Abstract
The Cacna1fnob2 mouse is reported to be a naturally occurring null mutation for the Cav1.4 calcium channel gene and the phenotype of this mouse is not identical to that of the targeted gene knockout model. We found two mRNA species in the Cacna1fnob2 mouse: approximately 90% of the mRNA represents a transcript with an in-frame stop codon within exon 2 of CACNA1F, while approximately 10% of the mRNA represents a transcript in which alternative splicing within the ETn element has removed the stop codon. This latter mRNA codes for full length Cav1.4 protein, detectable by Western blot analysis that is predicted to differ from wild type Cav1.4 protein in a region of approximately 22 amino acids in the N-terminal portion of the protein. Electrophysiological analysis with either mouse Cav1.4wt or Cav1.4nob2 cDNA revealed that the alternatively spliced protein does not differ from wild type with respect to activation and inactivation characteristics; however, while the wild type N-terminus interacted with filamin proteins in a biochemical pull-down experiment, the alternatively spliced N-terminus did not. The Cacna1fnob2 mouse electroretinogram displayed reduced b-wave and oscillatory potential amplitudes, and the retina was morphologically disorganized, with substantial reduction in thickness of the outer plexiform layer and sprouting of bipolar cell dendrites ectopically into the outer nuclear layer. Nevertheless, the spatial contrast sensitivity (optokinetic response) of Cacna1fnob2 mice was generally similar to that of wild type mice. These results suggest the Cacna1fnob2 mouse is not a CACNA1F knockout model. Rather, alternative splicing within the ETn element can lead to full-length Cav1.4 protein, albeit at reduced levels, and the functional Cav1.4 mutant may be incapable of interacting with cytoskeletal filamin proteins. These changes, do not alter the ability of the Cacna1fnob2 mouse to detect and follow moving sine-wave gratings compared to their wild type counterparts.
Introduction
Influx of calcium through voltage-gated calcium channels (VGCCs) leads to excitation-contraction coupling, excitation-transcription coupling, neurotransmitter release, and programmed cell death. Disorders of synaptic transmission are thought to be instrumental in two forms of human X-linked congenital stationary night blindness (CSNB): the “incomplete” form (iCSNB, or CSNB2), in which rod- and cone-driven electroretinogram (ERG) responses are reduced in amplitude, but oscillatory potentials (OPs) can be recorded; and the “complete” form (cCSNB, or CSNB1), in which rod-driven ERG responses are greatly reduced or absent but cone-driven responses are relatively well preserved, and OPs are rarely recorded. Mutations within the CACNA1F gene coding for Cav1.4 L-type calcium channels have been identified as one cause of CSNB2 [1], [2], as well as X-linked cone-rod dystrophy (CORDX3) [3] and Åland Island eye disease [4]. Over seventy CSNB2 nonsense and missense mutations have been identified (for example, [5]–[12], several of which have been shown to alter the biophysical properties of the channels [13]–[19]; reviewed in [20]).
Knockout of CACNA1F protein in mice following insertion of a self-excising Cre-lox-neo cassette into exon 7 results in an in-frame premature stop codon (G305X) in the Cav1.4 protein [21]. These Cacna1fG305X mice are characterized by complete loss of the b-wave and oscillatory potentials of the electroretinogram (ERG), absence of cone-driven visually-evoked activity in the superior colliculus, >90% reduction in calcium influx in photoreceptors, and disrupted retinal morphology with loss of photoreceptor synapses and sprouting of horizontal and bipolar cell dendrites into the outer nuclear layer [21]. Cacna1fG305X mice, therefore, resemble CSNB1 patients in their lack of cone-driven functions. More recently, a second mouse model said to be null for CACNA1F has been described [22]. This Cacna1fnob2 mouse arose by spontaneous insertion of a transposable element (ETn) into exon 2, which is predicted to produce an in-frame premature stop codon. Interestingly, while the Cacna1fnob2 mouse also displays disrupted retina morphology similar to that of the Cacna1fG305X mouse, the ERG of the Cacna1fG305X mouse is more similar to that of CSNB2 patients, being characterized by a reduced b-wave and oscillatory potentials while cone-driven responses are maintained.
The ETn element responsible for the Cacna1fnob2 mouse belongs to a family of early retrotransposons, approximately 5600 base pairs in length, which are transcribed during embryogenesis [23]–[25]. While they often are a source of mutations by insertion into the coding frame of genes, their characteristic long terminal repeat regions allow them to be alternatively spliced. This property can result in transcription of the gene into which they were inserted, at reduced levels [26], [27]. We hypothesized that the difference in phenotypes of the two mouse models was the result of alternative splicing of the ETn element, which would allow some transcription and synthesis of Cav1.4 protein. We have found two mRNA species in the Cacna1fnob2 mouse, one of which encodes an in-frame stop codon, and another in which the stop codon is missing as a result of splicing within the ETn; as a result, full-length protein was detectable by Western blotting using an antibody raised against the C-terminus of the α1F channel subunit (Cav1.4) protein. The alternatively spliced protein did not differ from the wild type protein with respect to activation and inactivation characteristics in an expression system; however, unlike the wild type protein N-terminus, the alternatively spliced N-terminus did not bind to cytoskeletal filamin proteins. Interestingly, while the outer retinal layers of the Cacna1fnob2 mouse were significantly disorganized, the optokinetic response was not dramatically different from that of wild type mice.
Results
Two mRNA species are detected in the Cacna1fnob2 mouse
Total RNA from the eyes of Cacna1fwt and Cacna1fnob2 mice was analyzed by RT-PCR. As shown in Figure 1.B, while only a single band was detected in Cacna1fwt mice with either primer combination (RR44+RR 45, or RR44+RR46), two bands were detected in Cacna1fnob2 mice (see table 1 for primers). Using the fluorescence intensity of the bands to assess relative quantity, we estimated that ∼90% of the mRNA is accounted for by the larger (Mr) band, and ∼10% by the smaller (Mr) band (consistent results from three separate reactions/gels). The cDNA bands were subsequently isolated and sequenced; the larger and more intense band corresponded to the CACNA1F-encoding sequence containing the ETn transposable element with an in-frame stop codon (Figure 2A), whereas the smaller and less intense band corresponded to the CACNA1F-encoding sequence containing the shorter ETn element that lacks the in-frame stop codon (Figure 2B). Genotyping of these mice, along with genomic DNA samples purchased from Jackson Laboratory, confirmed that our Cacna1fnob2 mice were identical in genotype to the original Cacna1fnob2 mouse line, as described previously [22]. An alignment of the predicted protein sequences corresponding to wild type Cav1.4 (hereafter referred to as Cav1.4wt) and Cav1.4 from the smaller size band (hereafter referred to as Cav1.4nob2) is provided in Figure 2C.
Figure 1. RT-PCR analysis of Cacna1fwt and Cacna1fnob2 mice.
A. Schematic representation of the location of PCR primers used. Primers RR44, 45, and 46 were used for RT-PCR reactions; primers RR50, 51, 52, and 53 were used for genomic PCR reactions. B. Agarose gel depicting RT-PCR reaction products for mRNA isolated from Cacna1fwt and Cacna1fnob2 mice. Regardless of the primer pair used, only a single band is detected using mRNA from Cacna1fwt mice. Using mRNA from Cacna1fnob2 mice, however, two bands are visible (see arrows). The relative intensities of the fluorescence signals indicate that the larger-Mr band accounts for ∼90%, and the smaller-Mr band for ∼10%, of the total mRNA.
Table 1. Primer sequences.
| Primer Name | Primer Sequence (5′→3′) | Primer Use |
| RR44 | atgtcggaatctgaagtcgg | RT-PCR (Figure 1A) |
| RR45 | caatgctgatgcaggaccg | |
| RR46 | gcagtgttggagtcgtcctc | |
| CDRR69 | ggatccaaatgtcggaatctgaagtcg | N-terminus amplification |
| CDRR70 | gaattctcacttccactctacaatgctcatgc |
Figure 2. Sequencing results of Cacna1fwt and Cacna1fnob2 mice.
A. Sequencing results from the larger-Mr, more intense band. Sequences are given for the DNA (small letters) and corresponding protein (capital letters); the sequence corresponding to wild type Cav1.4 protein is highlighted in grey. The ETn transposable element is inserted after the ninth Cav1.4 amino acid, and encodes an in-frame stop codon (highlighted in red) which is predicted to result in truncation of the Cav1.4 protein after only 25 amino acids. The underlined ETn sequence highlights a repetitive sequence (compare to the beginning of the ETn sequence, beginning at amino acid number 4). B. Same as A., but for the smaller-Mr, less intense band. Note that the inserted ETn element is shorter, and the in-frame stop codon is missing. C. Alignment of the predicted N-terminal amino acid sequences of wild type Cav1.4 protein and Cav1.4 protein encoded in the smaller-Mr band. A region of approximately 22 amino acids differs in the N-termini of the two clones.
