Abstract
Trans-splicing adeno-associated viral (AAV) vectors hold great promise in many gene therapy applications. We have shown that rational selection of the gene-splitting site in a therapeutic target gene can lead to extremely efficient trans-splicing vectors [Lai, Y., Yue, Y., Liu, M., Ghosh, A., Engelhardt, J.F., Chamberlain, J.S., and Duan, D. (2005). Nat. Biotechnol. 23, 1435–1439]. Our original strategy requires the screening of endogenous introns that are capable of overcoming the mRNA accumulation barrier. To further develop trans-splicing vectors, we have tested whether the use of a generic synthetic intron can bypass the labor-intensive intron-screening process. Two previously characterized exon/intron/exon junctions (60/60/61 and 63/63/64, respectively) in the 6 kb minidystrophin gene were used as templates to represent highly efficient (60/60/61) and relatively poor (63/63/64) gene-splitting sites. We compared RNA production from the reconstituted viral genome and transduction efficiency of the trans-splicing vectors in dystrophin-null mdx mouse skeletal muscle. Our results suggest that a synthetic intron can successfully overcome the mRNA accumulation barrier at the exon 63/64 junction. Furthermore, when the gene was split at the exon 63/64 junction, the synthetic intron-based vectors performed better than the endogenous intron-based vectors. When the gene was split at the exon 60/61 junction, we observed only nominal improvement in mRNA production. Nevertheless, vectors based on the exon 60/61 junction remain the best set in transduction efficiency. Taken together, our results suggest that optimizing intron sequence may boost the transduction efficiency of trans-splicing AAV vectors.
OVERVIEW SUMMARY
Successful transduction of trans-splicing AAV vectors depends on the efficient uptake of both donor and acceptor vectors, unidirectional head-to-tail viral genome recombination, and productive transcription and splicing of the reconstituted genome. In this study, we explored the strategy of using synthetic intron to overcome the transcription/splicing barrier. We hypothesized that substituting a less than optimal endogenous intron with a highly conserved synthetic intron will lead to enhanced mRNA production. On the basis of our previous studies of minidystrophin gene trans-splicing AAV vectors, we replaced endogenous introns with synthetic introns in artificially reconstituted viral genomes and in trans-splicing vectors, respectively. Consistent with our initial hypothesis, we observed significant improvement in mRNA production from constructs based on a poor gene-splitting site, namely, the exon 63/64 junction. Synthetic intron replacement also increased transduction efficiency in vectors based on this junction. However, it never reached the level of our best trans-splicing vectors, namely, those based on the exon 60/61 junction. In summary, we have developed a convenient method of using synthetic intron to overcome the mRNA accumulation barrier in trans-splicing vectors. Additional investigation of factors other than mRNA accumulation will likely help to develop better trans-splicing vectors.
INTRODUCTION
The single-stranded adeno-associated virus (AAV) has been adapted as one of the most effective gene delivery vehicles (Carter, 2004; Flotte and Berns, 2005; Gao et al., 2005). It shows persistent and high-level transduction in a variety of tissues in vivo and an outstanding safety profile in human patients. The rapid development in AAV technology is finally reaching the point of materializing gene therapy benefit in patients (Carter, 2005; Manno et al., 2006). Yet, there is one more hurdle in capitalizing AAV gene therapy in diseases involving large genes.
Wild-type AAV has a 4.7-kb genome consisting of a cap gene, a rep gene, and two flanking inverted terminal repeats (ITRs). In AAV vectors, all viral genes can be removed and replaced with a therapeutic and/or reporter expression cassette. Because of inherent limitations of the viral capsid, the maximal vector genome that can be assembled in a single AAV virion is less than 6 kb (Dong et al., 1996; Warrington et al., 2004; Grieger and Samulski, 2005). Delivering a large (≥6 kb) expression cassette has been a challenge in AAV gene therapy of muscular dystrophy, cystic fibrosis, hemophilia A, and a number of other diseases (Chamberlain, 2002; Flotte, 2005; Wang and Herzog, 2005).
Dual-vector strategies were developed to overcome the size limitation. In dual-vector approaches, two independent AAV particles are produced and each carries a portion of an intact expression cassette. Among these approaches, the trans-splicing method is particularly attractive. In this method, a large therapeutic gene is split into two arbitrarily designated “exons,” one carrying the 5′ expression cassette and the splicing donor and the other carrying the splicing acceptor and the 3′ expression cassette. Each part is packaged into one AAV virus. Expression is achieved by ITR-mediated recombination and subsequent splicing of the recombinant genome in coinfected cells (Nakai et al., 2000; Sun et al., 2000; Yan et al., 2000; Duan et al., 2001). Despite the promise of being able to deliver a large gene, earlier studies revealed modest transduction efficiency comparing with that of a single intact AAV vector. To improve gene expression from the trans-splicing vectors, we and others have systematically investigated several potential barriers including viral serotype, ITR-mediated recombination, mRNA accumulation, and the gene-splitting site (Reich et al., 2003; Xu et al., 2004; Lai et al., 2005; Yan et al., 2005). These results suggest that the gene-splitting site has a profound influence on mRNA accumulation and protein expression.
