Abstract
Lipin-1 deficiency in the mouse causes generalized lipodystrophy, characterized by impaired adipose tissue development and insulin resistance. Lipin-1 expression in differentiating preadipocytes is required for normal expression of adipogenic transcription factors, including peroxisome proliferator-activated receptor γ and CCAAT enhancer binding protein α, and for the synthesis of triacylglycerol. The requirement of lipin-1 for adipocyte differentiation can be explained, in part, by its activity as the sole adipocyte phosphatidic acid phosphatase-1 enzyme, which converts phosphatidate to diacylglycerol, the immediate precursor of triacylglycerol. Here we identify glucocorticoids as the stimulus for the induction of lipin-1 expression in differentiating adipocytes, and characterize a glucocorticoid response element (GRE) in the Lpin1 promoter. The Lpin1 GRE binds to the glucocorticoid receptor and leads to transcriptional activation in adipocytes and hepatocytes, as demonstrated by reporter gene transcription, electrophoretic mobility shift, and chromatin immunoprecipitation assays. This represents the first gene regulatory element identified to directly influence lipin-1 expression levels, and may modulate lipin-1 mRNA levels in adipose tissue and liver in conditions associated with increased local glucocorticoid concentrations in vivo, such as obesity and fasting.
Keywords: adipocyte differentiation, triacylglycerol, phosphatidate phosphatase
Lipin-1 was originally identified as the defective gene product that causes lipodystrophy in the fatty liver dystrophy (fld) mouse (1). Lipodystrophy in these animals is characterized by a lack of normal adipose tissue throughout the body and insulin resistance (2). Lipin-1 was subsequently identified as a phosphatidic acid phosphatase-1 (PAP1) enzyme, which catalyzes the conversion of phosphatidate to diacylglycerol, the immediate precursor of triacylglycerol and of zwitterionic phospholipids (3). The PAP1 activity is conferred by the active site motif DxDxT, which is present in the conserved C-terminal domain of the three mammalian lipins (lipin-1, lipin-2, and lipin-3) as well as all known lipin homologs in invertebrates and yeast. White and brown adipose tissue from fld mice completely lack PAP1 enzyme activity, indicating that lipin-1 is the sole PAP1 enzyme in adipose tissue, consistent with the failure of adipocytes in fld mice to accumulate triglyceride (4).
Variations in the lipin-1 mRNA levels in adipose tissue are associated with key metabolic traits, including adiposity, insulin sensitivity, energy expenditure, and gene expression. Thus, whereas lipin-1 deficiency produces lipodystrophy, enhanced lipin-1 expression in mouse models promotes obesity. Transgenic mice expressing constitutively elevated lipin-1 levels specifically in mature adipocytes exhibit increased triglyceride content per cell and increased expression of lipogenic genes (5). Interestingly, despite their increased adiposity, these adipose tissue-specific lipin-1 transgenic mice have increased insulin sensitivity compared with wild-type mice. A similar relationship between lipin-1 levels in adipose tissue and insulin sensitivity has been documented in several studies in humans (6–9). Although the mechanism is not known, possible contributions may be effects of lipin-1 on glucose uptake and oxidative gene expression in adipose tissue, and enhanced triglyceride storage in adipose tissue as protection against triglyceride accumulation in tissues such as muscle (4, 9, 10). It has long been known that PAP1 activity in liver is increased in diabetes, hypoxia, and in response to elevated glucocorticoid levels (11–14). In adipocytes, lipin-1 expression is enhanced by antidiabetic compounds of the thiazolidinedione (TZD) family and by harmine, both in vitro and in vivo (9, 15). The molecular mechanisms by which these compounds and conditions modulate lipin-1 expression and/or PAP1 activity in liver and adipose tissue have not been elucidated.
Lipin-1 is expressed at two stages during adipocyte differentiation, as assessed with the 3T3-L1 adipocyte cell line (16). Adipocyte differentiation occurs through an ordered cascade of gene expression changes (17, 18). The nuclear receptor peroxisome proliferator-activated receptor γ (PPARγ) is considered to be the key adipogenic transcriptional regulator. It is induced very early in the differentiation process and promotes the expression of numerous target genes involved in adipogenesis and lipid accumulation, such as those encoding CCAAT enhancer binding protein α, adipocyte fatty acid binding protein aP2, and lipoprotein lipase (19–21). Lipin-1 is initially expressed transiently between 10–20 h after induction of differentiation and then returns to baseline levels. Lipin-1 is then induced again at 2 days after initiation of differentiation, reaching peak levels in mature, lipid-loaded adipocytes. This biphasic pattern suggests that lipin-1 has a role in establishing the adipogenic gene expression program during early stages of differentiation, as well as a role in triglyceride accumulation in mature adipocytes. The latter function is likely to be directly attributable to lipin-1 PAP1 activity (4), but the function of lipin-1 in the early stages of adipocyte differentiation has not been elucidated.
Lipin-1 can localize to the nucleus or to the cytoplasm (1, 22). In mature adipocytes, but not in preadipocytes, the subcellular localization differs for two lipin-1 isoforms that are generated by alternative mRNA splicing (22). Lipin-1A and lipin-1B isoforms differ by the presence of an additional 33 amino acids in the lipin-1B isoform (22, 23). In mature adipocytes, lipin-1A localizes predominantly to the nucleus, whereas lipin-1B is largely cytoplasmic (22). The significance of the distinct localization of the two isoforms is not clear, but may influence lipin function. Since PAP1 enzyme activity is known to reside in the cytosol and to translocate to the endoplasmic reticulum in response to elevated fatty acid levels (24), cytoplasmic lipin-1 is likely to be involved primarily in glycerolipid synthesis. Nuclear lipin-1 could, however, serve a different function. In hepatocytes, Finck et al. (25) identified lipin-1 as a transcriptional coactivator working in conjunction with PPARγ coactivator-1α and PPARα to regulate hepatic gene expression during fasting.
As described above, increased lipin-1 expression in adipose tissue of both mouse and human is associated with enhanced insulin sensitivity. It is therefore important to identify factors that regulate lipin-1 expression and function in adipocytes. Here we characterize a molecular mechanism for the induction of lipin-1 expression during adipogenesis by a glucocorticoid receptor (GR)-dependent process. These findings suggest that lipin-1 may be a mediator of glucocorticoid effects in conditions such as fasting and obesity, and that genetic variations in this response could contribute to previously observed interindividual variations in lipin-1 expression levels.
MATERIALS AND METHODS
Animals
C57BL/6J and ob/ob mice were purchased from Jackson Laboratories (Bar Harbor, ME). For fasting and re-feeding experiments, C57BL/6J mice were fasted for 16 h, or fasted 16 h and re-fed for 4 h with standard rodent chow for fasted and re-fed samples, respectively. Diet-induced obesity was attained by feeding C57BL/6J mice a high-fat diet (35% fat and 33% carbohydrate; Diet F3282, Bio-Serve, Frenchtown, NJ) for 16 weeks beginning at 8 weeks of age. All animal studies were performed under approved institutional protocols and according to guidelines established in the “Guide for the Care and Use of Laboratory Animals.”
