Abstract
Bile salts, the major end metabolites of cholesterol, vary significantly in structure across vertebrate species, suggesting that nuclear receptors binding these molecules may show adaptive evolutionary changes. We compared across species the bile salt specificity of the major transcriptional regulator of bile salt synthesis, the farnesoid X receptor (FXR). We found that FXRs have changed specificity for primary bile salts across species by altering the shape and size of the ligand binding pocket. In particular, the ligand binding pockets of sea lamprey (Petromyzon marinus) and zebrafish (Danio rerio) FXRs, as predicted by homology models, are flat and ideal for binding planar, evolutionarily early bile alcohols. In contrast, human FXR has a curved binding pocket best suited for the bent steroid ring configuration typical of evolutionarily more recent bile acids. We also found that the putative FXR from the sea squirt Ciona intestinalis, a chordate invertebrate, was completely insensitive to activation by bile salts but was activated by sulfated pregnane steroids, suggesting that the endogenous ligands of this receptor may be steroidal in nature. Our observations present an integrated picture of the coevolution of bile salt structure and of the binding pocket of their target nuclear receptor FXR.
Keywords: fishes, lampreys, lithocholic acid, molecular evolution, molecular models, nuclear hormone receptors, sequence homology, structure-activity relationship, Urochordata, farnesoid X receptor
Bile salts are water-soluble, amphipathic end metabolites of cholesterol that facilitate intestinal absorption of lipids (1), enhance proteolytic cleavage of dietary proteins (2), and have potent antimicrobial activity in the small intestine (3). In addition, bile salt signaling via nuclear hormone receptors (NHRs) is important for bile salt homeostasis (4). Bile salts have not been detected in invertebrate animals. In contrast to steroid hormones and vitamins, whose structures tend to be strongly conserved, bile salts exhibit marked structural diversity across species (5–7).
Bile salts vary in three main ways: a) structure of the side chain, which determines the bile salt class; b) stereochemistry of the A/B juncture [cis (5β) or trans (5α)]; and c) hydroxylation pattern of the nucleus and/or side chain (e.g., 7α, 12α, 23R). We use the term “bile salts” to refer to the broad class of cholesterol end metabolites in all vertebrates. More specific subclasses of bile salts are “bile acids” (with a carboxylic acid group on the terminal carbon atom of the side chain) and “bile alcohols” (with a primary alcohol on the terminal carbon atom of the side chain). Examples of bile acids and alcohols are illustrated in Fig. 1. The evolutionarily “earliest” bile salts are most likely bile alcohol sulfates that retain all 27 carbon atoms of cholesterol and have a trans A/B ring juncture, leading to an overall planar structure (Fig. 1). Chenodeoxycholyltaurine (also called taurochenodeoxycholic acid) and cholyltaurine are examples of more evolutionarily recent 24 carbon atom (C24) bile acids that have a “bent” shape because of their cis A/B ring juncture (Fig. 1).
Fig. 1.
Bile salts for the model animals analyzed in this study. All bile salts are derived from cholesterol, illustrated with the four steroid rings labeled A, B, C, and D (topmost structure). Both sea lamprey and zebrafish utilize bile salts that have an A/B ring juncture that is trans, resulting in an overall planar and extended structure of the steroid rings (see representation of the A, B, and C rings on the right side). One of the two major primary bile salts in humans is chenodeoxycholyltaurine, which has an A/B ring juncture that is cis, resulting in a bent conformation of the steroid rings. Lithocholic acid is one of the smallest naturally occurring bile acids and results from bacterial enzyme-mediated deconjugation and 7-dehydroxylation of chenodeoxycholyltaurine. The sodium salt of lithocholic acid is insoluble in water at body temperature; the calcium salt has an extremely low-solubility product.
In mammals, bile salts are formed from cholesterol by at least two complex biosynthetic pathways involving multiple organelles in the hepatocyte (8, 9). In the neutral pathway, nuclear biotransformations occur before biotransformation of the side chain. In this pathway, cholesterol undergoes microsomal hydroxylation at C-7, followed by coordinated isomerization of the double bond from C-5/C-6 to C-3/C-4 and dehydrogenation at C-3. The product, 7α-hydroxy-4-cholest-3-one, is the substrate for microsomal hydroxylation at C-12 by cytochrome P450 8B1. Subsequent steps result in reduction of the 3-oxo group to a 3α-hydroxy group and reduction of the double bond to give a fully saturated A ring. In the acidic pathway, the initial step in bile acid biosynthesis is oxidation to a carboxyl group at C-27 followed by changes in the bile salt nucleus. In animals forming C24 bile acids, the C8 side chain undergoes oxidative decarboxylation in the peroxisomes. The enzymes involved in bile salt biosynthesis in basal vertebrates have not yet been characterized. Primary bile salts are defined as those synthesized directly by the liver. Secondary bile salts result from the modification of nuclear substituents of primary bile salts by host bacteria in the intestine (1, 10).
In considering the biochemistry of the bile salt biosynthetic pathway, 5α-bile alcohols are the simplest saturated bile salt structure that can be derived from cholesterol, in terms of the number of enzymatic steps likely required. An example of a 27 carbon (C27) 5α-bile alcohol sulfate is found in the Atlantic hagfish Myxine glutinosa (11). Agnatha (“jawless fish”) is a paraphyletic superclass of the phylogenetically most basal vertebrates and includes extant hagfish and lampreys (12). All Agnathan species examined by our group or others (11, 13) exclusively utilize 5α-bile alcohols. Based on biochemical considerations and the study of Agnathan bile salts, we conclude that the first ancestral bile salts were most likely 5α-C27 bile alcohol sulfates.
Three NHRs are activated by bile salts and regulate their synthesis and/or metabolism: farnesoid X receptor (FXR; NR1H4) (14–16), pregnane X receptor (PXR; NR1I2) (17), and vitamin D receptor (VDR; NR1I1) (18, 19). FXR serves as the major transcriptional regulator of bile salt synthesis, in part by controlling the rate-limiting enzyme in bile salt synthesis (4). Mammalian FXRs are activated best by primary bile salts such as chenodeoxycholic acid (CDCA; 3α,7α-dihydroxy-5β-cholan-24-oic acid) (14–16, 20). FXR is typically expressed at high levels in the liver, intestine, kidney, and adrenal glands (4). A second FXR, termed FXRβ (NR1H5), is found in some animal species (21). FXRβ does not appear to be involved with bile salt binding or regulation and is not discussed further in this work. Throughout this article, FXR refers to NR1H4, or what might be termed FXRα in species possessing two FXRs.
Multiple FXR isoforms generated by alternative promoter usage have been described (22, 23). The isoforms described so far contain identical ligand binding domains (LBDs) but may differ in their interaction with coregulators, heterodimerization with retinoid X receptors, or DNA binding properties (23). The studies of receptor activation in our study utilize GAL4/LBD chimeras and thus would not be affected by differences in FXR domains outside the LBD.
In contrast to the extensive study of FXRs in mammals, only a few nonmammalian FXRs have been characterized (24, 25). The African clawed frog (Xenopus laevis) expresses an unusual FXR (also termed FOR, for FXR-like orphan receptor) that has a 33 amino acid insert, not found in mammalian FXRs, in helix 7 of the LBD (25). Similar to mammalian FXRs, X. laevis FXR is highly expressed in liver and kidney of adults and also in liver and kidney of metamorphosing tadpoles. X. laevis FXR was insensitive to synthetic FXR ligands or 5β-bile acids like CDCA; however, the receptor was activated by a partially purified extract of frog gallbladder bile (25). An FXR cloned from the little skate (Leucoraja erinacea) was found to be insensitive to bile salts, even those from cartilaginous fish (24). The skate FXR showed significant differences in sequence from other vertebrate FXRs, including novel insertions, and there is the possibility that this receptor is actually orthologous to FXRβ.