The gene-splitting site is defined by the splicing signals in both intron and exons. Optimizing either one may improve the overall transduction efficiency of trans-splicing vectors. Altering DNA exon sequence may change protein coding and function. But intron sequence is more manipulable. In this study we tested whether intron optimization can augment gene expression from trans-splicing vectors. We replaced the endogenous intron in the minidystrophin gene with an optimized synthetic intron and measured mRNA accumulation from recombined genome and protein expression from the respective pairs of trans-splicing vectors. Our results suggest that synthetic introns can effectively replace endogenous introns in trans-splicing vectors. Furthermore, they can help to overcome the mRNA accumulation barrier at a poor junction. However, factors other than mRNA accumulation also contribute to the ultimate transduction efficiency in the trans-splicing vectors.
MATERIALS AND METHODS
Plasmids for comparing synthetic and endogenous intron splicing signals by RNase protection assay
Four plasmid constructs were used in this study: p60/60/61, p60/syn/61, p63/63/64, and p63/syn/64. These plasmids were named in the order of exon/intron/exon. Numeric numbers were used to indicate the respective endogenous exons and introns. Synthetic intron (syn) was obtained from pCI (Promega, Madison, WI). The splicing donor signal in synthetic intron derives from the first intron in the human β-globulin gene. The splicing acceptor signal in synthetic intron comes from the human immunoglobulin heavy chain gene. The double-D ITR sequence, a structure resembling the recombined ITR junction, is included in the center of the intron. All the constructs were under the transcriptional regulation of the cytomegalovirus (CMV) promoter and the simian virus 40 (SV40) polyadenylation signal. The detailed cloning strategy is provided elsewhere (see http://mmi.missouri.edu/pubs/duan/Hum-Apr-06-58-Supplementary.pdf).
Plasmids for generating RNase protection assay probe
RNase protection assay (RPA) probes were designed to hybridize with the 5′ exon and the intron donor sequence. Briefly, corresponding regions from p60/60/61, p60/syn/61, p63/63/64, and p63/syn/64 were amplified by polymerase chain reaction (PCR) and then inserted into the EcoRI/BamHI sites in pGEM-3Z (Promega). Primer sequences are provided elsewhere (see http://mmi.missouri.edu/pubs/duan/Hum-Apr-06-58-Supplementary.pdf). Probes for p60/60/61 and p63/63/64 have been described previously (Lai et al., 2005). All constructs were validated by sequencing.
Plasmids for generating minidystrophin trans-splicing vectors
A pair of cis plasmids, including a donor and an acceptor, was generated for making each set of trans-splicing vectors. Vectors developed from the endogenous intron were designated as pcisDonor.60endo and pcisAcceptor.60endo (for splitting the minigene at intron 60; these plasmids were previously reported as pcisDonor.60 and pcisAcceptor.60, respectively), and pcisDonor. 63endo and pcisAcceptor.63endo (for splitting the minigene at intron 63; these plasmids were previously reported as pcisDonor.63 and pcisAcceptor.63, respectively) (Lai et al., 2005). The vectors developed from the synthetic intron were designated as pcisDonor.60syn and pcisAcceptor.60syn (for splitting the minigene between exons 60 and 61), and pcisDonor.63syn and pcisAcceptor.63syn (for splitting the minigene between exons 63 and 64). All the donor plasmids carry the CMV promoter and all the acceptor plasmids carry the SV40 poly(A) sequence. Furthermore, the Kozak sequence was engineered into the translation starting site to facilitate minidystrophin gene expression (Lai et al., 2005). The detailed cloning strategy to make the synthetic intron-based vectors is provided elsewhere (see http://mmi.missouri.edu/pubs/duan/Hum-Apr-06-58-Supplementary.pdf).
RNase protection assay
The RPA was performed essentially as described previously (Xu et al., 2004; Lai et al., 2005). For each experimental construct (p60/60/61, p60/syn/61, p63/63/64, and p63/syn/64), 4 μg of plasmid and one-eighth (0.5 μg) of a control luciferase plasmid were cotransfected into 70–80% confluent 293 cells, using Lipofectamine and PLUS reagent (Invitrogen, Carlsbad, CA). Forty-eight hours after transfection, cells were harvested. One-fifth of the cells were used in a luciferase assay to normalize transfection efficiency. The remaining cells were lysed in 4 M guanidine isothiocyanate and total RNA was extracted by ultracentrifugation in 5.7 M CsCl.