Cell culture
Murine 3T3-L1 and Hepa 1-6 cell lines were obtained from American Type Culture Collection (Manassas, VA). 3T3-L1 preadipocytes were grown in DMEM supplemented with 10% calf serum, glutamine, sodium pyruvate, penicillin (100 U/ml), and streptomycin (100 μg/ml). Forty-eight hours after reaching confluence, cells were treated with a differentiation cocktail containing 0.25 μM dexamethasone (DEX), 1 μg/ml insulin, 0.5 mM 3-isobutyl-l-methylxanthine (MIX), and 10% FBS. After 48 h, the differentiation cocktail was replaced with DMEM containing 10% FBS and insulin. Adipocyte morphology was monitored by the appearance of cytoplasmic lipid droplets, which was closely correlated with the acquisition of mature adipocyte markers (26). Hepa 1-6 cells were propagated in DMEM plus 10% FBS, nonessential amino acids, sodium pyruvate, penicillin (100 U/ml), and streptomycin (100 μg/ml).
Cryopreserved subcutaneous human primary preadipocytes from female donors with a normal body mass index (22.24 kg/m2) were purchased from Zen-Bio, Inc. (Research Triangle Park, NC). Cells were maintained in Preadipocyte Medium (Zen-Bio, Inc.) and differentiated at passage 3 by treating confluent cells with Adipocyte Differentiation Medium (Zen-Bio, Inc.) for 7 days. Immediately prior to experiments with glucocorticoid treatment, cells were cultured overnight in DMEM containing 1% FBS.
Quantitative RT-PCR
Real-time PCR quantitation of gene expression was performed as we have reported previously (4, 5, 15, 16, 22). Briefly, total RNA was isolated from cultured cells with TRIzol reagent, and 2 μg of total RNA was reverse transcribed in a 20 μl reaction volume with Omniscript RT kit and oligo(dT) primers (Invitrogen, Carlsbad, CA). Real-time PCR was performed on the iCycler iQ Real-time Detection System (Bio-Rad; Hercules, CA) using SYBR Green PCR Quanti-Tect reagent kit (Qiagen; Valencia, CA). Threshold cycle numbers (Ct) were determined with Sequence Detector software (version 1.6; Applied Biosystems, Foster City, CA) and transformed using the ΔCt method as described by the manufacturer. Expression levels were normalized to the endogenous controls, TATA box binding protein, 18S rRNA, and hypoxanthine-guanine phosphoribosyltransferase. Results were expressed as fold induction or repression by normalizing to the control condition. To amplify splice variants of lipin-1 using a single PCR primer set (mlipin-1 f/r primers; see supplementary Table I), RNA samples obtained at various times after the addition of DEX were analyzed by semi-quantitative RT-PCR as described (22). Primers used for real-time PCR are provided in supplementary Table I.
PAP1 activity measurements
3T3-L1 adipocytes were harvested in 0.25 M sucrose containing 2 mM dithiothreitol, Phosphatase Inhibitor Cocktails I and II (Sigma Aldrich; St. Louis, MO), protease inhibitor cocktail (EDTA-free; Roche Diagnostics, Indianapolis, IN), and 0.15% Tween-20, and homogenized using a probe sonicator for 3–5 s. The protein concentration of lysates was determined using the bicinchoninic acid protein assay (Pierce Biotechnology; Rockford, IL). Analysis of PAP1 activity was performed in the presence and absence of N-ethylmaleimide with 2–8 μg of cell protein in a volume of 0.1 ml as described (4). Total activities for the N-ethylmaleimide-inhibitable PAP1 were calculated from measurements at three different protein concentrations to ensure the proportionality of the assay.
Western blot analysis
Cell lysates were prepared as for PAP1 measurements, and protein samples (30 μg per lane) were separated by NuPAGE Novex Tris acetate 3–8% gradient gel system (Invitrogen) and transferred to nitrocellulose filters. The blots were incubated with anti-lipin-1 antibody (4), followed by goat-anti-rabbit IgG (Santa Cruz Biotechnology, Inc.; Santa Cruz, CA), and developed with ECL Plus Western blotting detection system (Amersham Biosciences; Piscataway, NJ) and quantified by densitometry using Quantity One-4.5.2 software (Bio-Rad).
mRNA start site and construction of luciferase reporter constructs
The transcription initiation site was determined by 5′-rapid amplification of cDNA ends (RACE) from 3T3-L1 adipocytes with a SMART RACE cDNA Amplification kit (Clontech Laboratories, Inc., Mountain View, CA). One microgram total RNA from 3T3-L1 adipocytes 4 days postdifferentiation was converted into cDNA for RACE reactions. The resulting PCR product was gel purified and subcloned into pCR2.1TOPO-TA cloning vector (Invitrogen). An identical nucleotide sequence corresponding to the 5′ end of lipin-1 cDNA was obtained by sequence analysis of three independent clones.
Using the Expand High Fidelity PLUS PCR System (Roche Diagnostics), lipin-1 promoter constructs were generated by PCR amplification of appropriate DNA fragments from C57BL/6 mouse genomic DNA and ligated into the promoterless luciferase expression vector pGL3-basic (Promega; Madison, WI). Forward (F1–F7) and reverse (R1) gene-specific primers were engineered with the 5′ KpnI and 3′ BglII sites, respectively, for directional cloning into the KpnI/BglII sites of pGL3-basic. Primer sequences are provided in supplementary Table II.
Site-directed mutagenesis
Site-directed mutagenesis was performed using the QuickChange site-directed mutagenesis kit (Stratagene; La Jolla, CA) according the manufacturer's protocol, with the following oligonucleotide (boldface letters indicate GR binding regions, and lowercase letters show mutated bases): 5′ AGGGGAAGAACTAATTGccccCTGACATAAGGTCAAAA 3′ and its reverse complement. The plasmid Lip-823-Luc was used as a template to generate the Lip-823-Luc-GRE mutant.
Transient transfection and reporter assays
3T3-L1 and Hepa 1-6 cells were transfected with Effectene Transfection Reagent (Qiagen). Briefly, 80,000 cells were transfected with 150 ng of reporter plasmid, 80 ng phRL-TK Renilla control vector (Promega), and 100 ng rat GR expression vector pSG5-GR (27). Twenty-four hours after transfection, cells were treated with 1 μM DEX or the solvent DMSO in DMEM containing 1% FBS. Eighteen hours later, cells were washed in 1× PBS and lysed in Passive Lysis Buffer (Promega). Luciferase assays were performed with the Dual-Luciferase Reporter Assay System (Promega) following the protocol provided by the manufacturer. Firefly luciferase activity was normalized to Renilla luciferase activity to account for differences in transfection efficiency. In each experiment, samples were analyzed in quadruplicate, and each experiment was repeated at least twice.