Based on our studies of bile salt diversity across vertebrates, we selected five model animals for the study of FXR: human (h), mouse (m), African clawed frog (xl; X. laevis), zebrafish (zf; Danio rerio), and sea lamprey (lamp; Petromyzon marinus) as well as the sea squirt (ci; Ciona intestinalis), an invertebrate from Urochordata, a subphylum thought to contain the closest extant invertebrate relatives of vertebrates (26). As will be shown, these animals span a range of bile salt phenotypes: C24 5β-bile acids (human, mouse), a mixture of C24 and C27 bile acids and bile alcohols (X. laevis), C27 5α-bile alcohol sulfates (zebrafish), C24 and C27 5α-bile alcohol sulfates (sea lamprey), and the absence of bile salts (Ciona). In this study, we focus on determining in detail the bile salt selectivity for FXRs in vitro. We additionally utilize molecular modeling methods to rationalize the structural changes that have occurred during evolution to affect FXR ligand specificity and to compare these results with structural models for bile salt interactions with the related NHRs PXR and VDR.
EXPERIMENTAL PROCEDURES
Animals
Adult X. laevis were purchased from NASCO (Fort Atkinson, WI). The AB strain was used for the zebrafish experiments. Transformer-stage sea lampreys were obtained from Acme Lamprey Co. (Harrison, ME). Animal studies were approved by the University of Pittsburgh Institutional Animal Care and Use Committee and performed in conformity with the Public Health Service Policy on the Humane Care and Use of Laboratory Animals, incorporated in the Institute for Laboratory Animal Research Guide for the Care and Use of Laboratory Animals.
Chemicals
The sources of the chemicals were as follows: fexaramine, GW3965, GW4064, MS-222, phenobarbital, and 4-[(E)-2-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)-1-propenyl]benzoic acid were from Sigma (St. Louis, MO); T-0901317 was from Axxora (San Diego, CA); 5α-cholic acid, 5α-petromyzonol, 5α-petromyzonol 24-sulfate, and 3-keto-5α-petromyzonol sulfate were from Toronto Research Chemical, Inc. (North York, Ontario, Canada); 1α,25-(OH)2-vitamin D3 and the Nuclear Receptor Ligand Library (76 compounds known as ligands of various NHRs) were from BIOMOL International (Plymouth Meeting, PA). All other commercially purchased steroids and bile salts were obtained from Steraloids (Newport, RI).
Isolation and purification of animal bile
The bile salts of many nonmammalian species differ from the typical bile salts found in mammals and are generally unavailable from commercial sources. To allow for the study of these compounds, the following chemically diverse bile salts were isolated from animals by extraction and flash column chromatography (27): 5α-myxinol-(3β,7α,16α,27-tetrahydroxy-5α-cholestane)-3β,27-disulfate from the Atlantic hagfish (Myxine glutinosa); 5α-cyprinol-(3α,7α,12α,26,27-pentahydroxy-5α-cholestane)-27-sulfate from the Asiatic carp (Cyprinus carpio); 5β-scymnol-(3α,7α,12α,24,26,27-hexahydroxy-5β-cholestane)-27-sulfate from the spotted eagle ray (Aetobatus narinari); and taurine-conjugated 3α,7α,12α-trihydroxy-5β-cholestan-27-oic acid from the American alligator (Alligator mississippiensis). The isolation and purification of 5α-cyprinol and 5α-cyprinol sulfate have been described previously in detail (27). Bile alcohol sulfates were deconjugated by incubating in a solution of 2,2-dimethoxypropane and 1.0 N HCl (7:1, v/v) for 2 h at 37°C, followed by the addition of water and extraction into ether. Completeness of deconjugation and assessment of purity were performed by thin-layer chromatography using known standards.
Analysis of bile
ESI-MS
Biliary contents were dissolved and diluted in methanol (Burdick and Jackson, Muskegon, MI) and analyzed using ESI-MS on a PE Sciex API III (Perkin-Elmer, Alberta, Canada) triple-quadrupole tandem mass spectrometer modified with a nanoESI source from Protana A/S (Odense, Denmark) (28). When operating in the negative mode, the following voltages were used: ionspray voltage, 600; interface voltage, 110; and orifice voltage, 90 V. A curtain gas of ultrapure nitrogen was pumped into the interface at a rate of 0.6 l/min to aid the evaporation of solvent droplets and to prevent particulate matter from entering the analyzer. Medium-sized palladium-coated borosilicate glass capillaries from Protana A/S were used for sample delivery. Collision gas-induced fragmentation used for structural identification was performed with ultrapure argon as a collision gas. Precursor ion spectra were acquired by scanning the first quadrupole, while collisions with argon in the second quadrupole produced dissociated ions. The third quadrupole was used to mass select the fragment ion. Spectra were the result of averaging from 10 to 100 scans, depending on the scan time and the number of scans necessary to obtain a sufficient signal-to-noise ratio. ESI-MS used a Hewlett-Packard HP 1100 MSD operated in the negative or positive mode. The HPLC column was removed, and the injector output was coupled directly to the ESI inlet. Samples (2 μl) were injected in methanol-water (90:10, v/v) mobile phase running at a flow rate of 0.35 ml/min. The fragmenter was set to 200 V, and the capillary voltage was set to 5,000 V.
HPLC
Conjugated bile acids were analyzed by HPLC using a modification of a previously reported technique (29). An octadecylsilane column (RP C-18; Beckman Instruments, Fullerton, CA) was used with isocratic elution at 0.75 ml/min. The eluting solution was composed of a mixture of methanol and 0.01 M KH2PO4 (67.4%, v/v) adjusted to an apparent pH of 5.35 with H3PO4. Bile acids were quantified by measuring their absorbance at 205 nm. Bile acid amidates (taurine and glycine) have similar extinction coefficients. Bile acids were tentatively identified by matching their relative retention times with those of known standards.
GC-MS
Glycine and taurine conjugates of bile acids were deconjugated chemically using 1.0 N NaOH at 130°C for 4 h. Bile alcohol sulfates were deconjugated enzymatically. Unconjugated bile acids were isolated by acidification followed by solvent extraction into ethyl acetate. They were then analyzed by capillary GC-MS as methyl ester acetates (prepared using acetic anhydride in acetic acid with perchloric acid catalyst) or as methyl ester trimethylsilyl derivatives (prepared using Tri-Sil; Pierce Chemicals, Rockford, IL). GC was performed using a Hewlett-Packard 5890 gas chromatograph-5970 MSD, controlled by HP/UX Chem Station software. The column was a Supelco 30 m 0.25 mm inner diameter intermediate polarity SPB-35 of 35% phenyl methyl silicone (Supelco Co., Bellefonte, PA) operated at 277°C (isothermal). A splitless injection was used with an injection temperature of 290°C. Helium was used as the carrier gas with a 7 p.s.i. column head pressure. Relative retention times and fragmentation spectra of peaks obtained by GC-MS were compared with those of known standards for identification.