To make antisense RPA probe, the parental plasmids (see above) were first linearized at the EcoRI site. The probes were synthesized with the linearized plasmid as template and [α-32P]rUTP as substrate by in vitro transcription with SP6 RNA polymerase.
The RPA reaction was carried out with RNA (total, 10 μg) and an excess amount of radiolabeled probes. Unprotected RNA was digested away with RNase T1 and RNase A at final concentrations of 0.923 and 18.46 μg/ml, respectively. Protected RNA was electrophoresed in 8% denaturing polyacrylamide gels. The protected RNA signals were quantified with a Molecular Imager FX system and Quantity One imaging software (version 4.2.2) (Bio-Rad, Hercules, CA). Relative signal intensity was normalized for transfection efficiency (by luciferase activity), the number of 32P-labeled uridines in each protected band, and loading (by the internal human β-actin control).
Recombinant AAV stock preparation
The trans-splicing vectors were packaged in AAV serotype 6 capsid according to previously published protocols (Lai et al., 2005; Liu et al., 2005). Briefly, 70–80% confluent 293 cells were cotransfected with the respective cis-plasmid, pMTrep2 and pCMVcap6 (gifts from A. Dusty Miller, Fred Hutchinson Cancer Research Center, Seattle, WA), and an adenoviral helper plasmid (pHelper; Stratagene, La Jolla, CA) at a ratio of 1:1:3:3. Crude viral lysate was harvested 72 hr later and purified through three rounds of CsCl isopycnic ultracentrifugation. Before ultracentrifugation, crude lysate was sequentially treated by eight rounds of freeze–thawing, a 10-min sonication at a power output of 5.5 (Misonic Cell Disruptor S3000; Misonix, Inc., Farmingdale, NY), DNase I digestion (11,000 Kunitz units per 15 × 150 mm plate cell lysate), 1% sodium deoxycholate treatment, and 0.05% tissue culture-grade trypsin digestion. Viral titer determination and quality control were carried out as described previously (Lai et al., 2005; Liu et al., 2005).
Animal studies
All animal experiments were approved by the Animal Care and Use Committee at the University of Missouri and were in accordance with National Institutes of Health (NIH, Bethesda, MD) guidelines. All experimental mice were housed in a specific pathogen-free facility. Two-month-old male mdx mice were purchased from Jackson Laboratory (Bar Harbor, ME). Recombinant AAV-6 trans-splicing vectors were delivered to the tibialis anterior (TA) muscles according to a previously described protocol (Lai et al., 2005). For viruses generated with endogenous intron (AV.Donor.60endo and AV.Acceptor. 60endo, and AV.Donor.63endo, and AV.Acceptor.63endo), only donor and acceptor coinfection was performed. We have previously shown that single vector alone did not yield minidystrophin expression. For viruses generated with synthetic intron (AV.Donor.60syn and AV.Acceptor.60syn, and AV.Donor. 63syn and AV.Acceptor.63syn), we performed both single-vector infection and donor and acceptor coinfection. For TA muscle infection, 2 × 109 vector genome particles of each virus was delivered.
Morphometric quantification of minidystrophin expression in trans-splicing vector-infected muscle
Minidystrophin was detected by indirect immunofluorescence staining according to previously published protocols (Yue et al., 2003; Lai et al., 2005). Because the 6 kb minigene was derived from the human dystrophin gene and also contains the C-terminal domain, three antibodies were used to evaluate its expression. These included human dystrophin N-terminus-specific monoclonal antibody Dys-3 (1:10 dilution; Novocastra Laboratories, Newcastle upon Tyne, UK), dystrophin C-terminus-specific monoclonal antibody Dys-2 (1:30 dilution; Novocastra Laboratories), and an affinity-purified polyclonal anti-dystrophin N-terminus-specific antibody (1:600 dilution; a gift from J. Chamberlain, University of Washington, Seattle, WA). Transduction efficiency was determined by quantifying the total number of Dys-3-positive cells in the entire muscle section. At least two montage composites of entire cross-sections in the mid-belly were quantified for each muscle sample.
Statistical analysis
Data are presented as means ± standard error of mean. Statistical analysis was performed with SPSS software (SPSS, Chicago, IL). Statistical significance among all experimental groups was determined by one-way analysis of variance (ANOVA). Statistical significance in the same group (the same exon/exon junction but either endogenous or synthetic intron) was determined by Student t test. A difference was considered significant when p < 0.05.
