Abstract
StarD4 protein is a member of the StarD4 subfamily of steroidogenic acute regulatory-related lipid transfer (START) domain proteins that includes StarD5 and StarD6, proteins whose functions remain poorly defined. The objective of this study was to isolate and characterize StarD4's sterol binding and to determine in a hepatocyte culture model its sterol transport capabilities. Utilizing purified full-length StarD4, in vitro binding assays demonstrated a concentration-dependent binding of [14C]cholesterol by StarD4 similar to that of the cholesterol binding START domain proteins StarD1 and StarD5. Other tested sterols showed no detectable binding to StarD4, except for 7α-hydroxycholesterol, for which StarD4 demonstrated weak binding on lipid protein overlay assays. Subsequently, an isolated mouse hepatocyte model was used to study the ability of StarD4 to bind/mobilize/distribute cellular cholesterol. Increased expression of StarD4 in primary mouse hepatocytes led to a marked increase in the intracellular cholesteryl ester concentration and in the rates of bile acid synthesis. The ability and specificity of StarD4 to bind cholesterol and, as a function of its level of expression, to direct endogenous cellular cholesterol suggest that StarD4 plays an important role as a directional cholesterol transporter in the maintenance of cellular cholesterol homeostasis.
Keywords: liver, protein, metabolism, steroidogenic acute regulatory protein, steroidogenic acute regulatory-related lipid transfer domain
Cholesterol is a structural component of mammalian cell membranes and also serves as a precursor to bile acids (in the liver), steroid hormones (in the adrenal, testis, and ovaries), and vitamin D. Homeostasis of cholesterol within the body is maintained through the coordinated regulation of its cell-mediated uptake, transport/trafficking, sorting, biosynthesis, storage (i.e., esterification), secretion, and catabolism to bile acids (1). More specifically, the steroidogenic acute regulatory-related lipid transfer (START) domain superfamily of proteins has been shown to be involved in several pathways of intracellular trafficking of cholesterol (2–4). It has been predicted that all proteins with a START domain contain a similar binding pocket with modifications in that pocket that determine ligand binding specificity and function (5). The START domains are 200–210 amino acid motifs that appear in a wide range of proteins and have been implicated in several cellular functions, including lipid transport and metabolism, signal transduction, and transcriptional regulation (3, 4, 6). The START-related lipid transfer protein 4 (StarD4) belongs to the StarD4 subfamily, a START subfamily that also contains the proteins StarD5 and StarD6. The proteins in the StarD4 subfamily have been shown to contain 205–233 amino acid residues, sharing 26–32% identity with each other (7). Within the StarD4 subfamily, StarD5 has been shown in vitro to bind cholesterol and 25-hydroxycholesterol (4). StarD1 and MLN64/StarD3 (the nearest related proteins to the StarD4 subfamily) have been shown to bind only cholesterol (4, 8). Furthermore, full-length StarD1, StarD4, StarD5 and truncated MLN64/StarD3 have been reported to increase steroidogenesis and/or increase free cholesterol in the microsomes after overexpression in cell culture, representative evidence of their ability to transfer cholesterol (4, 9–12).
The first START domain protein crystal structure reported was the C-terminal portion of human MLN64/StarD3 (8), followed subsequently by the structure of mouse StarD4 (13). The protein structures of MLN64/StarD3 and StarD4 revealed similar secondary structural elements and a hydrophobic tunnel with a size consistent with the binding of one cholesterol molecule (8, 13). Less related to the StarD4 subfamily, but also containing a START domain with a known lipid ligand, is the phosphatidylcholine transfer protein (PCTP/StarD2) (14). The crystal structure of PCTP/StarD2 also shows a hydrophobic tunnel, but in contrast to MLN64/StarD3 and StarD4, it appears to selectively bind phosphatidylcholine (15). More recent modeling studies of the structure of StarD1 and MLN64 have shown that cholesterol appears to be bound by the predicted hydrophobic tunnel of both proteins and reveals changes in the loop at the entrance of the hydrophobic tunnel that may be sufficient for the uptake and release of cholesterol (16).
Although the roles of proteins like StarD1, PCTP/StarD2, and MLN64/StarD3 have been studied extensively with a better appreciation of their possible roles (9–11, 13–15, 17, 18), the roles of the StarD4 subfamily proteins remain unclear. In contrast to StarD1 and MLN64/StarD3, StarD4, StarD5, and StarD6 do not have N-terminal targeting sequences that should direct these proteins to specific cellular organelles. Therefore, they are predicted to be cytoplasmic proteins like PCTP/StarD2 (7, 19).
The expression of the StarD4 subfamily proteins within different tissues/cells is now starting to be revealed for StarD6 and StarD5. However, little is known about StarD4, although some preliminary studies have suggested a diffuse cytoplasmic and nuclear localization (expression studies utilizing a green fluorescent protein-StarD4 fusion protein in HeLa cells) (20). StarD6 has been localized in mouse testis but has not been found in the ovary, suggesting a special role for StarD6 during germ cell maturation in the adult testis (7, 21). StarD5 mRNA was first found in whole human liver (4), with StarD5 liver protein later localized to the Kupffer cells. Subsequent studies have shown the detection of StarD5 in high concentrations in other immune-related cells (macrophages, monocytes, basophils, etc.). Within the cell, StarD5 was found not only in the cytoplasm but also exhibited a strong association with Golgi membranes (22).
With respect to the regulation of StarD4, StarD5, and StarD6, there exist only relatively preliminary data about StarD6. StarD5 mRNA expression is induced in response to endoplasmic reticulum (ER) stress, either in free cholesterol-loaded mouse macrophages or in NIH-3T3 cells after being treated with different ER stressors (12). In contrast, StarD4 mRNA expression has been reported as responsive to sterols through sterol-regulatory element binding proteins (SREBPs), being sterol-repressed and predominantly activated by SREBP-2 activation (12). Most recently, new studies have shown a possible regulation of StarD4 mRNA expression under ER stress conditions, although limited to the early phase (23).
The objectives of this study were to purify human StarD4, to determine its sterol binding capabilities/specificities, and to alter its levels of expression in order to pursue the protein's function. The in vitro binding assays demonstrating StarD4's ability to selectively bind [14C]cholesterol, coupled with the observation of increased intracellular cholesteryl ester formation and bile acid synthesis following StarD4 overexpression, are supportive of StarD4 as a cholesterol carrier to different cellular compartments. Furthermore, under the conditions explored, it appears that cholesterol redistribution occurred as a function of StarD4 expression.
