Abstract
A critical requirement for integration of retroviruses, other than HIV and possibly related lentiviruses, is the breakdown of the nuclear envelope during mitosis. Nuclear envelope breakdown occurs during mitotic M-phase, the envelope reforming immediately after cell division, thereby permitting the translocation of the retroviral preintegration complex into the nucleus and enabling integration to proceed. In the oocyte, during metaphase II (MII) of the second meiosis, the nuclear envelope is also absent and the oocyte remains in MII arrest for a much longer period of time compared with M-phase in a somatic cell. Pseudotyped replication-defective retroviral vector was injected into the perivitelline space of bovine oocytes during MII. We show that reverse-transcribed gene transfer can take place in an oocyte in MII arrest of meiosis, leading to production of offspring, the majority of which are transgenic. We discuss the implications of this mechanism both as a means of production of transgenic livestock and as a model for naturally occurring recursive transgenesis.
Pronuclear injection has been widely used in attempts to produce transgenic cattle and other livestock. However, after a decade of use this technique has been unable to achieve transgenesis in more than 1% of injected embryos (1).
Retroviral infection, in which the genetic information is transferred as an RNA molecule, was the earliest method used for gene transfer into embryos (2). Repeated attempts over a number of years showed that the lack of control of gene dose and timing using this process results in nearly all of the animals born being genetic mosaics, with multiple and different gene insertion locations in different tissues (3). Recent studies placed retroviral packaging cells into the perivitelline space of bovine embryos and obtained several mosaic transgenic fetuses, but no live animals were produced (4). In general, the replication-defective retroviral vectors have seen only limited use in transduction of embryos (4–6), and no transgenic farm animals have been produced.
The extensive body of research in retroviral biology provides insights to how this highly effective biological system can be used to advantage in the production of transgenics. After entry of the retroviral RNA into the cell and reverse transcription into DNA, the integration of the DNA provirus into the host cell genome is mediated by the retroviral integrase and specific nucleotide sequences at the ends of the retroviral genome (7–9). Most retroviruses can infect only dividing cells (9–11). The critical requirement for integration of retroviruses, other than HIV and possibly related lentiviruses, is the breakdown of the nuclear envelope during mitosis. Nuclear envelope breakdown occurs during mitotic M-phase, the envelope reforming immediately after cell division. Envelope breakdown permits the translocation of the retroviral preintegration complex into the nucleus, enabling integration to proceed (7–10). Thus, the window of opportunity for access of the integration complex to the chromatin in somatic cells in mitosis is during M-phase. The genetic mosaics produced in the earlier embryo gene transfer using retroviruses arose because integration occurred late in development, so that only some cell lineages received the transgene. In the oocyte, during metaphase II (MII) of the second meiosis, the nuclear envelope is also absent and the oocyte remains in MII arrest for a much longer period of time compared with M-phase in a somatic cell (12). We hypothesized that, rather than targeting an embryo, retroviral vector gene introduction into a MII oocyte should result in a higher probability of preintegration complexes gaining access to the chromatin, and therefore should increase gene integration efficiency into the genome. Moreover, as the genes would be inserted before fertilization, the resulting offspring should not be mosaic. We show that reverse-transcribed gene transfer can take place in an oocyte in MII arrest of meiosis, leading to production of offspring, the majority of which are transgenic.
Not only do these results have the potential to greatly enhance the ability to produce transgenic animals, but they also provide an insight to an endogenous means of generation of genetic diversity. Chromatin is exposed for a prolonged period during MII arrest in an oocyte. During this time random insertion of DNA generated through reverse transcription and integration of transposable elements present as RNA provides a simple, but powerful, potential mechanism for generation of genetic diversity in progenitor cells as a prerequisite to evolutionary selection.
MATERIALS AND METHODS
Vector Production and Characterization.
To overcome the low titer and restricted host range of the commonly used vectors, we used a replication-defective vector based on Moloney murine leukemia virus, pseudotyped with the envelope glycoprotein of vesicular stomatitis virus (VSV-G) (13–15). In contrast to the native retroviral envelope proteins, absent from this vector, the VSV-G interacts with phospholipid components of the host cell plasma membrane (16–17). Retroviral vectors pseudotyped with VSV-G have an expanded range of infectivity and can be concentrated without significant loss of infectivity (13–16). Pseudotyped vectors were produced by using standard methodologies described in detail elsewhere (13–15). Titers of pseudotyped retroviral vectors were estimated in 208F rat fibroblast cells. Cells were exposed to G418 24 hr after infection, and the resistant colonies were counted after 12–14 days in selection. Plasmid pLRGeo and pLSRNL were used to construct the retroviral vectors LRgeoL-(VSV-G) and LSRNL-(VSV-G) as described (18–20). pLRGeo has a neomycin phosphotransferase (N) and β-galactosidase fusion gene expressed from Rous sarcoma virus (RSV) promoter (R) inserted downstream to the murine sarcoma virus long terminal repeat (LTR) (5′L). pLSRNL has hepatitis B surface antigen gene (HbsAg) (S) inserted downstream from the 5′L. The gene for neomycin phosphotransferase (N) is expressed from the RSV promoter (R).