RESULTS
Synthetic intron increases mRNA production from a relatively poor junction in the minidystrophin gene
To evaluate the influence of intron sequence in the reconstituted vector genome on mRNA accumulation, we replaced the endogenous intron in the minidystrophin gene with a commercially available synthetic intron (Duan et al., 2003). In the hope of further improving our minidystrophin trans-splicing vectors (Lai et al., 2005), the intron replacement experiment was performed on the 60/60/61 (exon/intron/exon) and 63/63/64 junctions, two previously characterized gene-splitting sites in the minidystrophin gene.
Both the 60/60/61 and 63/63/64 junctions have a relatively high consensus splicing value. Replacing the endogenous intron with synthetic intron further optimizes the splicing parameters. In particular, the 5′ splicing signals were matched perfectly with the conserved motif in both the 60/syn/61 and 63/syn/64 junctions (Table 1) (Shapiro and Senapathy, 1987). To compare mRNA production between constructs carrying either endogenous or synthetic intron, we generated two new minicassettes in which the double-D ITR junction is flanked by the synthetic splicing signals (Fig. 1A). We then measured total RNA, unspliced RNA, and mRNA by RPA. Representative RPA gels are shown in Fig. 1B. Normalized data (for transfection efficiency, RNA extraction, and loading) are presented in Table 1. Consistent with our previous report (Lai et al., 2005), the 60/60/61 junction resulted in approximately 2-fold higher total RNA and mRNA than did the 63/63/64 junction (Fig. 1B and Table 1).
Table 1.
Consensus Splicing Values and RPA Results
|
Exon/intron/exon combination |
||||
|---|---|---|---|---|
| 60/60/61 | 60/syn/61 | 63/63/64 | 63/syn/64 | |
| 5′ CV (%)a | 94.16 | 100.00 | 95.44 | 100.00 |
| 3′ CV (%)a | 98.13 | 96.56 | 89.70 | 90.79 |
| Total RNA | 338.5 ± 20.5 | 381.9 ± 128.6 | 143.6 ± 8.2 | 399.1 ± 34.8b |
| Unspliced RNA | 87.5 ± 5.8 | 81.7 ± 4.4 | 19.3 ± 1.3 | 36.7 ± 2.8b |
| Spliced RNA | 251.0 ± 25.1 | 300.2 ± 86.6 | 124.3 ± 8.2 | 362.4 ± 32.3b |
| S:U ratioc | 2.5 ± 0.4 | 6.4 ± 1.6 | 5.1 ± 0.5 | 7.7 ± 0.3b |
| % of splicing | 70.5 | 82.8 | 83.3 | 88.5 |
Consensus splicing value (CV) at the 5′ and 3′ ends of each indicated intron. It is presented as the percentage of homology to the most conserved motif in the primate gene. A higher value indicates a better match. Calculated according to Shapiro and Senapathy (1987).
Values are significantly different from that of p63/63/64 (p < 0.015).
The ratio of spliced (S) to unspliced (U) RNA transcript.
FIG. 1.
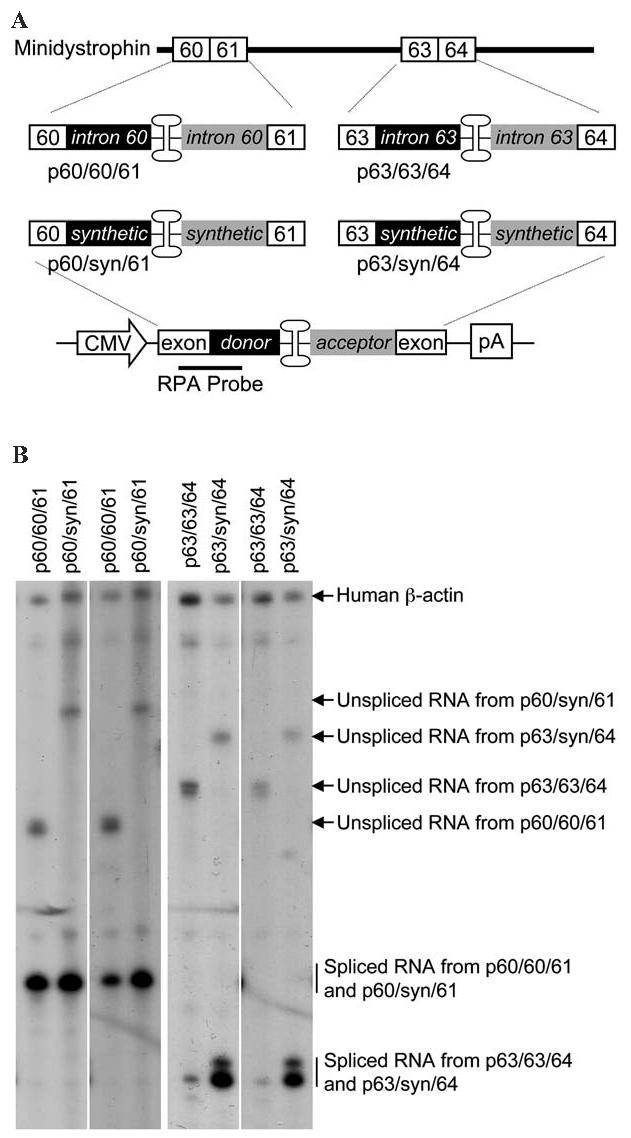
Effect of intron optimization on transcription and splicing in the reconstituted AAV genome. (A) Schematic diagram of the constructs used in RNase protection assays. Intron splicing signals and the double-D ITR junction are inserted either between exons 60 and 61 or between exons 63 and 64 in the minidystrophin gene to generate minicassettes for the RPA. Each minicassette consists of the exon, the splicing donor, the double-D ITR junction, the splicing acceptor, and the downstream exon. For the RPA study, the minicassette is under the transcriptional regulation of the CMV promoter and SV40 poly(A) signals. The antisense RPA probe is located