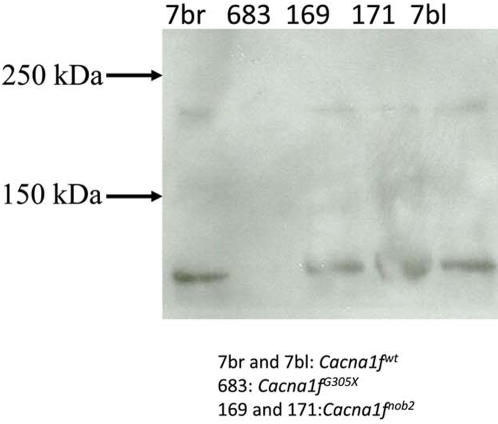
To test for Cav1.4 protein in the Cacna1fnob2 mouse, we probed lysates from spleen with a Cav1.4-specific antibody [16]. As shown in Figure 3, a band near 230 kDa was detected in samples from both Cacna1fwt and Cacna1fnob2 mice, but not from Cacna1fG305X mouse. A lower-Mr band (<150 kDa) was also detectable in extracts from all mouse strains tested, except Cacna1fG305X; this band likely is a truncated form of the wild type protein, as is characteristic of other L-type calcium channel proteins [28].
Figure 3. Western blot of spleen samples from Cacna1fwt, Cacna1fnob2, and Cacna1fG305X mice probed with a Cav1.4-specific antibody directed against the C-terminus of the channel.
Full-length protein is visible in all lanes except for Cacna1fG305X.
Cacna1fnob2 mouse has a selective b-wave defect
These results indicated that a full-length Cav1.4 channel (corresponding to the mRNA sequence from the smaller Mr, less intense band in Figure 1B) could be present in the Cacna1fnob2 mouse. This was surprising, since this mouse was reported to be null for the Cav1.4 channel protein [22]. Therefore, we next tested to ensure that the ERGs in our mice were identical to those previously published.
ERG recordings showed a selective b-wave defect in Cacna1fnob2, compared with that in age-matched Cacna1fwt mice (Figure 4), as previously reported [22], [29]. Under both scotopic and photopic adaptation conditions, the amplitude of the b-wave was selectively diminished. The intensity-response curves for the scotopic a-wave, as well as maximal a-wave amplitudes and thresholds (minimum luminance to reach criterion amplitude of 20 µV), were comparable in our mutant and wild type mice. In contrast, however, the b-waves in mutant and wild type mice were very different. The proportion of maximal b-wave remaining in Cacna1fnob2 compared with age-matched Cacna1fwt was statistically significantly greater (p<0.05, U-test) under scotopic (0.51±0.11) than under photopic (0.29±0.19) conditions. A typical way to document selective b-wave loss, clinically, is to compare the b/a ratios with values from control subjects: lower ratios mean selective b-wave defects. There was a statistically significant decrease in b/a ratios in mutant compared to wild type mice. The reductions in b/a ratios were more pronounced under photopic (0.99±0.19 in Cacna1fnob2 compared with 3.58±0.81 in Cacna1fwt; p<0.05, U-test) than scotopic conditions (0.88±0.05 in Cacna1fnob2 compared to 1.94±0.09 in Cacna1fwt; p<0.05, U-test). In addition, the threshold was higher in Cacna1fnob2 mice under scotopic as well as photopic conditions, and the photopic thresholds in Cacna1fnob2 mice were significantly higher than the scotopic thresholds. One b-wave property that was not affected in the Cacna1fnob2 mice compared with Cacna1fwt mice was the intensity at which maximal b-wave amplitudes were attained. Finally, while we did not systematically quantify the amplitudes of ERG oscillatory potentials, qualitatively they were depressed in Cacna1fnob2 as compared with those in Cacna1fwt mice.
Figure 4. ERG results showing selective inner retina defect in Cacna1fnob2 mice.
Representative ERG traces of responses obtained at various intensities under scotopic (panel A) and photopic (panel B) adaptation are given for a Cacna1fwt (left column) and an age-matched (52 days) Cacna1fnob2 mouse (right column); numbers on left of the traces correspond to the luminances (log cd/m2) of the flashes that elicit these responses; arrows point to the b-wave apex. Corresponding amplitudes of the a-wave and b-wave under scotopic conditions are shown in panels C and D, respectively; and of b-wave amplitudes under photopic conditions are shown in panel E. Finally, examples of comparisons of responses obtained from Cacna1fwt and Cacna1fnob2 are illustrated by superimposing the respective traces obtained from these two mouse types. Responses to high-intensity stimuli (1.89 cd/m2) are shown in panels F (scotopic adaptation) and G (photopic adaptation), and responses to low-intensity stimuli are shown in panels H (−0.81 cd/m2 under scotopic adaptation) and I (0.38 cd/m2 under photopic adaptation).
Upon histological examination, we found that the structure of retinas from Cacna1fnob2 was disorganized compared to that of Cacna1fwt mice, with substantially reduced outer plexiform layer thickness and dendritic sprouting of second-order neurons into the outer nuclear layer (data not shown). These findings are consistent with those reported previously in the original Cacna1fnob2 mouse strain [22], [30].
Cacna1fnob2 and Cacna1fwt optokinetic responses (OKR) are similar
Given that our Cacna1fnob2mice produced mRNA capable of encoding full-length Cav1.4 calcium channels (albeit at reduced levels, with only ∼10% of the mRNA encoding full-length Cav1.4 protein) but still had defects in ERG and retinal structure, it was important to test the consequences of these deficits for vision. Therefore we assessed the ability of the mouse to track moving sine-wave gratings.
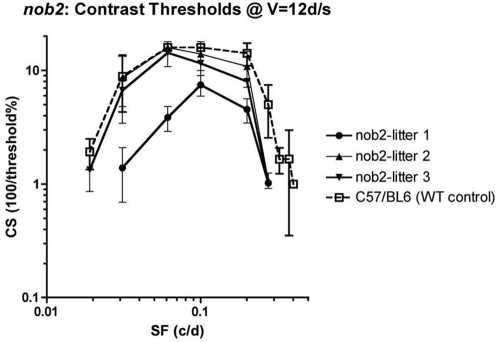
All Cacna1fnob2 mice tested showed robust optokinetic (more correctly, head-turning or optocollic) responses to moving sine-wave gratings, over a wide range of drift velocities and spatial frequencies (Table 2; Figure 5). Their contrast sensitivities (CS) were maximal at V = 12 degrees/second (d/s), at all spatial frequencies (data for other velocities not shown). The CS functions at V = 12 d/s were identical in litters 2 and 3; CS was maximal at 0.061 cycles/degree (c/d), and declined precipitously at spatial frequencies above 0.2 c/d but more gradually at spatial frequencies below 0.061 c/d. In litter 1, however, CS was maximal at 0.1 c/d and was substantially lower than that of litters 2 and 3 at most spatial frequencies tested (Figure 5). The lower limit of spatial frequencies that the animals could follow was 0.019–0.031 c/d, and the upper limit (“acuity”) was 0.275 c/d, in all three litters. There was no significant difference between the contrast sensitivities of litters 2 and 3 (P>0.05; Student-Neuman-Keuls Multiple Comparisons Test), whereas the contrast sensitivity of litters 2 and 3 (at V = 12 d/s) differed significantly from that of litter 1 (both P<0.001) at most spatial frequencies.
Table 2. Optokinetic Response Data for Cacna1fnob2 and Cacna1fwt mice.
| Group (n) | Optimum spatial freq. @ V = 12 d/s | Threshold Contrast (%) at Optimum | Contrast Sensitivity at Optimum | Mean Difference ( 0.061 c/d; V = 12 d/s) | P (2-tailed t-test) |
| nob2 - Litter#1 (6) | 0.1 c/d | 13.39% | 7.47 | N/A | —— |
| 0.061 c/d | 25.9% | 3.86 | −12.038 vs Litter #3; | <0.001 (highly significant) | |
| 10.458 vs Litter #2 | <0.001 (highly significant) | ||||
| nob2 – Litter#2 (4) | 0.061 c/d, | 6.3% | 15.9 | −1.58, vs Litter #3 | >0.05* |
| nob2 – Litter #3 (5) | 0.061 c/d, | 7.0% | 14.32 | See above | —— |
| nob2 -Combined Litters#(2+3) (9) | 0.061 c/d, | 6.7% | 15.08 (averaged) | N/A | —— |
| WT (C57/BL6) (6) | 0.061 c/d, | 6.3% | 15.9 | 0.88 vs Litters(2+3) | 0.3454 |
Figure 5. Optokinetic spatial contrast-sensitivity functions for three different Cacna1fnob2 litters and one Cacna1fwt litter.