In preparatory in vitro studies, we used high titer LRgeoL-(VSV-G) pseudotyped vector (21) to verify that the vector system was functional at 39°C, the critical operating temperature for in vitro maturation and fertilization of oocytes and culture of the early bovine embryo. Pseudotyped vector stability at 39°C was determined by titering with 208F cells after incubation of the vectors. The infectivity of the pseudotyped vectors decreased rapidly at 39°C to less than 10% at 4 hr, which gives a very narrow time window of efficient infection.
In Vitro Oocyte Maturation and Embryo Production.
In vitro production of bovine embryos was performed as described (22–25). In brief, oocytes collected by aspiration postmortem from cow ovaries were placed in maturation medium with 10% fetal bovine serum, 0.2 mM sodium pyruvate, 50 μg/ml of gentamycin sulfate, 5 μg/ml of luteinizing hormone, and 1 mg/ml of estradiol-17β in TC-199 at 39°C with 5% CO2 for 24 hr. The matured oocytes were mixed with thawed semen (acquired from a commercial bull stud), in which sperm concentration was adjusted to 1 × 106/ml. Sperm and oocytes were cocultured at 39°C in 5% CO2. Twenty-four hours postsemen addition, zygotes were further cultured in bovine embryo culture medium (25) with balanced salt solution supplemented with 5 mM lactate, 1 mM l-glutamine, 0.4 mM sodium pyruvate, 50 μg/ml of gentamycin sulfate, 6 mg/ml of BSA (fatty acid free), 10 μl/ml of 100× MEM amino acids solution, and 20 μl/ml of 50× basal medium Eagle amino acids solution at 39°C with 5% CO2.
Pronuclear Injection.
Pronuclear injection was performed at 18–22 hr postsemen addition. Pronuclei were visualized by centrifugation of zygotes at 12,000 × g for 6 min. DNA solution at 4 ng/μl was microinjected into one of the pronuclei by using an Eppendorf Transjector 5462.
Perivitelline Space Injection (PSI).
When PSI of vector was required, oocytes were recovered at 16 hr postincubation in maturation medium. Cumulus cells were removed mechanically, and oocytes were ready for injection. Oocytes then were coincubated with sperm at 24 hr postincubation in maturation medium. When PSI was applied to zygotes, the same procedure was followed at 12 hr postsemen addition. The estimated volume of the perivitelline space, lying between the zona pellucida and the plasma membrane of a bovine oocyte, is 2.8 × 10−7 cm3 (280 picoliters). Because of the limited volume of the perivitelline space, high titer vector stock is critical for successful PSI into early-stage embryos. Vector stocks were concentrated to 109 colony-forming units/ml (13), to permit injection of 10 picoliters containing at least a single infectious unit. We used two different variants of the same vector backbone containing an internal RSV promoter.
5-Bromo-4-Chloro-3-Indolyl β-d-Galactoside (X-gal) Staining.
To perform X-gal staining (5, 6), embryos were washed twice in 0.1 M Na-PBS supplemented with 0.3% polyvinylpyrrolidone (PVP) and 2 mM MgCl2 before fixing for 10 min at room temperature in 0.2% glutaraldehyde in the same buffer containing 5 mM EGTA. Fixed embryos were washed with PBS supplemented with 0.01% of sodium desoxycholate, 0.02% of Nonidet P-40, and 2 mM MgCl2, three times within 2 hr at room temperature. Embryos were incubated overnight in X-gal solution [20 mM K3Fe(CN)6, 20 mM K4Fe(CN)6-3H2O, 2 mM MgCl2, and 1 mg/ml of X-gal dissolved in dimethylformamide] at 37°C. X-gal staining of cultured cells was similar to that of embryos except the PBS lacked PVP and detergents.
Embryo Transfer.
Hormonal synchronization of recipient cattle was performed as described (26, 27). Recipients received i.m. injection of 100 μg of gonadotropin-releasing hormone (GnRH) on day 0 and 25 mg F2α on day 7. Two days after PGF2α injection, a second dose of GnRH (100 μg) was given; ovulation occurred between 24 and 32 hr later. Seven days after ovulation, in vitro produced experimental blastocysts, at 7 days postsemen addition, which were transferred to the uteri of synchronized recipient cattle.
Expression of HbsAg.
HbsAg detection was performed with a commercially available capture ELISA (Auzyme, Abbott).
PCR Analysis.
