTIP mass and cholesterol of column fractions were measured. LDL and HDL designations at the top of the figure indicate the elution peaks for these lipoproteins. Data are representative of more than five experiments.
The incubation-induced transfer of LTIP to LDL resulted in a reduced ability of LDL to participate in CETP-mediated lipid transfer reactions, showing that this LTIP is metabolically active (Table 3). From these data, a linear, inverse association between the LTIP content of LOL and CETP transfer activity was observed (r = −0.99). Collectively, these studies show that the association of LTIP with the inactive pool is dependent on the chemical status of plasma lipoproteins. Whether the loss of LTIP from the inactive complex and gain in the active LDL complex is driven by changes in LDL composition or 470 kDa composition, or both, remains to be defined.
TABLE 3.
Increased LTIP content of LDL reduces its reactivity with CETP
| Incubation Time | Plasma LTIP | CETP Activity | CETP Activity |
|---|---|---|---|
| % on LDL | % transfer | % control | |
| 0 h | 19.6 | 9.58 ± 0.43 | 100 |
| 24 h + prx | 40.0 | 7.41 ± 0.27 | 77.4 |
| 24 h | 60.8 | 5.47 ± 0.22 | 57.1 |
CETP, cholesteryl ester transfer protein. Fresh plasma was incubated at 37°C for the indicated times in the absence or presence of 1 mM paraoxon (prx) as indicated. The fraction of plasma LTIP associated with LDL was determined from the data shown in Fig. 6. Endogenous CETP activity on LDL was measured by adding [3H]CE-LDL (10 μg cholesterol) to incubated plasma samples (50 μl) and further incubating 4.5 h at 37°C. Subsequently, the extent of [3H]CE transfer from LDL to HDL was determined by precipitating apolipoprotein B-containing lipoproteins with manganese chloride in the presence of phosphate buffer (21), then quantifying the 3H content of the supernatant. CETP activities are the mean ± SD (n = 5).
DISCUSSION
In this study, we report the novel finding that a major portion of LTIP (apolipoprotein F) resides in a 470 kDa complex that has an electrophoretic mobility indistinguishable from HDL. LTIP in this complex was determined to be functionally inactive. The 470 kDa complex is larger than HDL2 in size, but isolates in the HDL3 fraction by density ultracentrifugation with a mean density of 1.134. This density translates into a particle composition of 49% lipid and 51% protein. Thus, the 470 kDa complex breaks from the commonly observed pattern for lipoproteins, in which particle density and size are inversely related (28). Mass spectrometry analysis of a fraction enriched in the 470 kDa complex revealed the presence of multiple proteins. While the stoichiometry of the complex remains to be determined, it is reasonable to assume that the very high levels of apolipoprotein A-I found in the enriched 470 kDa fraction partly reflect contamination from HDL3, because lipoproteins containing more than three to four copies of this protein have not been reported. Assuming four copies of apolipoprotein A-I, this protein could account for about 40% of the protein mass of the 470 kDa complex. This leaves room for multiple copies of several other proteins, including LTIP.
This bimodal distribution of LTIP mass in plasma differs from our earlier report that LTIP mass was found only on LDL (9). These previous studies used antibodies prepared against LTIP peptides, whereas the current study utilized antibodies prepared against whole LTIP. The most likely explanation for these discrepant results is that the LTIP epitopes recognized by the anti-peptide antibodies are not accessible in the 470 kDa complex under the near-native conditions of agarose electrophoresis used previously. Even under denaturing conditions, we have shown that these two antibody preparations react uniquely with LTIP, perhaps due to the variable carbohydrate content of plasma LTIP (15).
Sequestration of proteins into an inactive complex appears to be a common mechanism for regulation of lipoprotein metabolism. Several examples are LCAT, PLTP, and hepatic lipase. Krimbou et al. (25) reported that 40% of LCAT is bound to α(2) macroglobulin in an 18.5 nm-diameter particle. LCAT in this complex is inactive. Further, LCAT associated with α(2) macroglobulin is bound, internalized, and degraded by cells expressing the LDL receptor-related protein receptor, suggesting that this complex may also function to mediate clearance of plasma LCAT. Based on its large size, this LCAT complex is clearly distinct from the 470 kDa LTIP complex. Also, Oka et al. (26) have shown that about 70% of PLTP is contained in a 520 kDa inactive complex. Given the resolution of the gel filtration method used here, we cannot exclude the possibility that the 520 kDa PLTP complex and the 470 kDa LTIP complex are the same. However, this appears unlikely, because the PLTP complex is not stable under the high-salt and shear stress of ultracentrifugation (26), unlike the 470 kDa LTIP complex, and the distributions of PLTP and LTIP among gel filtration fractions overlap but are not the same (Fig. 3). Finally, hepatic lipase activity is regulated by its binding to HDL. Tighter binding of hepatic lipase to apolipoprotein A-II containing HDL particles renders it inactive (29).