Electrophoretic mobility shift assay
Double-stranded oligonucleotides corresponding to the native lipin-1 glucocorticoid response element (GRE) (5′ GGAAGAACTAATTGTAAACTGACA 3′) or mutated lipin-1 GRE (5′ GGAAGAACTAATTGccccCTGACA 3′, mutated bases shown in lowercase letters) were labeled using the 3′-biotin end-label kit (Pierce Biotechnology). An electrophoretic mobility shift assay (EMSA) was performed as described (27) by using the Lightshift kit from Pierce Biotechnology according to the manufacturer's protocol. Briefly, binding reactions containing 25 or 150 ng of purified human GR protein (Protein One; Bethesda, MD), 10 mM Tris, 50 mM KCl, 1 mM DTT, 10% glycerol, 5 mM MgCl2, 0.05% Nonidet P-40, and 2 pmol of oligonucleotide probe were incubated for 20 min at 4°C. Specific binding was confirmed by using a 50- to 250-fold excess of unlabeled probe as specific competitor. Protein DNA complexes were separated on a 4% nondenaturing acrylamide gel at 20 mA in 0.5× tris-borate-EDTA. Complexes were transferred to positively charged nylon membranes and ultraviolet cross-linked. Gel shifts were visualized with streptavidin-HRP followed by chemiluminescent detection.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were performed as described (28). Briefly, confluent 3T3-L1 cells were incubated with vehicle (DMSO) or DEX for 2 h. Chromatin was cross-linked with 1% formaldehyde for 10 min at 25°C. The reaction was stopped with glycine (125 mM final concentration) for 5 min and cells were harvested. Aliquots were removed and saved as the input control prior to precipitation with anti-GR or control IgG antibodies. Precipitated DNA was amplified by semi-quantitative PCR with primers for the Lpin1 upstream promoter region (−632/−169) and for Glut4 exon 10 (see supplementary Table I).
RESULTS
Glucocorticoid induces lipin-1 gene transcription during adipocyte differentiation
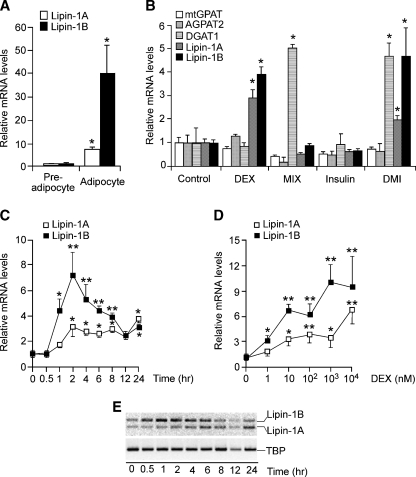
Lipin-1A and -B expression levels increased dramatically during differentiation of 3T3-L1 preadipocytes to mature adipocytes (Fig. 1A). This process occurred in response to a cocktail containing DEX, insulin, and MIX, indicating that one or more of these components is capable of inducing Lpin1 expression. To determine which component(s) of the differentiation cocktail induced lipin-1 expression, 2 day postconfluent 3T3-L1 preadipocytes were treated for 4 h with individual components or with the complete cocktail. As shown in Fig. 1B, both lipin-1 isoforms were significantly induced by DEX and were not affected by the other components. In contrast, DEX did not induce genes encoding other enzymes upstream or downstream of lipin-1 in the triglyceride biosynthetic pathway, including mitochondrial glycerol 3-phosphate acyltransferase (mtGPAT), acylglycerolphosphate acyltransferase 2 (AGPAT2), and diacylglycerol acyltransferase 1 (DGAT1). In contrast to lipin-1, DGAT1 was induced by MIX, whereas neither GPAT nor AGPAT2 was increased after a 4 h treatment with any of the components (Fig. 1B).
Fig. 1.
Glucocorticoid induces lipin-1 gene expression during adipocyte differentiation. A: Lipin-1 is dramatically induced in 3T3-L1 cells in response to adipocyte differentiation cocktail. Confluent 3T3-L1 cells (preadipocytes) were induced to differentiate with cocktail containing insulin, 3-isobutyl-l-methylxanthine (MIX), and dexamethasone (DEX) for 6 days (adipocytes). mRNA levels were determined by real-time RT-PCR and normalized to TATA box binding protein (TBP) mRNA levels. Values are expressed as the fold change compared with confluent cells (mean ± SD). B: Effects of differentiation cocktail components on expression of enzymes of triacylglycerol synthesis. Confluent 3T3-L1 preadipocytes were treated for 4 h with 10% FBS (control), DEX, MIX, insulin, or complete cocktail (DMI). mRNA levels for several enzymes in triacylglycerol biosynthesis were determined by real-time RT-PCR and expressed as fold difference compared with FBS-treated cells (mean ± SD). mtGPAT, mitochondrial glycerol-3-phosphate acyltransferase; AGPAT2, acylglycerolphosphate acyltransferase 2; DGAT1, diacylglycerol acyltransferase 1. C: Time course of DEX induction of lipin-1A and lipin-1B mRNA expression. Confluent 3T3-L1 preadipocytes were treated with DEX for the times indicated, and mRNA levels were expressed as fold difference compared with time 0 (mean ± SD). D: DEX induces lipin-1A and lipin-1B mRNA expression in a dose-dependent manner. Confluent 3T3-L1 preadipocytes were incubated with the indicated concentration of DEX in DMEM containing 10% FBS for 4 h, and mRNA levels were determined by real-time RT-PCR. E: Relative lipin-1A and -B isoform mRNA expression levels during DEX time course were determined by quantitative RT-PCR with primers that amplify both isoforms simultaneously. Products were resolved by agarose gel electrophoresis. Time of DEX treatment is indicated at bottom. TATA box binding protein was amplified as a control. For all studies in A–D, n = 3; * P < 0.05, ** P < 0.01 versus corresponding controls.
A time course study revealed that in response to 1 μM DEX treatment, lipin-1B mRNA levels increased as early as 1 h, and lipin-1A levels after 2 h (Fig. 1C). Lipin-1B levels peaked with a 7-fold induction at 2 h, and gradually declined afterward. Lipin-1A levels increased to a maximum of 3-fold above baseline and remained elevated near this level throughout 24 h. Visualization of the lipin-1A and -1B isoforms using PCR primers that amplify both forms simultaneously confirmed that lipin-1B is increased to a greater extent than lipin-1A between 1 and 8 h of treatment, suggesting that DEX may also influence alternative splicing of the lipin-1 mRNA (Fig. 1E). To evaluate the dose response to DEX, the cells were treated with DEX at concentrations ranging from 1 nM to 10 μM for 4 h. One nanomole/liter DEX induced expression of both lipin-1 isoforms, and levels generally increased as DEX concentration increased (Fig. 1D). These results indicate that DEX rapidly induces lipin-1 gene expression in a dose-dependent manner.
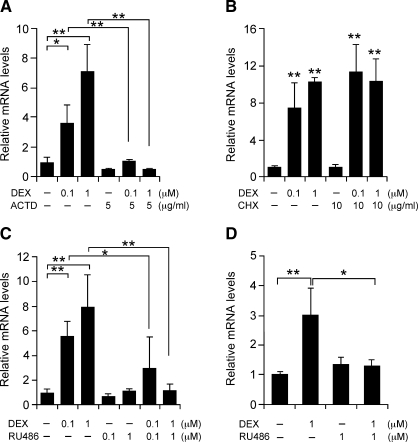
We next investigated whether the induction of lipin-1 mRNA expression by DEX is dependent on gene transcription and/or de novo protein synthesis. Two-day postconfluent 3T3-L1 cells were incubated with DEX in the presence or absence of actinomycin D, a transcription inhibitor, or cycloheximide (CHX), a protein synthesis inhibitor. Actinomycin D completely abolished the stimulatory effect of DEX on expression of both lipin-1 isoforms, indicating that DEX acts at the level of lipin-1 gene transcription (Fig. 2A and supplementary Figure IA for lipin-1B and lipin-1A, respectively). In contrast, incubation with CHX did not alter the DEX induction of lipin-1 mRNA expression, indicating that protein synthesis is not required (Fig. 2B and supplementary Figure IB). To determine whether regulation of lipin-1 mRNA expression by DEX is mediated by the GR, we studied the effects of RU486, a noncompetitive GR antagonist. One micromole/liter RU486 completely reversed the effects of DEX on lipin-1A and lipin-1B mRNA expression (Fig. 2C and supplementary Figure IC). These results demonstrate that the lipin-1 induction by DEX is dependent on the GR.