DNA was extracted and subjected to PCR analysis using two sets of primers. Amplification of the neor+ gene, primer set Neo-1 and Neo-2 yields a 349-bp amplicon (18). Amplification of HBsAg gene and primer set S-1 and S-3 yields a 334-bp amplicon (21). PCR was in a final volume of 50 μl. Cycles were 94°C for 2 min, 50°C for 2 min, and 72°C for 2 min. After 30 cycles the products were analyzed on 2% agarose gel.
Southern Hybridization.
Genomic DNA was digested with either the restriction enzyme HindIII (two digestion site in pLSRNL) or BamHI (single digestion site in pLSRNL). DNA fragments were separated by electrophoresis on a 0.8% agarose gel and transferred to Hybond-N+ nylon membranes. The blot was hybridized with a 32P-labeled PCR product of primer set S-1 and 3 on pLSRNL (334 bp) in rapid hybridization buffer (Amersham). After four washes at 65°C with high stringency buffer (2× standard saline citrate), the blot was exposed to x-ray film at −80°C.
RESULTS
In Vitro Generation and Evaluation of Transgenic Embryos.
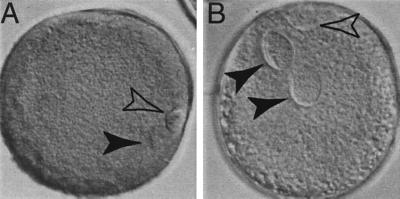
The distinctive nuclear configuration in bovine oocytes during maturation and after fertilization is shown in Fig. 1. Bovine oocytes and zygotes at the developmental stages shown in Fig. 1 were subjected to either pseudotyped vector LRgeoL-(VSV-G) infection by PSI or to conventional vector-free pronuclear injection of linear plasmid DNA of lacZ gene driven by simian virus 40 promoter (SV-lac). Perivitelline space-injected oocytes at 39°C were fertilized by using standard in vitro procedures 6–8 hr after PSI. Thus, the semen addition occurs at the normal stage of maturation 24 hr after placement in maturation medium. At the rate of loss of vector infectivity at 39°C found in control experiments (<10% remained after a 4-hr incubation period), we calculated that <1% of the initial vector titer remained by the time of fertilization. The timing of the process was designed to insure that any gene insertions have a very high probability of having occurred before fertilization.
Figure 1.
Bovine oocytes matured in vitro demonstrate distinctive nuclear configuration compared with zygotes at the pronuclear stage. Oocytes were fixed 18 hr after placing in maturation medium and zygotes were fixed 18 hr postsemen addition, in three parts ethanol/one part acetic acid for 24 hr, before acid-orcein staining (1% orcein stain in 40% acetic acid in H2O). (A) After induction of maturation, the oocyte undergoes germinal vesicle (4N) breakdown, the chromatin condenses, the first meiotic division occurs, and the first polar body, containing half of the genome (2N), is extruded. The remaining condensed, diploid chromatin is aligned at the second metaphase plate (arrowhead). Chromatin is not enclosed in a nuclear membrane and is located next to the first polar body. The polar body also has intense chromatin, which is enclosed in a plasma membrane during autosome segregation (open arrowhead). The oocyte remains arrested in MII until the completion of meiosis, which is heralded by the extrusion of the second polar body (1N) induced by fertilization. (B) After fertilization, the oocyte progresses from MII to interphase. Zygote contains both maternal and paternal pronuclei enclosed by a nuclear envelope (arrowhead). The second polar body is located next to the maternal pronucleus with distinctive chromatin staining (open arrowhead). In addition to the difference in nuclear configuration, oocytes have very condensed chromatin and intense chromatin staining, whereas zygotes have dispersed chromatin with less intense staining.
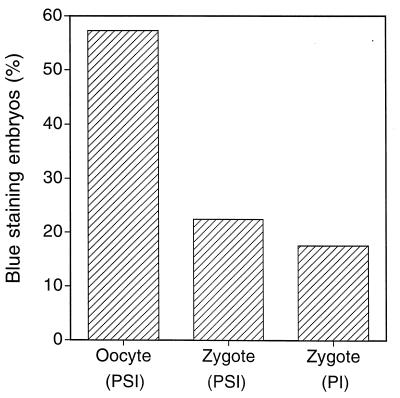
Embryos were stained at morula stage, and those with blue X-gal staining, indicating gene transfer and expression, were scored as positive. The morula stage embryos produced when MII oocytes were exposed to the pseudotyped vector showed the highest positive staining rate (178/316, 56%). Infection with pseudotyped vector at zygote stage resulted in stained morulae at less than half this efficiency (49/226, 22%) and conventional pronuclear injection of DNA showed the lowest rate of gene transfer (25/144, 17%) (Fig. 2). The lower rate observed in the zygotes may be because integration is delayed until the next cell cleavage after infection, decreasing the integration ability of the preintegration complex.
Figure 2.