A unique feature of LTIP activity regulation is the influence of lipoprotein status on the 470 kDa inactive complex. We observed that hypercholesterolemic subjects have an unusual distribution of LTIP between the LDL and 470 kDa forms, and that this cannot be replicated in vitro by supplementing normal plasma with sufficient LDL to match the levels present in hypercholesterolemic subjects. Further, 24 h incubation of normolipidemic plasma in vitro caused a marked redistribution of LTIP from the 470 kDa inactive to the active LDL fraction. Together, these data strongly suggest that lipoprotein composition influences the distribution of LTIP between these two fractions. These data are consistent with the hypothesis that LTIP is regulated by its reversible association with an inactive complex, and that this association is driven by metabolic events in plasma.
We have previously shown that LDL is the preferred substrate for LTIP, but LTIP is also capable of suppressing CETP activity on HDL in vitro. The hierarchy of CETP inhibition by LTIP with different lipoprotein donors is LDL > HDL2 >> HDL3 (30). Because LTIP activity is dependent on its binding to the lipoprotein surface (10), this implies that LTIP must be bound to these lipoproteins to some extent. The current data do not address LTIP binding to HDL, because the 470 kDa peak is not adequately resolved from the HDL subfractions by FPLC. Alternative methodologies will be required to verify the existence of these minor complexes, and to determine whether there is enhanced association of LTIP with other lipoproteins, especially HDL2, when LTIP redistribution is induced by 37°C incubation.
In summary, we have shown that LTIP exists in plasma associated with LDL, and in an inactive complex that is slightly larger than HDL2. The finding of inactive LTIP in the HDL fraction resolves a long-standing discrepancy between data showing LTIP activity only in the LDL fraction (9) and other studies showing that LTIP (apolipoprotein F) protein is primarily found in the HDL fraction (31, 32). Our data further clarify that LTIP present in the HDL fraction is associated with a specific lipoprotein subset that is distinct from the major HDL subfractions. The distribution of LTIP between these two pools appears to be dynamic, and can be modulated by changes in lipoprotein composition, such as those induced by 37°C incubation. Undoubtedly, during such incubations there are multiple proteins affecting lipoprotein remodeling. Although we present evidence that LCAT activity may promote a portion of the incubation-induced redistribution, further study is required to understand the factors that define the distribution of LTIP between its plasma forms. An obvious candidate in the LTIP redistribution process is the TG enrichment of LDL and HDL facilitated by CETP during the incubation. In this view, LTIP activity is being regulated by the actions of CETP, the protein that it, in turn, regulates. Higher LTIP activity results in reduced participation of LDL in lipid transfer events. One possible benefit of reducing the participation of LDL in lipid transfer events during times of elevated TG is that it provides for maximum CE flux from HDL to VLDL. This would have the effect of stimulating reverse cholesterol transport, because in humans, a major portion of LCAT-derived CE is transferred from HDL to other lipoproteins before being cleared from plasma, and VLDL has the shortest plasma lifetime of the possible CE recipients (33).
Abbreviations
CE, cholesteryl ester
CETP, cholesteryl ester transfer protein
FPLC, fast-protein liquid chromatography
LC-tandem MS, liquid chromatography-tandem mass spectrometry
LTIP, lipid transfer inhibitor protein
PLTP, phospholipid transfer protein
TG, triglyceride
Published, JLR Papers in Press, March 27, 2008.
Footnotes
This research was supported in part by Grant HL-60934 from the National Heart, Lung, and Blood Institute, National Institutes of Health.