Cell culture of HepG2, A6, and ZFL cell lines
The creation of a HepG2 (human liver) cell line stably expressing the human Na+-taurocholate cotransporter (NTCP; SLC10A1) has been reported previously (30). HepG2-NTCP cells were grown in modified Eagle's medium-α containing 10% fetal bovine serum and 1% penicillin/streptomycin. The cells were grown at 37°C in 5% CO2. The X. laevis A6 kidney cell line [American Type Culture Collection (ATCC), Manassas, VA] was grown in 75% NCTC 109 medium to which was added 15% distilled water and 10% fetal bovine serum at 26°C in 2% CO2. The zebrafish ZFL liver cell line (ATCC) was grown in 50% Leibovitz's L-15 medium with 2 mM l-glutamine, 35% Dulbecco's modified Eagle's medium with 4.5 g/l glucose and 4 mM l-glutamine, 15% Ham's F-12 with 1 mM l-glutamine supplemented with 0.15 g/l sodium bicarbonate, 15 mM HEPES, 10 μg/ml human insulin (Sigma), 50 ng/ml recombinant human epidermal growth factor (Sigma), and 5% fetal bovine serum. ZFL cells were grown at 28°C in room air. Except as noted above, all media and media supplements for the HepG2, A6, and ZFL cell lines were obtained from Invitrogen (Carlsbad, CA).
Molecular biology
Plasmids containing human organic anion-transporting polypeptide (Oatp1a1; Slco1a1), as well as the reporter construct tk-UAS-Luc and the “empty” vector PM2, were generously provided by S.A. Kliewer, J.T. Moore, and L.B. Moore (GlaxoSmithKline, Research Triangle Park, NC). To permit comparison between species and to avoid mismatching of nonmammalian receptors with mammalian retinoid X receptor, cofactors, and chromatin-remodeling factors, all receptors were studied as LBD/GAL4 chimeras. For the GAL4/LBD expression constructs, the reporter plasmid is tk-UAS-Luc, which contains GAL4 DNA binding elements driving luciferase expression.
The LBD of hFXR was cloned by PCR from IMAGE clone 4606016 (ATCC 10329524). The LBD of the xlFXR was cloned by PCR from IMAGE clone 4033203 (ATCC 10550277). The xlFXR characterized in this study corresponds to the LBD for the receptor referred to as FXR-like orphan receptor 1 (FOR1) in a previous report (25). zfFXR was cloned by RT-PCR from total RNA extracted from the ZFL liver cell line (ATCC CRL-2643). The D-domain and LBD of lampFXR were cloned by RT-PCR from total RNA extracted from liver of transformer-stage sea lamprey using sequence information from the partial draft of the sea lamprey genome (Genome Sequencing Center, Washington University, St. Louis, MO). ciFXR was cloned by RT-PCR from total RNA extracted from stomach, intestine, and pharynx of adult C. intestinalis. The LBDs of hFXR (amino acid residues 192–472), xlFXR (amino acid residues 196–513), zfFXR (amino acid residues 202–483), lampFXR, and ciFXR (amino acid residues 244–516) were inserted into the pM2-GAL4 vector to create GAL4/LBD chimeras.
Cotransfections and transactivation assays
The basic methodology for the luciferase reporter assays in 96-well format using HepG2-NTCP cells has been reported previously (30, 31). For the FXRs, 75 ng/well GAL4/LBD plasmid was cotransfected with 100 ng/well tk-UAS-Luc, 20 ng/well β-galactosidase, and 10 ng/well SLC21. Each compound concentration was performed at least in quadruplicate and repeated in separate experiments for a total of at least three times. For screening experiments, at least three concentrations of each compound were tested. On day 4, the cells were washed with Hanks' buffered salt solution and then exposed to 150 μl of lysis buffer (Reporter Lysis Buffer; Promega). Separate aliquots were taken for the measurement of β-galactosidase activity (Promega) and luciferase activity (Steady-Glo; Promega).
To facilitate more reliable cross-species comparisons, complete concentration-response curves for ligands were determined on the same microplate as the determination of response to a maximal activator. This allows for the determination of relative efficacy (ɛ), defined as the maximal response to the test ligand divided by the maximal response to a reference maximal activator (note than ɛ can exceed 1). The maximal activators and their concentrations were as follows: hFXR, 20 μM CDCA; xlFXR and zfFXR, 150 μM 5α-cyprinol 27-sulfate; lampFXR, 50 μM 5α-petromyzonol 24-sulfate; ciFXR, 10 μM AM-580 (BIOMOL). All comparisons with maximal activators were done on the same microplate. Luciferase data were normalized to the internal β-galactosidase control and represent means ± SD of the assays.
NHR sequences
Sequences for NHR genes were downloaded from the National Center for Biotechnology Information http://www.ncbi.nlm.nih.gov/entrez) and Ensembl http://www.ensembl.org) public databases. Complete listings of all genes, species, and accession numbers are provided in supplementary Table I. Sequences were aligned using ClustalW (http://www.ebi.ac.uk/clustalw) and Tcoffee http://tcoffee.vital-it.ch/cgi-bin/Tcoffee/tcoffee_cgi/index.cgi) software and manually adjusted as needed. For supplementary Fig. I, ligand binding residues for FXR were identified from published crystal structures for human FXR (20) and rat FXR (32) and are highlighted in boldface type. A comparison of the amino acid sequences of human VDR, FXR, and PXR is provided in supplementary Fig. II.
Molecular modeling studies
Lithocholic acid was docked into the crystal structure for human PXR [Protein Data Bank (PDB) accession number 1ILG] (33) and human VDR (PDB accession number 1DB1) (34) using the GOLD docking program (35). During the docking process, the protein was held fixed, while full conformational flexibility was allowed for the ligands. For each ligand, 30 independent docking runs were performed to achieve the consensus orientation in the ligand binding pocket (LBP). Structural homology models of the LBDs of hFXR, xlFXR, zfFXR, lampFXR, and ciFXR were constructed using the Insight II Homology Module (Accelrys) with the published crystal structure of rat FXR in complex with 6α-ethyl-CDCA (PDB accession number 1OSV) (32) as the modeling template (this structure was chosen because it has bound bile acid, unlike available crystal structures of hFXR). In addition, a homology model of zfPXR was constructed using a human PXR crystal structure (PDB accession number 1ILG) as the template. Several energy minimization-based refinement procedures were implemented on the initial models, and the quality of the final model was confirmed by the WHATIF-Check program. The homology model for hFXR was compared with the previously solved crystal structure with the synthetic ligand fexaramine (PDB accession number 1OSH) (20). Molecular docking studies were performed with CDCA and fexaramine in the various FXR homology models using GOLD. Estimation of the volume of the LBP for the crystallographic structures and homology models described above was determined using CASTp http://sts.bioengr.uic.edu/castp/calculation.php). For each receptor, when possible, the mean ± SD was calculated (36).
