1. Introduction
1.1. Overview of Direct Repair of DNA Alkylation Damage
Cellular DNA is constantly subjected to modifications by intracellular and extracellular chemicals, which can result in covalent changes. 1,2 Alkylating agents are one group of such chemicals that can lead to DNA damage.3 These agents are prevalent in the environment and are used as anticancer compounds in the clinical setting.4–10 Alkylating agents also exist endogenously inside cells; for instance, S-adenosylmethionine, a methyl donor for many cellular reactions, has been shown to produce methylation damage.11,12 The attack on DNA by these alkylating agents can lead to various types of lesions on the heterocyclic bases or backbone.3,13–15 Most of these resulting adducts are mutagenic or toxic, and cells have evolved various proteins to detect and repair them.9,16,17 Interestingly, many of these alkylation lesions are repaired through the direct removal of the adduct. Other than the photolyase that catalyzes direct reversal of the thymine dimer created by UV light,18,19 all known direct DNA repair proteins are engaged in alkylation DNA damage repair. These are the N-terminal domain of the Escherichia coli (E. coli) Ada protein, the O6-alkylguanine-DNA alkyltransferase family, and the AlkB family.9
1.1.1. Alkylation of DNA
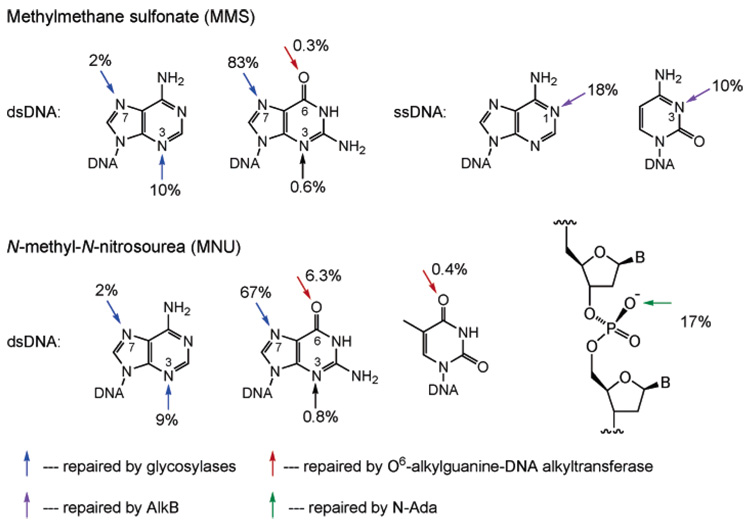
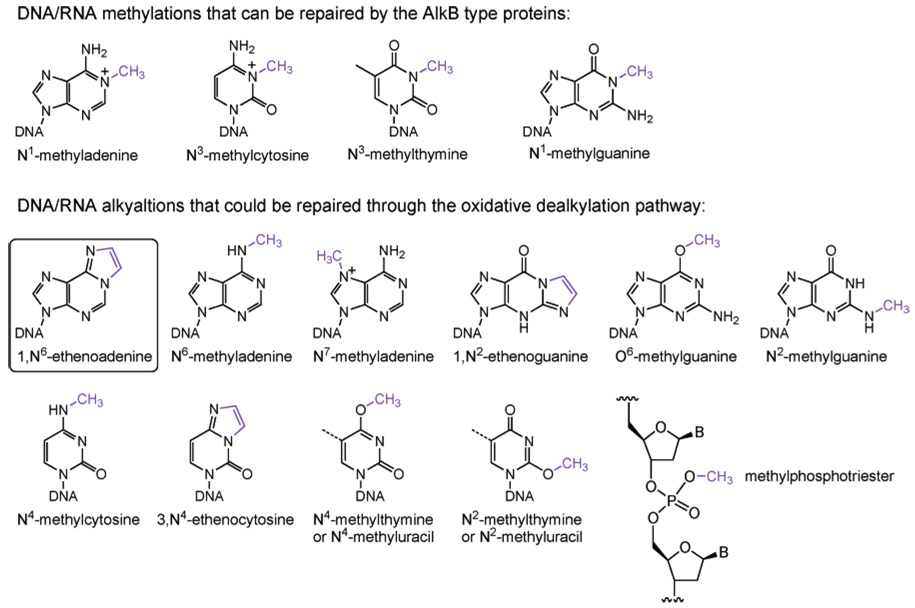
Alkylating reagents can be divided into SN1 and SN2 types based on the mechanism of the alkylation attack. The alkylation susceptibility of each site on the bases or backbone varies depending on the reagent used (Figure 1); the resulting lesions also have different mutagenic and cytotoxic effects. The N7-position of guanine is the most vulnerable site on DNA; unsurprisingly, it also serves as the best ligand on the DNA for metal ions such as platinum(II).20 Treating double-stranded DNA (dsDNA) with methylating agents such as methylmethane sulfonate (MMS, an SN2 type methylating agent) or N-methyl-N′-nitrosourea (MNU, an SN1 type methylating agent) typically results in 70–80% of the methylation occurring on the N7-position of guanine. Despite being the most abundant product of alkylation damage, N7-methylguanine is relatively innocuous and is removed mostly through spontaneous depurination.21 The resulting abasic site is toxic and repaired enzymatically.22 The N3-methyladenine is the second most abundant alkylation lesion formed in dsDNA. This lesion can block DNA replication and is removed by AlkA in E. coli and 3-methyladenine-DNA-glycosylases.23–25
Figure 1.
Methylation patterns of the DNA bases and phosphate backbone with MMS and MNU. The blue arrows indicate methylation sites that are repaired by glycosylases; the red arrows are for sites repaired by O6-alkylguanine-DNA alkyltransferases; the purple arrows are for sites repaired by the AlkB proteins; and the green arrow is the site repaired by N-Ada.