Mean contrast sensitivities±standard deviation, at drift velocity (V) = 12 degrees per second (d/s) and various spatial frequencies (SF) in cycles per degree (c/d). See Table 2 for further details and statistics.
The contrast sensitivities of Cacna1fwt mice were maximal at V = 12 d/s, at all spatial frequencies tested (data for other velocities not shown). The CS function at V = 12 d/s was similar to that of Cacna1fnob2 litters 2 and 3 at 0.031–0.1 c/d, but contrast sensitivity remained high at 0.2 c/d and declined precipitously only at spatial frequencies ≥0.275 c/d (Figure 5). The upper limit of spatial frequencies that could be followed by Cacna1fwt mice was 0.4 c/d, or ∼1.5× the acuity of Cacna1fnob2 mice. The diminished contrast sensitivity seen in the one litter (#1) of Cacna1fnob2 mice has not been seen in any WT mice, regardless of litter or age (S.P. Bonfield, unpublished results).
Biophysical properties of Cav1.4wt and Cav1.4nob2 channels are statistically indistinguishable
Next, we tested whether the Cav1.4nob2 channel could support ionic currents. Standard whole-cell electrophysiological recordings were obtained from tsA-201 cells transfected with mouse Cav1.4wt or Cav1.4nob2 (along with rat β2a and α2–δ1) constructs. With 20 mM Ba2+ as charge carrier, the average activation and inactivation parameters for Cav1.4nob2 were not significantly different from those of Cav1.4wt (p>0.05, t-test; Table 3; Figure 6). Also, with 2 mM Ca2+ as the charge carrier and using ramp protocols [31], [32], no differences were observed in the activation properties between the two clones (see inset, Fig 6).
Table 3. Summary of biophysical properties of mouse Cav1.4wtand Cav1.4nob2 constructs recorded with 20 mM Ba2+ as charge carrier.
| Vact (mV) | Gmax (nS) | S (mV) | Erev (mV) | Vinact (mV) | z | |
| Cav1.4wt | −3±4 (11) | 4±3 (11) | 9±1 (11) | 44±6 (11) | −18±11 (11) | 1.9±0.6 (11) |
| Cav1.4nob2 | −1±4 (13) | 3±1 (13) | 8.2±0.8 (13) | 45±6 (13) | −22±10 (12) | 3±3 (12) |
| Cav1.4wt (s.s.) | −1±6 (4) | 4±3 (4) | 9±1 (4) | 44±7 (4) | −22±9 (4) | 2.0±0.6 (4) |
| Cav1.4wt (s.s.+PD98059) | 1±2 (4) | 3.4±0.8 (4) | 8.4±0.8 (4) | 46±4 (4) | −17±9 (4) | 1.8±0.4 (4) |
| Cav1.4nob2 (s.s) | −4±3 (4) | 2.1±0.3 (4) | 8.7±0.3 (4) | 41±6 (4) | −28±6 (4) | 2.4±0.7 (4) |
| Cav1.4nob2 (s.s.+PD98059) | −5±2 (5) | 2.0±0.9 (5) | 8.4±0.3 (5) | 41±6 (5) | −18±5 (5) | 1.7±0.5 (4) |
s.s. denotes serum starvation for 2 hours; no statistical difference is observed between mouse Cav1.4wt and Cav1.4nob2, or between the constructs when currents were recorded in the absence or presence of 20 µM PD98059 following two hours of serum starvation (p>0.05, one-way ANOVA).
Figure 6. Biophysical properties of Cav1.4wt and Cav1.4nob2 channels, coexpressed with β2a and α2–δ1 subunits in tSA-201 cells.
A. Representative current waveforms for Cav1.4wt (left) and Cav1.4nob2 (right) recorded with 20 mM Ba2+ external saline. Horizontal scale bars denote 25 ms, and vertical scale bars 25 pA. B. Average activation (filled symbols) and inactivation (hollow) symbols for Cav1.4wt (squares) and Cav1.4nob2 (circles) recorded with 20 mM Ba2+ external saline. Average activation parameters from 11 Cav1.4wt cells and 13 Cav1.4nob2 cells are: Vact, wt = −3±4 mV, Vact, nob2 = −1±4 mV (n = 13); Gmax, wt = 4±3 nS and Gmax, nob2 = 3±1 nS; Swt = 9±1 mV and Snob2 = 8.2±0.8 mV. These values are stastically identical, and are summarized in Table 3. Average inactivation parameters from these cells are Vinact, wt = −18±11 mV and Vinact, nob2 = −22±10 mV, with a large fraction of non-inactivating current for both channels. These values statistically identical and are summarized in Table 3. C. Average half-inactivation potentials for channels recorded with 2 mM Ca2+ as charge carrier. Currents were substantially smaller than with 20 mM Ba2+, but were distinguishable from background noise, and were obtained using a ramp protocol identical to that previously reported [31], obtained by ramping voltage from −100 mV to +100 mV over 500 ms. Values were Vact, wt = −17±8 mV (average peak current size −9±4 pA) and Vact, nob2 = −17±6 mV (average peak current size −9±3 pA). The shift observed with switching from 20 mM Ba2+ to 2 mM Ca2+ as external charge carrier is similar to that we have previously reported for the human Cav1.4 channels [16].
The Cav1.4wt N-terminus contains a putative site for phosphorylation by mitogen-activated protein (MAP) kinase that is not present in Cav1.4nob2 (serine residue in PEPSPAN region of wild type clone (Figure 2C) identified by ELM – Functional Sites in Proteins, http://www.elm.eu.org). To test whether this predicted phosphorylation site is differentially phosphorylated, thereby causing a functional difference between the two clones, we blocked MAP kinase activity in serum-starved cells by treating them for 2 hours with 20 µM PD98059 (a specific inhibitor for MAP kinase kinase; e.g., [33], [34]). PD98059 did not significantly alter the biophysical properties of either Cav1.4wt or Cav1.4nob2 channels (p>0.05) in comparison with those that were recorded in the absence of the inhibitor (Table 3).
N-terminus from Cav1.4wt and not Cav1.4nob2 can interact with filamin proteins
To test whether Cav1.4wt and Cav1.4nob2 constructs differentially interact with cellular proteins, we utilized a biochemical pull-down assay. As shown in Figure 7A, a band of approximately 37 kDa was pulled down by Cav1.4wt but not Cav1.4nob2 N-termini. This band was excised, and the protein was subsequently identified as filamin A by means of LC/MS/MS.
Figure 7. A. Representative Coomassie Brilliant Blue-stained 10% PAGE gel from three separate experiments.
N-termini from Cav1.4wt or Cav1.4nob2 were fused to glutathione S-transferase and purified on glutathione beads. Lysates from eye (lanes D–F) or spleen (G–I) were then incubated with the beads, and subsequently washed. Unpurified eye or spleen lysates (5 µL) or purified beads+lysates (40 µL) were loaded onto the gel. Lanes are as follows: A: unpurified eye, B: unpurified spleen, C: protein ladder, D: glutathione-sepharose beads+eye lysates, E: glutathione-sepharose beads+eye lysates+GST-Cav1.4 N-terminus, F: glutathione-sepharose beads+eye lysates+GST-Cav1.4nob2 N-terminus, G: glutathione-sepharose beads+spleen lysates, H: glutathione-sepharose beads+spleen lysates+GST-Cav1.4 N-terminus, I: glutathione-sepharose beads+spleen lysates+GST-Cav1.4nob2 N-terminus. A prominent band of Mr slightly larger than 37 kDa is observed in lanes E and H, but is absent from lanes F and I. This band was interpreted as a filamin A protein fragment. B. GST-fused N-termini from Cav1.4wt or Cav1.4nob2 were incubated with HA-tagged C-termini of filamin A or filamin B. Only GST-Cav1.4wt N-terminus was capable of interacting with either filamin A or filamin B. Lanes 1-3 correspond to filamin A (with GST-Cav1.4wt, GST-Cav1.4nob2, and GST, respectively); lanes 4–6 correspond to filamin B (with GST-Cav1.4wt, GST-Cav1.4nob2, and GST, respectively); lanes 7 and 8 to filamin A and B, respectively (no GST construct); lanes 9–11 GST-Cav1.4wt, GST-Cav1.4nob2, and GST, respectively (with no filamin constructs); lane 12 is unpurified GST-Cav1.4wt lysate; lane 13 is unpurified GST-Cav1.4nob2 lysate.