RESULTS
Bile salt profiles of sea lamprey, zebrafish, and African clawed frog
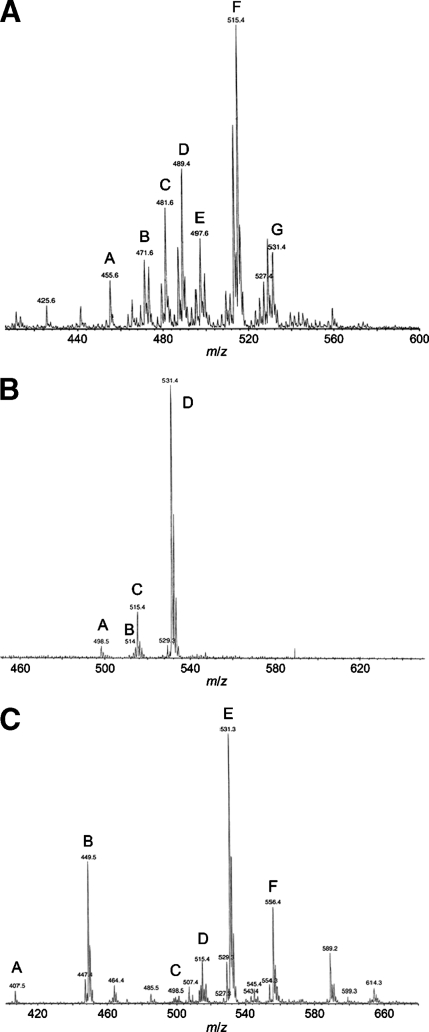
Analyses of the bile salts of African clawed frog (37) and sea lamprey (13) were reported over three decades ago. Using more detailed analytical methods (e.g., ESI-MS), we resolved more completely the bile salt profiles of these two animals as well as that of the zebrafish (Fig. 2). Previous studies have shown that sea lamprey biliary bile contains a unique C24 bile alcohol sulfate, 5α-petromyzonol 24-sulfate (13, 38). We additionally identified two C27 5α-bile alcohol sulfates (tetrahydroxylated and pentahydroxylated) as the major constituent of bile from the transformer-stage sea lamprey (Fig. 2A). 5α-Petromyzonol 24-sulfate was also identified (peak D of Fig. 2A), as were dihydroxylated and trihydroxylated C24 and C27 bile alcohols with a single double bond. The bile profile revealed is more complex than that originally reported by Haslewood and Tökés in 1969 (13). This may result from our use of ESI-MS but also possibly from the differences in age or developmental stage of the lampreys analyzed by us compared with Haslewood and Tökés (13). The dihydroxylated and trihydroxylated unsaturated C24 and C27 bile alcohols may represent synthetic precursors to 5α-petromyzonol 24-sulfate and the two major C27 bile alcohol sulfates. The major bile salt in zebrafish bile was 5α-cyprinol 27-sulfate (39) (Fig. 2B), a C27 bile alcohol sulfate that is also the major bile salt of Asiatic carp (27), another member of the family Cyprinidae. In both sea lamprey and zebrafish, at least 98% of the bile salts in biliary bile were in the 5α (A/B trans) configuration, as determined by GC-MS analysis.
Fig. 2.
ESI-MS analysis of biliary bile salts from sea lamprey, zebrafish, and African clawed frog. From an extensive library generated from the analysis of many bile specimens, the major ions are annotated with probable matches indicating the number of carbon atoms (24 or 27), whether the compound is a bile acid or bile alcohol, the number of hydroxyl groups, the presence of a double bond (if any), and the type of conjugation. Additional studies (data not shown) included GC-MS, thin-layer chromatography, and high-performance liquid chromatography. The orientation of the hydrogen atom on carbon 5 was generally resolved by GC-MS. A: ESI-MS analysis of sea lamprey biliary bile. The labeled peaks in the spectra are as follows: peak A, C24 bile alcohol-(OH)2-SO4 with one double bond; peak B, C24 bile alcohol-(OH)3-SO4 with one double bond; peak C, C27 bile alcohol-(OH)2-SO4 with one double bond; peak D, C24 bile alcohol-(OH)4-SO4; peak E, C27 bile alcohol-(OH)3-SO4 with one double bond; peak F, C27 bile alcohol-(OH)4-SO4; peak G, C27 bile alcohol-(OH)5-SO4. Peak D corresponds to 5α-petromyzonol 24-sulfate. B: ESI-MS analysis of zebrafish biliary bile. The labeled peaks in the spectra are as follows: peak A, C24 bile acid-(OH)2-taurine; peak B, C24 bile acid-(OH)3-taurine; peak C, C27 bile alcohol-(OH)4-SO4; peak D, C27 bile alcohol-(OH)5-SO4. The most abundant ion (peak D) corresponds to 5α-cyprinol 27-sulfate. C: ESI:MS analysis of bile salts from African clawed frog biliary bile. The labeled peaks in the spectra are as follows: peak A, C24 bile acid-(OH)3; peak B, C27 bile acid-(OH)3; peak C, C24 bile acid-(OH)2-taurine; peak D, C24 bile acid-(OH)3-taurine; peak E, C27 bile alcohol-(OH)5-SO4; peak F, C27 bile acid-(OH)3-taurine. The most abundant ion (peak E) corresponds to 5β-cyprinol 27-sulfate, the A/B cis isomer to the main zebrafish bile salt.
The African clawed frog has a complicated biliary bile salt profile, with the presence of C24 bile acids, C27 bile acids, and C27 bile alcohol sulfates (Fig. 2C). This bile profile is quite similar to our prior analysis of biliary bile from Silurana tropicalis (Western clawed frog, also known as Xenopus tropicalis), a related frog species (40). GC-MS analysis (data not shown) showed that most of the bile salts from African clawed frog were in the 5β (A/B cis) orientation, with only a minor fraction (<10%) in the 5α orientation. Although the presence of 5β-bile acids as a component (albeit minor) of X. laevis primary bile salts suggested the possibility of secondary bile acid conversion, we did not detect 7-deoxy bile acids in either biliary bile or intestinal contents of adult frogs. The primary bile salts of humans and mice have been well described and are C24 5β-bile acids: cholic acid (3α,7α,12α-trihydroxy-5β-cholan-24-oic acid) and CDCA in humans, and β-muricholic acid (3α,6β,7β-trihydroxy-5β-cholan-24-oic acid), α-muricholic acid (3α,6β,7α-trihydroxy-5β-cholan-24-oic acid), and cholic acid in mice (6).
Structure-activity studies for the activation of FXRs
Using luciferase-based reporter assays based on GAL4/LBD chimeras, we determined detailed structure-activity relationships for activation of the various FXRs using an extensive panel of 49 naturally occurring bile salts, bile salt precursors, and synthetic analogs (Fig. 3; see supplementary Table II). It should be pointed out that it is not known whether NTCP and Oatp1a1 are capable of transporting all of the bile salts and bile salt precursors tested. An absence of transactivation may, in principle, also be due to inefficient uptake by the HepG2 cells. In general, the vertebrate FXRs showed selectivity for the major species-specific primary bile salt(s), while the putative FXR from the invertebrate C. intestinalis was not activated by any bile salt. Both lampFXR and zfFXR were activated nearly exclusively by 5α-bile alcohol sulfates, while hFXR and mFXR were activated best by 5β-bile acids and only weakly or not at all by 5α-bile salts (see supplementary Table II). xlFXR was activated by both 5α- and 5β-bile alcohols but poorly by bile acids (see supplementary Table II), consistent with the findings of a previous study that 5β-bile acids did not activate xlFXR but extracts from frog bile (which would contain mostly bile alcohols) did activate xlFXR (25). Conjugation by glycine or taurine had minimal effects on FXR activation. Overall, these results show a shift in ligand selectivity from evolutionarily early to more recent ligands, and this is most clearly illustrated by comparing lampFXR and zfFXR with mFXR and hFXR.
Fig. 3.