Fig. 2.
Lipin-1 regulation by glucocorticoid occurs at the transcriptional level. Results in panels A–C are for lipin-1B; the analogous experiments for lipin-1A are presented in supplementary Figure I, panels A–C. A: Induction of lipin-1B expression by DEX is inhibited by the RNA synthesis inhibitor actinomycin D (ACTD). Confluent 3T3-L1 preadipocytes were treated with the indicated concentration of DEX in the absence or presence of ACTD for 4 h. B: Induction of lipin-1B was not inhibited by the protein synthesis inhibitor cycloheximide (CHX). Confluent 3T3-L1 preadipocytes were treated with the indicated concentration of DEX in the absence or presence of CHX for 4 h. C: Induction of lipin-1B was inhibited by the glucocorticoid receptor (GR) antagonist RU486. Confluent 3T3-L1 preadipocytes were treated with the indicated concentration of DEX in the absence or presence of RU486 for 4 h. D: Human primary preadipocytes were treated with 1 μM DEX in the absence or presence of 1 μM RU486 for 4 h and exhibit the same response of lipin-1 gene expression as observed in 3T3-L1 cells. The analogous experiment in mature human primary adipocytes is presented in supplementary Figure ID. For all studies, values represent the mean ± SD for three samples. * P < 0.05, **P < 0.01 versus controls or for comparisons indicated.
To rule out the possibility that the results described above were limited to the mouse 3T3-L1 cell line, we performed similar studies in primary human preadipocytes and adipocytes. Total lipin-1 mRNA expression was significantly induced by DEX in human preadipocytes and differentiated adipocytes (Fig. 2D and supplementary Figure ID, respectively). Furthermore, RU486 completely reversed the effects of DEX, confirming the requirement for the GR. These results establish that lipin-1 gene expression is regulated in a GR-dependent manner in both mouse and human adipocytes.
Glucocorticoid-induced Lpin1 regulation leads to increased protein and PAP1 activity
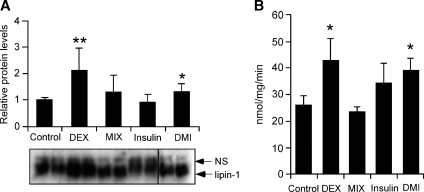
We next investigated whether induction of lipin-1 mRNA expression by DEX led to parallel increases in lipin-1 protein expression and PAP1 activity in adipocytes. Mature 3T3-L1 adipocytes (4 days postdifferentiation) were incubated with DEX, MIX, or insulin for 8 h. As observed previously in rat adipose tissue (23), lipin-1 in 3T3-L1 adipocyte extracts was detected as a heterogeneous band that migrates at ∼140 kDa (Fig. 3A). The heterogeneity has been attributed to multiple phosphorylation sites on the lipin-1 protein (23, 29). Consistent with its effects on lipin-1 mRNA levels, DEX treatment increased lipin-1 protein levels 2-fold, compared with treatment with control culture medium containing only FBS (Fig. 3A). Moreover, DEX significantly increased PAP1 activity by 70% (Fig. 3B). Neither insulin nor MIX affected lipin-1 protein or PAP1 activity levels, consistent with the previous findings that insulin and epinephrine do not affect lipin-1 PAP1-specific activity (29). However, insulin and MIX did alter lipin-1 electrophoretic mobility (Fig. 3A). As reported previously for rat adipose tissue (23), insulin treatment caused a proportion of the protein to shift to slower mobility, indicative of increased phosphorylation. In contrast, protein isolated from MIX-treated cells had increased mobility, suggestive of reduced phosphorylation. The MIX effect appeared to be dominant over the insulin effect, as indicated by the rapid mobility of lipin-1 protein bands in samples treated with the complete differentiation cocktail. These results establish that induction of lipin-1 transcription by DEX is reflected in increased lipin-1 protein synthesis and PAP1 activity.
Fig. 3.
Glucocorticoid-induced lipin-1 mRNA expression leads to increased lipin-1 protein and phosphatidic acid phosphatase-1 (PAP1) activity. 3T3-L1 adipocytes (differentiated for 8 days) were incubated in 1% FBS overnight, then treated for 8 h with DEX, MIX, insulin, or DMI mixture. A: Cell lysates were analyzed for lipin-1 protein levels by Western blotting with rabbit polyclonal anti-lipin-1 antibody and bands quantitated (n = 8). A representative gel is shown at bottom with duplicate samples from each treatment. Note that the lanes labeled DMI are from the same gel, but are shown as a separate box because of removal of irrelevant lanes in the figure. A nonspecific band is indicated (NS). B: N-ethylmaleimide-inhibitable PAP1 activity in cell lysates from samples in A. PAP1 activity was normalized to total cell protein. n = 6; * P < 0.05, ** P < 0.01 versus control treatment. Values presented represent mean ± SD.
DEX activation of the Lpin1 promoter is mediated by a GRE motif
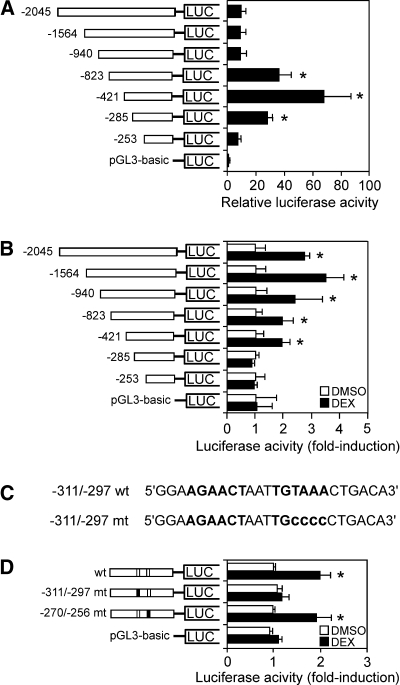
We hypothesized that the DEX induction of lipin-1 gene transcription occurs through a GRE in the Lpin1 5′ flanking sequence. We first verified the position of the lipin-1 transcription initiation site in 3T3-L1 cells using 5′-RACE (30). This confirmed the initiation site at Chr12:16615246, which was very close to the major site reported previously (Chr12:16615250) based on sequencing of lipin-1 cDNA and expressed sequence tag clones (1, 31). Sequence analysis revealed several putative GREs between −940 and −253 relative to the Lpin1 transcription start site. To evaluate these putative regulatory elements, we cloned a Lipin1 promoter fragment encompassing sequences from −2,045 to +70 into a luciferase reporter plasmid. From this parent construct, a series of 5′ deletions of the region were also generated.