Gene transfer efficiency of LRgeoL-(VSV-G) in bovine oocytes and zygotes. Gene transfer efficiency was evaluated by X-gal staining of embryos 4 days postsemen addition. Pronuclear injection of linearized SV-lac into zygotes was used as a control. Infection with LRgeoL-(VSV-G) was achieved by PSI at the indicated time points. (A) Oocytes at 20 hr after placing in maturation medium were infected by LRgeoL-(VSV-G) PSI. Four hours after infection, oocytes were incubated with thawed semen. X-gal staining was observed in 56% (178/316) of infected embryos. Zygotes at 18 hr postsemen addition were infected by LRgeoL-(VSV-G) PSI. X-gal staining was observed in 22% (49/226) of infected embryos. Zygotes at 18 hr postsemen addition were pronuclear injected with SV-lac, and 17% (25/144) of microinjected embryos were stained with X-gal. Cytoplasmic expression, in the case of pronuclear microinjection, does not differentiate between integration and extrachromosomal expression.
These results demonstrate that a meiotic bovine oocyte is highly susceptible to VSV-G-pseudotyped vector infection. There was no apparent morphological difference among the embryos derived either from PSI of VSV-G-pseudotyped vector into oocytes and zygotes, or pronuclear injection of DNA solution into zygotes.
Embryos for Transfer to Recipients.
For the production of animals to be carried to term, we selected a vector that has a similar backbone structure to that used in the LRgeo-(VSV-G). The reason for changing to a different vector was that expression of lacZ-neor+ fusion protein from LRgeo-(VSV-G) treatment leads to abnormal morphology of blastocysts, suggesting a detrimental effect on the cells. Treatment of embryos with the LSRNL vector, containing the same internal promoter driving neor+, did not lead to abnormal embryo morphology. LSRNL can be prepared at the high titer required (≈109 colony-forming units/ml). Preliminary experiments showed no reduction in the number of embryos developing to blastocyst during the use of this vector. Normal mechanisms blocking multiple retroviral infections are not operative with the pseudotyped system. When the pseudotyped vectors are used multiple insertional events frequently occur (21). For our purposes multiple insertions would complicate subsequent analysis. Therefore, we chose a combination of injection volume and titer that would provide a multiplicity of infection of approximately one.
To assess the utility of pseudotyped vectors for germ-line transformation, we injected the perivitelline spaces of oocytes (n = 836) and zygotes (n = 584) with high titer pseudotype [LSRNL-(VSV-G)]. Injection volumes were calibrated to provide one colony-forming unit per 10 picoliter injection. At this vector concentration, Poisson probabilities indicate that a portion of embryos likely will have multiple insertions, whereas some injections will contain no vector, making transgenesis impossible. After PSI, 174/836 (21%) oocytes and 193/584 (33%) zygotes developed to the blastocyst stage. Control experiments showed that the lower efficiency of blastocyst formation is not caused by PSI. Disturbance of the cumulus cells surrounding the oocyte and the resulting polyspermy are possible causes. Embryos for transfer to recipients were selected from the groups at random. They were not preselected in any way for the presence of transgenes but were merely selected based on their normal morphological appearance. They were transferred to recipient cows by using embryo transfer procedures used commercially in cattle. Seven days after fertilization in vitro, 10 blastocysts from the oocyte PSI group were transferred into five hormonally synchronized recipient cows, and 12 blastocysts from the zygote PSI group were transferred into six recipient cows. The results are summarized in Table 1. The blastocysts not transferred were pooled at random in groups of five and frozen at −20°C for subsequent analysis. Six groups of blastocysts were prepared from the nontransferred embryos from each of the zygote and oocyte treatments and subsequently were analyzed by PCR. Although it was not possible to determine the percentage of individual transgenic embryos by this method, all groups were positive for the transgene for HbsAg protein.
Table 1.
Embryo transfer of randomly selected experimental blastocysts
| Vector treatment stage
|
||
|---|---|---|
| 17-hr mature oocyte | 12-hr postfertilization zygote | |
| Number of oocytes | 836 | 584 |
| Number developing to blastocyst (% of oocytes) | 174 (21) | 193 (33) |
| Number of embryos transferred to recipients | 10 | 12 |
| Number of recipients | 5 | 6 |
| Number of pregnancies | 3 | 4 |
| Number of calves | 4 | 4* |
| Transgenic offspring | 4 | 1 |
Includes one pregnancy that terminated in late-stage abortion of twins, neither of which was transgenic.