Bile salt activation of farnesoid X receptors (FXRs) from different species. Frog and zebrafish FXRs are activated by the early bile salt 5α-cyprinol sulfate (A) but are insensitive to the recent primary bile salt 5β-chenodeoxycholyltaurine (TCDCA; B) and its corresponding deconjugated and dehydroxylated secondary bile salt 5β-lithocholic acid (C). In contrast, human and mouse FXRs are activated by 5β-bile acids (B, C) but not by 5α-cyprinol sulfate (A). The C. intestinalis FXR is insensitive to all three bile salts (A–C). Data shown are means ± SD.
We also determined the selectivity of the FXRs for synthetic ligands. Fexaramine and GW4064, both previously described as selective hFXR agonists (20), activated hFXR at submicromolar concentrations. GW4064 but not fexaramine activated zfFXR, while neither fexaramine nor GW4064 activated xlFXR or lampFXR (see supplementary Table III). T-0901317, originally thought to be a selective liver X receptor agonist (41) but now known to also activate mammalian FXRs and PXRs (42–44), activated hFXR and zfFXR but not xlFXR or lampFXR. The selective liver X receptor agonist GW3965 did not activate any of the FXRs (see supplementary Table III).
Ligand specificity of the Ciona FXR
Analysis of the C. intestinalis genome revealed a single putative ortholog to vertebrate FXRs (45). Examination of the C. intestinalis genome also revealed a single putative ortholog to vertebrate NR1I (VDR, PXR, and CAR) genes, which we have provisionally classified as a VDR (31, 46), although the sequence similarity of this gene to any of the vertebrate NR1I genes is low, especially in the LBD. The pharmacological properties of the “Ciona VDR” are quite different from any of the characterized NR1H and NR1I receptors, so more accurate phylogenetic assignment requires further study. From the Ghost database of C. intestinalis genomic and cDNA resources http://ghost.zool.kyoto-u.ac.jp/indexr1.html), cDNA clones cidg803k09 and cicl102e19 correspond to the ciFXR analyzed in our study. These cDNAs, based on expressed sequence tag counts, show expression in eggs, cleaving embryo, tailbud embryo, young adult animals, and mature animals. Within adult animals, expression was seen in the digestive gland.
Neither bile salts nor synthetic FXR agonists such as fexaramine, GW4064, or T-0901317 were capable of activating ciFXR (Fig. 3; see supplementary Tables II, III). Screening of the 76 compound BIOMOL Nuclear Receptor Ligand Library revealed that 6-formylindolo-[3,2]carbazole, a tryptophan photoproduct that is a high-affinity agonist of the aryl hydrocarbon receptor (47), and AM-580 both activated ciFXR in the low micromolar range. Screening of an additional 85 compounds comprising steroid hormones, vitamins, and xenobiotics revealed that some sulfated steroids activated ciFXR (see supplementary Table III). The most efficacious activators were those sulfated at the 3 position, including 5-pregnen-3β-ol-20-one sulfate (pregnanolone sulfate), 5β-pregnan-3α-ol-20-one sulfate (pregnenolone sulfate), 5β-pregnan-3β-ol-20-one sulfate (epipregnanolone sulfate), and 5α-pregnan-3β-ol-20-one sulfate (epiallopregnanolone sulfate). These findings suggest that the endogenous ligand(s) of ciFXR may be steroidal in nature.
Docking studies comparing interactions of FXR, VDR, and PXR with bile salts
We generated novel homology models of hFXR, xlFXR, zfFXR, lampFXR, ciFXR, and zfPXR, which were used along with available crystal structures (rat FXR, hPXR, hVDR, and zfVDR) for computational docking studies with bile salts (Fig. 4). These studies revealed a common binding mechanism for bile salt interactions with hVDR, hFXR, and hPXR. Particularly key interactions are hydrogen bonding with a histidine residue (hVDR His-397, hFXR His-444, hPXR His-407), hydrophobic interactions with a phenylalanine (hVDR Phe-422, hPXR Phe-429) or tryptophan (hFXR Trp-466) residue, and an arginine charge clamp (hVDR Arg-274, hFXR Arg-328, hPXR Arg-287) (see supplementary Fig. III). However, beyond the similar binding interactions described above, the disparate architectures of the LBPs markedly influence the specificity of human PXR, VDR, and FXR for bile salts.
Fig. 4.
Docking of ligands into structural models derived from X-ray crystallography or homology modeling. A: The curved binding pocket of human FXR accommodates the bent conformation of 5β-lithocholic acid well. B: The narrow binding pocket of sea lamprey FXR is suited to binding of the evolutionary early, planar bile salt 5α-cyprinol 27-sulfate. C: The binding pocket of Ciona FXR is smaller than that of any of the vertebrate FXRs analyzed and was not predicted to be capable of binding any bile salts but does bind the synthetic ligand AM-580. D: The large binding pocket of human pregnane X receptor (PXR) readily accommodates lithocholic acid. E: Similar to lamprey FXR, zebrafish PXR has a flat binding pocket suited to binding of the planar bile salt 5α-cyprinol 27-sulfate. F: Human vitamin D receptor has a relatively small, slightly curved binding pocket that can barely accommodate the small secondary bile acid lithocholic acid. See Experimental Procedures for details on the structural models.
Structural changes in FXRs across species
The shape of the hFXR LBP has a major impact on bile salt selectivity and clearly favors A/B cis 5β-bile acids (Fig. 4A). The models predict that the more extended, planar A/B trans bile salts cannot achieve favorable complementarity to residues within the hFXR LBP and, therefore, have limited or no ability to activate hFXR, consistent with our structure-activity studies (see supplementary Table II) and those of others (48). In contrast, homology models and docking studies of zfFXR and lampFXR predict narrow LBPs permitting binding of planar 5α-bile salts like 5α-cyprinol sulfate (a major zebrafish bile salt) (Fig. 4B) but not evolutionarily more recent 5β-bile salts that have a bent structure. The homology model for ciFXR revealed a significantly smaller LBP (648 Å3) compared with that for hFXR (814 Å3); docking analysis also suggested that the ciFXR LBP should not bind bile salts. Docking studies did show that ciFXR could favorably bind AM-580 (Fig. 4C), a synthetic compound from the BIOMOL Nuclear Receptor Ligand Library found to activate ciFXR (see supplementary Table III).
Structural models for PXR activation by bile salts
We and others have demonstrated in previous studies that hPXR has broad specificity for bile salt structures and is activated by conjugated, unconjugated, A/B trans, and A/B cis bile salts (30, 49, 50). In contrast, zfPXR is chiefly activated by 5α-bile alcohol sulfates (46, 49). We concluded previously that the sea lamprey genome likely lacks a PXR gene, based on several lines of evidence: a) the absence of any PXR-like sequence in the sea lamprey draft genome; b) our inability to clone a sea lamprey PXR; and c) our studies of cultured lamprey primary hepatocytes that failed to show any evidence of PXR-mediated functions (31). Homology modeling of zfPXR predicted a LBP well suited to binding of planar bile salts such as 5α-cyprinol 27-sulfate (Fig. 4E). The differences in the interactions of hPXR and zfPXR with bile salts are attributable to two main factors. First, the estimated LBP volume of zfPXR (∼1,000 Å3) is smaller than that of hPXR (∼1,300 Å3). Second, zfPXR differs from hPXR in multiple ligand binding residues identified in the hPXR X-ray crystal structures that interact with ligands (33, 44, 51). Such residues include Arg-287 and His-407 discussed above (comparable to His-283 and tyrosine-404, respectively, in zfPXR).