To characterize Lpin1 promoter activity, the reporter constructs carrying deletions of the gene 5′-flanking region were transfected into 3T3-L1 preadipocytes, in combination with a plasmid expressing recombinant GR. Luciferase activity was measured 36 h after transfection. As shown in Fig. 4A, the inclusion of Lpin1 5′-flanking sequence from −2,045 to −253 bp conferred activity several-fold above that of the pGL3 vector sequences. There was a 4-fold increase in luciferase activity upon deletion of sequences between −940 and −823, and a further 2-fold increase upon deletion to −421, suggesting the presence of negative regulatory elements in these regions. Subsequent deletion to −285 led to a reduction in luciferase activity, probably due to loss of elements that enhance expression.
Fig. 4.
Dexamethasone activation of the Lpin1 promoter is mediated by a glucocorticoid response element (GRE) motif. A: Transcriptional activity of Lpin1 promoter-luciferase constructs under basal conditions (10% FBS). 3T3-L1 preadipocytes were cotransfected with Lpin1 promoter luciferase constructs, renilla-luciferase control vector, and rat GR expression vector (pSG5-GR). Lpin1 promoter activity was normalized to renilla luciferase activity. Data represent the mean ± SE of four samples, expressed as the ratio of the Lpin1 promoter segment to that of the pGL3-basic vector. B: Effect of 1 μM DEX on the activity of the Lpin1-luciferase constructs in 3T3-L1 cells. Luciferase activity was measured 24 h after treatment with vehicle (DMSO) or DEX. Data are shown as fold difference (mean ± SE) between DEX and the vehicle-treated groups. n = 4; * P < 0.05 versus vehicle treated. C: Sequence of the Lpin1 promoter region containing the GRE (−311 to −297). The wild-type (wt) putative GRE motif and the mutant (mt) GRE (−311/−297) used in subsequent studies are in boldface. D: Effect of 1 μM DEX on Lpin1 promoter-luciferase constructs containing wild-type and mutant versions of the GRE. n = 4; * P < 0.05 versus vehicle-treated.
To identify functional GREs, cells transfected with the reporter constructs were treated with DEX or vehicle (DMSO), and relative expression levels were determined. Sequences between −2,045 and −421 conferred DEX responsiveness, whereas deletion of more-proximal sequences abolished this response. This was true in both 3T3-L1 (Fig. 4B) and Hepa 1-6 hepatoma cells (see supplementary Figure IIA), consistent with the previous report that glucocorticoids upregulate lipin-1 expression in mouse liver (25). These results suggest that the region between −421 and −285 is sufficient to transduce the glucocorticoid effect in adipocyte and hepatocyte cell lines.
Sequence analysis of the Lpin1 −421/−285 region revealed the presence of an imperfect palindromic 15 mer sequence characteristic of GREs located at −311 to −297 (Fig. 4C). Although not a perfect match, this putative GRE has high similarity to the functional GRE of the 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase (PFK2) gene (22). To evaluate function of this element, mutations were generated within the putative GRE at −311/−297, and in a negative control sequence located at −270/−256 that had some similarities to the putative GRE (Fig. 4D). As expected, the wild-type sequence promoted a 2-fold induction in luciferase activity in response to DEX. Mutation of the GRE residing at −311/−270 abolished DEX responsiveness in 3T3-L1 cells. Mutation of the irrelevant sequences at −270/−256 did not alter the DEX response (Fig. 4D). An identical effect was conferred by the GRE in Hepa 1-6 cells (see supplementary Figure IIB).
The Lpin1 promoter directly binds the GR in a hormone-dependent manner
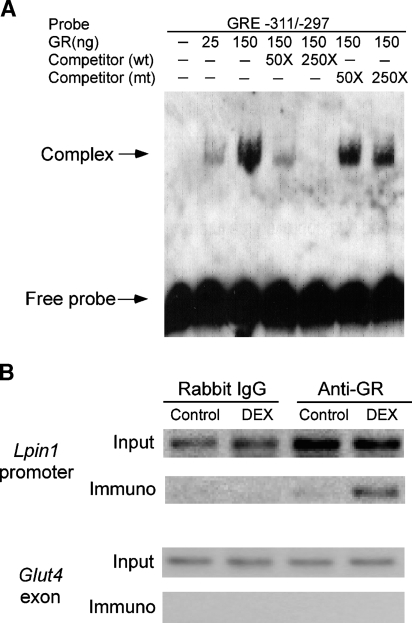
To determine whether GR binds directly to the lipin-1 GRE, we employed an EMSA. Double-stranded oligonucleotides corresponding to the native or mutant lipin-1 GRE labeled at the 3′ end with biotin were incubated with purified GR, and analyzed by nondenaturing electrophoresis and chemiluminescent detection. As shown in Fig. 5A, GR bound specifically to the biotinylated lipin-1 GRE. Unlabeled wild-type oligonucleotide effectively competed for binding in a dose-dependent manner, whereas mutant oligonucleotide did not block binding. These results establish that GR protein binds to the lipin-1 GRE in a sequence-specific manner.
Fig. 5.
The lipin-1 GRE binds GR. A: Analysis of lipin-1 GRE binding to GR by electrophoretic mobility shift assay (EMSA). An EMSA was performed using purified human GR and a biotinylated oligonucleotide probe containing the lipin-1 GRE and surrounding sequences (−311 to −297; see Fig. 4C). Competition assays were performed by preincubating the reaction for 25 min with 50- or 250-fold molar excess of unlabeled oligonucleotides containing wild-type (wt) or mutant (mt) GRE sequences. Arrows indicate position of unbound oligonucleotide probe and probe complexed with GR. Results are representative of three independent experiments. B: Chromatin immunoprecipitation (ChIP) assay to detect the binding of GR to the Lpin1 promoter in chromatin from 3T3-L1 adipocytes. Cells were treated with vehicle (DMSO) or DEX for 4 h, and association of the GR with Lpin1 promoter sequences (−632/−169) was detected by immunoprecipitation with anti-GR antibody followed by PCR amplification of Lpin1 promoter sequences. Input lanes show presence of Lpin1 sequence before immunoprecipitation in all samples. Rabbit IgG was used as a negative control for specificity of the immunoprecipitation, and an exon of the Glut4 gene was used as a negative control for the PCR amplification.
To determine whether GR binds to the native Lpin1 promoter in intact adipocytes, we performed ChIP assays using anti-GR antibodies and chromatin isolated from mature 3T3-L1 adipocytes. GR was found to associate poorly with the Lpin1 promoter under basal conditions, but was enhanced substantially in response to DEX treatment (Fig. 5B). In contrast, no binding was observed when experiments were performed with a nonspecific antibody (rabbit IgG), or when binding to an unrelated sequence (an exon within the Glut4 gene) was ascertained.