To investigate these interactions further, both -N-termini and filamin proteins were used as biochemical pull-down ligands. Filamin A and filamin B constructs with an HA tag were mixed with the GST- Cav1.4wt and GST-Cav1.4nob2 lysates. As shown in Figure 7B, the N-terminus of the Cav1.4wt channel protein interacted with the C-termini of both filamin A and filamin B, whereas the N-terminus of the Cav1.4nob2 channel protein did not.
Discussion
Taken together, our results suggest that alternative splicing within the ETn element inserted into exon 2 of the CACNA1F gene of the Cacna1fnob2 mouse allows for full-length Cav1.4 protein to be produced, and therefore this mouse model is not null for the Cav1.4 calcium channel. These results contrast with those previously published [22].
Because of their highly repetitive sequence, transposable elements have previously been shown to undergo alternative splicing; this allows for full-length protein to be produced, albeit at reduced levels relative to those in WT controls [26], [27]. In agreement with these previous findings (depicted in Figure 1B), we detected differentially spliced mRNA isoforms in the Cacna1fnob2 mouse, with approximately 90% of the mRNA expected to be transcribed to Cav1.4 protein having a premature stop codon in exon 2, and 10% of the mRNA expected to be transcribed to full-length channel protein with a mutated N-terminus (Figure 2C). Full-length protein was detected in spleen samples with Western blots (Figure 3), confirming that the alternatively spliced mRNA species is capable of producing Cav1.4 channel proteins with a molecular mass of ∼230 kDa. We attempted to detect Cav1.4 channel protein in sections of mouse retina; however our antibody is directed against the C-terminus of the human protein, which shares only ∼50% sequence homology with the mouse protein (rendering the antibody only weakly cross-reactive and therefore unsuitable for immunohistochemistry in mouse). Therefore, it was necessary to use concentrated protein lysates from spleen on Western blots, where Cav1.4 protein appears to be greatly enriched [16]. While it is possible no functional Cav1.4 protein is present in the Cacna1fnob2 mouse retina, the reduced protein levels in the retina may have accounted for the apparent background labeling in previous reports (for example, the faint labeling in Figure 1b from Chang et al., 2006). Thus, the CSNB2 phenotype in the Cacna1fnob2 mouse could arise simply from reduced Cav1.4 protein levels, rather than complete knockout of protein. This hypothesis is supported by previous studies, which have shown that R508Q and L1364H mutations of CACNA1F linked to CSNB2 reduce the amount of functional protein in the membrane, without significantly altering the biophysical properties of the channel [15].
Since our findings differ substantially from those reported previously [22], it was necessary to ensure that the CACNA1F gene mutation in the Cacna1fnob2 mouse line that we were testing was the same as that originally described. Therefore, we sequenced genomic DNA, obtained both from our in-house colony and as supplied by the Jackson Laboratory. In both cases the genomic sequence confirmed the presence of the ETn element inserted into exon 2 of the CACNA1F gene. Additionally, the mice we tested showed alterations in the ERGs (Figure 4) that were consistent with previous findings [22], [30]. Taken together, these results suggest that the CACNA1F gene mutation in the mice we were testing was identical to that in the line originally described, and that the inherited defect in our colony was not the result of a random genetic mutation or a deletion of the ETn element from exon 2.
As shown in Figure 4, the Cacna1fnob2 mouse retina is characterized by a functional defect that selectively affects the light-driven activation of neurons in the inner retinal layers; this is supported by the depression of the ERG b-wave (reflecting diminished modulation of activity in depolarizing bipolar cells), while the a-wave amplitudes are unaffected (indicating that photoreceptor activity is relatively unaffected). The partial preservation of the b-wave in these mice is reminiscent of the incomplete form of CSNB (CSNB2 due to CACNA1F mutations), in contrast to the complete absence of a recordable b-wave in the complete form of CSNB (CSNB1; [35], [36]. As previously reported in Cacna1fnob2 mice, CACNA1F mutations in humans cause ERG b-wave reductions under both dark-adapted (scotopic) and light-adapted (photopic) conditions. Another similarity between Cacna1fnob2 mice and CSNB2 patients is the depression of the OPs under scotopic conditions. Finally, both humans with CACNA1F mutations and Cacna1fnob2 mice have normal photopic and scotopic a-wave amplitudes [35], [36], (Figure 5). There is evidence that decreases in b-wave amplitude might lead to increases in a-wave amplitudes. For instance, pharmacological blockade of mGluR6 receptors (with intravitreal injections of L-glutamate or APB) does completely abolish the b-wave (also known as PII) and reveal photoreceptor activity (also known as PIII) over its full time course, and does increase the amplitude of the a-wave/PIII. However, in cases of complete b-wave loss due to nyctalopin mutations, a-wave amplitudes are normal under both scotopic and photopic adaptation. So the outcome, whether b-wave reductions affect a-wave amplitudes, might depend on the mechanism by which the b-wave is abolished or decreased in amplitude. The possibility that the mutated Cav1.4 channel might lead to reduced a-wave amplitudes, and that these reductions might be compensated by the indirect effect of increased a-wave amplitudes due to reduced b-wave amplitudes, does exist, but we have no experimental evidence to verify whether this is the case.
Previously, Cacna1fnob2 mice were found to have a reduced b-wave, and morphological studies of the Cacna1fnob2 retina showed a reduced OPL thickness and sprouting of second-order neurons into the ONL. Therefore, we expected that the visual performance of Cacna1fnob2 mice would be greatly reduced or absent, as in Cacna1fG305X mice [37]. Quite unexpectedly, our Cacna1fnob2 mice responded robustly to a full range of sine-wave gratings, showing only slight differences from the optokinetic performance of WT-C57/BL6 mice. The major difference in visual performance of the best-performing Cacna1fnob2 mice was that their CS functions cut off more sharply than those of Cacna1fwt mice at spatial frequencies ≥0.2 c/d, resulting in an optokinetic acuity of 0.275 c/d (Figure 6; Table 2). As a result, the spatial acuity of Cacna1fnob2 mice is about two-thirds of the acuity of C57BL6 mice, which is 0.4 c/d [37].
Thus, our behavioral results show that the Cacna1fnob2 mouse has good vision (at least for the optokinetic response), despite its severe ERG phenotype and morphologically abnormal outer retina. The relatively good vision of Cacna1fnob2 mice contrasts starkly with the lack of optokinetic responses in a targeted CACNA1F knockout, the G305X mutant [37], suggesting that the Cacna1fnob2 phenotype is not due solely to a truncation or loss-of-function mutation. Since our Cacna1fnob2 animals perform nearly as well as wild type controls, it is clear that in spite of the histological and electroretinographic evidence of severe retinal dysfunction, the Cacna1fnob2 mouse maintains at least some nearly normal visual processing in proximal levels of the retina. Our finding that about 10% of whole-retinal Cacna1fnob2 mRNA is expected to encode a full-length channel protein similar to the wild type (whereas 90% carries the in-frame stop codon) raises the question whether every photoreceptor cell makes 10% WT-like protein, or 10% of photoreceptor cells make 100% wild type-like protein (or some combination of these). On the one hand, the loss of synapses in the OPL and the sprouting of dendrites into the ONL point to significant defects in the synthesis of Cav1.4 protein, and consequently of transmitter release, in the vast majority of photoreceptor cells; this suggests that nearly all Cacna1fnob2 photoreceptor cells are making insufficient amounts of functional channel protein. On the other hand, the retention of normal contrast sensitivity and near-normal optokinetic acuity under photopic conditions indicates that cone pathways are only minimally affected. We suggest, therefore, that the wild type-like transcript is expressed mainly or exclusively in cone photoreceptors, accounting for the relative sparing of cone transmission as seen in the optokinetic response, and that the mutant transcript is expressed mainly or exclusively in rod photoreceptors, accounting for the striking loss of synaptic terminals and consequent thinning in the OPL. The discrepancy, between retention of cone function in the optokinetic response and loss of cone signals in the ERG, may be due to differential processing of cone information or mutation-dependent changes in the retinal circuits responsible for the OKR and ERG. The experiments that would be required to test this hypothesis are beyond the scope of the present study.