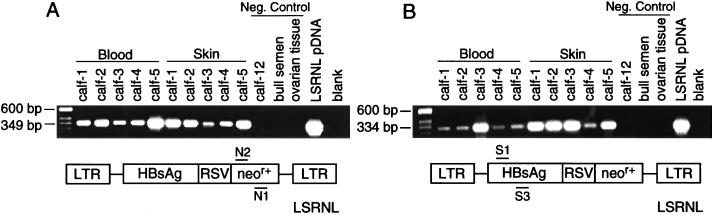
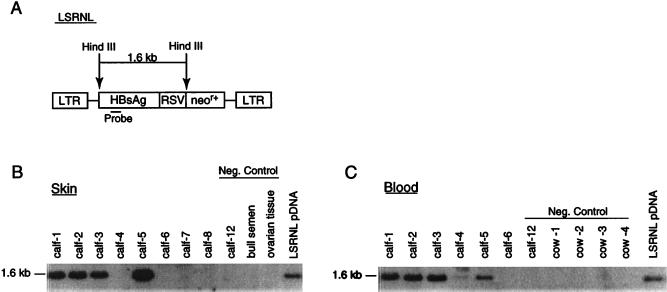
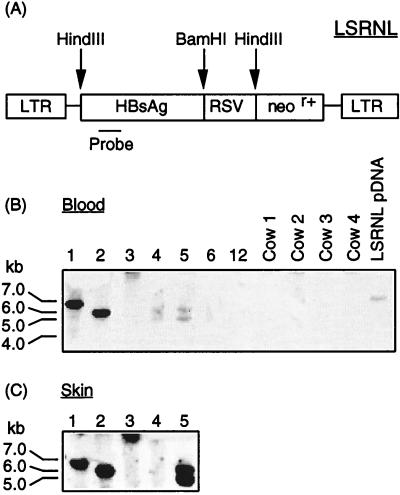
Overall, the embryo survival rate, gestation length, and animal size were normal and consistent with that observed in commercial bovine embryo transfer. Four calves (three female, one male) were born from the oocyte treatment group, and two calves (one female, one male) were born from the zygote treatment group. One pregnancy from the zygote group terminated in late-stage abortion of normal-size twins. All four calves derived from the oocyte treatment group were transgenic, and one of the two calves derived from the zygote treatment group was transgenic. The aborted twins from the zygote treatment group were nontransgenic. In every transgenic calf, DNA extracts of whole blood (mesoderm) and skin (ectoderm) showed the presence of the transgene by PCR (Fig. 3). Calves 1, 2, 3, and 5 showed the presence of the transgene by Southern blot analysis of both skin and blood (Fig. 4). Calf 4 showed inconsistent Southern blot results from skin; some preparations from skin demonstrated a band, whereas that shown in Fig. 4 does not. Southern blot analysis of blood of calf 4 did consistently show a band. This finding implies that there may be an uneven distribution of the transgene in calf 4. This observation may arise because the distribution of the gene is mosaic within the ectodermal lineage or it may simply reflect inconsistencies in preparation of DNA from the skin of this animal, which is a Zebu crossbred with a very different consistency to its skin. We have not been able to exclude the possibility that calf 4, a twin, may be a leucochimera. In cattle, the chorionic membranes of twins usually fuse early in embryonic development, allowing blood to be exchanged between the two fetuses. Calves 1, 2, 3, and 5 also each showed identity of integration sites in their blood and skin (Fig. 5), and thus that the genes are integrated into the genome. Three of the calves had a single integration site; calves 4 and 5 had two integration sites. As expected, different flanking regions were noted between calves (Fig. 5).
Figure 3.
PCR analysis to detect neor+ (A) and HBsAg (B) transgenes in DNA extracts of whole blood (mesoderm) and skin (ectoderm) of calves derived from embryos infected with VSV-G-pseudotyped vector. (A and B) Lanes 1–5 are DNA from blood samples and lanes 6–10 are DNA from skin tissue of transgenic calves (lane 1: zygote treatment group, male; lane 2: oocyte treatment group, female; lane 3: oocyte treatment group, female; lane 4: oocyte reatment group, female; lane 5; oocyte treatment group, male.). Lanes 4 and 5 were from twins resulting from implantation of two embryos and the birth of phenotypically distinct calves of different breeds. The negative controls in lanes 11–13 comprise DNA from a blood sample from calf 12, which was a naturally conceived nontransgenic calf, commercial bull semen and ovarian tissue from a cow. Lane 14 contains the plasmid DNA of LSRNL, and lane 15 is a water control. Tissue samples from blood and skin show positive signal with both primer sets, but none of the negative controls show the presence of transgene. Differences in intensity of signal do not reflect copy numbers of the transgene inserted.
Figure 4.
Southern blot analysis of HindIII-digested genomic DNA (A) to detect the 1.6-kb fragment from the HBsAg gene in blood (B) and skin (C) tissue of calves. (B) Lanes 1–5, transgenic calves. Lane 6, nontransgenic calf born after VSV-G-pseudotyped vector injection. Lanes 7 and 8, nontransgenic aborted fetuses. Negative controls include calf 12, which was a naturally conceived nontransgenic calf, ovarian tissue from a cull cow and commercial bull semen. (C) Lanes 1–5, transgenic calves. Lane 6, nontransgenic calf. Negative controls, calf 12 (naturally conceived nontransgenic). Cows 1–4 were the recipients carrying the transgenic calves. HindIII-digested pLSRNL DNA was included as a positive control.