Adipose tissue lipin-1 expression is increased in conditions associated with increased local glucocorticoid concentrations in vivo
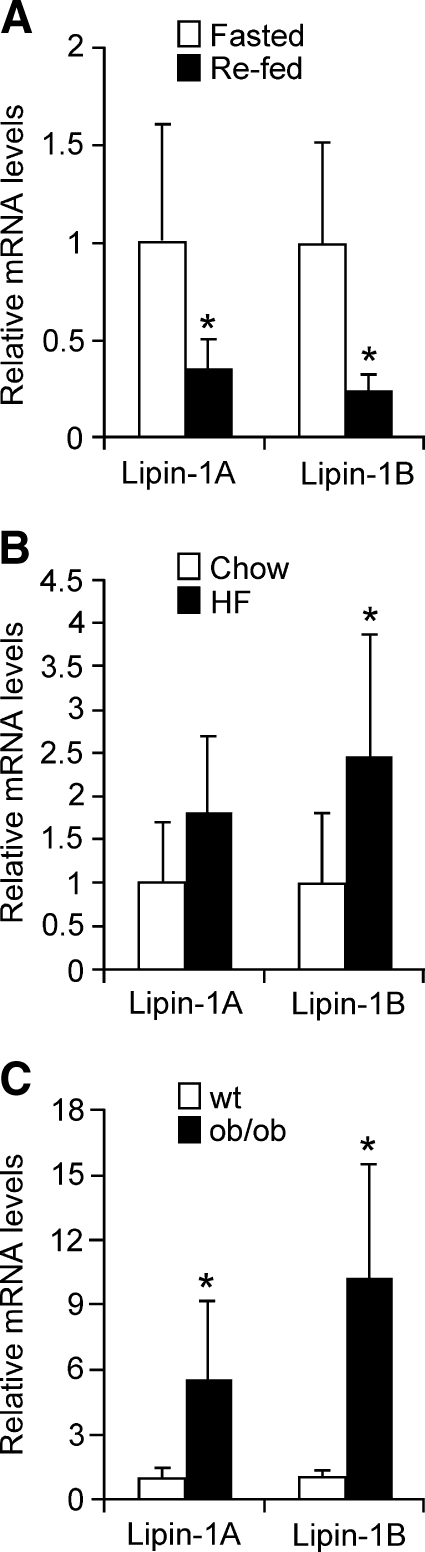
As demonstrated here, Lpin1 transcription is induced by glucocorticoids during adipocyte differentiation, and acts through binding of the GR to specific promoter sequences. This suggests that physiological conditions characterized by enhanced glucocorticoid levels should lead to increased lipin-1 mRNA levels in vivo. To test this, we quantitated mouse adipose tissue lipin-1 mRNA levels in the fasted condition, which is known to increase circulating glucocorticoid levels, and in obesity, which is characterized by increased local glucocorticoid levels due to the action of 11β-hydroxysteroid dehydrogenase 1 (32). Compared with re-fed mice, lipin-1A and -1B expression in white adipose tissue of fasted C57BL/6J mice was increased by approximately 3-fold (Fig. 6A). Lipin-1 expression was also increased in adipose tissue of mice made obese by genetic manipulation in the leptin-deficient ob/ob mouse (Fig. 6B), and in diet-induced obesity (Fig. 6C).
Fig. 6.
Adipose tissue lipin-1 expression is increased in conditions associated with increased local glucocorticoid levels. A: Lipin-1 mRNA levels in epididymal adipose tissue from C57BL/6J mice after a 16 h fast (fasted) or after a 16 h fast plus a 4 h re-feeding (re-fed). mRNA levels were determined by real-time RT-PCR and normalized to TBP mRNA levels. n = 6; * P < 0.05 versus fasted samples. B: Lipin-1 levels in epididymal adipose tissue from C57BL/6J mice fed a chow or high-fat diet for 16 weeks. n = 6; * P < 0.05 high-fat diet versus chow diet. C: Lipin-1 levels in epididymal adipose tissue of ob/ob and wild-type (wt) littermates. The value of wt was set to 1, and levels in ob/ob mice were expressed relative to wt. n = 5; * P < 0.05. Data are shown as mean ± SEM.
DISCUSSION
Adipose tissue plays an important role in metabolic homeostasis, inasmuch as both too little and too much adipose tissue can promote insulin resistance and hyperlipidemia (33, 34). Lipodystrophy in the lipin-1-deficient mouse provides compelling evidence for the critical role of lipin-1 in adipocyte differentiation and maturation. There is also substantial evidence that lipin-1 levels in human adipose tissue are correlated with insulin sensitivity, energy expenditure, and expression of lipid metabolism genes (6–9). These observations provided the rationale for our investigation of Lpin1 gene regulation. We determined that the synthetic glucocorticoid DEX induces lipin-1 mRNA, protein expression, and PAP1 activity in a GR-dependent manner in mouse adipocytes. The effects of DEX on lipin-1 expression were also observed in primary human adipocytes and mouse hepatocytes, demonstrating that glucocorticoids are a general regulatory stimulus for lipin-1 expression. We further identified a functional GRE in the Lpin1 upstream sequence that confers DEX induction, and demonstrated that inactivation of this element abolishes the response.
Our findings provide a mechanism for numerous observations made prior to isolation of the lipin-1 gene regarding the effect of glucocorticoids on PAP1 activity in vivo. More than 20 years ago, Pittner, Fears, and Brindley (13) reported that DEX increases the activity of PAP1 in rat hepatocytes. Recently, glucocorticoid administration was shown to strongly induce lipin-1 gene expression in mouse liver (25), and this effect is regulated at the mRNA level (35). In addition, lipin-1 exhibits a circadian pattern of expression in liver and adipose tissue of mice, and the rhythmicity of lipin-1 mRNA expression is lost in adrenal ectomized mice, suggesting control by glucocorticoids (36). Also, in rats, the maximum PAP1 activity in the liver occurs about 4 h after the diurnal peak in corticosterone concentrations (37). The ethanol-induced increase in PAP1 in the livers of rats is also strongly attenuated by adrenalectomy, implicating glucocorticoids in observed regulation in response to ethanol (12).
Glucocorticoids are major stimulators of adipose tissue development and fat accumulation, especially in combination with insulin (38). Our conclusion that lipin-1 expression is increased by glucocorticoids in adipose tissue in obesity is compatible with this and with the observed increase in PAP1 activity in adipose tissue in obesity associated with hyperinsulinemia (39). The finding that lipin-1 expression is also increased in adipose tissue of fasted mice is expected from the glucocorticoid induction, although this is probably not related to adipogenesis. Increased lipin-1 levels could be a mechanism to prevent an accumulation of unesterified fatty acids in adipocytes as a consequence of high rates of lipolysis. Indeed, there is precedence for a futile cycle acting in adipocytes under conditions such as fasting, in which the products of triglyceride hydrolysis can be recycled into triglyceride within the cell (40). Another possible function of the lipin-1 induction in adipose tissue during fasting is in the regulation of genes involved in fatty acid oxidation. Lipin-1 exhibits a strong positive correlation with PPARα and medium-chain acylCoA dehydrogenase expression levels in human adipose tissue (6). This relationship was also seen in lipin-1 transgenic adipose tissue, suggesting that the increase in fatty acid oxidation gene expression is a direct or indirect consequence of the increased lipin-1 levels (6). Thus, it is possible that lipin-1 plays a role in the activation of fatty acid oxidation through PPARα in adipose tissue analogous to that observed in liver (25). Increased fatty acid oxidation would provide an alternative mode of energy production to glucose oxidation in fasting as well as some protection for the adipocyte against fatty acid accumulation.