While full-length Cav1.4 protein is produced (albeit at presumably reduced levels) in the Cacna1fnob2 mouse, the encoded amino acid sequence differs from that in the wild type by approximately 20 residues in the N-terminus of the protein (Figure 2C). As shown in Figure 6, the electrophysiological properties of Cav1.4nob2 are not significantly different from those of Cav1.4wt when expressed in tsA-201 cells, with either 20 mM Ba2+ or 2 mM Ca2+ as charge carrier, suggesting that the differences between Cacna1fnob2 and Cacna1fwt mice do not arise from changes in the activation or inactivation characteristics of the Cav1.4 channels. Interestingly, within the span of amino acids that differ between the mutant and wild type proteins, the Cav1.4nob2 channel sequence lacks a putative MAP kinase phosphorylation site that is present in the wild type protein. PD98059, a specific inhibitor of MAPKK, did not have a differential effect on the biophysical properties of either Cav1.4nob2 or Cav1.4wt, suggesting that the difference in biophysical properties of these channels is not due to a difference in phosphorylation within these 20 amino acids.
However, as shown in Figure 7, the mutant and wild type N-terminal regions do differ in that they interact with different proteins, since the N-terminal GST-fusion protein for the Cav1.4wt sequence pulls down filamin protein, whereas the Cav1.4nob2 peptide does not. Filamins are a family of cytoskeleton proteins (200–300 kDa) that regulate and crosslink filamentous actin. They are highly sensitive to proteolysis, and numerous fragments of varying size, from 10 kDa to 280 kDa, have been identified [38]; reviewed in [39]. Calpain 3 has been shown to cleave full-length filamin A (∼260 kDa), yielding one identified product of ∼220 kDa [40]; the other cleavage product, which is expected to have a molecular mass of ∼40 kDa, thus may account for the filamin fragment identified in Figure 7A. Additionally, several previous studies have demonstrated that interactions of ion channels with filamin proteins result in enhanced targeting of the ion channels to the plasma membrane accompanied by increased current densities, specifically for Kir2.1 (binds to C-terminus of FLNA; [41]), Kv4.2 and Kv4.3 [42], and CFTR [43]–[46]. In the case of Kv4.2, a proline-rich region of the channel was identified as necessary for the interaction, with only four amino acids (PTPP) required for binding [42]. In our case, the wild type sequence of the 20 amino-acid region unique to the N-terminus of Cav1.4wt contains six proline residues, while that of Cav1.4nob2 only contains one; furthermore, at the boundary of this region with the downstream N-terminal sequence, the wild type clone has a PGPP motif (recall PTPP motif for Kv4.2), while the Cav1.4nob2 construct has an NRPP motif. While we did not map the exact residues in this 20 amino acid region necessary for this interaction, these data suggest that the CSNB2-like phenotype of the Cacna1fnob2 mouse may result not only from lower Cav1.4-mediated calcium-current densities, due to the diminished production of full-length protein, but also to a failure of targeting of the mutant protein to the plasma membrane, due to its inability to interact with filamin. Since filamin proteins are present in most cell types, any effects on current densities may be masked in our overexpression (transfected cell) system, but such effects could be profound in the highly regulated environment of photoreceptor cells.
Methods
Cacna1fnob2 (AXB6/pgnJ, stock #001678) mice were purchased from Jackson Laboratory (Bar Harbor, ME, USA). A colony was established by breeding Cacna1fnob2 mice with in-house C57BL/6 mice at the University of Calgary Health Sciences Animal Resource Centre. One male Cacna1fG305X mouse was obtained from Dr. N.T. Bech-Hansen (University of Calgary). PCR was used to confirm the genotype of all the mice used in this study. All experimental protocols were approved by University of Calgary Animal Care Committee, in accordance with guidelines established by the Canadian Council of Animal Care and the ARVO Statement for the Use of Animals in Ophthalmic and Visual Research.
RT-PCR, splice variant generation, and genomic analysis
Total RNA was isolated from a whole mouse eye (minus lens), using Trizol (Invitrogen). To synthesize cDNA, 5 µL of isolated RNA was added to oligo-dT primers along with Superscript II reverse transcriptase (Invitrogen) and incubated according to the manufacturer's instructions. To probe for CACNA1F transcripts, corresponding primers were synthesized flanking the ETn element in exon 1 (RR44) and exon 2 (RR45) or exon 3 (RR46) (Table 1; Figure 1A). For amplification, 2 µL of cDNA and Hot Start Taq (Qiagen) were used according to the manufacturer's instructions, and the reaction product was analyzed on a 1.5% agarose gel; DNA bands were isolated using a gel extraction kit (Qiagen), cloned into pGEM-T-easy (Stratagene), and then sequenced to confirm identity.
Full length mouse Cav1.4 cDNA (Cav1.4wt; [47] was subcloned into pCDNA3.1zeo, and the Cav1.4nob2 splice variant was synthesized using site-directed mutagenesis and Not I and Spe I restriction enzymes. After the sequences had been confirmed, constructs were transfected into an expression system for electrophysiological analysis (see below).
Biochemistry
For analysis of Cav1.4wt or Cav1.4nob2 expression by polyacrylamide gel electrophoresis, protein concentrations in spleen lysates were determined using a DC protein assay kit (BioRad), and equal total protein was loaded into each lane. Cav1.4 proteins were detected using 0.7 µg of an affinity purified rabbit polyclonal anti-Cav1.4 antibody (1∶1000 dilution; [16]). The membranes were then washed, incubated with secondary HRP-conjugated anti-rat IgG antibody (1∶5000 dilution, GE Healthcare) and detected using standard ECL methods. Spleen lysate from a Cacna1fG305X mouse, which does not make complete Cav1.4 channel protein [21], was prepared similarly as a negative control. For the Western blots, 4 separate experiments (n = 4) were performed and revealed the same results.
PCR was used to amplify the N-termini of mouse Cav1.4wt and Cav1.4nob2 from their appropriate cDNA templates, using primers CDRR69 (with incorporated BamHI restriction site; Table 1) and CDRR70 (with incorporated EcoRI site, Table 1). The fragment was ligated into the T-easy vector system (Promega). After the PCR product sequence had been verified, the N-terminal fragments were excised (BamHI, EcoRI) and subcloned into the pGEX-5X-1 GST Fusion System (Pharmacia). The resulting GST-fusion proteins were purified using glutathione sepharose beads (Amersham) according to the manufacturer's instructions. A 50 µL aliquot of beads was mixed with 20 µL eye or spleen lysate overnight at 4°C (whole eyes minus lens, from 7 C57/BL6 mice, were pooled and homogenized in 200 µL total volume; spleens from these mice were pooled and homogenized in 1 mL total volume), beads washed, and then proteins resolved on a 10% polyacrylamide gel stained with Coomassie Brilliant Blue. These experiments were performed 3 separate times (n = 3) resulting in identical results. Protein bands of interest were excised from the gel, and the protein sequence was determined using LC/MS/MS (Southern Alberta Mass Spectrometry Centre).
Mouse filamin cDNA was obtained from OpenBiosystems. PCR was used to amplify (past repeat 23) and add an HA tag onto the C-terminal regions of filamins A and B. Each fragment was ligated into the T-easy vector system (Promega), the sequence was confirmed, and the fragment was subcloned into the expression vector pCDNA3.1neo. The resulting construct was transfected into tsA-201 cells (transfection details below). After a two-day incubation the cells were lysed on ice with lysis buffer (300 mM NaCl, 50 mM Tris pH 7.5, 0.1% Triton X-100) containing a protease-inhibitor cocktail (complete Mini EDTA-free, Roche). The lysate was incubated with sepharose G beads (Amersham) and Anti-HA High Affinity antibody (Roche) at 4°C for 4 hours. The beads were then washed and incubated with 1 ml of GST- Cav1.4wt or GST- Cav1.4nob2 cell lysate, overnight at 4°C. The beads were washed again, and then proteins were resolved using PAGE (12% gel), transferred to a PVDF membrane, and probed with Anti-GST antibody (1∶2000 dilution, GE Healthcare). Finally the membranes were washed, incubated with HRP-conjugated anti-rat-IgG antibody (1∶5000 dilution, GE Healthcare) and detected using standard ECL methods. These experiments were performed 3 independent times (n = 3) and had the identical results.
Tissue culture and transfection
Culturing and transfection of tsA-201 cells via the calcium phosphate method have been previously described by us in detail [16]. Briefly, tsA-201 cells were maintained in DMEM supplemented with 10% FBS and 50 U/mL penicillin-streptomycin. Cells were grown to 80% confluency (37°C, humidified, 5% CO2), dissociated enzymatically (trypsin-EDTA), and then plated at 10% confluency on glass cover slips. After recovering for 8 hours, cells were transfected using standard calcium phosphate techniques (in all cases 6 µg each of cDNA encoding mouse α1, rat β2a, and rat α2–δ1 subunits, and 1 µg pIRES transfection marker). Twelve hours later, cells were washed with fresh media and then moved to 29°C (5% CO2, humidified) for two to three days before evaluation by the whole-cell patch clamp technique.