EXPERIMENTAL PROCEDURES
Materials
The QIAprep-Spin Miniprep Kit, QIAquick PCR Purification Kit, and QIAquick Gel Extraction Kit for nucleotide purification and extraction and nickel-nitrilotriacetic acid agarose resin for cholesterol binding assays were purchased from Qiagen (Valencia, CA). The following materials were purchased from Novagen (Madison, WI): His-Bind resin, 8× Charge buffer, and 4× Strip buffer, Escherichia coli strain Novablue (recA, endA, lacIq) used for construction of clones, and pET-30a(+) vector. Glutathione Sepharose 4B, reduced glutathione, and pGEX-4T-3 vector were purchased from Amersham (Piscataway, NJ). Antibiotics used for bacterial cultures were purchased from Sigma (St. Louis, MO). Enzymes used for cloning procedures were purchased from Promega (Madison, WI). The labeled sterols [14C]cholesterol and [3H]25-hydroxycholesterol were purchased from Perkin-Elmer (Boston, MA); 7α-hydroxycholesterol, 7-keto-cholesterol, 20-hydroxycholesterol, 24-hydroxycholesterol, 27-hydroxycholesterol, and 24,25-hydroxycholesterol were purchased from Steraloids (Newport, RI); stigmasterol was from ICN Biomedicals (Costa Mesa, CA). [14C]27-hydroxycholesterol was made in our laboratory as described previously (4). SuperSignal® West Pico Chemiluminiscent Substrate was purchased from Pierce (Rockford, IL). The E. coli strain BL21 was used. All oligonucleotides were synthesized at the Sigma-Genosys facility.
Expression and purification
PCR procedure
PCR experiments were carried out in a total volume of 0.1 ml. PCR performed with the StarD5 cDNA contained PCR buffer (1×), 1.5 mM MgCl2, 250 μM deoxynucleoside triphosphates, 20 ng of template, 1 μM of each primer, and 1 unit of Taq polymerase. Amplifications were performed using the following reaction conditions: 1 cycle of 2 min at 95°C, followed by 30 cycles of 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C. The reaction was finished with 1 cycle of 10 min at 72°C. The PCR products were then analyzed by agarose gel electrophoresis.
Cloning and purification of His-tag/StarD5, His-tag/(N62)StarD1, and His-tag/StarD4 fusion proteins
The fusion proteins His-tag/StarD5 and His-tag/(N62)StarD1 were obtained as described previously (4). The cDNA corresponding to the human StarD4 protein was obtained by PCR using the plasmid pZEROTG-CMV/hStAR-D4 as template and the following primers: 5′-GGATACGTATGAATTCATGGAAGGCCTGTCTGATGTT-3′ and 5′-TCAGGCTACGGAATTCTCATAAAGCTTTTCGTAAATCAC-3′ (the EcoRI restriction site is underlined). The PCR product was purified and then digested with EcoRI. After digestion, the fragment was ligated into pET-30a(+) vector, previously digested with EcoRI and purified, creating a C-His-tag/StarD4 DNA, which provided a protease (enterokinase) cleavage site between the histidine (His) tag and the sequence of StarD4. The ligation mixture was used to transform Novablue competent cells. The single colonies grew on Luria-Bertani (LB) agar plates and were screened for the 618 nucleotide fragment by digestion with EcoRI, corresponding to the StarD4 coding region. For further confirmation, the plasmid with the StarD4 coding region was sequenced and also confirmed the right direction of the cloned cDNA. Finally, the plasmid was used to transform BL21 competent cells for expression of the recombinant protein. Purification, Western blot analysis, and protein sequencing via Edman chemistry were performed as described previously for StarD5 (4).
For protein sequencing and circular dichroism (CD) spectroscopy analysis, the fusion protein was digested with enterokinase, following the manufacturer's instructions, to obtain the native StarD4 protein.
Cloning and purification of glutathione S-transferase protein and glutathione S-transferase/StarD4 fusion protein
The cloning of the human StarD4 cDNA into the vector pGEX-4T-3, creating a glutathione S-transferase (GST)-StarD4 DNA, was performed following a strategy similar to the one described above but using primers 5′-GTTCCGCGTGGATCCATGGAAGGCCTGTCTGATGTT-3′ (the BamHI restriction site is underlined) and 5′-TCATTATTACTCGAGTAAAGCTTTTCGTAAATCACC-3′ (the XhoI restriction site is underlined). The single colonies grew on LB agar plates and were screened for the 618 nucleotide fragment by digestion with BamHI and XhoI, corresponding to the StarD4 coding region. For further confirmation, the plasmid with the StarD4 coding region was sequenced using the commercially available primers pGEX 5′ and pGEX 3′ (Amersham) and also confirmed the right direction of the cloned cDNA. The plasmid was used to transform BL21 competent cells for the expression of the recombinant protein. To purify GST and GST/StarD4, cells BL21(DE3) containing pGEX-4T-3 or pGEX-4T-3/StarD4 plasmids were grown in 10 ml of LB broth with 100 μg/ml ampicillin at 37°C overnight. One liter of LB broth containing 0.36% glucose was inoculated with 6.25 ml of the overnight culture and was grown at 37°C with shaking until the optical density at 600 nm reached 0.3–0.6 (∼3 h). At this point, 1 ml of a 1 M isopropylthio-β-galactoside solution was added and incubated for an additional 24 h at 25°C with shaking. Cells were harvested by centrifugation (8,000 rpm for 10 min at 4°C), resuspended in 50 ml of BugBuster 1X, and incubated at room temperature for 30 min for complete bacterial lysis. Bacterial lysates were centrifuged at 14,000 rpm for 20 min at 4°C. The supernatant was incubated with Glutathione Sepharose 4B overnight at 4°C. The GST protein and GST/StarD4 fusion protein were purified following a batch method as described by the manufacturer (Amersham). Fractions from each step of the purification were analyzed by SDS-PAGE, under reducing conditions on a 12% gel, following the method described by Laemmli (24).
Preparation of chimeric pZEROTG-CMV/hStarD4 and CMV-StarD1 constructs and propagation
pZEROTG-CMV/hStarD4 was prepared by placing a PCR fragment containing the coding region for the human StarD4 into the pZEROTG-CMV expression vector opened with EcoRV and dephosphorylated with calf intestinal alkaline phosphatase. The primers used for the reaction were 5′-ATGGAAGGCCTGTCTGATGTTGC-3′ and 5′-TCATAAAGCTTTTCGTAAATCACC-3′. Five micrograms of human liver total RNA in a final volume of 20 μl were used as template in an room temperature reaction using 100 pmol of random hexamers as primers and 1.5 mM Mg2+ for 10 min at 25°C and 1 h at 42°C. Five microliters of the room temperature reaction were used as template in the PCR. The PCR conditions were as follows: 5 min at 98°C, 5 min at 72°C (1 cycle), followed by 35 cycles of 1 min at 94°C, 1 min at 62°C, and 1.5 min at 72°C. The reaction was finished with one cycle of 10 min at 72°C. The pZEROTG-CMV expression vector was obtained from Dr. Kris Valerie's laboratory at Virginia Commonwealth University. The CMV-StarD5 and CMV-StarD1 adenoviruses were constructed as described previously (4, 25).
Recombinant virus CMV-StarD4 was transfected into human embryonic kidney 293 cells (American Type Culture Collection, Manassas, VA). Adenovirus DNA from the resulting plaques was further screened by Southern blotting for the presence of the inserts. To purify the recombinant virus, the crude supernatant was carefully layered over a two-step CsCl gradient as described previously (25).
Isolation of primary mouse hepatocytes
Hepatocytes were isolated from male C57BL mice as described previously (26) using the collagenase perfusion technique of Bissell and Guzelian (27).