It has recently been demonstrated that lipin-1 gene expression is induced in adipose tissue in vivo by two classes of compounds with antidiabetic effects, the TZD class of drugs, and the small molecule harmine. Thus, adipose tissue samples from individuals treated with the TZD pioglitazone exhibited a 2-fold induction of lipin-1B, whereas no effect on lipin-1 expression levels occurred in subjects treated with the non-TZD anti-diabetic, metformin (9). A similar induction of lipin-1B expression by TZD occurred in 3T3-L1 adipocytes, suggesting that the induction was a direct rather than a secondary effect, and that TZD may also promote splicing of the lipin-1B isoform. Interestingly, TZDs activate GR nuclear translocation (41), raising the possibility that the observed effect of TZDs on lipin-1B expression may occur through the same mechanism as the glucocorticoid induction shown here. Notably, whereas DEX increased both lipin-1A and -1B mRNA levels, there appeared to be greater magnitude of induction of the lipin-1B isoform, similar to that observed with TZD treatment. This suggests that glucocorticoids may influence both Lpin1 transcription and isoform-specific mRNA splicing in adipocytes. Lipin-1A and -1B mRNA levels are induced by harmine treatment both in vitro and in vivo, but the molecular mechanism remains to be determined (15). Harmine also stimulates adipocyte differentiation, and it is likely that the induction of lipin-1 expression by harmine contributes to both the expression of adipogenic genes and the triglyceride accumulation that occurs with harmine treatment.
In conclusion, we have identified a molecular mechanism for lipin-1 regulation by glucocorticoids during adipocyte differentiation. This represents the first gene regulatory element identified to directly influence lipin-1 levels, and may determine the glucocorticoid-induced increase in PAP1 activity that occurs in liver or adipose tissue following fasting and ethanol consumption and during the circadian cycle. These results raise the possibility that variations in glucocorticoid levels, or genetic polymorphisms in the GRE, might contribute to differences in lipin-1 expression levels, which have been associated with insulin sensitivity and adiposity in human subjects (6, 7, 9, 42).
Supplementary Material
Acknowledgments
The authors thank Jay Dewald and Darryl Kong for excellent technical assistance, Dr. Laurent Vergnes for RNA samples from fasted/re-fed mice, Dr. Keith Yamamoto for the gift of plasmid pSG5-GR, and Dr. Miklós Péterfy for valuable discussions.
Abbreviations
AGPAT2, acylglycerolphosphate acyltransferase 2
ChIP, chromatin immunoprecipitation
CHX, cycloheximide
DEX, dexamethasone
DGAT1, diacylglycerol acyltransferase 1
EMSA, electrophoretic mobility shift assay
GPAT, glycerol 3-phosphate acyltransferase
GR, glucocorticoid receptor
GRE, glucocorticoid response element
MIX, 3-isobutyl-l-methylxanthine
PAP1, phosphatidic acid phosphatase-1
PPARγ, peroxisome proliferator-activated receptor γ
RACE, rapid amplification of cDNA ends
TZD, thiazolidinedione
Published, JLR Papers in Press, March 24, 2008.
Footnotes
These studies were supported by National Institutes of Health Grant HL-28481 (K.R.) and by the Canadian Institutes of Health Research (MOP 81137) and the Canadian Heart and Stroke Foundation (D.N.B.).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two tables and two figures.
References
- 1.Péterfy M., J. Phan, P. Xu, and K. Reue. 2001. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat. Genet. 27 121–124. [DOI] [PubMed] [Google Scholar]
- 2.Reue K., P. Xu, X. P. Wang, and B. G. Slavin. 2000. Adipose tissue deficiency, glucose intolerance, and increased atherosclerosis result from mutation in the mouse fatty liver dystrophy (fld) gene. J. Lipid Res. 41 1067–1076. [PubMed] [Google Scholar]
- 3.Han G. S., W. I. Wu, and G. M. Carman. 2006. The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281 9210–9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donkor J., M. Sariahmetoglu, J. Dewald, D. N. Brindley, and K. Reue. 2007. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J. Biol. Chem. 282 3450–3457. [DOI] [PubMed] [Google Scholar]
- 5.Phan J., and K. Reue. 2005. Lipin, a lipodystrophy and obesity gene. Cell Metab. 1 73–83. [DOI] [PubMed] [Google Scholar]
- 6.Donkor J., L. M. Sparks, H. Xie, S. R. Smith, and K. Reue. 2008. Adipose tissue lipin-1 expression is correlated with PPARalpha gene expression and insulin sensitivity in healthy young men. J. Clin. Endocrinol. Metab. 93 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miranda M., M. R. Chacon, J. Gomez, A. Megia, V. Ceperuelo-Mallafre, S. Veloso, M. Saumoy, L. Gallart, C. Richart, J. M. Fernandez-Real, et al. 2007. Human subcutaneous adipose tissue LPIN1 expression in obesity, type 2 diabetes mellitus, and human immunodeficiency virus-associated lipodystrophy syndrome. Metabolism. 56 1518–1526. [DOI] [PubMed] [Google Scholar]
- 8.Suviolahti E., K. Reue, R. M. Cantor, J. Phan, M. Gentile, J. Naukkarinen, A. Soro-Paavonen, L. Oksanen, J. Kaprio, A. Rissanen, et al. 2006. Cross-species analyses implicate Lipin 1 involvement in human glucose metabolism. Hum. Mol. Genet. 15 377–386. [DOI] [PubMed] [Google Scholar]
- 9.Yao-Borengasser A., N. Rasouli, V. Varma, L. M. Miles, B. Phanavanh, T. N. Starks, J. Phan, H. J. Spencer III, R. E. McGehee, Jr., K. Reue, et al. 2006. Lipin expression is attenuated in adipose tissue of insulin-resistant human subjects and increases with peroxisome proliferator-activated receptor gamma activation. Diabetes. 55 2811–2818. [DOI] [PubMed] [Google Scholar]
- 10.van Harmelen V., M. Ryden, E. Sjolin, and J. Hoffstedt. 2007. A role of lipin in human obesity and insulin resistance: relation to adiopcyte glucose transport and GLUT4 expression. J. Lipid Res. 48 201–206. [DOI] [PubMed] [Google Scholar]
- 11.Brindley, D. N. 1988. Phosphatidate phosphohydrolase activity in the liver. In Phosphatidate Phosphohydrolase: Its Role in Glycerolipid Synthesis. CRC Press, Inc., Boca Raton. 21–77.