between the upstream exon and the splicing donor signal. Black boxes, intron donor signal; gray boxes, intron acceptor signal. (B) Representative photomicrographs of RPA results. Endogenous human β-actin RNA (the control for RPA and loading) was detected by an independent probe added at the same time. The expected locations for unspliced and spliced RNAs are indicated by arrows and vertical lines, respectively. Irrespective of the intron used in the constructs (endogenous or synthetic), only one species of the spliced RNA is revealed by the RPA probe for each exon/exon junction.
The ratio of spliced to unspliced RNA (S:U ratio) and the percentage of spliced RNA are relatively lower with the 60/60/61 junction (Table 1) (Lai et al., 2005). Replacing intron 60 with synthetic intron improved both (Table 1). Interestingly, the increment in splicing ratio and splicing percentage did not lead to a significant increase (although there was a trend) in the mRNA level. A much more dramatic improvement was observed when intron 63 was replaced with a synthetic intron. The levels of total RNA and mRNA produced from p63/syn/64 reached those from p60/60/61 and p60/syn/61 (Fig. 1B and Table 1). The S:U ratio and the splicing percentage were also enhanced (Table 1). Taken together, the inclusion of a synthetic intron enhanced splicing and mRNA production (moderately for the 60/60/61 junction and significantly for the 63/63/64 junction).
Synthetic intron-based trans-splicing vectors mediate efficient minidystrophin expression in mdx skeletal muscle
An important goal of our study was to develop the best strategy for making highly efficient trans-splicing vectors for gene therapy. Considering the beneficial effect of the synthetic intron at the mRNA level, we hypothesized that replacing endogenous intron in the trans-splicing AAV vectors might also improve transduction efficiency. To evaluate the in vivo performance of synthetic intron-based vectors, we engineered two additional sets of trans-splicing viruses: the AV.Donor.60syn/AV.Acceptor.60syn pair and the AV.Donor.63syn/AV.Acceptor.63syn pair. Except for the difference in intron splicing sequences, these vectors are identical to our published AV. Donor.60endo/AV.Acceptor.60endo pair and AV.Donor.63endo/AV.Acceptor.63endo pair, respectively (Lai et al., 2005).
For synthetic intron-based vectors, we first evaluated background transduction from each vector by infecting adult mdx mouse TA muscle with either donor or acceptor virus alone. Similar to our results with endogenous intron-based vectors, none of the single-virus infections yielded any minidystrophin-positive cells (data not shown). We next performed coinfection experiments. We have previously shown that the transduction efficiency of AV.Donor.63endo/AV.Acceptor.63endo coinfection reached only 10–20% compared with AV.Donor.60endo/AV.Acceptor.60endo coinfection (Lai et al., 2005). This result was confirmed (Fig. 2). In the RPA study, both p60/60/61 and p60/syn/61 were efficient in mRNA production (Fig. 1B and Table 1). This was also reflected in the trans-splicing vectors. Both AV.Donor.60endo/AV.Acceptor.60endo coinfection and AV.Donor.60syn/AV.Acceptor.60syn coinfection yielded high-level minidystrophin expression (Fig. 2).
FIG. 2.