Figure 5.
Detection of chromosomal integration of HBsAg gene by Southern blot hybridization of the BamHI-digested genomic DNA from blood and skin tissue of calves. (A) Digestion sites of pLSRNL. Various-sized fragments were produced from different calves that were different from the linearized plasmid DNA control, demonstrating the successful unique insertion of the transgene into the host cell genome of each calf. (B) Blood—lanes 1–5, transgenic calves. Lane 6, nontransgenic calves born after VSV-G-pseudotyped vector injection. The negative controls in lanes 7–11 comprise calf 12, a naturally conceived nontransgenic calf, and blood samples from recipient cows (nos. 1–4) that carried the transgenic embryos. Lane 12 is the plasmid DNA of LSRNL digested by BamHI. (C) Skin—lanes 1–5, transgenic calves that show the same integration pattern as the corresponding blood samples.
Semen was collected from the two males when they reached sexual maturity. PCR analysis of semen indicated the presence of both transgenes, neor+ and HBsAg. This finding demonstrated that the genes were in the germ line, as well as in mesoderm and ectoderm. The transgenic semen from the bull derived from oocyte treatment (calf 5) was used in the in vitro fertilization protocol. Five of the nine embryos produced were shown by PCR to be transgenic. Thus, the transgene is transmitted via the germ line with apparently Mendelian segregation frequency.
Gene Expression.
These studies were conducted to test the hypothesis that nuclear membrane breakdown at MII would facilitate retroviral transduction and to evaluate this system as a means of production of transgenic animals. Therefore, the vectors we used were not constructed to provide a specific expression location within the whole animal but, rather, were useful laboratory models. From their use in cell culture systems, retroviral promoters and enhancers are thought of as strong gene control elements, but little is known about their activities other than in cell culture systems.
After birth of the transgenic animals, we measured the level of HBsAg in their serum. One animal (calf 5) showed a moderate level of HBsAg (reactive in the nonquantitative Auzyme test) in the precolostral serum sample drawn just after birth, but no HBsAg has been detected in subsequent samples. The animals now are approximately 2 years old and are normal in all physical respects.
DISCUSSION
These studies demonstrate that VSV-G-pseudotyped RNA-based vectors can genetically transform bovine oocytes or zygotes and lead to stable integration of the provirus in the host cell genome. Both infection of oocytes and zygotes produce transgenics at rates that greatly exceed existing pronuclear microinjection methods (1).
The enhancement we have seen in integration efficiency in MII stage oocytes compared with zygotes underscores the importance of nuclear envelope breakdown for retroviral integration (10). Given the broad infectivity of VSV-G pseudotypes, we anticipate that this method can be used to produce transgenic animals of any species for which either oocytes can be matured in vitro or MII oocytes are otherwise accessible. Combined with the use of internal promoters that confer tissue specificity of gene expression and incorporation of other engineering features, this gene transfer system should substantially facilitate transgenic animal production.
Recent data has shown very restricted spatial/temporal expression of genes under the control of LTR elements (28, 29). For example, in transgenic mice the intracisternal A-particle retrotransposon LTR has been shown to be active only in a very restricted period of male germ-line development, that of gonocytes and premeiotic undifferentiated spermatogonia (29).
On a per-copy basis, retroviral vectors integrated by their normal biological integration system exhibit significantly higher levels of expression than other means of genetic transformation (30). Pronuclear injection (as well as standard DNA transfection techniques) leads to insertion of large tandem arrays of DNA. These tandem arrays are unstable and subject to rearrangements and deletions in subsequent cell divisions (31). Currently, transgenic technology relies on breeding rare founder animals while retaining the desired phenotype. Because of the DNA structure at the insertion site, the phenotype is likely to be unstable during breeding. The system we have described permits single-copy gene insertions, potentially at many sites in the genome, that animal breeding experience suggests should behave predictably during breeding. It creates challenges and opportunities for new transgenic strategies: (a) efficient production of transgenic founders would enable evaluation of phenotypic expression in large numbers of founder animals, using standard population genetics procedures; (b) efficient production of transgenic founders will make it possible to do the genetic design necessary to obtain constructs with appropriate temporal-spatial in vivo expression patterns; and (c) efficient production of transgenic animals carrying the genetic constructs throughout their bodies will provide a potentially useful method for evaluation of gene therapy constructs.
Although we have examined directed genetic change, our approach suggests processes that may be operating naturally. Retroviruses and retrovirus-like elements are naturally inherited elements in the germ line of many organisms, where they show relatively stable Mendelian inheritance (32). It has been estimated that as much as 10% of the mammalian genome has been introduced by mechanisms involving reverse transcription (33, 37). The extended period of coevolution of these elements with the mammalian genome suggests either an evolutionary advantage or a special, potentially symbiotic, relationship to the remainder of the genome. Recent studies have shown that retroviral elements select a scaffold or matrix-attached region of the chromatin flanked by DNA with a high binding potential, possibly enhancing potential for recombination and transcription (34, 35) and implicated in the efficient expression of tandem transgene arrays in transgenic animals (36).