Comparison of FXR and VDR binding of bile acids
Previous studies have shown that hVDR and mVDR are activated by a limited number of C24 5β-bile acids, especially unconjugated lithocholic acid and derivatives (18, 19), but are not activated by 5α-bile acids or alcohols (30, 31, 49). Our modeling studies show that hVDR has a tight, “curved” binding pocket that can accommodate bent A/B cis bile acids like lithocholic acid but not the more extended, planar A/B trans bile salts (Fig. 4F). We propose that the smaller size of the LBP of hVDR relative to hFXR and hPXR, together with the relatively short distance between the key histidine and arginine residues (see supplementary Fig. III), renders hVDR responsive only to small A/B cis bile acids. The docking orientation of lithocholic acid in the hVDR LBP determined by our analyses using the docking algorithm GOLD is ∼180° opposite that of two previous studies that used FlexX as the docking algorithm (18, 52). We believe that the model we present here better explains the available structure-activity data on VDR interactions with bile salts (18, 30, 49, 52).
DISCUSSION
There is increasing interest in ligand-nuclear receptor coevolution, as illustrated by steroid hormone interactions with NHRs (53, 54). Our results with FXRs identify a shift in selectivity from early, “ancestral” ligands (5α-bile alcohols) to evolutionarily recent ligands (5β-bile acids). The FXR results are consistent with vertebrate FXRs controlling specificity for bile salts by increasing the size and changing the topology of the LBP, compared with an ancestral invertebrate receptor. Study of additional invertebrate FXRs would help support or refute this hypothesis. This hypothesis would fit with a major function of FXRs being to monitor hepatocyte concentrations of conjugated bile salts and then appropriately regulate their biosynthesis, conjugation, and transport (4). As primary bile salt structures changed across species, FXRs adapted to the new structures, losing the sensitivity to early bile salts in the process. It is also worth noting that the variability of FXRs across vertebrate species contrasts markedly with the liver X receptors, closely related NHRs whose endogenous ligands are oxysterols (4). Liver X receptors α and β (NR1H3 and NR1H2, respectively) are much more highly conserved in sequence and pharmacology across vertebrates than FXRs (30, 40).
The evolutionary changes in FXRs contrast somewhat with cross-species differences in bile salt recognition by PXRs. PXR, also known as the steroid and xenobiotic receptor, is activated by a wide range of endogenous and exogenous molecules, including steroid hormones, bile salts, and xenobiotics. PXR regulates the transcription of enzymes and transporters involved in metabolism and the elimination of structurally diverse compounds, including detoxification of toxic bile acids via sulfation pathways (55). Previous studies have shown significant cross-species differences in selectivity for bile salts by PXRs (56), with mammalian PXRs being activated by a broad range of bile salt structures and, in contrast, zebrafish PXR being activated by only a narrow range of bile salts (30, 49, 50). Whereas human and mouse FXRs have “lost” the ability to be activated by evolutionarily early bile acids, human PXR is activated by a wide range of bile salt structures, in effect retaining the ability to be activated by the ancestral ligands (30, 49). The PXR results may be explained by observations that PXRs also bind a wide range of chemical structures other than bile salts as part of a broader function to detect potentially toxic compounds (17). The evolution of PXRs would thus be driven by at least two factors: adaptation to changes in bile salts structures and function as a xenobiotic sensor. Expansion of the size and alteration in topology of the hPXR LBP, relative to nonmammalian PXRs, permits recognition of the evolutionarily recent 5β-bile acids typical of terrestrial vertebrates, while also retaining sensitivity to evolutionarily early bile salt structures.
In contrast to FXRs, which evidently became sensitive to bile salts early in vertebrate evolution, VDRs likely acquired the ability to be activated by bile salts later in evolution, based on our previous studies comparing mammalian and nonmammalian VDRs (30, 31, 49). The complete lack of sensitivity of VDRs to 5α-bile salts suggests that VDRs “gained” the ability to recognize bile acids in animals that had already made the evolutionary shift from 5α- to 5β-bile salts, possibly as an adaptive response to help detoxify lithocholic acid (19, 30). The evolution of secosteroid sensitivity of VDRs is also not known. Our study of a VDR-like receptor from C. intestinalis revealed no activation by vitamin D compounds or by vertebrate steroids or bile salts (31, 46). This receptor was activated, however, by a small number of planar synthetic molecules. Better resolution of the VDR phylogeny requires the study of additional invertebrates and basal vertebrates.
Our studies with the putative Ciona FXR reveal a receptor with quite different pharmacology from vertebrate FXRs, which is perhaps not surprising given that invertebrates are not thought to produce bile salts. However, this receptor was activated by sulfated steroids and several compounds (AM-580 and 6-formylindolo-[3,2]carbazole) known to activate other NHRs. The sensitivity of Ciona FXR to sulfated steroids raises the possibility that an ancestral FXR selective for invertebrate steroids or structurally related ligands adapted during vertebrate evolution to recognize bile salts. Sulfated endogenous compounds, including steroids, are common in marine invertebrate animals (57). One report identified 3α,4β,7α,27-tetrahydroxycholestane-3α,27-disulfate as a chemoattractant for Ciona sperm (58). This compound, termed sperm-activating and -attracting factor, bears close resemblance to early vertebrate bile salts such as 5α-myxinol 3β,27-disulfate from the Atlantic hagfish (11). Overall, our results are consistent with a complex evolutionary history for FXRs, with the acquisition of sensitivity to bile salts from a bile salt-insensitive ancestral receptor and then adaptation within vertebrates to cross-species differences in biliary bile salt composition.
Supplementary Material
Acknowledgments
The authors thank Drs. Anna Di Rienzo (University of Chicago), George Michalopoulos, Alan Wells, and Harry Blair (University of Pittsburgh) for helpful advice and comments on the manuscript.
Abbreviations
CDCA, chenodeoxycholic acid
FXR, farnesoid X receptor
LBD, ligand binding domain
LBP, ligand binding pocket
NHR, nuclear hormone receptor
PXR, pregnane X receptor
VDR, vitamin D receptor
Published, JLR Papers in Press, March 24, 2008.
Footnotes
M.D.K. is supported by Grant K08 GM-074238 from the National Institutes of Health and the Competitive Medical Research Fund from the University of Pittsburgh Medical Center. W.J.W., S.E., and N.A. acknowledge support for this work provided by the Environmental Protection Agency-funded Environmental Bioinformatics and Computational Toxicology Center, under STAR Grant GAD R 832721-010 (this work has not been reviewed by and does not represent the opinions of the funding agency). A.F.H. and L.R.H. are supported by National Institutes of Health Grant DDK-64891 (to A.F.H.).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three figures and three tables.