The SN1 type methylating agents such as MNU and N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) are highly mutagenic because they attack the oxygen atoms on DNA bases to give a significant amount of O6-methylguanine (O6-meG) and a small amount of O4-methylthymine (Figure 1).13,14 O6-meG mispairs with thymine during DNA replication, which gives rise to a transition mutation of G:C to A:T.26–29 Thus, this lesion must be rapidly located and removed in order to maintain the integrity of the genome. The O6-alkylguanine-DNA alkyltransferase family of proteins performs this important task in almost all organisms.29–33
The SN2 type methylating agents such as MMS and methyl halides can react with single-stranded DNA (ssDNA) to generate large portions of N1-methyladenine and N3-methylcytosine (Figure 1).3,9,13,14 These two positions are protected by hydrogen bonding in dsDNA but are quite nucleophilic when exposed in ssDNA or replication forks. When these sites are exposed, they are vulnerable to nucleophilic attack; the pKa’s of these two nitrogen sites are 4.1 and 4.5 in ssDNA, which are higher than that of N7-guanine.34–38 The resulting lesions prevent formation of Watson–Crick base pairs which could be toxic for cells. The protein involved in the repair of these lesions has been revealed only very recently. A family of iron(II)-dependent dioxygenases was found to catalytically remove these alkylation lesions.9,39,40
The phosphodiester DNA backbone is also subject to alkylation damage. For instance, 17% of the total methylation occurs on the backbone to yield methylphosphotriesters when dsDNA is treated with MNU (Figure 1). The neutral phosphotriester can be hydrolyzed by water much faster than the diester, which leads to cleavage of the backbone. The Sp-methylphosphotriester is repaired by the N-terminal domain of the Ada protein (N-Ada) in E. coli.17,41 This repair serves mostly as a signaling pathway to induce expression of methylation resistance genes, as will be discussed below. The other diastereomer, Rp-methylphosphotriester, cannot be repaired by N-Ada. There is no homologue of N-Ada found in eukaryotes. It is unclear whether methylphosphotriester is repaired in eukaryotes.
1.1.2. Direct Removal of the Alkyl Damage by Nucleophilic Cys Residues
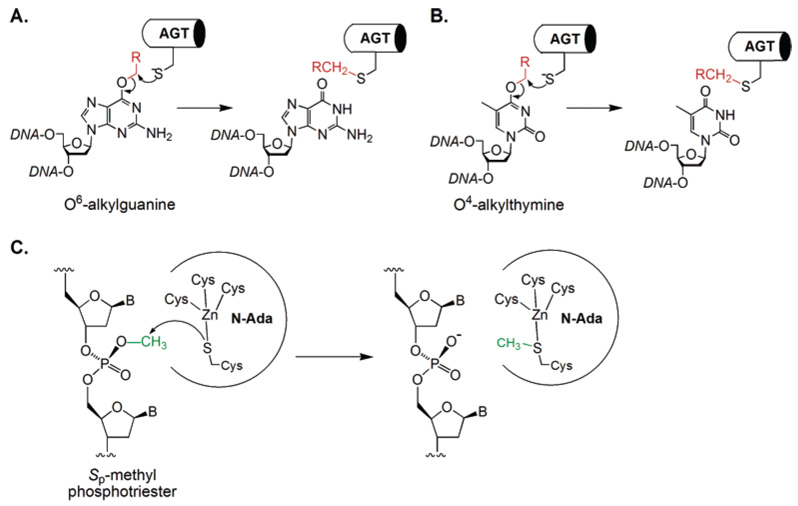
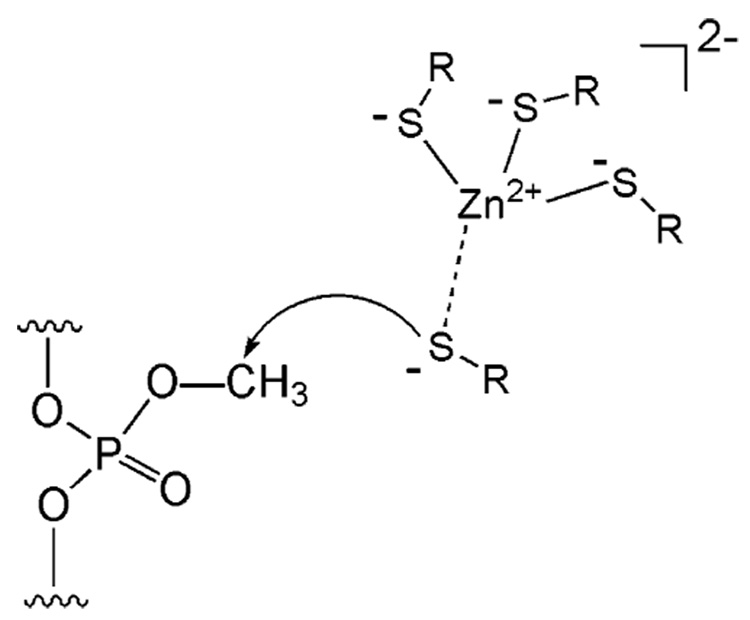
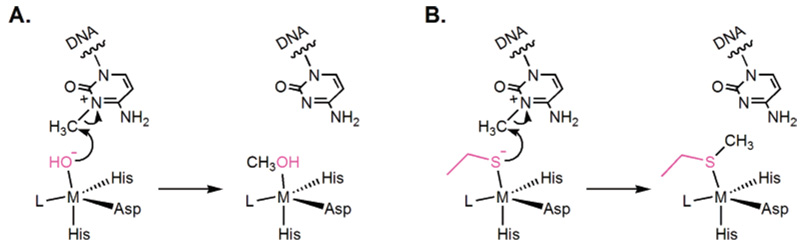
A common strategy used by nature to directly repair alkylation damage is to irreversibly transfer the alkyl groups to nucleophilic Cys residues in repair proteins. O6-Alkylguanine and O4-alkylthymine are repaired in such a manner by the sacrificial protein O6-alkylguanine-DNA alkyltransferase (AGT or MGMT).9,17,42–45 A nucleophilic Cys residue is utilized to receive the alkyl lesion in a SN2 manner (Figure 2A and B). The alkylated protein cannot be regenerated and is degraded after the repair. In this case, the protein serves as an alkyl transfer reagent instead of as a strictly defined enzyme. The Sp-methylphosphotriester is also repaired by a direct transfer of the methyl group from methylphosphotriester to a reactive Cys residue in E. coli N-Ada.41 N-Ada is the prototype of proteins that catalyze zinc(II)-mediated alkyl transfer to thiols. The reactive Cys residue in N-Ada is a ligand of an active site zinc(II) ion (Figure 2C).46,47 The methyl transfer process mediated by N-Ada is irreversible as well. The methylated Ada is converted into a potent transcriptional activator.17
Figure 2.
A Cys residue is used by the AGT proteins to remove the alkyl adducts on the O6-position of guanine (A) and the O4-position of thymine (B). A zinc-bound Cys residue is also used by the N-terminal domain of the E. coli Ada protein to displace a methyl group from Sp-methylphosphotriester (C). All these alkyl transfers from DNA lesions to Cys residues are irreversible.