References
- 1.Morton R. E. 1990. Interaction of lipid transfer protein with plasma lipoproteins and cell membranes. Experientia. 46 552–560. [DOI] [PubMed] [Google Scholar]
- 2.Tall A. 1995. Plasma lipid transfer proteins. Annu. Rev. Biochem. 64 235–257. [DOI] [PubMed] [Google Scholar]
- 3.Morton R. E. 1999. Cholesteryl ester transfer protein and its plasma regulator: lipid transfer inhibitor protein. Curr. Opin. Lipidol. 10 321–327. [DOI] [PubMed] [Google Scholar]
- 4.Klerkx A. H. E. M., K. E. Harchaoui, W. A. van der Steeg, S. M. Boekholdt, E. S. G. Stroes, J. J. P. Kastelein, and J. A. Kuivenhoven. 2006. Cholesteryl ester transfer protein (CETP) inhibition beyond raising high-density lipoprotein cholesterol levels: pathways by which modulation of CETP activity may alter atherogenesis. Arterioscler. Thromb. Vasc. Biol. 26 706–715. [DOI] [PubMed] [Google Scholar]
- 5.Barter P. J., H. B. Brewer, Jr., M. J. Chapman, C. H. Hennekens, D. J. Rader, and A. R. Tall. 2003. Cholesteryl ester transfer protein: a novel target for raising HDL and inhibiting atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 23 160–167. [DOI] [PubMed] [Google Scholar]
- 6.Gautier T., D. Masson, A. Athias, P. Gambert, D. Aunis, M. H. Metz-Boutigue, and L. Lagrost. 2000. Human apolipoprotein C-I accounts for the ability of plasma high density lipoproteins to inhibit the cholesteryl ester transfer protein activity. J. Biol. Chem. 275 37504–37509. [DOI] [PubMed] [Google Scholar]
- 7.Gautier T., D. Masson, M. C. Jong, L. Duverneuil, N. Le Guern, V.Deckert, J. P. Pais de Barros, L. Dumont, A. Bataille, Z. Zak, et al. 2002. Apolipoprotein CI deficiency markedly augments plasma lipoprotein changes mediated by human cholesteryl ester transfer protein (CETP) in CETP transgenic/ApoCI-knocked out mice. J. Biol. Chem. 277 31354–31363. [DOI] [PubMed] [Google Scholar]
- 8.Dumont L., T. Gautier, J. P. de Barros, H. Laplanche, D. Blache, P.Ducoroy, J. Fruchart, J. C. Fruchart, P. Gambert, D. Masson, et al. 2005. Molecular mechanism of the blockade of plasma cholesteryl ester transfer protein by its physiological inhibitor apolipoprotein CI. J. Biol. Chem. 280 38108–38116. [DOI] [PubMed] [Google Scholar]
- 9.Wang X., D. M. Driscoll, and R. E. Morton. 1999. Molecular cloning and expression of lipid transfer inhibitor protein reveals its identity with apolipoprotein F. J. Biol. Chem. 274 1814–1820. [DOI] [PubMed] [Google Scholar]
- 10.Morton R. E. 1985. Binding of plasma-derived lipid transfer protein to lipoprotein substrates. The role of binding in the lipid transfer process. J. Biol. Chem. 260 12593–12599. [PubMed] [Google Scholar]
- 11.Morton R. E., and D. J. Greene. 1994. Regulation of lipid transfer between lipoproteins by an endogenous plasma protein: selective inhibition among lipoprotein classes. J. Lipid Res. 35 836–847. [PubMed] [Google Scholar]
- 12.Mann C. J., F. T. Yen, A. M. Grant, and B. E. Bihain. 1991. Mechanism of plasma cholesteryl ester transfer in hypertriglyceridemia. J.Clin. Invest. 88 2059–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murakami T., S. Michelagnoli, R. Longhi, G. Gianfranceschi, F. Pazzucconi, L. Calabresi, C. R. Sirtori, and G. Franceschini. 1995. Triglycerides are major determinants of cholesterol esterification/transfer and HDL remodeling in human plasma. Arterioscler. Thromb. Vasc. Biol. 15 1819–1828. [DOI] [PubMed] [Google Scholar]
- 14.Serdyuk A. P., and R. E. Morton. 1999. Lipid transfer inhibitor protein defines the participation of lipoproteins in lipid transfer reactions: CETP has no preference for cholesteryl esters in HDL versus LDL. Arterioscler. Thromb. Vasc. Biol. 19 718–726. [DOI] [PubMed] [Google Scholar]
- 15.Morton R. E., H. M. Gnizak, D. J. Greene, K-H. Cho, and V. M. Paromov. 2008. Lipid transfer inhibitor protein (apolipoprotein F) concentration in normolipidemic and hyperlipidemic subjects. J. Lipid Res. 49 127–135. [DOI] [PubMed] [Google Scholar]