References
- 1.Hofmann A. F. 1999. The continuing importance of bile acids in liver and intestinal disease. Arch. Intern. Med. 159 2647–2658. [DOI] [PubMed] [Google Scholar]
- 2.Gass J., H. Vora, A. F. Hofmann, G. M. Gray, and C. Khosla. 2007. Enhancement of dietary protein digestion by conjugated bile acids. Gastroenterology. 133 16–23. [DOI] [PubMed] [Google Scholar]
- 3.Hofmann A. F., and L. Eckmann. 2006. How bile acids confer gut mucosal protection against bacteria. Proc. Natl. Acad. Sci. USA. 103 4333–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalaany N. Y., and D. J. Mangelsdorf. 2006. LXRs and FXR: the yin and yang of cholesterol and fat metabolism. Annu. Rev. Physiol. 68 159–191. [DOI] [PubMed] [Google Scholar]
- 5.Haslewood G. A. D. 1967. Bile salt evolution. J. Lipid Res. 8 535–550. [PubMed] [Google Scholar]
- 6.Moschetta A., F. Xu, L. R. Hagey, G. P. van Berge Henegouwen, K. J. van Erpecum, J. F. Brouwers, J. C. Cohen, M. Bierman, H. H. Hobbs, J. H. Steinbach, et al. 2005. A phylogenetic survey of biliary lipids in vertebrates. J. Lipid Res. 46 2221–2232. [DOI] [PubMed] [Google Scholar]
- 7.Une M., and T. Hoshita. 1994. Natural occurrence and chemical synthesis of bile alcohols, higher bile acids, and short side chain bile acids. Hiroshima J. Med. Sci. 43 37–67. [PubMed] [Google Scholar]
- 8.Norlin M., and K. Wikvall. 2007. Enzymes in the conversion of cholesterol into bile acids. Curr. Mol. Med. 7 199–218. [DOI] [PubMed] [Google Scholar]
- 9.Russell D. W. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72 137–174. [DOI] [PubMed] [Google Scholar]
- 10.Hofmann A. F. 2004. Detoxification of lithocholic acid, a toxic bile acid: relevance to drug hepatotoxicity. Drug Metab. Rev. 36 703–722. [DOI] [PubMed] [Google Scholar]
- 11.Haslewood G. A. D. 1966. Comparative studies of bile salts. Myxinol disulphate, the principal bile salt of hagfish (Myxinidae). Biochem. J. 100 233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forey P., and P. Janvier. 1993. Agnathans and the origin of jawed vertebrates. Nature. 361 129–134. [Google Scholar]
- 13.Haslewood G. A. D., and L. Tökés. 1969. Comparative studies of bile salts: bile salts of the lamprey Petromyzon marinus L. Biochem. J. 114 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H., J. Chen, K. Hollister, L. C. Sowers, and B. M. Forman. 1999. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell. 3 543–553. [DOI] [PubMed] [Google Scholar]
- 15.Parks D. J., S. G. Blanchard, R. K. Bledsoe, G. Chandra, T. G. Consler, S. A. Kliewer, J. B. Stimmel, T. M. Willson, A. M. Zavacki, D. D. Moore, et al. 1999. Bile acids: natural ligands for an orphan nuclear receptor. Science. 284 1365–1368. [DOI] [PubMed] [Google Scholar]
- 16.Makishima M., A. Y. Okamato, J. J. Repa, H. Tu, R. M. Learned, A. Luk, M. V. Hull, K. D. Lustig, D. J. Mangelsdorf, and B. Shan. 1999. Identification of a nuclear receptor for bile acids. Science. 284 1362–1365. [DOI] [PubMed] [Google Scholar]
- 17.Kliewer S. A., and T. M. Willson. 2002. Regulation of xenobiotic and bile acid metabolism by the nuclear pregnane X receptor. J. Lipid Res. 43 359–364. [PubMed] [Google Scholar]
- 18.Adachi R., A. I. Shulman, K. Yamamoto, I. Shimomura, S. Yamada, D. J. Mangelsdorf, and M. Makishima. 2004. Structural determinants for vitamin D responses to endocrine and xenobiotic signals. Mol. Endocrinol. 18 43–52. [DOI] [PubMed] [Google Scholar]
- 19.Makishima M., T. T. Lu, W. Xie, G. K. Whitfield, H. Domoto, R. M. Evans, M. R. Haussler, and D. J. Mangelsdorf. 2002. Vitamin D receptor as an intestinal bile acid sensor. Science. 296 1313–1316. [DOI] [PubMed] [Google Scholar]
- 20.Downes M., M. A. Verdecia, A. J. Roecker, R. Hughes, J. B. Hogenesch, H. R. Kast-Woelbern, M. E. Bowman, J-L. Ferrer, A. M. Anisfeld, P. A. Edwards, et al. 2003. A chemical, genetic, and structural analysis of the nuclear bile acid receptor FXR. Mol. Cell. 11 1079–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otte K., H. Kranz, I. Kober, P. Thompson, M. Hoefer, B. Haubold, B. Remmel, H. Voss, C. Kaiser, M. Albers, et al. 2003. Identification of farnesoid X receptor β as a novel mammalian nuclear receptor sensing lanosterol. Mol. Cell. Biol. 23 864–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber R. M., K. Murphy, B. Miao, J. R. Link, M. R. Cunningham, M. J. Rupar, P. L. Gunyuzlu, T. F. Haws, A. Kassam, F. Powell, et al. 2002. Generation of multiple farnesoid-X-receptor isoforms through the use of alternative promoters. Gene. 290 35–43. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y., H. R. Kast-Woelbern, and P. A. Edwards. 2003. Natural structural variants of the nuclear farnesoid X receptor affect transcriptional activation. J. Biol. Chem. 278 104–110. [DOI] [PubMed] [Google Scholar]
- 24.Cai S. Y., L. Xiong, C. G. Wray, N. Ballatori, and J. L. Boyer. 2007. The farnesoid X receptor, FXRα/NR1H4, acquired ligand specificity for bile salts late in vertebrate evolution. Am. J. Physiol. Regul. Integr. Comp. Physiol. 293 R1400–R1409. [DOI] [PubMed] [Google Scholar]
- 25.Seo Y-W., S. Sanyal, H-J. Kim, D. H. Won, J-Y. An, T. Amano, A. M. Zavacki, H-B. Kwon, Y-B. Shi, W-S. Kim, et al. 2002. FOR, a novel orphan nuclear receptor related to farnesoid X receptor. J. Biol. Chem. 277 17836–17844. [DOI] [PubMed] [Google Scholar]
- 26.Delsuc F., H. Brinkmann, D. Chourrout, and H. Philippe. 2006. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 439 965–968. [DOI] [PubMed] [Google Scholar]
- 27.Goto T., F. Holzinger, L. R. Hagey, C. Cerrè, H-T. Ton-Nu, C. D. Schteingart, J. H. Steinbach, B. L. Shneider, and A. F. Hofmann. 2003. Physicochemical and physiological properties of 5α-cyprinol sulfate, the toxic bile salt of cyprinid fish. J. Lipid Res. 44 1643–1651. [DOI] [PubMed] [Google Scholar]
- 28.Chatman K., T. Hollenbeck, L. R. Hagey, M. Vallee, R. Purdy, F. Weiss, and G. Suizdak. 1999. Nanoelectrospray mass spectrometry and precursor ion monitoring for quantitative steroid analysis and attomole sensitivity. Anal. Chem. 71 2358–2363. [DOI] [PubMed] [Google Scholar]
- 29.Rossi S. S., J. L. Converse, and A. F. Hofmann. 1987. High pressure liquid chromatography analysis of conjugated bile acids in human bile: simultaneous resolution of sulfated and unsulfated lithocholyl amidates and the common conjugated bile acids. J. Lipid Res. 28 589–595. [PubMed] [Google Scholar]