Infection of primary mouse hepatocytes with adenovirus
Primary mouse hepatocytes were isolated and plated as described previously at 15–20% of normal density in Williams medium with insulin and dexamethasone, on six-well culture plates, with a coverslip at the bottom of each well and incubated at 37°C and 5% CO2. The cells, 24 h after plating, were infected with purified recombinant adenovirus encoding StarD4, StarD1, StarD5, or Ad-CMV control virus. After 2 h of infection, the medium was removed and replaced with fresh medium.
RNA isolation and real-time RT-PCR
Total RNA was isolated from cultured cells infected with recombinant adenovirus encoding StarD4 using the SV Total RNA Isolation System (Promega). Two and one-half micrograms of RNA were reverse-transcribed in 20 μl using oligo(dT) and Moloney murine leukemia virus reverse transcriptase (Invitrogen) following the manufacturer's instructions. Relative quantification of StarD4 gene expression was performed in 20 μl reactions using 25 ng of cDNA, forward and reverse primers at 2 μM, and LightCycler® 480 SYBR Green I Master (Roche, Indianapolis, IN). Primer pairs for human StarD4 (5′-CAAAGCCCAAGGTGTTATAGATGAC-3′ and 5′-ACAGCAATTCTCTTCAAAGTTCTCC-3′), StarD1 (5′-AGCTCTCTACTCGGTTCTCGGCTA-3′ and 5′-CTGCTGACTCTCCTTCTTCCAGCC-3′), and StarD5 (5′-CCAAAGCATCACTGACACCCT-3′ and 5′-GGTCCCATCCTCATATCTCTTGAC-3′) were designed with Beacon Designer 2 (Premier Biosoft). Primer pair sequences used for mouse β-actin were 5′-TCTACGAGGGCTATGCTCTCC-3′ and 5′-TCTTTGATGTCACGCACGATTTC-3′. A 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA) was used with the default thermal cycling profile of 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 1 min. Carboxy-X-rhodamine (Roche) was the passive reference dye for normalization. The threshold was set at 0.2 unit of normalized fluorescence. Relative standard curves were plotted for StarD4, StarD1, and StarD5, and the mean threshold cycle for each cDNA sample was expressed as an arbitrary value relative to the standard. For each cDNA, values for StarD4, StarD1, and StarD5 were normalized to the corresponding value for β-actin and expressed as a ratio.
Biochemical characterization
CD spectroscopy
Far-ultraviolet (UV; 195–250 nm) CD measurements were carried out in a 1.0 mm path length cuvette at 200 μg protein/ml purified protein (9 μM for StarD4) in 0.5× PBS buffer, pH 7.4, at 20°C in a Olis CD module at a scan speed of 2.7 nm/min; three spectra were averaged. Also, far-UV (195–250 nm) CD spectra were obtained with StarD4 protein at 200 μg/ml (9 μM) alone and in the presence of cholesterol at 1, 3, 6, 9, 12, and 30 μM final concentrations.
Sterol binding assays
Binding assays were performed according to the method described previously by Rodriguez-Agudo et al. (4). The sterols analyzed in these binding studies were [14C]cholesterol, [3H]25-hydroxycholesterol, and [14C]27-hydroxycholesterol. When labeled sterols were not available, competition assays were performed with unlabeled sterols (7α-hydroxycholesterol, 24-hydroxycholesterol, and 24,25-hydroxycholesterol) as described previously by Rodriguez-Agudo et al. (4).
Lipid protein overlay assays
Lipid protein overlay(LPO) assays were performed according to the method described by Dowler, Kular, and Alessi (28) with small modifications. Briefly, sterols were diluted in a 2:1:0.8 solution of methanol-chloroform-water to different concentrations ranging between 10 and 0.1 mM. Nitrocellulose membranes were soaked in TBS buffer (50 mM Tris-HCl, pH 7.5, and 150 mM NaCl) before 1 μl of the solutions was spotted and allowed to dry for 1 h at room temperature. Then, the membranes were incubated with gentle rocking in TBS buffer + 2 mg/ml BSA for 1 h at room temperature. The membranes were incubated overnight at 4°C with gentle rocking in TBS buffer + 2 mg/ml BSA containing 0.6 μg/ml GST protein or GST/StarD4 fusion protein and washed three times over 30 min with TBS buffer + 0.1% Tween 20 (TBST). The membranes were then incubated for 1 h at room temperature with TBST + 2 mg/ml BSA containing a 1:2,000 dilution of anti-GST monoclonal antibody produced in mouse (Sigma) and washed three times over 30 min with TBST. The membranes were finally incubated for 1 h at room temperature with TBST + 2 mg/ml BSA containing a 1:5,000 dilution of the HRP-conjugated anti-mouse secondary antibody and washed three times over 30 min with TBST. Protein bound to the lipids on the membrane was visualized using SuperSignal® West Pico Chemiluminiscent Substrate with a LAS-1000 camera (FujiFilm) in Intelligent Dark Box (FujiFilm) using Image Reader LAS-1000 Pro software.
Functional studies
Filipin staining of cholesterol in hepatocytes
Primary mouse hepatocytes were prepared and infected as mentioned above with purified recombinant adenovirus encoding StarD4, StarD5, StarD1, or Ad-CMV control virus. The next day, media were removed and the cells were washed twice with PBS and fixed with 3.7% formaldehyde in PBS for 10 min at 4°C. The cells were then washed three times, 5 min each, with PBS while rocking gently at room temperature. Cells were permeabilized with 0.1% Triton, in PBS, for 3 min at 4°C, then stained with 5 mg/ml Filipin in PBS plus 0.5% BSA for 30 min at 37°C, and washed three times, 5 min each, with PBS while rocking gently at room temperature in the dark. The coverslips containing the cells were then taken from the wells and mounted onto glass slides. The cells were allowed to dry for at least 45 min before being placed on a fluorescence microscope stage at excitation filter 360/40 nm, emission filter 460/50 nm, and beam splitter 400 nm.
Integrated optical density determination
Fluorescence was quantified via Image-Pro® Plus analysis software and expressed as integrated optical density.
Oil Red O staining
Primary mouse hepatocytes were prepared and infected as mentioned above with purified recombinant adenovirus encoding StarD4, StarD1, or Ad-CMV control virus. After 48 h, media were removed and the cells were washed twice with PBS and fixed with 3.7% formaldehyde in PBS for 10 min at room temperature. The cells were washed three times, 5 min each, with PBS while rocking gently at room temperature and then stained with a solution of 0.3% Oil Red O in PBS/isopropanol (1:1) and washed three times, 5 min each, with PBS while rocking gently at room temperature. Finally, the coverslips containing the cells were taken from the wells and mounted onto glass slides before being placed on a bright-field microscope stage.
Quantification of bile acid synthesis rates and cholesteryl ester formation
Bile acid synthesis rates were determined by the addition of 2.5 mCi of [14C]cholesterol to each 150 mm plate of confluent primary mouse hepatocyte cultures (2.5 × 107 cells) at 24 h after plating. The medium and the cells were harvested at 48 h after viral infection with recombinant adenovirus encoding StarD4, StarD1, StarD5, or Ad-CMV control virus. Conversion of [14C]cholesterol to [14C]methanol-water-soluble products was determined by scintillation counting after Folch extraction (29) with chloroform-methanol (2:1, v/v) of the culture medium. The rates of bile acid biosynthesis following recombinant adenovirus infection were calculated as the ratio of [14C]methanol-water-soluble counts to the sum of chloroform-methanol-water-soluble counts. The rates of cholesteryl ester formation were determined from the chloroform phase by TLC. To determine cholesteryl ester formation from newly synthesized cholesterol, 5 μCi of (1) acetate was added instead of [14C]cholesterol.