Electrophysiology
Electrophysiological data were acquired using an Axopatch 200B amplifier (Axon Instruments, Union City, CA) linked to a personal computer with Digidata 1322A interface, using pClamp 9.1 (Axon) software. In all cases, pipette capacitance and series resistance were compensated (series resistance by 80%). Currents were filtered at 1 kHz and digitized at 2 kHz. Prior to recording, individual cover slips were transferred to a 3 cm culture dish filled with bath recording solution (in mM: 20 BaCl2 (or 2 CaCl2 for ramp protocols), 65 CsCl, 40 TEA-Cl, 1 MgCl2, 10 glucose, 10 HEPES, pH 7.20 with TEA-OH). Pipettes (BF-150-86 borosilicate glass, Sutter Instruments, Novato CA) were pulled on a P-87 microelectrode puller (Sutter Instruments) and fire-polished with an MF-830 microforge (Narishige, Japan) to a resistance of 1–4 MΩ when filled with recording solution (in mM: 108 CsCH3SO3, 4 MgCl2, 9 EGTA, 9 HEPES pH 7.20 with CsOH).
Current-voltage relations were obtained by holding cells at −100 mV before stepping to various test potentials for 150 ms, typically at a frequency of 0.2 Hz. Ramp experiments were obtained by changing membrane voltage from −100 mV to +100 mV over 500 ms. Whole-cell current-voltage relations were fitted with Equation 1, where I denotes peak current amplitude, V is the test potential, V act is the half-activation potential, S is a slope factor, G max is maximum chord/slope (whole-cell) conductance, and E rev is reversal potential.
| (1) |
Inactivation-voltage relations were obtained by holding cells at −100 mV, depolarizing to a test potential of +20 mV for 50 ms, stepping back to −100 mV for 1 ms before initiating a 10 s conditioning pulse to various potentials, stepping back to −100 mV for 1 ms, and initiating a second test pulse to +20 mV for 50 ms. The degree of inactivation was determined as the ratio of the second test pulse to the first test pulse. Inactivation-voltage relations were fitted with Equation 2, where I is the degree of inactivation, V is the conditioning potential, V inact is the half-inactivation potential, z is a slope factor reflecting effective gating charge, and x represents the non-inactivating fraction of current. Since experiments were typically carried out at room temperature, R (universal gas constant), F (Faraday constant), and T (absolute temperature) simplify to a value of 25.6 mV in this equation (ie. RT/F = 25.6 mV).
| (2) |
Electrophysiological data were analyzed using Clampfit9.1 software (Axon Instruments, Union City, CA) and SigmaPlot2000 (Jandel Scientific, Chicago, IL).
PD98059, a mitogen-activated protein (MAP) kinase inhibitor, (Tocris Biosciences) was dissolved in DMSO at a stock concentration of 25 mM, and then diluted into the final recording solution immediately before recording. Control and test cells were serum-starved for 2 hours in the presence or absence of inhibitor (see for example, [34], [48]–[50]) prior to electrophysiological analysis. Control (with DMSO vehicle) and drug (20 µM) saline solutions were delivered by a gravity-driven microperfusion system.
ERG recordings
ERGs were recorded from Cacna1fnob2 (n = 6) and age-matched Cacna1fwt (n = 5) mice, as previously described [51]. Briefly, after overnight dark-adaptation, mice were prepared for bilateral recordings under dim red light. While under anesthesia (xylazine 10 mg/kg i.p; ketamine 150 mg/kg i.p.) the mouse body temperature was monitored with a rectal probe and maintained at 38°C using a homeothermic blanket. Both pupils were dilated using 1% tropicamide. A drop of methylcellulose, applied on each cornea, prevented dehydration and allowed electrical contact with the recording electrode (gold wire loop). A pair of 25-gauge platinum needles inserted subdermally behind each eye served as reference electrodes. Amplification (1–1000 Hz bandpass), stimulus presentation, and data acquisition were provided by the Espion E 2 ERG system (Diagnosys LLC, Lowell, MA). First, scotopic intensity-response functions were determined using single flashes (6500K, 10 µs duration) presented to dark-adapted animals at nineteen increasing intensity steps from −5.22 to 2.86 log cds/m2. The inter-stimulus-interval (ISI) was increased progressively from 5 sec (at the lowest stimulus intensity) to 2 minutes (at the highest stimulus intensity), so as to minimize rod photopigment bleaching and desensitization. The amplitude of the b-wave was measured from the a-wave negative peak to the b-wave positive apex, and not to the peak of oscillatory potentials (OPs), which can exceed the b-wave apex [52]. After 10 min photopic adaptation (30 cd/m2 background), cone-driven intensity-response functions were obtained, using single flashes (6500K, 10 µs duration) presented at eleven increasing intensity steps from −1.63 to 2.86 log cds/m2. The time interval between steps was 10 seconds, and each stimulus was presented 6 times at 5 sec intervals. Responses were averaged for the six flashes at each intensity. For data analysis, responses from both eyes were considered, therefore the values in the graphs represent average±standard deviations for n = 12 Cacna1fnob2 and n = 10 Cacna1fwt eyes.
Optokinetic (Head-Turning or Optocollic) Responses
Contrast sensitivity functions were obtained for optokinetic responses to moving sine-wave gratings, using the virtual optomotor apparatus, OptoMotry™ (Cerebral Mechanics, Lethbridge, AB, Canada; [53], [54]). Briefly, three sets of Cacna1fnob2 littermates of either sex (litter #1: n = 6; litter #2: n = 4; litter #3: n = 5), or their wild type background littermates (n = 6), aged 90–110 days, were placed individually on a 5 cm platform mounted in the middle of a closed testing chamber enclosed by four 17-inch LCD computer monitors (model 1703FP; Dell, Phoenix, AZ). We observed head-following responses by means of a digital video camera mounted above the platform in the lid of the chamber. Horizontally drifting sine-wave gratings, drifting to either the left or right, were presented at various spatial frequencies (0.019 cycles/degree (c/d) to 0.3 c/d) and drift velocities (6, 12, 18, 24, 30 and 36 degrees/second (d/s), for most spatial frequencies). Contrast threshold, defined as the lowest contrast at which the animal could follow the moving grating reliably (Michelson contrast: Equation 3, where Lmax and Lmin are the maximum and minimum luminances of the monitors), was determined by a modified staircase procedure and a two-alternative forced-choice paradigm. Contrast sensitivity was defined as the inverse of contrast threshold (100/threshold % contrast), and acuity was defined as the highest spatial frequency at which the animal could follow the drifting grating at 100% contrast.
| (3) |
Contrast, spatial frequency, and rotation velocity are expressed here as the computer settings. True contrast and luminance at the center of the animal's viewing platform were measured with a Minolta LS-110 Luminance Meter, operating as a spot photometer with a 1 degree acceptance angle. The mean luminance was 55 cd/m2 for a 1.0 c/d sine-wave grating of maximum contrast moving at 12 d/s, and the mean luminances of “black” and “white” (i.e., the maximum and minimum luminances), determined with a stationary 0.011 c/d grating at 100% contrast, were 3.98 cd/m2 and 104.2 cd/m2, respectively. Thus, the actual Michelson contrast range was 3.8–100%; contrast thresholds, and the related contrast sensitivities reported in this paper, were not corrected for this small discrepancy from the instrument's contrast settings of 0–100%. Mean luminance of the gratings varied from 27.5 to 110 cd/m2 with distance from the monitor screens, as the animal moved freely on the platform.
Statistics
Statistical analyses were performed using either SigmaStat2.03 (SSI, Richmond, CA), or Prism™ or InStat™ (GraphPad Software Inc., San Diego, CA, USA). Unless otherwise stated, numbers shown are mean±standard deviation, and numbers in parentheses denote the number of experiments ( = number of animals tested). Significant differences are denoted by * (p<0.05). For optokinetic responses, a one-way ANOVA (InStat™) was used to determine whether significant differences existed among the data gathered for the three litters.