- 30.Krasowski M. D., K. Yasuda, L. R. Hagey, and E. G. Schuetz. 2005. Evolutionary selection across the nuclear hormone receptor superfamily with a focus on the NR1I subfamily (vitamin D, pregnane X, and constitutive androstane receptors). Nucl. Recept. 3 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reschly E. J., A. C. D. Bainy, J. J. Mattos, L. R. Hagey, N. Bahary, S. R. Mada, J. Ou, R. Venkataramanan, and M. D. Krasowski. 2007. Functional evolution of the vitamin D and pregnane X receptors. BMC Evol. Biol. 7 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mi L-Z., S. Devarakonda, J. M. Harp, Q. Han, R. Pellicciari, T. M. Willson, S. Khorasanizadeh, and F. Rastinejad. 2003. Structural basis for bile acid binding and activation of the nuclear receptor FXR. Mol. Cell. 11 1093–1100. [DOI] [PubMed] [Google Scholar]
- 33.Watkins R. E., G. B. Wisely, L. B. Moore, J. L. Collins, M. H. Lambert, S. P. Williams, T. M. Willson, S. A. Kliewer, and M. R. Redinbo. 2001. The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science. 292 2329–2333. [DOI] [PubMed] [Google Scholar]
- 34.Rochel N., J. M. Wurtz, A. Mitschler, B. Klaholz, and D. Moras. 2000. The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol. Cell. 5 173–179. [DOI] [PubMed] [Google Scholar]
- 35.Jones G., P. Willett, R. C. Glen, A. R. Leach, and R. Taylor. 1997. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 267 727–748. [DOI] [PubMed] [Google Scholar]
- 36.Liang J., H. Edelsbrunner, and C. Woodward. 1998. Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci. 7 1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson I. G., G. A. D. Haslewood, R. S. Oldham, B. Amos, and L. Tökés. 1974. A more detailed study of bile salt evolution, including techniques for small-scale identification and their application to amphibian biles. Biochem. J. 141 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li W., A. P. Scott, M. J. Siefkes, H. Yan, Q. Liu, S-S. Yun, and D. A. Gage. 2002. Bile acid secreted by male sea lamprey that acts as a sex pheromone. Science. 296 138–141. [DOI] [PubMed] [Google Scholar]
- 39.Anderson I. G., T. Briggs, and G. A. D. Haslewood. 1964. Comparative studies of ‘bile salts.’ 18. The chemistry of cyprinol. Biochem. J. 90 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reschly, E. J., N. Ai, W. J. Welsh, S. Ekins, L. R. Hagey, and M. D. Krasowski. 2008. Ligand specificity and evolution of liver X receptors. J. Steroid Biochem. Mol. Biol. In press. [DOI] [PMC free article] [PubMed]
- 41.Schultz J. R., H. Tu, A. Luk, J. J. Repa, J. C. Medina, L. Li, S. Schwendner, S. Wang, M. Thoolen, D. J. Mangelsdorf, et al. 2000. Role of LXRs in control of lipogenesis. Genes Dev. 14 2831–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitro N., L. Vargas, R. Romeo, A. Koder, and E. Saez. 2007. T0901317 is a potent PXR ligand: implications for the biology ascribed to LXR. FEBS Lett. 581 1721–1726. [DOI] [PubMed] [Google Scholar]
- 43.Houck K. A., K. M. Borchert, C. D. Hepler, J. S. Thomas, K. S. Bramlett, L. F. Michael, and T. P. Burris. 2004. T0901317 is a dual LXR/FXR agonist. Mol. Genet. Metab. 83 184–187. [DOI] [PubMed] [Google Scholar]
- 44.Xue Y., E. Chao, W. J. Zuercher, T. M. Willson, J. L. Collins, and M. R. Redinbo. 2007. Crystal structure of PXR-T1317 complex provides a scaffold to examine the potential for receptor antagonism. Bioorg. Med. Chem. 15 2156–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yagi K., Y. Satou, F. Mazet, S. M. Shimeld, B. Degnan, D. Rokhsar, M. Levine, Y. Kohara, and N. Satoh. 2003. A genomewide survey of developmentally relevant genes in Ciona intestinalis. III. Genes for Fox, ETS, nuclear receptors and NFκB. Dev. Genes Evol. 213 235–244. [DOI] [PubMed] [Google Scholar]
- 46.Ekins S., E. J. Reschly, L. R. Hagey, and M. D. Krasowski. 2008. Evolution of pharmacologic specificity in the pregnane X receptor. BMC Evol. Biol. 8 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rannug A., U. Rannug, H. S. Rosenkranz, L. Winqvist, R. Westerholm, E. Agurell, and A-K. Grafström. 1987. Certain photooxidized derivatives of tryptophan bind with very high affinity to the Ah receptor and are likely to be endogenous signal substances. J. Biol. Chem. 262 15422–15427. [PubMed] [Google Scholar]
- 48.Nishimaki-Mogami T., Y. Kawahara, N. Tamehiro, T. Yoshida, K. Inoue, Y. Ohno, T. Nagao, and M. Une. 2006. 5α-Bile alcohols function as farnesoid X receptor antagonists. Biochem. Biophys. Res. Commun. 339 386–391. [DOI] [PubMed] [Google Scholar]
- 49.Krasowski M. D., K. Yasuda, L. R. Hagey, and E. G. Schuetz. 2005. Evolution of the pregnane X receptor: adaptation to cross-species differences in biliary bile salts. Mol. Endocrinol. 19 1720–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moore L. B., J. M. Maglich, D. D. McKee, B. Wisely, T. M. Willson, S. A. Kliewer, M. H. Lambert, and J. T. Moore. 2002. Pregnane X receptor (PXR), constitutive androstane receptor (CAR), and benzoate X receptor (BXR) define three pharmacologically distinct classes of nuclear receptors. Mol. Endocrinol. 16 977–986. [DOI] [PubMed] [Google Scholar]
- 51.Chrencik J. E., J. Orans, L. B. Moore, Y. Xue, L. Peng, J. L. Collins, G. B. Wisely, M. H. Lambert, S. A. Kliewer, and M. R. Redinbo. 2005. Structural disorder in the complex of human PXR and the macrolide antibiotic rifampicin. Mol. Endocrinol. 19 1125–1134. [DOI] [PubMed] [Google Scholar]
- 52.Adachi R., Y. Honma, H. Masuno, K. Karana, I. Shimomura, S. Yamada, and M. Makishima. 2005. Selective activation of vitamin D receptor by lithocholic acid acetate, a bile acid derivative. J. Lipid Res. 46 46–57. [DOI] [PubMed] [Google Scholar]
- 53.Ortlund E. A., J. T. Bridgham, M. R. Redinbo, and J. W. Thornton. 2007. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 317 1544–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bridgham J. T., S. M. Carroll, and J. W. Thornton. 2006. Evolution of hormone-receptor complexity by molecular exploitation. Science. 312 97–101. [DOI] [PubMed] [Google Scholar]
- 55.Sonoda J., W. Xie, J. M. Rosenfeld, J. L. Barwick, P. S. Guzelian, and R. M. Evans. 2002. Regulation of a xenobiotics sulfonation cascade by nuclear pregnane X receptor (PXR). Proc. Natl. Acad. Sci. USA. 99 13801–13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reschly E. J., and M. D. Krasowski. 2006. Evolution and function of the NR1I nuclear hormone receptor subfamily (VDR, PXR, and CAR) with respect to metabolism of xenobiotics and endogenous compounds. Curr. Drug Metab. 7 349–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kornprobst J-M., C. Sallenave, and G. Barnathan. 1998. Sulfated compounds from marine organisms. Comp. Biochem. Physiol. B. 119 1–51. [DOI] [PubMed] [Google Scholar]
- 58.Yoshida M., M. Murata, K. Inaba, and M. Morisawa. 2002. A chemoattractant for ascidian spermatozoa is a sulfated steroid. Proc. Natl. Acad. Sci. USA. 99 14831–14836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.