TLC analysis
Sterol samples (chloroform phase) from primary mouse hepatocytes were analyzed by TLC as described previously (25). 14C-labeled sterols were visualized with a PhosphorImager (FujiFilm).
Statistics
Data from sterol binding, filipin staining, bile acid synthesis rates, and real-time quantitative RT-PCR in primary mouse hepatocytes are reported as means ± SEM of three separate experiments.
RESULTS
Purification of the recombinant human StarD4
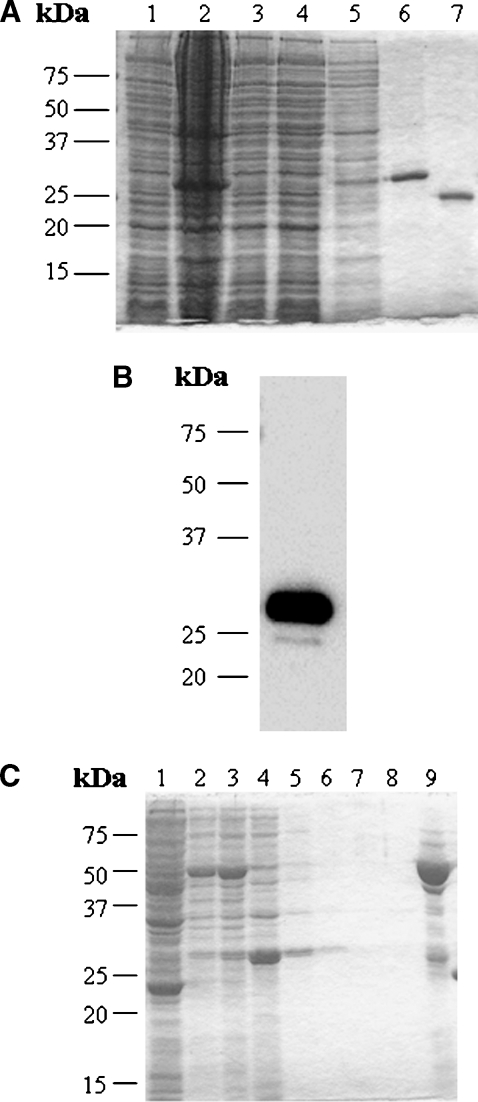
Protein expression using the human StarD4 cDNA cloned into pET-30a(+) resulted in a 27 kDa major protein band by SDS-PAGE analysis (Fig. 1A, lane 6). From 575 mg of total soluble cell protein derived from 1 liter of culture, 6.3 mg of His-tag/protein was purified. Recombinant His-tag/protein accounted for 7% of the total soluble cell protein, which corresponds to 40 mg of His-tag/StarD4. The yield was 15.75%, calculated as milligrams of pure His-tag/protein obtained divided by the total His-tag/protein in the soluble cell fraction. The pure protein obtained was immunoreactive to a monoclonal antibody against polyhistidine as determined by Western blot analysis (Fig. 1B). After digestion of the His tag with recombinant enterokinase (Fig. 1A, lane 7), the purified protein was analyzed by N-terminal sequencing, and after six cycles of Edman degradation, the sequence obtained was Met-Glu-Gly-Leu-Ser-Asp, which is identical to the N-terminal sequence of the human StarD4 protein.
Fig. 1.
Purification and identification of recombinant human StarD4 protein. A: SDS-PAGE analysis and Coomassie blue staining of human StarD4 overexpressed in BL21 cells at each step of the purification. Lane 1, total soluble protein before induction (30 μg); lane 2, total soluble protein after induction (50 μg); lane 3, total soluble protein after incubation with the resin (30 μg); lane 4, proteins eluted with wash buffer I (30 μg); lane 5, proteins eluted with wash buffer II (10 μg); lane 6, protein eluted with lysis buffer plus 1 M imidazole (20 μg); lane 7, StarD4 protein after cleavage of the His tag with recombinant enterokinase. A major protein band was found around 27 kDa. B: Western blot analysis of human StarD4 with monoclonal anti-polyhistidine antibody. Protein eluted with lysis buffer plus 1 M imidazole (200 ng). C: SDS-PAGE analysis and Coomassie blue staining of GST-StarD4 fusion protein overexpressed in BL21 cells at each step of the purification. Lane 1, total soluble protein before induction (50 μg); lane 2, total soluble protein after induction (30 μg); lane 3, total soluble protein before incubation with Glutathione Sepharose 4B (30 μg); lane 4, proteins not bound by the resin (30 μg); lanes 5 to 8, proteins eluted with PBS (10 μg); lane 9, protein eluted with reduced glutathione (20 μg).
In order to confirm StarD4 lipid binding studies by LPO assays, a recombinant StarD4 with a GST tag was required. Protein expression using the human StarD4 cDNA resulted in a 50 kDa major protein band by SDS-PAGE analysis (Fig. 1C, lane 9). The pure protein obtained was immunoreactive to a monoclonal antibody against GST, as determined by Western blot analysis (data not shown). To confirm the presence of StarD4 in the fusion protein, the protein was digested with thrombin and loaded again into Glutathione Sepharose 4B to separate GST from StarD4 protein. StarD4 purified protein was analyzed by N-terminal sequencing as described above.
StarD4 CD spectroscopy
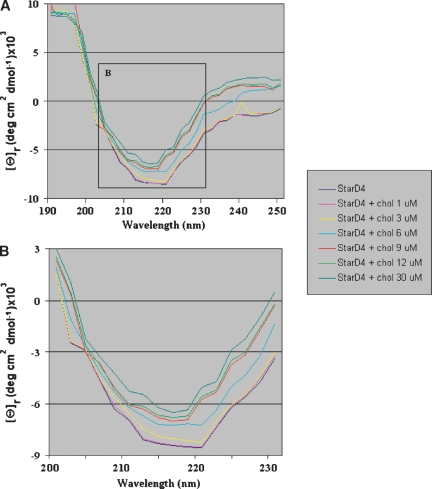
The far-UV CD spectrum of the StarD4 protein gave a minimum at 220 nm (Fig. 2), suggestive of the presence of a large number of β-sheet structures as opposed to other secondary structures (i.e., random coils and α-helices). The minimum at 210, however, suggests the presence of some α-helical structures in the protein (30). Far-UV CD spectra of the StarD4 protein following incubation with cholesterol (Fig. 2) showed changes in the ellipticity at 220 nm, consistent with StarD4 binding of cholesterol, as shown in cholesterol binding assays and LPO assays. Addition of cholesterol at 1 and 3 μM to the StarD4 solution did not change the CD spectrum of the protein. At 6 μM cholesterol, the spectrum still shows the minimum at 220 nm and another one at 216 nm, which indicates the binding of cholesterol. At equimolar amounts of StarD4 and cholesterol, the 220 nm minimum of StarD4 completely shifts to a minimum at 216 nm. At greater concentrations of cholesterol (12 and 30 μM), the spectrum remained similar to the one observed at 9 μM cholesterol, showing a minimum at 216 nm, only shifting the spectra because the addition of greater amounts cholesterol to the solution diluted the protein.