1.1.3. Oxidative Dealkylation
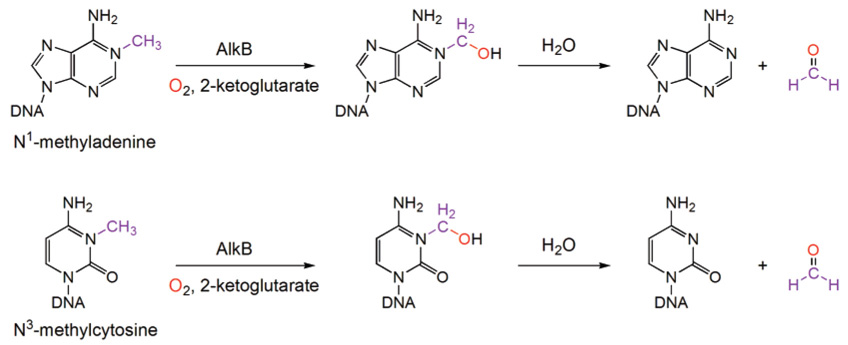
The transfer of alkyl adducts to Cys residues was the only known mechanism for direct alkylation repair before the function of AlkB was elucidated. In 2002, two groups simultaneously demonstrated that E. coli AlkB performs an unprecedented oxidative dealkylation repair of N1-methyladenine (1-meA) and N3-methylcytosine (3-meC) lesions in ssDNA.39,40 The AlkB protein belongs to the family of α-ketoglutarate/iron(II)-dependent dioxygenases.48 It activates dioxygen to oxidize the methyl group. The resulting oxidized product decomposes spontaneously in aqueous solution to give formaldehyde and the repaired base (Figure 3). Human homologues have been identified and exhibit similar functions.49,50 This exciting discovery opens new frontiers in DNA repair and cancer research.
Figure 3.
Novel oxidative dealkylation mechanism used by the AlkB proteins to remove methyl or other alkyl groups on the N1-position of adenine and the N3-position of cytosine.
1.2. Scope of Review
This review covers the three direct dealkylation repair protein families: N-Ada, O6-alkylguanine-DNA alkyltransferase, and AlkB. Excellent reviews and articles have been written on these subjects in the past; due to space constraints, we apologize for any references that we may have omitted.3,9,16,51–53 We hope to emphasize the chemical aspects of these repair functions and focus on the recent advances made toward understanding the structure, function, and mechanism of these proteins. Inhibition of human AGT (hAGT) as a potential strategy to improve the efficacy of anticancer alkylation chemotherapies will be discussed, as well as recent strategies to utilize this protein for in vivo imaging and protein-immobilization applications. The function of AlkB was revealed only three years ago. We will try to cover most of the advances in studying this family of proteins.
2. Direct Repair of Sp-Methylphosphotriesters by E. Coli N-Ada
2.1. The Ada-Regulated Adaptive Response in E. Coli
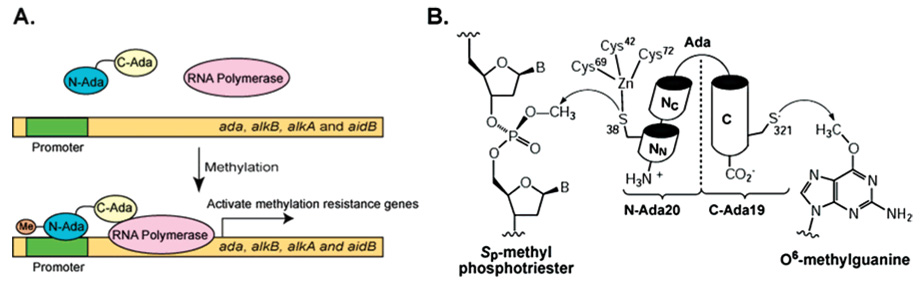
Many bacteria can defend against abnormally high levels of environmental methylating agents by mounting an inducible response.3,9,16,17,54–56 This so-called adaptive response has been studied extensively in E. coli, in which expression of four genes, ada, alkA, alkB, and aidB is activated through a post-translational modification of the Ada protein.57 The three gene products, Ada, AlkA, and AlkB, are all engaged in repair of DNA alkylation damage; the function of AidB is still unknown. The Ada protein, a protein with only 354 amino acids, possesses three different functions (Figure 4)! It has two distinct domains: a 20 kDa N-terminal domain (N-Ada) repairs the Sp-configurated methylphosphotriester (Figure 4B),41,58 and a 19 kDa C-terminal domain (C-Ada) repairs one of the most mutagenic DNA base lesions, O6-meG, as well as O4-methylthymine (Figure 4B).31 The two domains can be separated and still retain their corresponding repair activities. Ada is unique in that it is also a transcriptional activator that regulates the methylation resistance adaptive response pathway (Figure 4A).17,57,59
Figure 4.
The E. coli Ada protein is a multiple function protein that protects E. coli against methylation challenge. (A) Ada is a transcriptional activator. Methylation of N-Ada increases Ada’s affinity to DNA and converts it into a potent transcriptional activator. N-Methylated Ada binds several promoter sites and recruits RNA polymerase to initiate transcriptional activation of the ada regulon that includes the methylation resistance genes ada, alkB, alkA, and aidB. (B) Ada possesses two repair activities: the N-terminal domain of Ada utilizes a zinc-activated Cys residue to repair Sp-methylphosphotriester; the C-terminal domain contains a nucleophilic Cys residue to remove alkyl groups on the O6-position of guanine and the O4-position of thymine.
N-Ada performs a direct, irreversible transfer of the methyl group from the Sp-methylphosphotriester to one of its Cys residues.41 This simple modification converts N-Ada into a much tighter DNA binder; it was estimated that methylation of N-Ada increases its DNA affinity by 100–1000-fold!58,60 After this methylation, Ada utilizes its methylated N-terminus to recognize the promoter regions of the ada regulon and recruits RNA polymerase to initiate transcriptional activation of the methylation resistance genes (Figure 4A).17,56,61–63 Thus, Ada is a chemosensor for DNA methylation damage in E. coli cells.