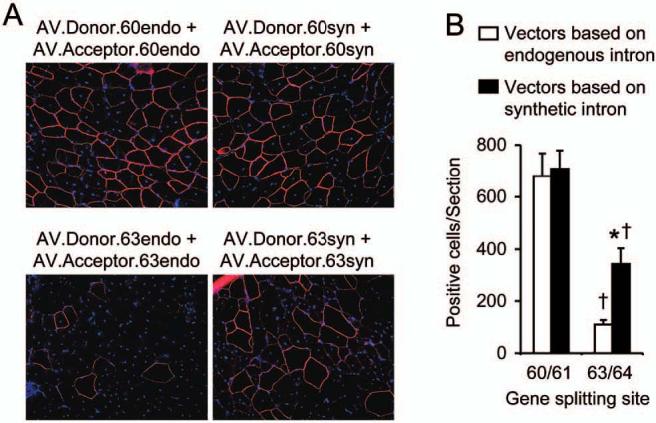
Quantitative evaluation of minidystrophin expression from the trans-splicing vectors in the mdx TA muscle. (A) Representative indirect immunofluorescence staining photomicrographs of muscle infected with each indicated pair of trans-splicing vectors. Minidystrophin is shown as continuous peripheral staining of the sarcolemma. Nuclei are highlighted by DAPI staining. (B) Quantification of minidystrophin-positive myofibers. Numerical numbers in the x axis refer to the exon/exon junction. n = 4 to 9 samples for each group. Open box, minidystrophin expression from vectors based on endogenous intron; closed box, minidystrophin expression from vectors based on synthetic intron; asterisk, when the minigene was split between exons 63 and 64, the difference between the endogenous intron-based vector set and the synthetic intron-based vector set was statistically significant; cross, minidystrophin expression from vectors based on the exons 63/64 junction is significantly different from that of vectors based on the exons 60/61 junction.
Because synthetic intron overcame the mRNA accumulation barrier at the exon 63/64 junction, we initially hypothesized that the AV.Donor.63syn/AV.Acceptor.63syn set should perform better than the AV.Donor.63endo/AV.Acceptor.63endo set, and ideally should reach an efficiency level comparable with that from vectors based on the exon 60/61 junction. In support of this hypothesis, transduction efficiency (as quantified by minidystrophin-positive cell number) of the AV.Donor.63syn/AV.Acceptor.63syn set was 3-fold higher than that of the AV.Donor.63endo/AV.Acceptor.63endo set (Fig. 2). Surprisingly, the transduction efficiency of the AV.Donor.63syn/AV.Acceptor.63syn pair reached only 50% of that based on the exon 60/61 junction (Fig. 2).
DISCUSSION
The development of trans-splicing vectors has significantly raised the hope of expanding AAV gene therapy to many diseases that need a large therapeutic gene (Reich et al., 2003; Xu et al., 2004; Lai et al., 2005; Yan et al., 2005). The success of the trans-splicing vectors hinges on efficient coinfection, viral genome recombination, transcription, and splicing. We have demonstrated that mRNA accumulation, rather than viral coinfection, is a critical rate-limiting factor in this approach (Xu et al., 2004). Furthermore, rational selection of the gene-splitting site overcomes the mRNA accumulation barrier.
Using the 6-kb minidystrophin gene as a template, we achieved therapeutic level protein expression in dystrophic muscle when the gene was split at intron 60 but not when it was split at intron 63 (Lai et al., 2005). Endogenous intron was used in our original study. One problem with endogenous intron is that the splicing signals are often less than optimal. To determine whether optimizing splicing signals can surpass the mRNA accumulation barrier, we replaced the endogenous intron with a highly efficient synthetic intron. Others have shown that site-directed mutagenesis can strengthen a weak intron (Carothers et al., 1993). However, this approach is labor intensive and time-consuming. Furthermore, specific mutations must be introduced into each intron. To develop a generic strategy for the trans-splicing AAV vectors, we explored the possibility of using synthetic introns in the trans-splicing vectors. We hypothesized that an optimized intron should augment mRNA production and therefore enhance the overall transduction efficiency of the trans-splicing AAV vectors. As expected, synthetic intron at the exon 63/64 junction significantly increased mRNA accumulation. The mRNA level from p63/syn/64 was doubled compared with that from p63/63/64 (Fig. 1 and Table 1). Interestingly, we observed only a nominal improvement in mRNA level at the exon 60/61 junction. Because synthetic intron increased the overall consensus splicing value at both the exon 60/61 and 63/64 junctions, it is currently not clear why enhancement was seen only at the exon 63/64 junction. One possibility is that the mRNA production from p60/60/61 may have already reached the maximal level for this transcription unit. Replacing endogenous intron 60 with a synthetic intron would therefore not yield further improvement in p60/syn/61.