Retrotransposons, retroviruses, and other like genetic elements are widely distributed (32, 36, 38) and have particular utility as a means of assessing evolutionary changes. Recent findings by Agrawal et al. (39) provide an in vitro example of transposon-mediated evolutionary change. The approach we have implemented provides a model for how such a system may operate in nature, through recursive transgenesis. One can speculate the retrotransposons present in high copy number in genomes are very efficient at carrying out the same steps that we have implemented. During the prolonged MII arrest in an oocyte, large amounts of highly stable maternal mRNAs are found in the cell cytoplasm. Reverse transcription and random integration during MII arrest of meiosis would provide a powerful mechanism for generating evolutionary genetic diversity in progenitor cells. By occurring in meiosis, the system operates so that any useful change can be captured and can offer selective advantage. In this scenario, the nuclear membrane is the effective gatekeeper for the process, guarding the genome against rogue transposable elements except in meiosis. Operating repeatedly through many generations this system would provide a mechanism for periodic, dramatic quantum changes in phenotype. Other genetic changes brought about by recombination and crossing over during meiosis, because they normally act within a restricted area of the genome, are more likely to lead to smaller incremental changes. Movement of a retroelement to a new location in the genome of the organism introduces a stochastic event to the process.
Our goals were quite pragmatic. We were seeking a more efficient means of production of transgenic animals and testing a simple hypothesis. Although conventional retroviral vectors are limited in the size of inserted gene(s) that they carry, they have adequate capacity to carry coding sequences, internal promoters, and other necessary elements to produce proteins well in excess of 100 kDa, which includes the vast majority of proteins.
The efficiency of the process we designed was remarkable and provides a method of studying the generation of genetic diversity and the generation of transgenic animals. The use of vectors with appropriate expression characteristics, in combination with specific marker genes that could be monitored through successive generations, would allow the system we describe to be used as a model for understanding the coevolution of retroviral-like elements in the genome of animals. It also provides a model in which to study the continual generation of genetic diversity as the prerequisite to evolutionary selection.
Acknowledgments
Dr. Atsushi Miyanohara generously provided 293 gp/LSRNL producer cells. We thank Dr. Kurt Eakle for technical support and critical review on the manuscript, David Northey and Jerry Guenther for technical support in embryo transfer, and Drs. J. Been and M. Kieler for veterinary supervision of births. This work was supported in part by the College of Agricultural and Life Sciences of the University of Wisconsin-Madison and by Gala Design LLC.
ABBREVIATIONS
- MII
metaphase II
- VSV-G
vesicular stomatitis virus envelope glycoprotein G
- RSV
Rous sarcoma virus
- LTR
long terminal repeat
- HbsAg
hepatitis B surface antigen
- PSI
perivitelline space injection
- X-gal
5-bromo-4-chloro-3-indolyl β-d-galactoside
References
- 1.Wall R J. Theriogenology. 1996;45:57–68. [Google Scholar]
- 2.Jaenisch R. Proc Natl Acad Sci USA. 1976;73:1260–1264. doi: 10.1073/pnas.73.4.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaenisch R. Cell. 1980;19:181–188. doi: 10.1016/0092-8674(80)90399-2. [DOI] [PubMed] [Google Scholar]
- 4.Haskell R E, Bowen R A. Mol Reprod Dev. 1995;40:386–390. doi: 10.1002/mrd.1080400316. [DOI] [PubMed] [Google Scholar]
- 5.Kim T, Leibfried-Rutledge M L, First N L. Anim Biotechnol. 1993;4:53–69. [Google Scholar]
- 6.Kim T, Leibfried-Rutledge M L, First N L. Mol Reprod Dev. 1993;35:105–113. doi: 10.1002/mrd.1080350202. [DOI] [PubMed] [Google Scholar]
- 7.Goff V. Annu Rev Genet. 1992;26:527–544. doi: 10.1146/annurev.ge.26.120192.002523. [DOI] [PubMed] [Google Scholar]