Acknowledgments
The authors thank Dr. Francoise J Heseleer for the mouse Cav1.4wt cDNA clone and Dr. Torben Bech-Hansen for the Cacna1fG305X mouse tissue.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: CJD and RR are supported by Studentships from AHFMR. CJD is also supported by NSERC CGS, and a Killam Fellowship from the University of Calgary. WKS and SB are supported by a Discovery Grant from NSERC to WKS, and the OptoMotry system was purchased with a grant from the Lions Sight Centre Fund (University of Calgary) to WKS. YS is supported by a Canadian Institutes of Health Research Grant (151145), an Alberta Heritage Foundation for Medical Research equipment grant, and grants from the Canadian National Institute for the Blind (CNIB) and The Lena McLaughlin Foundation (Mona & Rod McLennan); he is a recipient of the Barbara Tuck/MacPhee Family Vision Research Award in Macular Degeneration. SCM was supported by the Foundation Fighting Blindness Canada. JEM is supported by an Operating Grant from CIHR.
References
- 1.Bech-Hansen NT, Naylor MJ, Maybaum TA, Pearce WG, Koop B, et al. Loss-of-function mutations in a calcium-channel alpha1-subunit gene in Xp11.23 cause incomplete X-linked congenital stationary night blindness. Nat Genet. 1998;19:264–267. doi: 10.1038/947. [DOI] [PubMed] [Google Scholar]
- 2.Strom TM, Nyakatura G, Apfelstedt-Sylla E, Hellebrand H, et al. An L-type calcium-channel gene mutated in incomplete X-linked congenital stationary night blindness. Nat Genet. 1998;19:260–263. doi: 10.1038/940. [DOI] [PubMed] [Google Scholar]
- 3.Jalkanen R, Mantyjarvi M, Tobias R, Isosomppi J, Sankila EM, et al. X linked cone-rod dystrophy, CORDX3, is caused by a mutation in the CACNA1F gene. J Med Genet. 2006;43:699–704. doi: 10.1136/jmg.2006.040741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jalkanen R, Bech-Hansen NT, Tobias R, Sankila EM, Mantyjarvi M, et al. A novel CACNA1F gene mutation causes Aland Island eye disease. Invest Ophthalmol Vis Sci. 2007;48:2498–2502. doi: 10.1167/iovs.06-1103. [DOI] [PubMed] [Google Scholar]
- 5.Boycott KM, Pearce WG, Bech-Hansen NT. Clinical variability among patients with incomplete X-linked congenital stationary night blindness and a founder mutation in CACNA1F. Can J Ophthalmol. 2000;35:204–213. doi: 10.1016/s0008-4182(00)80031-9. [DOI] [PubMed] [Google Scholar]
- 6.Boycott KM, Maybaum TA, Naylor MJ, Weleber RG, Robitaille J, et al. A summary of 20 CACNA1F mutations identified in 36 families with incomplete X-linked congenital stationary night blindness, and characterization of splice variants. Hum Genet. 2001;108:91–97. doi: 10.1007/s004390100461. [DOI] [PubMed] [Google Scholar]
- 7.Hope CI, Sharp DM, Hemara-Wahanui A, Sissingh JI, Lundon P, et al. Clinical manifestations of a unique X-linked retinal disorder in a large New Zealand family with a novel mutation in CACNA1F, the gene responsible for CSNB2. Clin Experiment Ophthalmol. 2005;33:129–136. doi: 10.1111/j.1442-9071.2005.00987.x. [DOI] [PubMed] [Google Scholar]
- 8.Jacobi FK, Hamel CP, Arnaud B, Blin N, Broghammer M, et al. A novel CACNA1F mutation in a french family with the incomplete type of X-linked congenital stationary night blindness. Am J Ophthalmol. 2003;135:733–736. doi: 10.1016/s0002-9394(02)02109-8. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura M, Ito S, Terasaki H, Miyake Y. Novel CACNA1F mutations in Japanese patients with incomplete congenital stationary night blindness. Invest Ophthalmol Vis Sci. 2001;42:1610–1616. [PubMed] [Google Scholar]
- 10.Wutz K, Sauer C, Zrenner E, Lorenz B, Alitalo T, et al. Thirty distinct CACNA1F mutations in 33 families with incomplete type of XLCSNB and Cacna1f expression profiling in mouse retina. Eur J Hum Genet. 2002;10:449–456. doi: 10.1038/sj.ejhg.5200828. [DOI] [PubMed] [Google Scholar]
- 11.Zeitz C, Minotti R, Feil S, Matyas G, Cremers FP, et al. Novel mutations in CACNA1F and NYX in Dutch families with X-linked congenital stationary night blindness. Mol Vis. 2005;11:179–183. [PubMed] [Google Scholar]
- 12.Zito I, Allen LE, Patel RJ, Meindl A, Bradshaw K, et al. Mutations in the CACNA1F and NYX genes in British CSNBX families. Hum Mutat. 2003;21:169. doi: 10.1002/humu.9106. [DOI] [PubMed] [Google Scholar]
- 13.Hemara-Wahanui A, Berjukow S, Hope CI, Dearden PK, Wu SB, et al. A CACNA1F mutation identified in an X-linked retinal disorder shifts the voltage dependence of Cav1.4 channel activation. Proc Natl Acad Sci U S A. 2005;102:7553–7558. doi: 10.1073/pnas.0501907102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoda JC, Zaghetto F, Koschak A, Striessnig J. Congenital stationary night blindness type 2 mutations S229P, G369D, L1068P, and W1440X alter channel gating or functional expression of Cav1.4 L-type Ca2+ channels. J Neurosci. 2005;25:252–259. doi: 10.1523/JNEUROSCI.3054-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoda JC, Zaghetto F, Singh A, Koschak A, Striessnig J. Effects of congenital stationary night blindness type 2 mutations R508Q and L1364H on Cav1.4 L-type Ca2+ channel function and expression. J Neurochem. 2006;96:1648–1658. doi: 10.1111/j.1471-4159.2006.03678.x. [DOI] [PubMed] [Google Scholar]
- 16.McRory JE, Hamid J, Doering CJ, Garcia E, Parker R, et al. The CACNA1F gene encodes an L-type calcium channel with unique biophysical properties and tissue distribution. J Neurosci. 2004;24:1707–1718. doi: 10.1523/JNEUROSCI.4846-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wahl-Schott C, Baumann L, Cuny H, Eckert C, Griessmeier K, Biel M. Switching off calcium-dependent inactivation in L-type calcium channels by an autoinhibitory domain. Proc Natl Acad Sci U S A. 2006;103:15657–15662. doi: 10.1073/pnas.0604621103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peloquin JB, Rehak R, Doering CJ, McRory JE. Functional analysis of congenital stationary night blindness type-2 CACNA1F mutations F742C, G1007R, and R1049W. Neuroscience. 2007;150:335–345. doi: 10.1016/j.neuroscience.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 19.Singh A, Hamedinger D, Hoda JC, Gebhart M, Koschak A, et al. C-terminal modulator controls Ca2+-dependent gating of Cav1.4 L-type Ca2+ channels. Nat Neurosci. 2006;9:1108–1116. doi: 10.1038/nn1751. [DOI] [PubMed] [Google Scholar]
- 20.Doering CJ, Peloquin JB, McRory JE. The Cav1.4 calcium channels: more than meets the eye. Channels. 2007;1:3–10. [PubMed] [Google Scholar]
- 21.Mansergh F, Orton NC, Vessey JP, Lalonde MR, Stell WK, et al. Mutation of the calcium channel gene Cacna1f disrupts calcium signaling, synaptic transmission and cellular organization in mouse retina. Hum Mol Genet. 2005;14:3035–3046. doi: 10.1093/hmg/ddi336. [DOI] [PubMed] [Google Scholar]
- 22.Chang B, Heckenlively JR, Bayley PR, Brecha NC, Davisson MT, et al. The nob2 mouse, a null mutation in Cacna1f: anatomical and functional abnormalities in the outer retina and their consequences on ganglion cell visual responses. Vis Neurosci. 2006;23:11–24. doi: 10.1017/S095252380623102X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgiev GP. Mobile genetic elements in animal cells and their biological significance. Eur J Biochem. 1984;145:203–220. doi: 10.1111/j.1432-1033.1984.tb08541.x. [DOI] [PubMed] [Google Scholar]