2.2. A Zinc(II)-Mediated Methyl Transfer in N-Ada
2.2.1. Identification of the Active Cys Residues: Only One Cys Residue Is Selectively Activated
An early work showed that the methylphosphotriester repair activity of Ada is contained in the N-terminal domain.64,65 A Cys residue in N-Ada was assigned as the potential methyl acceptor which is converted into a thioether after the methyl transfer step.41,58 The repair reaction was found to be stoichiometric, and the methylation could not be reversed. Only the Sp-configurated methylphosphotriester is repaired, leaving the other diastereomers intact. The mechanism underlying this selectivity was not clear at the time. N-Ada contains six Cys residues; the correct assignment of the active Cys residue in N-Ada was only achieved very recently through biochemical analysis of the methylated protein and structural characterization of the methylated N-Ada bound to DNA.58 Although the reactive residue was previously considered to be Cys69, Cys38 now has been identified as the methyl acceptor.58 This Cys residue is selectively activated, because treating N-Ada with methyl iodide (MeI) resulted in almost exclusive methylation of Cys38.66 Apparently, Cys38 in N-Ada possesses an intrinsically higher nucleophilicity than the other Cys residues. This residue turns out to be a ligand to an active site zinc(II) ion that is also bound by three other Cys residues.46,64,67
2.2.2. N-Ada Utilizes a Zinc(II)-Activated Cys Residue
The overexpression of soluble N-Ada20 (residue 1–179) was discovered to be dependent on the availability of excess of zinc(II) ions in the growth medium.64 This was the first indication that N-Ada might be a zinc(II)-containing protein. An experiment was performed to replace zinc(II) ion with cadmium(II) ion by using minimum medium containing cadmium(II). The resulting Cd(II)-substituted N-Ada retained the ability to repair Sp-methylphosphotriester, but the rate was one-fourth that of the native Zn(II)-containing protein.46 This experiment suggested that metal is directly involved in the rate-determining step of the repair. To determine the ligands around the metal ion, 113Cd-substituted N-Ada20 was prepared for 113Cd nuclear magnetic resonance (NMR) investigations.46 A single peak at 684 ppm was observed from the spectrum, which indicates a tetrahedral geometry with four Cys residues as ligands. Further NMR studies, together with biochemical experiments, assigned the four Cys ligands to zinc(II) as Cys38, Cys42, Cys69, and Cys72 with the active Cys residue bound to the zinc(II). The presence of this zinc(II) ion is also essential for the folding of N-Ada.
Prior to this discovery, Cys4-ligated zinc(II) sites were known for their structural roles in numerous transcriptional factors. The binding of zinc(II) to a Cys residue gives a deprotonated thiolate at neutral pH. This thiolate is typically inert due to its interaction with the zinc(II) ion; however, with four thiolates bound to a zinc(II) ion, the accumulation of negative charges on the whole cluster could induce a transient dissociation of one thiolate which becomes a good nucleophile (Figure 5).68,69 Most structural Cys4Zn sites have evolved to suppress this activity. Hydrogen-bonding interactions from the protein backbone amides to the sulfur atoms of cysteines are frequently observed in these clusters, which shield the Cys residues and alleviate the accumulation of negative charges on the clusters.70,71 However, the Cys4Zn site in N-Ada possesses a unique mechanism to selectively activate one Cys residue, Cys38, as a nucleophile. In fact, N-Ada is the prototype for the zinc-mediated biological alkyl transfer to thiol groups. Since the discovery of this function for N-Ada, similar activities were found for cobalamin-dependent and -independent methionine synthase, protein farnesyltransferase (FTase), geranylgeranyltransferase (GTase), and possibly methanogenic methyl transferases and epoxide carboxylase.47,72–74 All of these proteins utilize zinc(II) to generate a nucleophilic thiolate at neutral pH. N-Ada has provided an opportunity to understand the general principles of this process.
Figure 5.
A Cys4Zn cluster accumulates negative charges. A Cys residue, which may transiently dissociate from the zinc(II) center while still kept as the deprotonated form, can attack a methylphos-photriester to transfer the methyl group.
2.2.3. NMR Structure of N-Ada10
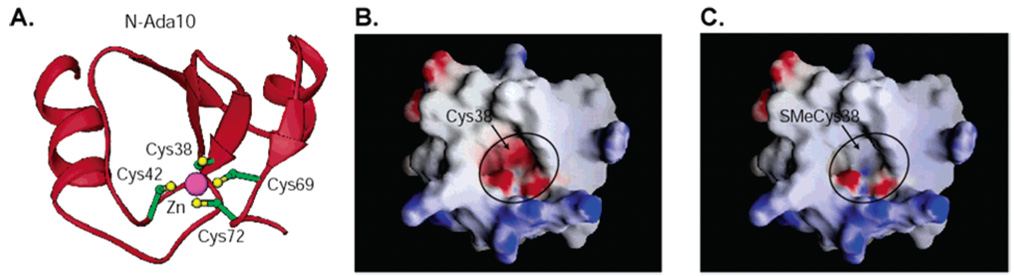
N-Ada can be further divided into two separate subdomains: N-AdaN and N-AdaC (Figure 4B). The zinc(II) cluster and the methyl transfer activity are contained in the 10 kDa N-AdaN subdomain (N-Ada10, residues 1–92).64,65 N-AdaC has a helix–turn–helix motif, which contributes to the DNA binding of the protein.75 A low-resolution structure of N-Ada10 solved by NMR showed that it consists of a β-sheet sandwiched between two α-helices, with the zinc site stabilizing the overall fold at the edge of the β-sheet (Figure 6A).67,76 The zinc atom is tetrahedrally ligated by four Cys residues, as suggested by 113Cd NMR experiments. This metal site is located at the bottom of a cavity on the protein surface. Cys38, Cys42, and Cys69 are all fairly exposed to solvents; however, how Cys38 is selectively activated cannot be explained by this low-resolution structure, nor could the structure shed any light on the enhanced DNA binding affinity of the protein after methylation of Cys38.
Figure 6.
NMR structure of N-Ada10 (PDB accession code 1EYF). (A) The overall fold and the Cys4Zn cluster. (B) An electrostatic surface (GRASP) representation of N-Ada10 in the unmethylated state. Red, negatively charged surfaces; blue, positively charged surfaces; white, neutral. (C) An electrostatic surface (GRASP) representation of N-Ada10 with a methyl group added to the sulfur atom of Cys38 (modeled from the NMR structure). Note the change in charge state of the Cys4Zn pocket that interacts with DNA.
X-ray absorption spectroscopy of N-Ada and its methylated form indicates that the SMeCys38 thioether is still ligated to the zinc(II) center.76 There is no ligand exchange process after methylation, which rules out the possibility of inducing a protein conformation change to enhance the protein’s affinity toward DNA.