Fig. 2.
Far-ultraviolet CD spectrum of StarD4. A: The data shown are averages of three scans of the recombinant human protein StarD4 alone at 200 μg/ml (9 μM) and in the presence of cholesterol at different concentrations. B: The inset from A showing a closer look at the spectra between 200 and 230 nm of StarD4 protein alone and in the presence of cholesterol at different concentrations.
StarD4-containing START domain binds cholesterol
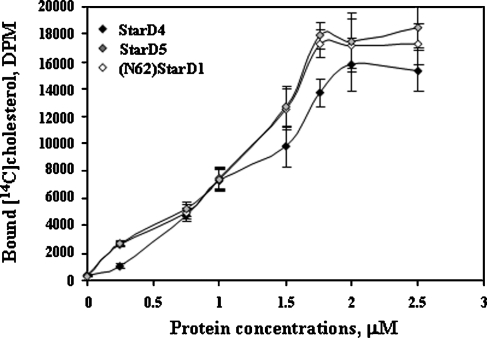
Rodriguez-Agudo et al. (4) and Tsujishita and Hurley (8) have reported data showing direct binding of cholesterol by a His-tagged StarD1 (full-length protein), (N62)StarD1 (a functional cholesterol binding StarD1 protein that lacks the mitochondrial targeting sequence), and StarD5. To determine whether StarD4 His-tagged protein was capable of binding cholesterol, the same method was used. At 2 μM protein, all [14C]cholesterol added was bound to the protein, at a stoichiometry of 1:0.9 (StarD4/cholesterol) (Fig. 3), similar to (N62) StarD1 and StarD5 (Fig. 3). Direct binding assays utilizing StarD4 with [3H]25-hydroxycholesterol and [14C]27-hydroxycholesterol or competition assays utilizing StarD4 between [14C]cholesterol and 7α-hydroxycholesterol, 24-hydroxycholesterol, and 24,25-hydroxycholesterol (data not shown) did not show any evidence of binding.
Fig. 3.
Characterization of StarD4 sterol binding specificities. Different concentrations of His-tagged StarD5, (N62)StarD1 (a completely functional StarD1 protein, as described in Results), and StarD4 were incubated with 1.76 μM [14C]cholesterol for 1 h at 37°C. Data are presented as means ± SEM of three independent experiments.
As a negative control, the assay was performed with StarD4 that had been heat-inactivated. Heat-inactivated StarD4 did not bind [14C]cholesterol (data not shown). As reported previously, the assay carried out with bovine albumin at different concentrations showed no evidence of nonspecific binding of [14C]cholesterol to the nickel resin (4).
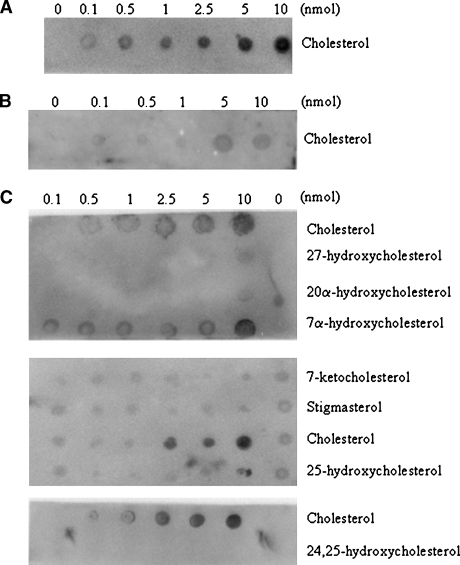
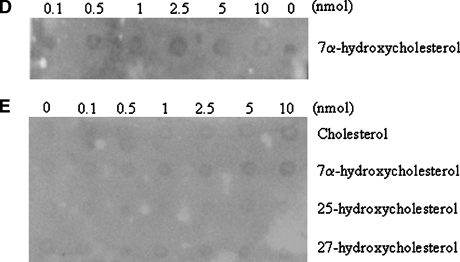
The binding assays described above are useful but with the limitation of the availability of radiolabeled sterols. When radiolabeled sterols were not available, competition assays were performed, but again with the limitation of possible false negatives because of the higher affinity of StarD4 for cholesterol. The LPO assay solves this problem, is less expensive, and is confirmatory of an existing standardized assay. The results with the LPO assay, however, are only qualitative. The results confirmed the ability of StarD4 to bind cholesterol (Fig. 4A), as shown with the lipid binding assays (Fig. 3). When GST-StarD4 fusion protein was incubated with cholesterol prior to the LPO assay, only a weak signal was obtained on the membrane where 5 and 10 nmol amounts were spotted (Fig. 4B). All of the other sterols tested were negative (Fig. 4C), with the exception of 7α-hydroxycholesterol, which was positive at 10 nmol (Fig. 4C). When GST-StarD4 fusion protein was incubated with cholesterol prior to the LPO assay with 7α-hydroxycholesterol, no signal was obtained on the membrane at any of the amounts spotted (Fig. 4D). As a negative control, purified GST protein was used on LPO assays with different sterols (Fig. 4E).
Fig. 4.
Lipid protein overlay (LPO) assays of the steroidogenic acute regulatory-related lipid transfer (START) domain containing protein StarD4 with sterols. The ability of the StarD4 protein to bind various sterols was analyzed. A: Serial dilutions (0–10 nmol) of cholesterol were spotted onto nitrocellulose membranes, which were then incubated with GST-StarD4 protein. B: Binding affinity for cholesterol by GST-StarD4 fusion protein was reduced when the protein was preincubated with cholesterol for 1 h at 37°C previous to the LPO assay. C: Serial dilutions (0–10 nmol) of the indicated sterols were spotted onto nitrocellulose membranes, which were then incubated with GST-StarD4 protein. Cholesterol was included as a positive control in all membranes. D: Binding affinity for 7α-hydroxycholesterol by GST-StarD4 fusion protein was annulled when the protein was preincubated with cholesterol for 1 h at 37°C previous to the LPO assay. E: No nonspecific binding of GST protein was observed when serial dilutions of the indicated sterols were spotted onto nitrocellulose membranes, which were then incubated with GST protein.
Overexpression of StarD4 increases bile acid synthesis rates in hepatocytes
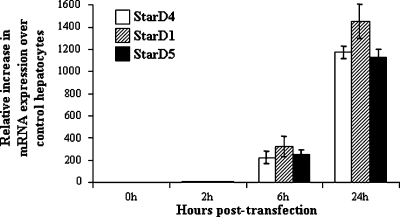
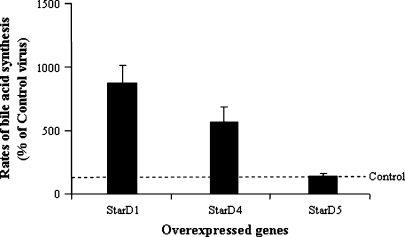
Overexpression of StarD4, StarD1, and StarD5 in primary mouse hepatocytes was assayed by quantitative real time PCR. Shown in Fig. 5 is the increase in mRNA levels at differing incubation times following virus infection. Overexpression of StarD4 following StarD4 virus infection increased, up to 7-fold, the rates of bile acid synthesis in 1° mouse hepatocytes, rates similar to that observed after StarD1 overexpression (10-fold increase). In contrast, StarD5 did not increase bile acid synthesis levels over control virus levels (Fig. 6).