- 16.Vaisar T., S. Pennathur, P. S. Green, S. A. Gharib, A. N. Hoofnagle, M. C. Cheung, J. Byun, S. Vuletic, S. Kassim, P. Singh, et al. 2007. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J. Clin. Invest. 117 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Havel R. J., H. A. Eder, and J. H. Bragdon. 1955. The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J. Clin. Invest. 34 1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morton R. E., and D. B. Zilversmit. 1981. A plasma inhibitor of triglyceride and cholesteryl ester transfer activities. J. Biol. Chem. 256 11992–11995. [PubMed] [Google Scholar]
- 19.Morton R. E., and D. B. Zilversmit. 1982. Purification and characterization of lipid transfer protein(s) from human lipoprotein-deficient plasma. J. Lipid Res. 23 1058–1067. [PubMed] [Google Scholar]
- 20.Pattnaik N. M., A. Montes, L. B. Hughes, and D. B. Zilversmit. 1978. Cholesteryl ester exchange protein in human plasma: isolation and characterization. Biochim. Biophys. Acta. 530 428–438. [DOI] [PubMed] [Google Scholar]
- 21.Morton R. E., and D. B. Zilversmit. 1981. The separation of apolipoprotein D from cholesteryl ester transfer protein. Biochim. Biophys. Acta. 663 350–355. [DOI] [PubMed] [Google Scholar]
- 22.Burstein M., H. R. Scholnick, and R. Morfin. 1970. Rapid method for the isolation of lipoproteins from human serum by precipitation with polyanions. J. Lipid Res. 11 583–595. [PubMed] [Google Scholar]
- 23.Towbin H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA. 76 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myers G. L., G. R. Cooper, C. L. Winn, and S. J. Smith. 1989. The Centers for Disease Control-National Heart, Lung and Blood Institute Lipid Standardization Program. An approach to accurate and precise lipid measurements. Clin. Lab. Med. 9 105–135. [PubMed] [Google Scholar]
- 25.Krimbou L., M. Marcil, J. Davignon, and J. Genest, Jr. 2001. Interaction of lecithin:cholesterol acyltransferase (LCAT).alpha 2-macroglobulin complex with low density lipoprotein receptor-related protein (LRP). Evidence for an alpha 2-macroglobulin/LRP receptor-mediated system participating in LCAT clearance. J. Biol. Chem. 276 33241–33248. [DOI] [PubMed] [Google Scholar]
- 26.Oka T., T. Kujiraoka, M. Ito, T. Egashira, S. Takahashi, M. N. Nanjee, N. E. Miller, J. Metso, V. M. Olkkonen, C. Ehnholm, et al. 2000. Distribution of phospholipid transfer protein in human plasma: presence of two forms of phospholipid transfer protein, one catalytically active and the other inactive. J. Lipid Res. 41 1651–1657. [PubMed] [Google Scholar]
- 27.Morton R. E., V. Nunes, L. Izem, and E. Quintão. 2001. Markedly elevated lipid transfer inhibitor protein in hypercholesterolemic subjects is mitigated by plasma triglyceride levels. Arterioscler. Thromb. Vasc. Biol. 21 1642–1649. [DOI] [PubMed] [Google Scholar]
- 28.Shen B. W., A. M. Scanu, and F. J. Kézdy. 1977. Structure of human serum lipoproteins inferred from compositional analysis. Proc. Natl. Acad. Sci. USA. 74 837–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boucher J., T. A. Ramsamy, S. Braschi, D. Sahoo, T. A. M. Neville, and D. L. Sparks. 2004. Apolipoprotein A-II regulates HDL stability and affects hepatic lipase association and activity. J. Lipid Res. 45 849–858. [DOI] [PubMed] [Google Scholar]
- 30.Paromov V. M., and R. E. Morton. 2003. Lipid transfer inhibitor protein defines the participation of high density lipoprotein subfractions in lipid transfer reactions mediated by cholesteryl ester transfer protein (CETP). J. Biol. Chem. 278 40859–40866. [DOI] [PubMed] [Google Scholar]
- 31.Olofsson S. O., W. J. McConathy, and P. Alaupovic. 1978. Isolation and partial characterization of a new acidic apolipoprotein (apolipoprotein F) from high density lipoproteins of human plasma. Biochemistry. 17 1032–1036. [DOI] [PubMed] [Google Scholar]
- 32.Day J. R., J. J. Albers, T. L. Gilbert, T. E. Whitmore, W. J. McConathy, and G. Wolfbauer. 1994. Purification and molecular cloning of human apolipoprotein F. Biochem. Biophys. Res. Commun. 203 1146–1151. [DOI] [PubMed] [Google Scholar]
- 33.Barter P. J., G. J. Hopkins, and G. D. Calvert. 1982. Transfers and exchanges of esterified cholesterol between plasma lipoproteins. Biochem. J. 208 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]