2.2.4. Repair and Transcriptional Activation Mechanism of N-Ada
N-Ada presents several quite interesting chemical problems: (i) how is one Cys residue selectively activated among all four Cys ligands? (ii) how does methylation of one Cys residue attenuate the protein’s affinity to DNA so dramatically? (iii) why is only the Sp-configurated methylphosphotriester repaired? These questions could only be answered with a high-resolution structure of the protein bound to DNA. Very recently, a crystal structure of methylated N-Ada bound to DNA has been obtained.58 This structure and subsequent biochemical studies reassigned Cys38 as the active residue. The structure also showed that the sulfur atoms of the other three residues, Cys42, Cys69, and Cys72, are “masked” by hydrogen bonding to the protein main chain amide protons. Their nucleophilicity is suppressed due to these interactions. Cys38 is selectively activated since it is the only Cys ligand that is not engaged in any hydrogen-bonding interaction. The stereoselectivity of the N-Ada’s repair function can be explained as well. N-Ada aligns the Sp oxygen atom of the nearby DNA phosphodiester backbone toward the sulfur atom of Cys38; the other oxygen atom points in the opposite direction, which cannot be accessed by Cys38.
How does one methyl transfer significantly increase the affinity of N-Ada to DNA? The structures of the protein and the protein/DNA complex do not give the answer to this question at first glance since there is no significant conformational rearrangement of the protein caused by methylation. However, an inspection of the protein surface that interacts with DNA indicates the presence of a negatively charged area localized on the zinc site for the wild type N-Ada (Figure 6B).58,76 The Cys4Zn cluster has a total charge of −2 with each sulfur atom of the Cys residues, including the surface exposing ones, bearing a negative charge of −½. This locally negatively charged surface will repulse the negatively charged DNA backbone, which will result in weak binding between protein and DNA, as would be expected. Methylation of Cys38 neutralizes this negatively charged surface of N-Ada and reduces the charge–charge repulsion between the protein and the DNA backbone (compare part C of Figure 6 with part B). This elimination of unfavorable charge–charge repulsion gives rise to enhanced DNA affinity for the methylated N-Ada. In essence, N-Ada has evolved a methylation-dependent electrostatic switching mechanism to control its DNA binding affinity and transcriptional regulation.
The same electrostatic argument also explains N-Ada’s ability to selectively recognize the methylphosphotriester damage on the DNA backbone. A methylphosphotriester renders this part of the DNA backbone neutral, which will not exhibit repulsion to the negatively charged Cys4Zn surface on N-Ada. While the undamaged phosphodiester backbone is repulsed away from this surface, the neutral triester is selectively recognized due to reduced unfavorable repulsion.58 This methylphosphotriester recognition serves as a convenient signal to trigger overexpression of other repair functions which are crucial for the survival of cells.
The adaptive response toward the methylation challenge does not occur in eukaryotes. Not surprisingly, no N-Ada homologues have been found in humans, and there has been no evidence that methylphosphotriesters are repaired in human cells.
2.3. Converting N-Ada into A Catalytic Enzyme
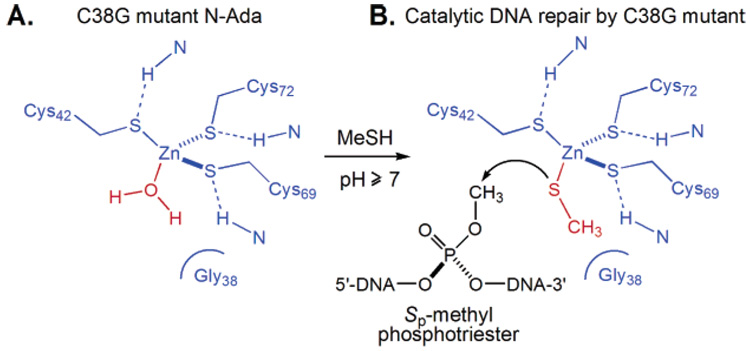
The sacrificial DNA repair protein N-Ada repairs the Sp-methylphosphotriester through an irreversible methyl transfer to a reactive Cys residue, Cys38 (Figure 4B).58 The methylated Cys38 cannot be regenerated, and each Ada can only repair one methylphosphotriester lesion. A strategy to convert N-Ada into a catalytic enzyme through protein engineering was employed. Cys38 was mutated to Gly, and the mutant protein was produced and purified.77 A water may occupy the fourth coordination site on zinc(II) in this mutant protein (Figure 7A). Addition of methanethiol, an external thiol, to the solution of the protein may replace the bound water molecule and generate a zinc(II)-bound thiolate (Figure 7B). Due to the accumulation of negative charge on the cluster, this newly formed thiolate should be nucleophilic since it is not “masked” by hydrogen bonding. When an ssDNA substrate containing a methylphosphotriester was added to this solution, repair activity was observed only when both the mutant protein and methanethiol are present.77
Figure 7.
Strategy to convert N-Ada into a catalytic enzyme. (A) The active Cys38 residue is mutated to Gly to open a coordination site on the zinc(II) center. (B) A methanethiol molecule can bind to zinc(II) and perform methyl transfer from Sp-methylphosphotriester.
Since external thiols are used as the methyl acceptor in this case, turnovers were observed, as expected. The repair is protein-based because the deprotonated methanethiol alone could not remove the methyl group from the triester. Furthermore, only the Sp-diastereomer was repaired, leaving the other diastereomer intact, which confirms the requirement of the protein to place the two reactants together for a stere-oselective methyl transfer. Rarely is a stoichiometric reagent protein converted into a catalytic enzyme. The strategy may work for other protein systems that use similar mechanisms.