Fig. 5.
Quantitative real-time PCR of StarD4, StarD1, and StarD5 mRNA in primary mouse hepatocytes. Following a 2 h transfection with Ad-CMV StarD4, StarD1, or StarD5, levels of the mRNAs were increased in a time-dependent manner over control Ad-CMV transfected hepatocytes. Data are presented as percentage increase over controls (means ± SEM of three independent experiments).
Fig. 6.
Effects of StarD1, StarD4, and StarD5 overexpression on the rates of bile acid synthesis in primary mouse hepatocytes. StarD1 and StarD4 overexpression led to an increase in the rates of bile acid synthesis, while StarD5 overexpression did not affect the rates of bile acid synthesis. Bile acid synthesis levels are expressed as percentages of Ad-CMV control transfected cells. Data are presented as means ± SEM of three experiments.
Overexpression of StarD4 increases intracellular cholesteryl esters in hepatocytes
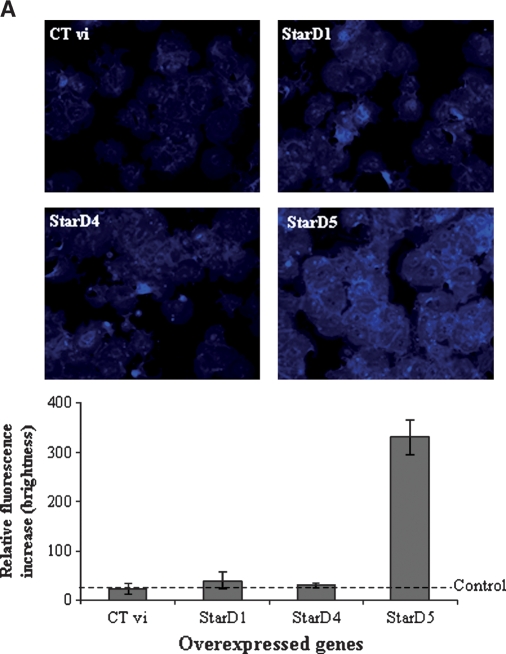
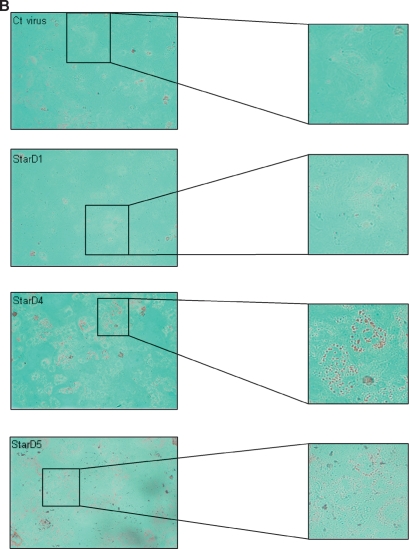
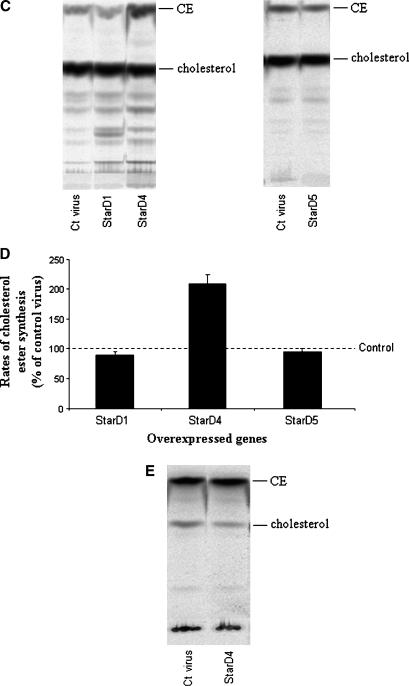
Free cholesterol content in the cells was analyzed by filipin staining. Primary mouse hepatocytes overexpressing human StarD4 did not show any increase of intracellular free cholesterol with respect to hepatocytes infected with adenovirus encoding StarD1 or Ad-CMV control virus, while cells infected with the StarD5 virus showed a 12-fold increase over controls (Fig. 7A), as previously reported in rat hepatocytes (4). Cholesteryl esters were increased in cells overexpressing StarD4, as shown by Oil Red O staining of primary mouse hepatocytes (Fig. 7B). No increase was observed following StarD1, StarD5, or control virus overexpression (Fig. 7B). Supportive results were obtained by TLC analysis of steroid products in chloroform-extractable phase from mouse hepatocytes incubated with [14C]cholesterol (Fig. 7C, D), in which cholesteryl esters increased up to 2-fold over control levels following StarD4 overexpression. To determine whether newly synthesized cholesterol can also serve as a substrate for StarD4 in increasing cholesteryl ester formation, [1-14C]acetate was added to the culture medium as a precursor for cholesterol synthesis. The results obtained after TLC analysis of steroid products in the chloroform-extractable phase did not show any difference in the levels of cholesteryl ester formation between cells overexpressing StarD4 and control cells (Fig. 7E).
Fig. 7.
Effects of the overexpression of different StAR proteins on the levels of free cholesterol and cholesteryl esters in primary mouse hepatocytes. A: Filipin staining for localization of free cholesterol in mouse hepatocytes. Overexpression of StarD1 and StarD4 did not affect the intracellular levels of free cholesterol over control virus, while overexpression of StarD5 increased the levels of free cholesterol up to 12-fold. B: Oil Red O staining of primary mouse hepatocytes overexpressing StarD1, StarD4, StarD5, or control virus. StarD4-overexpressing cells showed a noticeable increase in Oil Red O staining of neutral lipids. C: TLC analysis of sterols in primary mouse hepatocytes after overexpression of StarD1, StarD4, or StarD5. Overexpression of StarD4 in primary hepatocytes led to an increase in cholesteryl ester (CE) levels when medium was supplemented with [14C]cholesterol. No detectable differences were observed in free cholesterol levels following StarD4 overexpression. D: StarD4 overexpression led to increased cholesteryl ester formation, while StarD1 or StarD5 overexpression did not affect cholesteryl ester formation rates. Cholesteryl ester levels are expressed as percentages of Ad-CMV control virus. E: TLC analysis of sterols in primary mouse hepatocytes supplemented with [1-14C]acetate (as substrate for cholesterol synthesis) after overexpression of control StarD4. Overexpression of StarD4 in primary hepatocytes did not increase cholesteryl ester levels from newly synthesized cholesterol. Data are presented as means ± SEM of three independent experiments.