The ultimate goal is to improve protein-level gene expression from the trans-splicing AAV vectors. To determine whether intron optimization was sufficient to enhance transduction efficiency, we generated AAV-6 trans-splicing vectors corresponding to each respective intron (endogenous versus synthetic) and the exon/exon junction (60/61 versus 63/64) and evaluated their transduction efficiency in dystrophin-null mdx muscle.
Consistent with the RPA results, the highest expression was achieved with vectors based on the exon 60/61 junction. There is no significant difference between vectors based on endogenous intron or synthetic intron. This result suggests that for a gene-splitting site that is already at maximal efficiency, there is limited room for further improvement. Nevertheless, synthetic intron does not have a negative impact on transduction. The exon 63/64 junction was a poor site when endogenous intron was used (Lai et al., 2005). Optimizing the splicing signals with synthetic intron successfully overcame the mRNA accumulation barrier (Fig. 1 and Table 1). Consequently, transduction efficiency was improved in vectors based on synthetic intron (Fig. 2). Surprisingly, the expression level in AV.Donor.63syn/AV.Acceptor. 63syn-coinfected muscle did not reach that of muscle infected with vectors based on the exon 60/61 junction. Apparently, there are additional barriers besides mRNA accumulation that might also have contributed to the suboptimal expression from the AV.Donor.63syn/AV.Acceptor.63syn pair. Some possibilities may include the efficiency of head-to-tail intermolecular recombination and the relative stability of the recombined genome.
In summary, we have demonstrated a readily applicable approach of using synthetic intron to overcome the mRNA accumulation barrier in trans-splicing vectors. However, identification of the optimal gene-splitting site remains a critical parameter in developing trans-splicing vectors.
ACKNOWLEDGMENTS
The authors thank Drs. Shi-jie Chen and Song Cao for calculating consensus splicing value, and Dr. Jeffrey Chamberlain for providing the 6 kb ΔH2-R19 minidystrophin gene. The authors also thank Dr. A. Dusty Miller for the AAV-6 packaging plasmids. This work was supported by grants from the National Institutes of Health (AR-49419; D.D.), the Muscular Dystrophy Association (D.D.), and the Cystic Fibrosis Foundation (D.D.).
REFERENCES
- CAROTHERS AM, URLAUB G, GRUNBERGER D, CHASIN LA. Splicing mutants and their second-site suppressors at the dihydrofolate reductase locus in Chinese hamster ovary cells. Mol. Cell. Biol. 1993;13:5085–5098. doi: 10.1128/mcb.13.8.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CARTER BJ. Adeno-associated virus and the development of adeno-associated virus vectors: A historical perspective. Mol. Ther. 2004;10:981–989. doi: 10.1016/j.ymthe.2004.09.011. [DOI] [PubMed] [Google Scholar]
- CARTER BJ. Adeno-associated virus vectors in clinical trials. Hum. Gene Ther. 2005;16:541–550. doi: 10.1089/hum.2005.16.541. [DOI] [PubMed] [Google Scholar]
- CHAMBERLAIN JS. Gene therapy of muscular dystrophy. Hum. Mol. Genet. 2002;11:2355–2362. doi: 10.1093/hmg/11.20.2355. [DOI] [PubMed] [Google Scholar]
- DONG JY, FAN PD, FRIZZELL RA. Quantitative analysis of the packaging capacity of recombinant adeno-associated virus. Hum. Gene Ther. 1996;7:2101–2112. doi: 10.1089/hum.1996.7.17-2101. [DOI] [PubMed] [Google Scholar]
- DUAN D, YUE Y, ENGELHARDT JF. Expanding AAV packaging capacity with trans-splicing or overlapping vectors: A quantitative comparison. Mol. Ther. 2001;4:383–391. doi: 10.1006/mthe.2001.0456. [DOI] [PubMed] [Google Scholar]
- DUAN D, YUE Y, ENGELHARDT JF. Dual vector expansion of the recombinant AAV packaging capacity. Methods Mol. Biol. 2003;219:29–51. doi: 10.1385/1-59259-350-x:29. [DOI] [PubMed] [Google Scholar]
- FLOTTE TR. Adeno-associated virus-mediated gene transfer for lung diseases. Hum. Gene Ther. 2005;16:643–648. doi: 10.1089/hum.2005.16.643. [DOI] [PubMed] [Google Scholar]