3. O6-Alkylguanine-DNA Alkyltransferases
3.1. Direct Repair of O6-Alkylguanine and O4-Alkylthymine
The C-terminal end of Ada is responsible for the second repair function of the protein, the repair of O6-alkylguanine and O4-alkylthymine (Figure 2 and Figure 4). The first lesion is produced in low amounts by alkylating agents but is especially detrimental to all organisms due to its direct mutagenicity.29 If the damaged strand is used as the template for DNA replication, a thymine base will be preferentially incorporated opposite O6-alkylguanine, which when followed by a second round of replication gives rise to a G:C to A:T mutation.32 This is the most common mutation seen after cells are exposed to alkylating agents. C-Ada removes an alkyl lesion on the O6-position of a guanine when it is opposite a cytosine; however, it is unclear if C-Ada is capable of efficiently repairing O6-alkylguanine when it forms a stable base pair with an opposite thymine.78 Such a function can be performed by the human homologue of C-Ada, hAGT;79 the resulting G:T base pair is recognized and converted back into G:C by mismatch repair systems.80
Like N-Ada, repair by the C-Ada domain is also carried out by a direct transfer of the alkyl group from the damaged base to a reactive cysteine residue, Cys321, via a SN2 mechanism (Figure 2A). This action is irreversible and therefore inactivates this domain of the protein in a stoichiometric fashion; one C-Ada is depleted for each repaired base.17 C-Ada is actually capable of removing a variety of alkyl groups from guanine in DNA including ethyl78 and benzyl adducts,81 although the latter is repaired very slowly and is not repaired at all as the free base.81 While early studies indicated that the O6-methylguanine was repaired better than the O4-methylthymine,78,82 a more recent study avers that they are repaired with equal efficiency.83 The protein prefers lesions in dsDNA over ssDNA.29
3.2. E. Coli C-Ada
3.2.1. Structure of C-Ada
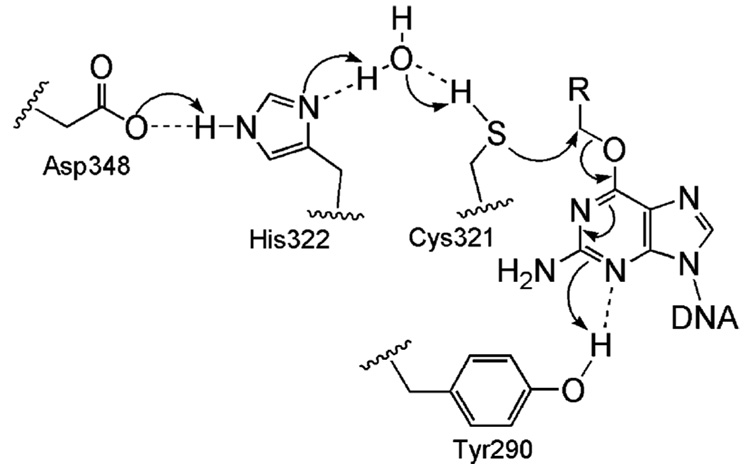
The crystal structure of C-Ada was solved in 1994.84 This domain of the Ada protein is divided into two subdomains. The first subdomain (residues 176–260) contains a β-sheet sandwiched between two helices. The second subdomain contains the conserved active site residues of the AGT family and folds into a previously unseen structure comprised primarily of helices and coils including a helix–turn–helix motif that is known to bind DNA. The structure suggested a mechanism by which Cys321 is activated for a nucleophilic attack of the lesion. At biological pH, alkanethiol groups should be predominately protonated (pKa ~ 8.4). In this protonated form, Cys321 would be unlikely to displace the alkyl group of the lesioned base. However, within the protein a hydrogen bonding network was observed that involves His322, Glu348, and a structured water to help deprotonate the sulfur atom of Cys321 to generate a nucleophilic thiolate anion that could perform the alkyl transfer function (Figure 8).
Figure 8.
A conserved hydrogen bond network is present in the active site of the AGT proteins to facilitate the repair.
The structure did present an interesting problem: the reactive Cys321 residue is buried in a pocket within the protein. Its position in the absence of a bound substrate indicates that either the protein or the DNA must make a conformational change for the alkyl group to come into contact with Cys321. At the time, circular dichroism studies and fluorescence analysis of hAGT indicated that the protein may change its conformation upon binding to DNA.85,86 Based on these results it was proposed that part of C-Ada would twist away to present the active cysteine to the bound DNA. While no crystal structures of the C-Ada/DNA complex have been solved, hAGT/DNA complex structures obtained recently make this hypothesis unlikely.
3.2.2. Other Homologues
Since O6-alkylguanine is one of the most harmful kinds of mutagenic damage that all cells must avoid, O6-alkylguanine-DNA alkyltransferase is widespread among almost all organisms. The family is easily recognized by the conserved sequence of its active site, –(I/V)PCHR(V/I)–. In addition to the inducible Ada, E. coli possesses a second copy of O6-alkylguanine-DNA alkyltransferase known as Ogt which is expressed constitutively as a house-keeping protein. Ogt shows a preference for repairing adducts larger than the methyl group,81 and it also prefers O6-alkylguanine over O4-alkylthymine although it is reasonably capable with both.82,87 Several archaea versions have been identified, including one from Pyrococcus kodakaraensis for which the crystal structure was solved.88 The agt genes in many eukaryotes have been cloned and expressed as well.89–95 Aside from the active site and some other residues in the C-terminal subdomain, the sequences of these proteins show rather low similarity, although most contain a N-terminal domain that does not seem essential for the alkyl transfer reaction. In fact, in the case of the yeast homologue, this domain does not even have the same predicted secondary structure as that of C-Ada.89 The function of this domain remains unknown. Two homologues have been found that contain only the C-terminal domain of AGT with an extra domain following. This domain in Caenorhabditis elegans is homologous to histone 1C96 while in Ferroplasma acidarmanus it has been proved to be an active endonuclease V domain.97
3.3. Human O6-Alkylguanine-DNA Alkyltransferase (hAGT)
3.3.1. Substrate Preferences
hAGT, encoded by the human agt (mgmt) gene, is more homologous to E. coli Ogt than C-Ada.84 It contains 207 amino acids (MW= 22 kDa) with Cys145 as the reactive cysteine. It also shares the substrate preferences of Ogt, as it is able to repair O6-BG as a free base and actually prefers it over O6-meG81 in oligonucleotides as well as repairing O6-meG more efficiently than O4-methylthymine.83 As for the case of C-Ada, the preferred substrate is the B-form dsDNA. Little neighboring sequence specificity has been found for the targeting of specific damage,98,99 and it was found that hAGT is able to repair O6-meG paired with any other base.100,101
3.3.2. Structure of hAGT
Several forms of the hAGT structure are now available. The first to be published showed it to have a similar fold to C-Ada even in the N-terminal domain where the two proteins share lower sequence similarity.102 A second native structure published shortly after, along with the structure of the benzylated form, revealed a surprising finding that the N-terminus contains a zinc(II) site bound by Cys5, Cys24, His29, and His85 in a tetrahedral geometry.103 These four residues are conserved in all mammalian sequences of the protein but are missing from both S. cerevisiae and bacterial homologues.104 Further biochemical studies showed that although the zinc(II) site is far from the reaction center, binding a zinc(II) ion serves to lower the pKa of the Cys145 even further than that is in the apo protein, which increases the reactivity of the protein.105,106 A detailed Cys145 activation and repair mechanism was proposed based on the stcuture.103 A hydrogen bonding network is used to activate the Cys145 residue, and a key Tyr residue, Tyr114, is employed to recognize the damaged guanine and provide a proton to facilitate the removal of the alkyl adduct (Figure 8). The increased preference of hAGT for large aromatic substrates is highlighted by the benzylated structure in which the benzyl group, covalently attached to the active site cysteine, is packed between Pro140, Ser159, and Tyr158 through hydrophobic interactions.103 In C-Ada the proline is replaced with an alanine, and a glycine, Gly160, on the other side of the hAGT pocket is replaced with a tryptophan. Thus, C-Ada has a narrow substrate binding pocket, which helps explain its preferences for small lesions.104
3.3.3. DNA Binding
Further studies of the AGT proteins, including NMR work,107 shifted the preferred hypothesis for AGT binding to DNA from protein conformational change to one in which the damaged base is flipped out and inserted into the active site to be repaired. The exact mode of damage-recognition and DNA binding by the AGT proteins has only been recently revealed from the structures of hAGT/DNA complexes (Figure 9). These structures show an unprecedented base flipping from the minor groove of the DNA duplex (Figure 9A and B).108,109 The protein interacts with DNA predominantly through phosphate backbone contacts. The base is inserted into the active site pocket while an Arg residue from the protein, Arg128, invades into the DNA helical stack to fill the space left by the flipped base. The side chain of this residue forms hydrophobic interactions with neighboring bases and hydrogen bonds to the opposite base. Tyr114 was found to disrupt the normal curve of the DNA backbone by occupying the space normally taken by the phosphate group 3′ of the flipped base (Figure 9). The DNA is bent away from the protein in all the structures; these distortions may facilitate base flipping.