DISCUSSION
To pursue functional studies on the role of StarD4 in lipid binding and transport, the human StarD4 protein was purified (Fig. 1) and sequenced and underwent CD spectrum analysis prior to being utilized in in vitro sterol binding studies. Once cholesterol binding with isolated full-length StarD4 was determined and found to be similar to StarD1 and StarD5, the effect of altering StarD4 expression was explored in hepatocyte cell culture. Increasing StarD4 expression in hepatocyte culture using a recombinant adenoviral vector system was found to mobilize/direct cellular cholesterol to increased storage (increased cellular cholesteryl ester concentration) and degradation (increased bile acid formation). These findings suggest that under the conditions explored in mouse hepatoctyes, cholesterol delivery, and not enzymatic metabolism of cholesterol, is rate-determining for cholesterol esterification and bile acid synthesis.
The StarD4 protein gives name to the StarD4 subfamily of proteins, which also includes StarD5 and StarD6. StarD4 does not exhibit N-terminal domains that could direct the protein to specific cellular organelles. Therefore, StarD4 has been predicted to be a cytoplasmic protein (7). Although it lacks an N-terminal targeting sequence, its START domain is predicted to bind cholesterol and/or other sterols. The binding assays reported here show the ability of StarD4 to bind cholesterol with a similar stoichiometry to those reported for StarD1 and StarD5 (4, 8), a closely related protein also able to bind 25-hydroxycholesterol (4). Furthermore, StarD4's ability to bind cholesterol was confirmed by LPO assays and confirmed that variation in the structure of the cholesterol molecule prevented StarD4 binding (Fig. 4C). The only other sterol for which StarD4 showed limited binding was 7α-hydroxycholesterol. The binding of 7α-hydroxycholesterol, however, was completely abolished when the recombinant protein was preincubated with cholesterol, demonstrating StarD4's higher affinity for cholesterol and bringing into question the physiologic relevance of the observed 7α-hydroxycholesterol binding. These results indicate that StarD4 is a selective cholesterol transporter.
To address a possible misfolding of the protein after purification, far-UV CD spectra were obtained. The StarD4 protein gave a minimum at 220 nm, suggesting primarily β-sheet character for this protein, which matches the structure of mouse StarD4 protein, which has a major number of β-sheets (13), suggesting that the structures of the mouse and human StarD4 proteins most likely share similar secondary structures and folding. These data suggested that the StarD4 protein adopted normal conformation after purification. The far-UV spectra of StarD4 incubated with cholesterol (Fig. 2) showed a shift at the 220 nm minimum and lost the minimum at 210 nm obtained with the protein alone, suggesting conformational changes of the protein upon cholesterol binding.
To assay for the ability of StarD4 to transport cholesterol within cells, primary mouse hepatocytes were infected with a recombinant StarD4 adenovirus. Mouse hepatocytes were chosen because of our laboratory's expertise with this model and because hepatocytes not only have sterol uptake, storage, and secretion capabilities but also exhibit an easily measurable pathway of sterol catabolism to bile acids. In initial studies, changes in cholesterol (free and esters) levels in the cells were examined. Filipin staining [a well-described marker for staining intracellular non-membrane-bound free cholesterol (31)] of hepatocytes overexpressing StarD4 did not detect changes in the levels of free cholesterol (Fig. 7A). Similar results were obtained with StarD1. As shown previously in rat hepatocytes, overexpression of StarD5 increased free cholesterol levels in mouse hepatocytes (4). By comparison, both TLC analysis and Oil Red O staining showed an increase in cellular cholesteryl esters (Fig. 7B–D). These data demonstrate the ability of StarD4 protein to bind, transport, position, and effect cholesterol esterification from exogenous cholesterol. Interestingly, cholesteryl ester formation from newly synthesized cholesterol was not altered in the presence of StarD4 overexpression (Fig. 7E). These observations indicate that StarD4 appears able to increase cholesteryl ester formation only from exogenous sources of cholesterol. Furthermore, the findings suggest that substrate availability for ACAT is a key determinant of the rate of cholesterol esterification. In preliminary studies in human macrophage culture and primary cultures of human hepatocytes, rates of cholesterol esterification were similarly increased following increased StarD4 expression (data not shown). A similar substrate availability phenomenon was observed previously following increased expression of StarD1, a mitochondrial cholesterol delivery protein (25).
The ability of StarD4 to mobilize/transport cholesterol to or in the ER differs from the recent observations with StarD5 in which the cytosolic StarD5 was found to have an association with the Golgi and appeared to participate in cholesterol delivery to the Golgi. Interestingly, in the presence of increased cholesterol esterification, the rates of bile acid synthesis also increased following increased StarD4 expression (Fig. 6). Under the culture conditions utilized, only the “acidic” or alternative pathway of bile acid synthesis is functional (hepatocytes do not express CYP7A1 in culture), that is, a pathway initiated by mitochondrial CYP27A1. Therefore, in addition to StarD4's ability to mobilize cholesterol to the ER, these findings indicate the ability of StarD4 to present cholesterol for further metabolism to the mitochondria as well. Whether this mitochondrial transport of cholesterol represents a physiologic phenomenon or not is still uncertain.
Although the physiologic role of StarD4 remains uncertain, the selectivity of StarD4 for cholesterol indicates differences from other members of the StarD4 subfamily, such as StarD5, which binds not only cholesterol but 25-hydroxycholesterol as well (4, 7). Those differences are further emphasized by the ability of StarD4 to transport cholesterol to locations that differ from those reported for StarD5 and StarD1 (4, 22, 25). Studies including protein crystallization, protein localization by Western analysis, cellular immunohistochemistry/immunocytochemistry, and the generation of StarD4 knockout conditions will be key in elucidating the physiologic role and regulation of this novel cholesterol binding protein.
Acknowledgments
The authors thank Dr. D. Peterson for his help with CD experiments.
Published, JLR Papers in Press, April 9, 2008.
Footnotes
This work was supported by grants from the Veterans Administration (Merit Review), the National Institutes of Health (P01 DK-38030 and R01 HL-078898), and the Jeffress Research Grant. D.R-A. is the recipient of an American Heart Association Postdoctoral Fellowship award.
References
- 1.Hylemon, P. B., W. M. Pandak, and Z. R. Vlahcevic. 2001. The liver: biology and pathobiology. In The Liver. I. M. Arias, J. L. Boyer, F. V. Chisari, et al., editors. Lippincott Williams & Wilkins, Philadelphia, PA. 231–247.