- 8.Brown P O, Bowerman B, Varmus H E, Bishop J M. Proc Natl Acad Sci USA. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulkosly J, Skalka A M. Pharmacol Ther. 1994;61:185–203. doi: 10.1016/0163-7258(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 10.Roe T Y, Reynolds T C, Yu G, Brown P O. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller D G, Adam M A, Miller V. Mol Cell Biol. 1990;10:4239–4242. doi: 10.1128/mcb.10.8.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray A, Hunt T. In: The Cell Cycle. Murray D G, Hunt T, editors. New York: Freeman; 1993. pp. 3–22. [Google Scholar]
- 13.Yee J K, Friedmann T, Burns J C. Methods Cell Biol. 1994;43:99–112. doi: 10.1016/s0091-679x(08)60600-7. [DOI] [PubMed] [Google Scholar]
- 14.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J K. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yee J K, Miyanohara A, LaPorte P, Bouic K, Burns J C, Friedmann T. Proc Natl Acad Sci USA. 1994;91:9564–9568. doi: 10.1073/pnas.91.20.9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedmann T, Yee J K. Natural Med. 1995;1:275–277. doi: 10.1038/nm0395-275. [DOI] [PubMed] [Google Scholar]
- 17.Mastromarino P, Conti C, Goldoni P, Hauttecoeur B, Orsi N. J Gen Virol. 1987;68:2359–2369. doi: 10.1099/0022-1317-68-9-2359. [DOI] [PubMed] [Google Scholar]
- 18.Lu J K, Chen T T, Allen S K, Matsubara T, Burns J, C. Proc Natl Acad Sci USA. 1996;93:3482–3486. doi: 10.1073/pnas.93.8.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emi N, Friedmann T, Yee J K. J Virol. 1991;65:1202–1207. doi: 10.1128/jvi.65.3.1202-1207.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyanohara A, Elam R L, Witstum J L, Friedmann T. New Biol. 1992;4:261–267. [PubMed] [Google Scholar]
- 21.Burns J C, Matsubara T, Lozinski G, Yee J K, Friedmann T, Washabaugh C H, Tsonis P A. Dev Biol. 1994;165:285–289. doi: 10.1006/dbio.1994.1253. [DOI] [PubMed] [Google Scholar]
- 22.Parrish J J, Susko-Parrish J, Winer M A, First N L. Theriogenology. 1986;25:591–600. doi: 10.1016/0093-691x(86)90143-3. [DOI] [PubMed] [Google Scholar]
- 23.Leibfried M L, First N L. J Anim Sci. 1979;48:76–86. doi: 10.2527/jas1979.48176x. [DOI] [PubMed] [Google Scholar]
- 24.Leibfried-Rutledge M L, Critser E S, Eyestone W H, Northey D L, First N L. Biol Reprod. 1987;36:376–393. doi: 10.1095/biolreprod36.2.376. [DOI] [PubMed] [Google Scholar]
- 25.Rosenkrans C F, Jr, First N L. J Anim Sci. 1994;72:434–437. doi: 10.2527/1994.722434x. [DOI] [PubMed] [Google Scholar]
- 26.Pursley J R, Kosorok M R, Wiltbank M C. J Dairy Sci. 1997;80:301–306. doi: 10.3168/jds.S0022-0302(97)75938-1. [DOI] [PubMed] [Google Scholar]
- 27.Pursley J R, Wiltbank M C, Stevenson J S, Ottobre J S, Garverick H A, Anderson L L. J Dairy Sci. 1997;80:295–300. doi: 10.3168/jds.S0022-0302(97)75937-X. [DOI] [PubMed] [Google Scholar]
- 28.Hopkin K. J NIH Res. 1997;9:41–42. [Google Scholar]
- 29.Dupressoir A, Heidmann T. Mol Cell Biol. 1996;16:4495–4503. doi: 10.1128/mcb.16.8.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schubeler D, Meilke C, Maass K, Bode J. Biochemistry. 1996;35:11160–11169. doi: 10.1021/bi960930o. [DOI] [PubMed] [Google Scholar]
- 31.Colman A. Am J Clin Nutr. 1996;63:639S–645S. [Google Scholar]
- 32.Doolittle R, Feng D, McClure M, Johnson M. Curr Top Microbiol Immunol. 1990;157:1–18. doi: 10.1007/978-3-642-75218-6_1. [DOI] [PubMed] [Google Scholar]
- 33.Temin H M. Cell Biophys. 1986;9:9–16. doi: 10.1007/BF02797372. [DOI] [PubMed] [Google Scholar]
- 34.Mielke C, Maass K, Tummler M, Bode J. Biochemistry. 1996;35:2239–2252. doi: 10.1021/bi952393y. [DOI] [PubMed] [Google Scholar]
- 35.Taruscio D, Manuelidis L. Chromosoma. 1991;101:141–156. doi: 10.1007/BF00355364. [DOI] [PubMed] [Google Scholar]
- 36.McKnight R A, Spencer M, Wall R J, Hennighausen L. Mol Reprod Dev. 1996;44:179–184. doi: 10.1002/(SICI)1098-2795(199606)44:2<179::AID-MRD6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 37.Nouvel P. Genetica. 1994;93:191–201. doi: 10.1007/BF01435251. [DOI] [PubMed] [Google Scholar]
- 38.Patience C, Wilkinson D A, Weiss R A. Trends Genet. 1997;13:116–120. doi: 10.1016/s0168-9525(97)01057-3. [DOI] [PubMed] [Google Scholar]
- 39.Agrawal A, Eastman Q M, Schatz D G. Nature (London) 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]