Figure 9.
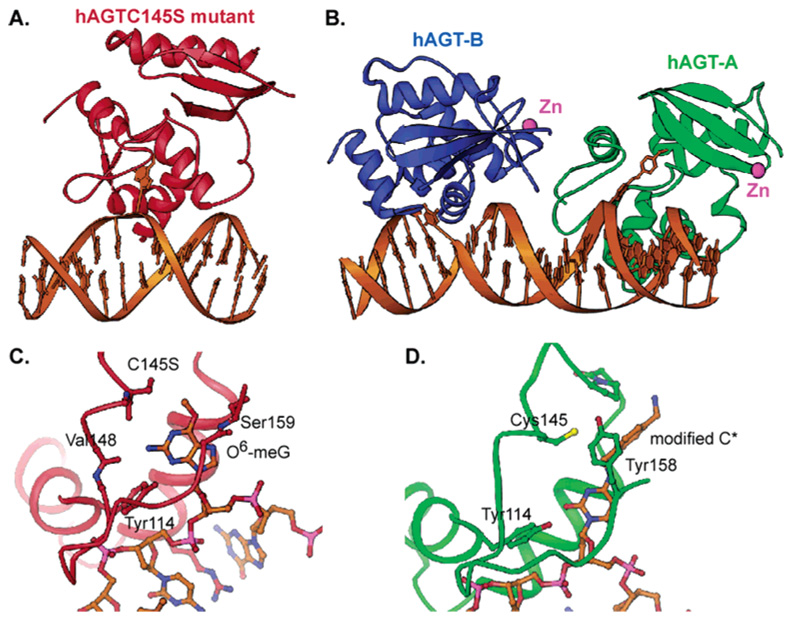
Structures of the hAGT/DNA complexes. (A) Overall structure of an inactive mutant hAGTC145S bound to a dsDNA containing a damaged base O6-meG. The lesioned base is flipped out of the minor groove of the DNA and inserted into the active site of the protein (PDB accession code 1T38). (B) Overall structure of the native hAGT bound to two different regions of DNA: hAGT-A flips a modified cytosine in DNA, and hAGT-B binds two ends of the DNA and recognizes a terminal overhanging T (1YFH). (C) Recognition of the O6-meG in the active site of the hAGTC145S mutant. (D) Binding of the modified cytosine in the active site of the native hAGT-A.
The preference for DNA as opposed to RNA repair is caused by the Cα of Gly131 which is too close to the sugar ring to allow for the extra hydroxyl group. In the first structure published with the inactive hAGTC145S mutant bound to an O6-meG-containing oligonucleotide (Figure 9A),108 several residues are found to recognize the flipped O6-meG. Tyr114 forms a hydrogen bond to the N3 nitrogen atom which is the main contact that may select O6-alkylguanine over the other bases (Figure 9C). The carbonyl oxygen atoms of Val148 and Ser145 are positioned to hydrogen bond to the N2-amino group of the guanine, and the main chain amide of Ser159 forms a hydrogen bond to the O6 atom. Some of these interactions may also help stabilize the transition state of the repair reaction.
The structure of the wild type protein bound to a modified cytosine having a hydrophobic group attached to the N4 nitrogen atom reveals contacts which may be important for the recognition of damaged thymine (hAGT-A in Figure 9B).109 In this case the phenol group of Tyr114 forms a hydrogen bond to the exocyclic O2 atom of the base (Figure 9D). No interaction was observed that may help protonate the N3 nitrogen atom of the base. The hydrophobic group is packed between Tyr158 and Pro140, confirming the prediction from the benzylated protein structure.103
3.3.4. Fate of the Alkylated hAGT
After alkylation it is known that hAGT is quickly degraded by the ubiquitin pathway.110 It has been postulated that the signal for this degradation is the disruption of a salt bridge between Asn137 and the carbonyl oxygen of Met134 due to steric clash of the new S-alkylcysteine.103 Crystal structures of the benzylated form of hAGT show a shift in the position of the guanine-binding loop 0.5–1.5 Å away from the protein core when compared to the native structure.103 While this seems to have little effect on the binding of the protein to DNA,111 it does lead to increased proteolysis and susceptibility to urea unfolding in the presence and absence of DNA.
A recent discovery suggested that methylated hAGT hijacks the estrogen receptor, the crucial transcription factor that regulates cell proliferation, from its transcriptional complex.112 Thus, the alkylated hAGT may briefly perform a signaling role after repairing the damage similar to that of Ada in E. coli. In this case it may act as a repressor of cell proliferation. This new discovery opens up possibilities for further investigation of mammalian responses to alkylation damage.