- 2.Strauss J. F., III, T. Kishida, L. K. Christenson, T. Fujimoto, and H. Hiroi. 2003. START domain proteins and the intracellular trafficking of cholesterol in steroidogenic cells. Mol. Cell. Endocrinol. 202 59–65. [DOI] [PubMed] [Google Scholar]
- 3.Ponting C. P., and L. Aravind. 2004. START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem. Sci. 24 130–132. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Agudo D., S. Ren, P. Hylemon, K. Redford, R. Natarajan, A. Del Castillo, G. Gil, and W. Pandak. 2005. Human StarD5, a cytosolic StAR-related lipid binding protein. J. Lipid Res. 46 1615–1623. [DOI] [PubMed] [Google Scholar]
- 5.Iyer L. M., E. V. Koonin, and L. Aravind. 2001. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins. 43 134–144. [DOI] [PubMed] [Google Scholar]
- 6.Schultz J., F. Milpetz, P. Bork, and C. P. Ponting. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA. 95 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soccio R. E., R. M. Adams, M. J. Romanowski, E. Sehayek, S. K. Burley, and J. L. Breslow. 2002. The cholesterol-regulated StarD4 gene encodes a StAR-related lipid transfer protein with two closely related homologues, StarD5 and StarD6. Proc. Natl. Acad. Sci. USA. 99 6943–6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsujishita Y., and J. H. Hurley. 2000. Structure and lipid transport mechanism of a StAR-related domain. Nat. Struct. Biol. 7 408–414. [DOI] [PubMed] [Google Scholar]
- 9.Lin D., T. Sugawara, J. F. Strauss, III, B. J. Clark, D. M. Stocco, P. Saenger, A. Rogol, and W. L. Miller. 1995. Role of steroidogenic acute regulatory protein in adrenal and gonadal steroidogenesis. Science. 267 1828–1831. [DOI] [PubMed] [Google Scholar]
- 10.Moog-Lutz C., C. Tomasetto, C. H. Regnier, C. Wendling, Y. Lutz, D. Muller, M. P. Chenard, P. Basset, and M. C. Rio. 1997. MLN64 exhibits homology with the steroidogenic acute regulatory protein (STAR) and is over-expressed in human breast carcinomas. Int. J. Cancer. 71 183–191. [DOI] [PubMed] [Google Scholar]
- 11.Bose H. S., R. M. Whittal, M. C. Huang, M. A. Baldwin, and W. L. Miller. 2000. N-218 MLN64, a protein with StAR-like steroidogenic activity, is folded and cleaved similarly to StAR. Biochemistry. 39 11722–11731. [DOI] [PubMed] [Google Scholar]
- 12.Soccio R. E., R. M. Adams, K. N. Maxwell, and J. L. Breslow. 2005. Differential gene regulation of StarD4 and StarD5 cholesterol transfer proteins: activation of StarD4 by SREBP-2 and StarD5 by endoplasmic reticulum stress. J. Biol. Chem. 280 19410–19418. [DOI] [PubMed] [Google Scholar]
- 13.Romanowski M. J., R. E. Soccio, J. L. Breslow, and S. K. Burley. 2002. Crystal structure of the Mus musculus cholesterol-regulated START protein 4 (StarD4) containing a StAR-related lipid transfer domain. Proc. Natl. Acad. Sci. USA. 99 6949–6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen D. E., R. M. Green, M. K. Wu, and D. R. Beier. 1999. Cloning, tissue-specific expression, gene structure and chromosomal localization of human phosphatidylcholine transfer protein. Biochim. Biophys. Acta. 1447 265–270. [DOI] [PubMed] [Google Scholar]
- 15.Roderick S. L., W. W. Chan, D. S. Agate, L. R. Olsen, M. W. Vetting, K. R. Rajashankar, and D. E. Cohen. 2002. Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nat. Struct. Biol. 9 507–511. [DOI] [PubMed] [Google Scholar]
- 16.Murcia M., J. D. Faraldo-Gomez, F. R. Maxfield, and B. Roux. 2006. Modeling the structure of the StART domains of MLN64 and StAR proteins in complex with cholesterol. J. Lipid Res. 47 2614–2630. [DOI] [PubMed] [Google Scholar]
- 17.Horton J. D., N. A. Shah, J. A. Warrington, N. N. Anderson, S. W. Park, M. S. Brown, and J. L. Goldstein. 2003. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA. 100 12027–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren S., P. Hylemon, D. Marques, E. Hall, K. Redford, G. Gil, and W. Pandak. 2004. Effect of increasing the expression of cholesterol transporters (StAR, MLN64, and SCP-2) on bile acid synthesis. J. Lipid Res. 45 2123–2131. [DOI] [PubMed] [Google Scholar]
- 19.Feng L., W. W. Chan, S. L. Roderick, and D. E. Cohen. 2000. High-level expression and mutagenesis of recombinant human phosphatidylcholine transfer protein using a synthetic gene: evidence for a C-terminal membrane binding domain. Biochemistry. 39 15399–15409. [DOI] [PubMed] [Google Scholar]
- 20.Alpy F., and C. Tomasetto. 2005. Give lipids a START: the StAR-related transfer (START) domain in mammals. J. Cell Sci. 118 2791–2801. [DOI] [PubMed] [Google Scholar]
- 21.Gomes C., S. D. Oh, J. W. Kim, S. Y. Chun, K. Lee, H. B. Kwon, and J. Soh. 2005. Expression of the putative sterol binding protein StarD6 gene is male germ specific. Biol. Reprod. 72 651–658. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez-Agudo D., S. Ren, P. B. Hylemon, R. Montanez, K. Redford, R. Natarajan, M. A. Medina, G. Gil, and W. M. Pandak. 2006. Localization of StarD5 cholesterol binding protein. J. Lipid Res. 47 1168–1175. [DOI] [PubMed] [Google Scholar]
- 23.Yamada S., T. Yamaguchi, A. Hosoda, T. Iwawaki, and K. Kohno. 2006. Regulation of human STARD4 gene expression under endoplasmic reticulum stress. Biochem. Biophys. Res. Commun. 343 1079–1085. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227 6573–6576. [DOI] [PubMed] [Google Scholar]
- 25.Pandak W. M., S. Ren, D. Marques, E. Hall, K. Redford, D. Mallonee, P. Bohdan, D. Heuman, G. Gil, and P. Hylemon. 2002. Transport of cholesterol into mitochondria is rate-limiting for bile acid synthesis via the alternative pathway in primary rat hepatocytes. J. Biol. Chem. 277 48158–48164. [DOI] [PubMed] [Google Scholar]
- 26.Hylemon P. B., E. C. Gurley, R. T. Stravitz, J. S. Litz, W. M. Pandak, J. Y. Chiang, and Z. R. Vlahcevic. 1992. Hormonal regulation of cholesterol 7 alpha-hydroxylase mRNA levels and transcriptional activity in primary rat hepatocyte cultures. J. Biol. Chem. 267 16866–16871. [PubMed] [Google Scholar]
- 27.Bissell D. M., and P. S. Guzelian. 1980. Phenotypic stability of adult rat hepatocytes in primary monolayer culture. Ann. N. Y. Acad. Sci. 349 85–98. [DOI] [PubMed] [Google Scholar]
- 28.Dowler S., G. Kular, and D. R. Alessi. 2002. Protein lipid overlay assay. Sci. STKE. 129 PL6. [DOI] [PubMed] [Google Scholar]
- 29.Folch J., M. Lees, and G. H. Sloane Stanley. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226 497–509. [PubMed] [Google Scholar]
- 30.Johnson W. C., Jr. 1988. Secondary structure of proteins through circular dichroism spectroscopy. Annu. Rev. Biophys. Biophys. Chem. 17 145–166. [DOI] [PubMed] [Google Scholar]
- 31.Kruth H. S., and D. L. Fry. 1984. Histochemical detection and differentiation of free and esterified cholesterol in swine atherosclerosis using filipin. Exp. Mol. Pathol. 40 288–294. [DOI] [PubMed] [Google Scholar]