- 24.Maksakova IA, Mager DL. Transcriptional regulation of early transposon elements, an active family of mouse long terminal repeat retrotransposons. J Virol. 2005;79:13865–13874. doi: 10.1128/JVI.79.22.13865-13874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonigo P, Wain-Hobson S, Bougueleret L, Tiollais P, Jacob F, Brulet P. Nucleotide sequence and evolution of ETn elements. Proc Natl Acad Sci U S A. 1987;84:3768–3771. doi: 10.1073/pnas.84.11.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu JL, Drappa J, Parnassa A, Elkon KB. The defect in Fas mRNA expression in MRL/lpr mice is associated with insertion of the retrotransposon, ETn. J Exp Med. 1993;178:723–730. doi: 10.1084/jem.178.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi S, Hirano T, Kakinuma M, Uede T. Transcriptional repression and differential splicing of Fas mRNA by early transposon (ETn) insertion in autoimmune lpr mice. Biochem Biophys Res Commun. 1993;191:617–624. doi: 10.1006/bbrc.1993.1262. [DOI] [PubMed] [Google Scholar]
- 28.Gomez-Ospina N, Tsuruta F, Barreto-Chang O, Hu L, Dolmetsch R. The C terminus of the L-type voltage-gated calcium channel Cav1.2 encodes a transcription factor. Cell. 2006;127:591–606. doi: 10.1016/j.cell.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu M, Peachey NS. Attenuation of oscillatory potentials in nob2 mice. Doc Ophthalmol. 2007 doi: 10.1007/s10633-007-9058-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bayley PR, Morgans CW. Rod bipolar cells and horizontal cells form displaced synaptic contacts with rods in the outer nuclear layer of the nob2 retina. J Comp Neurol. 2007;500:286–298. doi: 10.1002/cne.21188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doering CJ, McRory JE. Effects of extracellular pH on neuronal calcium channel activation. Neuroscience. 2007;146:1032–1043. doi: 10.1016/j.neuroscience.2007.02.049. [DOI] [PubMed] [Google Scholar]
- 32.Peloquin JB, Doering CJ, Rehak R, McRory JE. Temperature dependence of Cav1.4 calcium channel gating. Neuroscience. 2008;151:1066–1083. doi: 10.1016/j.neuroscience.2007.11.053. [DOI] [PubMed] [Google Scholar]
- 33.Ekinci FJ, Malik KU, Shea TB. Activation of the L voltage-sensitive calcium channel by mitogen-activated protein (MAP) kinase following exposure of neuronal cells to beta-amyloid. MAP kinase mediates beta-amyloid-induced neurodegeneration. J Biol Chem. 1999;274:30322–30327. doi: 10.1074/jbc.274.42.30322. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt H, Schulz S, Klutzny M, Koch T, Handel M, Hollt V. Involvement of mitogen-activated protein kinase in agonist-induced phosphorylation of the mu-opioid receptor in HEK 293 cells. J Neurochem. 2000;74:414–422. doi: 10.1046/j.1471-4159.2000.0740414.x. [DOI] [PubMed] [Google Scholar]
- 35.Miyake Y, Yagasaki K, Horiguchi M, Kawase Y, Kanda T. Congenital stationary night blindness with negative electroretinogram. A new classification. Arch Ophthalmol. 1986;104:1013–1020. doi: 10.1001/archopht.1986.01050190071042. [DOI] [PubMed] [Google Scholar]
- 36.Tremblay F, Laroche RG, De BI. The electroretinographic diagnosis of the incomplete form of congenital stationary night blindness. Vision Res. 1995;35:2383–2393. doi: 10.1016/0042-6989(95)00006-l. [DOI] [PubMed] [Google Scholar]
- 37.Bonfield S, Tejedor J, Sauve Y, Doering CJ, McRory JE, et al. Spatiotemporal contrast sensitivity characteristics of optokinetic responses in chicks and in normal and Cacna1f-mutant (CSNB2) mice. ARVO 2007 [Google Scholar]
- 38.Umeda T, Kouchi Z, Kawahara H, Tomioka S, Sasagawa N, et al. Limited proteolysis of filamin is catalyzed by caspase-3 in U937 and Jurkat cells. J Biochem (Tokyo) 2001;130:535–542. doi: 10.1093/oxfordjournals.jbchem.a003016. [DOI] [PubMed] [Google Scholar]
- 39.van der Flier A, Sonnenberg A. Structural and functional aspects of filamins. Biochim Biophys Acta. 2001;1538:99–117. doi: 10.1016/s0167-4889(01)00072-6. [DOI] [PubMed] [Google Scholar]
- 40.Taveau M, Bourg N, Sillon G, Roudaut C, Bartoli M, Richard I. Calpain 3 is activated through autolysis within the active site and lyses sarcomeric and sarcolemmal components. Mol Cell Biol. 2003;23:9127–9135. doi: 10.1128/MCB.23.24.9127-9135.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sampson LJ, Leyland ML, Dart C. Direct interaction between the actin-binding protein filamin-A and the inwardly rectifying potassium channel, Kir2.1. J Biol Chem. 2003;278:41988–41997. doi: 10.1074/jbc.M307479200. [DOI] [PubMed] [Google Scholar]
- 42.Petrecca K, Miller DM, Shrier A. Localization and enhanced current density of the Kv4.2 potassium channel by interaction with the actin-binding protein filamin. J Neurosci. 2000;20:8736–8744. doi: 10.1523/JNEUROSCI.20-23-08736.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cantiello HF. Role of actin filament organization in CFTR activation. Pflugers Arch 443 Suppl. 2001;1:S75–S80. doi: 10.1007/s004240100649. [DOI] [PubMed] [Google Scholar]
- 44.Prat AG, Xiao YF, Ausiello DA, Cantiello HF. cAMP-independent regulation of CFTR by the actin cytoskeleton. Am J Physiol. 1995;268:C1552–C1561. doi: 10.1152/ajpcell.1995.268.6.C1552. [DOI] [PubMed] [Google Scholar]
- 45.Prat AG, Cunningham CC, Jackson GR, Jr, Borkan SC, Wang Y, et al. Actin filament organization is required for proper cAMP-dependent activation of CFTR. Am J Physiol. 1999;277:C1160–C1169. doi: 10.1152/ajpcell.1999.277.6.C1160. [DOI] [PubMed] [Google Scholar]
- 46.Thelin WR, Chen Y, Gentzsch M, Kreda SM, Sallee JL, et al. Direct interaction with filamins modulates the stability and plasma membrane expression of CFTR. J Clin Invest. 2007;117:364–374. doi: 10.1172/JCI30376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haeseleer F, Imanishi Y, Maeda T, Possin DE, Maeda A, et al. Essential role of Ca2+-binding protein 4, a Cav1.4 channel regulator, in photoreceptor synaptic function. Nat Neurosci. 2004;7:1079–1087. doi: 10.1038/nn1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dudley DT, Pang L, Decker SJ, Bridges AJ, Saltiel AR. A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proc Natl Acad Sci U S A. 1995;92:7686–7689. doi: 10.1073/pnas.92.17.7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herrera R, Hubbell S, Decker S, Petruzzelli L. A role for the MEK/MAPK pathway in PMA-induced cell cycle arrest: modulation of megakaryocytic differentiation of K562 cells. Exp Cell Res. 1998;238:407–414. doi: 10.1006/excr.1997.3847. [DOI] [PubMed] [Google Scholar]
- 50.Meriane M, Duhamel S, Lejeune L, Galipeau J, Annabi B. Cooperation of matrix metalloproteinases with the RhoA/Rho kinase and mitogen-activated protein kinase kinase-1/extracellular signal-regulated kinase signaling pathways is required for the sphingosine-1-phosphate-induced mobilization of marrow-derived stromal cells. Stem Cells. 2006;24:2557–2565. doi: 10.1634/stemcells.2006-0209. [DOI] [PubMed] [Google Scholar]
- 51.Sauve Y, Pinilla I, Lund RD. Partial preservation of rod and cone ERG function following subretinal injection of ARPE-19 cells in RCS rats. Vision Res. 2006;46:1459–1472. doi: 10.1016/j.visres.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 52.Nusinowitz S, Nguyen L, Radu R, Kashani Z, Farber D, Danciger M. Electroretinographic evidence for altered phototransduction gain and slowed recovery from photobleaches in albino mice with a MET450 variant in RPE65. Exp Eye Res. 2003;77:627–638. doi: 10.1016/s0014-4835(03)00217-3. [DOI] [PubMed] [Google Scholar]
- 53.Douglas RM, Alam NM, Silver BD, McGill TJ, Tschetter WW, Prusky GT. Independent visual threshold measurements in the two eyes of freely moving rats and mice using a virtual-reality optokinetic system. Vis Neurosci. 2005;22:677–684. doi: 10.1017/S0952523805225166. [DOI] [PubMed] [Google Scholar]
- 54.Prusky GT, Alam NM, Beekman S, Douglas RM. Rapid quantification of adult and developing mouse spatial vision using a virtual optomotor system. Invest Ophthalmol Vis Sci. 2004;45:4611–4616. doi: 10.1167/iovs.04-0541. [DOI] [PubMed] [Google Scholar]