- 12.Brindley D. N., J. Cooling, S. L. Burditt, P. H. Pritchard, S. Pawson, and R. G. Sturton. 1979. The involvement of glucocorticoids in regulating the activity of phosphatidate phosphohydrolase and the synthesis of triacylglycerols in the liver. Effects of feeding rats with glucose, sorbitol, fructose, glycerol and ethanol. Biochem. J. 180 195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pittner R. A., R. Fears, and D. N. Brindley. 1985. Effects of cyclic AMP, glucocorticoids and insulin on the activities of phosphatidate phosphohydrolase, tyrosine aminotransferase and glycerol kinase in isolated rat hepatocytes in relation to the control of triacylglycerol synthesis and gluconeogenesis. Biochem. J. 225 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pittner R. A., R. Fears, and D. N. Brindley. 1985. Interactions of insulin, glucagon and dexamethasone in controlling the activity of glycerol phosphate acyltransferase and the activity and subcellular distribution of phosphatidate phosphohydrolase in cultured rat hepatocytes. Biochem. J. 230 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waki H., K. W. Park, N. Mitro, L. Pei, R. Damoiseaux, D. C. Wilpitz, K. Reue, E. Saez, and P. Tontonoz. 2007. The small molecule harmine is an antidiabetic cell-type-specific regulator of PPARgamma expression. Cell Metab. 5 357–370. [DOI] [PubMed] [Google Scholar]
- 16.Phan J., M. Péterfy, and K. Reue. 2004. Lipin expression preceding peroxisome proliferator-activated receptor-gamma is critical for adipogenesis in vivo and in vitro. J. Biol. Chem. 279 29558–29564. [DOI] [PubMed] [Google Scholar]
- 17.Farmer S. R. 2006. Transcriptional control of adipocyte formation. Cell Metab. 4 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosen E. D., and O. A. MacDougald. 2006. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 7 885–896. [DOI] [PubMed] [Google Scholar]
- 19.Rosen E. D., C. H. Hsu, X. Wang, S. Sakai, M. W. Freeman, F. J. Gonzalez, and B. M. Spiegelman. 2002. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev. 16 22–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoonjans K., J. Peinado-Onsurbe, A. M. Lefebvre, R. A. Heyman, M. Briggs, S. Deeb, B. Staels, and J. Auwerx. 1996. PPARalpha and PPARgamma activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO J. 15 5336–5348. [PMC free article] [PubMed] [Google Scholar]
- 21.Tontonoz P., E. Hu, R. A. Graves, A. I. Budavari, and B. M. Spiegelman.1994. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 8 1224–1234. [DOI] [PubMed] [Google Scholar]
- 22.Péterfy M., J. Phan, and K. Reue. 2005. Alternatively spliced lipin isoforms exhibit distinct expression pattern, subcellular localization, and role in adipogenesis. J. Biol. Chem. 280 32883–32889. [DOI] [PubMed] [Google Scholar]
- 23.Huffman T. A., I. Mothe-Satney, and J. C. Lawrence, Jr. 2002. Insulin-stimulated phosphorylation of lipin mediated by the mammalian target of rapamycin. Proc. Natl. Acad. Sci. USA. 99 1047–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cascales C., E. H. Mangiapane, and D. N. Brindley. 1988. Oleic acid promotes the activation and translocation of phosphatidate phosphohydrolase from the cytosol to particulate fractions of isolated rat hepatocytes. Biochem. J. 219 911–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finck B. N., M. C. Gropler, Z. Chen, T. C. Leone, M. A. Croce, T. E. Harris, J. C. Lawrence, Jr., and D. P. Kelly. 2006. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 4 199–210. [DOI] [PubMed] [Google Scholar]
- 26.Patel Y. M., and M. D. Lane. 1999. Role of calpain in adipocyte differentiation. Proc. Natl. Acad. Sci. USA. 96 1279–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darimont B. D., R. L. Wagner, J. W. Apriletti, M. R. Stallcup, P. J. Kushner, J. D. Baxter, R. J. Fletterick, and K. R. Yamamoto. 1998. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12 3343–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herzog B., R. K. Hall, X. L. Wang, M. Waltner-Law, and D. K. Granner. 2004. Peroxisome proliferator-activated receptor gamma coactivator-1alpha, as a transcription amplifier, is not essential for basal and hormone-induced phosphoenolpyruvate carboxykinase gene expression. Mol. Endocrinol. 18 807–819. [DOI] [PubMed] [Google Scholar]
- 29.Harris T. E., T. A. Huffman, A. Chi, J. Shabanowitz, D. F. Hunt, A. Kumar, and J. C. Lawrence, Jr. 2007. Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J. Biol. Chem. 282 277–286. [DOI] [PubMed] [Google Scholar]
- 30.Scotto-Lavino E., G. Du, and M. A. Frohman. 2006. 5′ end cDNA amplificaiton using classic RACE. Nat. Protocols. 1 2555–2562. [DOI] [PubMed] [Google Scholar]
- 31.Peterfy M., J. Phan, G. M. Oswell, P. Xu, and K. Reue. 1999. Genetic, physical, and transcript map of the fld region on mouse chromosome 12. Genomics. 62 436–444. [DOI] [PubMed] [Google Scholar]
- 32.Putignano P., F. Pecori Giraldi, and F. Cavagnini. 2004. Tissue-specific dysregulation of 11beta-hydroxysteroid dehydrogenase type 1 and pathogenesis of the metabolic syndrome. J. Endocrinol. Invest. 27 969–974. [DOI] [PubMed] [Google Scholar]
- 33.Garg A. 2006. Adipose tissue dysfunction in obesity and lipodystrophy. Clin. Cornerstone. 8 (Suppl.): 7–13. [DOI] [PubMed] [Google Scholar]
- 34.Reue K., and J. Phan. 2006. Metabolic consequences of lipodystrophy in mouse models. Curr. Opin. Clin. Nutr. Metab. Care. 9 436–441. [DOI] [PubMed] [Google Scholar]
- 35.Manmontri B., M. Sariahmetoglu, J. Donkor, M. Bou Khalil, M. Sundaram, Z. Yao, K. Reue, R. Lehner, and D. N. Brindley. 2008. Glucocorticoids and cAMP selectively increase hepatic lipin-1 expression, and insulin acts antagonistically. J. Lipid Res. 49 1056–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oishi K., N. Amagai, H. Shirai, K. Kadota, N. Ohkura, and N. Ishida. 2005. Genome-wide expression analysis reveals 100 adrenal gland-dependent circadian genes in the mouse liver. DNA Res. 12 191–202. [DOI] [PubMed] [Google Scholar]
- 37.Knox A. M., R. G. Sturton, J. Cooling, and D. N. Brindley. 1979. Control of hepatic triacylglycerol synthesis. Diurnal variations in hepatic phosphatidate phosphohydrolase activity and in the concentrations of circulating insulin and corticosterone in rats. Biochem. J. 180 441–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brindley D. N. 1992. Neuroendocrine regulation and obesity. Int. J. Obes. Relat. Metab. Disord. 16 (Suppl.): 73–79. [PubMed] [Google Scholar]
- 39.Jamal Z., A. Martin, A. Gomez-Munoz, P. Hales, E. Chang, J. C. Russell, and D. N. Brindley. 1992. Phosphatidate phosphohydrolases in liver, heart and adipose tissue of the JCR:LA corpulent rat and the lean genotypes: implications for glycerolipid synthesis and signal transduction. Int. J. Obes. Relat. Metab. Disord. 16 789–799. [PubMed] [Google Scholar]
- 40.Guan H. P., T. Ishizuka, P. C. Chui, M. Lehrke, and M. A. Lazar. 2005. Corepressors selectively control the transcriptional activity of PPARgamma in adipocytes. Genes Dev. 19 453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ialenti A., G. Grassia, P. Di Meglio, P. Maffia, M. Di Rosa, and A. Ianaro. 2005. Mechanism of the anti-inflammatory effect of thiazolidinediones: relationship with the glucocorticoid pathway. Mol. Pharmacol. 67 1620–1628. [DOI] [PubMed] [Google Scholar]
- 42.Croce M. A., J. C. Eagon, L. L. LaRiviere, K. M. Korenblat, S. Klein, and B. N. Finck. 2007. Hepatic lipin 1beta expression is diminished in insulin-resistant obese subjects and is reactivated by marked weight loss. Diabetes. 56 2395–2399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.