- FLOTTE TR, BERNS KI. Adeno-associated virus: A ubiquitous commensal of mammals. Hum. Gene Ther. 2005;16:401–407. doi: 10.1089/hum.2005.16.401. [DOI] [PubMed] [Google Scholar]
- GAO G, VANDENBERGHE LH, WILSON JM. New recombinant serotypes of AAV vectors. Curr. Gene Ther. 2005;5:285–297. doi: 10.2174/1566523054065057. [DOI] [PubMed] [Google Scholar]
- GRIEGER JC, SAMULSKI RJ. Packaging capacity of adeno-associated virus serotypes: Impact of larger genomes on infectivity and postentry steps. J. Virol. 2005;79:9933–9944. doi: 10.1128/JVI.79.15.9933-9944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAI Y, YUE Y, LIU M, GHOSH A, ENGELHARDT JF, CHAMBERLAIN JS, DUAN D. Efficient in vivo gene expression by trans-splicing adeno-associated viral vectors. Nat. Biotechnol. 2005;23:1435–1439. doi: 10.1038/nbt1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU M, YUE Y, HARPER SQ, GRANGE RW, CHAMBERLAIN JS, DUAN D. Adeno-associated virus-mediated microdystrophin expression protects young mdx muscle from contraction-induced injury. Mol. Ther. 2005;11:245–256. doi: 10.1016/j.ymthe.2004.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANNO CS, ARRUDA VR, PIERCE GF, GLADER B, RAGNI M, RASKO J, OZELO MC, HOOTS K, BLATT P, KONKLE B, DAKE M, KAYE R, RAZAVI M, ZAJKO A, ZEHNDER J, NAKAI H, CHEW A, LEONARD D, WRIGHT JF, LESSARD RR, SOMMER JM, TIGGES M, SABATINO D, LUK A, JIANG H, MINGOZZI F, COUTO L, ERTL HC, HIGH KA, KAY MA. Successful transduction of liver in hemophilia by AAV-factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- NAKAI H, STORM TA, KAY MA. Increasing the size of rAAV-mediated expression cassettes in vivo by intermolecular joining of two complementary vectors [see comments] Nat. Biotechnol. 2000;18:527–532. doi: 10.1038/75390. [DOI] [PubMed] [Google Scholar]
- REICH SJ, AURICCHIO A, HILDINGER M, GLOVER E, MAGUIRE AM, WILSON JM, BENNETT J. Efficient trans-splicing in the retina expands the utility of adenoassociated virus as a vector for gene therapy. Hum. Gene Ther. 2003;14:37–44. doi: 10.1089/10430340360464697. [DOI] [PubMed] [Google Scholar]
- SHAPIRO MB, SENAPATHY P. RNA splice junctions of different classes of eukaryotes: Sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUN L, LI J, XIAO X. Overcoming adeno-associated virus vector size limitation through viral DNA heterodimerization. Nat. Med. 2000;6:599–602. doi: 10.1038/75087. [DOI] [PubMed] [Google Scholar]
- WANG L, HERZOG RW. AAV-mediated gene transfer for treatment of hemophilia. Curr. Gene Ther. 2005;5:349–360. doi: 10.2174/1566523054065048. [DOI] [PubMed] [Google Scholar]
- WARRINGTON KH, JR., GORBATYUK OS, HARRISON JK, OPIE SR, ZOLOTUKHIN S, MUZYCZKA N. Adeno-associated virus type 2 VP2 capsid protein is nonessential and can tolerate large peptide insertions at its N terminus. J. Virol. 2004;78:6595–6609. doi: 10.1128/JVI.78.12.6595-6609.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XU Z, YUE Y, LAI Y, YE C, QIU J, PINTEL DJ, DUAN D. Trans-splicing adeno-associated viral vector-mediated gene therapy is limited by the accumulation of spliced mRNA but not by dual vector coinfection efficiency. Hum. Gene Ther. 2004;15:896–905. doi: 10.1089/hum.2004.15.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAN Z, ZHANG Y, DUAN D, ENGELHARDT JF. From the cover: Trans-splicing vectors expand the utility of adeno-associated virus for gene therapy. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6716–6721. doi: 10.1073/pnas.97.12.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAN Z, ZAK R, ZHANG Y, ENGELHARDT JF. Inverted terminal repeat sequences are important for intermolecular recombination and circularization of adeno-associated virus genomes. J. Virol. 2005;79:364–379. doi: 10.1128/JVI.79.1.364-379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YUE Y, LI Z, HARPER SQ, DAVISSON RL, CHAMBERLAIN JS, DUAN D. Microdystrophin gene therapy of cardiomyopathy restores dystrophin–glycoprotein complex and improves sarcolemma integrity in the mdx mouse heart. Circulation. 2003;108:1626–1632. doi: 10.1161/01.CIR.0000089371.11664.27. [DOI] [PMC free article] [PubMed] [Google Scholar]