3.4. Potential Damage-Searching Modes for the AGT Proteins
3.4.1. Proposed Mechanisms of Detection
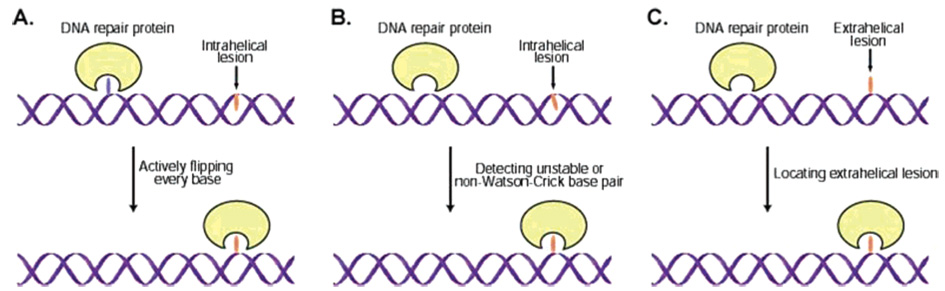
The genomes of mammalian cells contain billions of base pairs. A base-flipping repair protein such as AGT may have to search through millions of base pairs to find a single point of damage. How these proteins efficiently locate base lesions has been an interesting and long-standing question. Assuming a repair protein first binds DNA nonspecifically and then moves in one dimension, there are three potential damage-locating mechanisms (Figure 10): (i) every base is actively flipped and inserted into the active site by the protein until the damage is located (Figure 10A); (ii) a repair protein selectively detects an unstable/non-Watson–Crick base pair that contains the damaged base (Figure 10B); (iii) a transiently extrahelical base lesion is captured by the repair protein (Figure 10C). Studies on base-flipping glycosylases have been performed to help provide insights.113–117 The AGT proteins can also be used as models to probe this intriguing problem.
Figure 10.
Three potential damage-searching mechanisms for DNA repair proteins that recognize and process damaged bases extrahelically. (A) An active damage-searching mechanism. In this mechanism, the protein flips every base out and checks it in its active site pocket until the lesion is located. (B) A repair protein selectively detects the unstable/non-Watson–Crick base pair that contains the damaged base. (C) A mechanism of simply capturing a transiently extrahelical base lesion by the repair protein.
3.4.2. Study of C-Ada and hAGT
A disulfide cross-linking strategy was designed to probe DNA base flipping by the AGT proteins.118 In this strategy, a chemically modified base, which can trap the DNA/protein complex in the flipped position, was incorporated into stable and unstable base pairs in DNA probes. The probes were used to cross-link with both E. coli C-Ada and human AGT. The result indicates that C-Ada only detects unstable base pairs and cannot actively flip bases in stable, matched base pairs in DNA. hAGT, however, can efficiently locate damaged bases in unstable base pairs, but hAGT is also capable of extruding base lesions that are stabilized intrahelically.118 A recent kinetic and fluorescent analysis of hAGT repair indicates, however, that only O6-alkylguanine in an “unstable” base pair is flipped by hAGT.119 Normal G:C or A:T base pairs do not seem to be affected by the protein.
The structure study of hAGT bound to DNA suggests that the protein imposes a strain on DNA and searches for DNA regions that have already been destabilized.108,109 While this explains the efficient recognition of unstable alkylG:C base pairs, it does not address the ability of hAGT to break stable base pairs.100,101,118 More intriguingly, hAGT binds DNA from the minor groove while O6-alkylation is in the major groove. How does the protein locate the lesions buried in stable base pairs? Further investigations are required to understand the search modes of AGT proteins, especially the differences between homologues.
3.4.3. Directional Bias of ssDNA Repair by hAGT
Recent work from Pegg and Tainer et al. has suggested an interesting directionality of ssDNA repair by hAGT.108 It appears that hAGT may preferentially locate damage near the 5′ end of ssDNA. The protein does not show this bias with dsDNA. It was proposed that perhaps this phenomenon arises from a cooperative mechanism: hAGT recruits another monomer of itself faster on the 5′ side than on the 3′ side, and the repair is a cooperative event. More work is required to shed further light onto this observation.
3.5. Inhibition of hAGT in Anticancer Alkylation Chemotherapy
3.5.1. Alkylation Chemotherapy
The cytotoxic effects of alkylation damage on the O6-position of guanine can be used as a pathway for battling cancer. To this end many alkylating agents have been developed and are used clinically as antitumor drugs. Those which are known to attack the O6-position of guanine are the family of nitrosoureas and O6-(2-chloroethylating) agents such as carmustine (BCNU).10 Cyclophosphamide is also believed to attack this site through one of its metabolites.120 In this case the function of hAGT is undesired since it repairs the damage caused by the chemotherapeutic agents. Moreover, increased production of hAGT is one of the leading causes of resistance to such therapies.121 Therefore, inhibition of hAGT as a strategy to improve the efficacy of alkylation agents in chemotherapies has been a goal for some time.
3.5.2. Inhibition of hAGT by O6-Benzylguanine
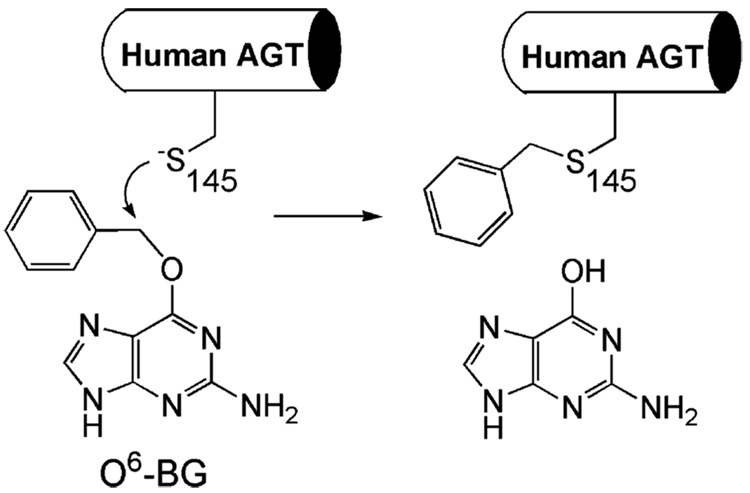
Due to its lack of turnover, the most obvious way to inhibit hAGT is to alkylate its active site cysteine. For some time the canonical substrate O6-meG as a free base was thought to be a possible compound to achieve this,122,123 but the insolubility of this pseudosubstrate and its failure in in vivo studies started the search for more efficacious inactivators. O6-BG was quickly found to be much more effective than the methylated form and has been tested extensively since then (Figure 11). Preincubation of cancer cell lines with this small molecule was repeatedly found to increase the cell’s sensitivity to subsequent treatment with the CNU type agents.124 Phase I clinical trials of O6-BG and BCNU set the maximum-tolerated dose for human patients at 100 and 40 mg/m2, respectively.125 The study also proved that O6-BG is quickly converted to 8-oxo-O6-BG in vivo, which is still a good inactivator for hAGT. Phase II trials of the same drug combination (120 mg/m2 O6-BG dose) did not produce tumor regression in nitrosourea-resistant malignant glioma patients; however, some patients did exhibit stable disease for up to 18 weeks with the treatment.126 These trials highlight the need for more advanced inhibitors with increased potency, bioavailability, and cancer cell targeting abilities.
Figure 11.
O6-BG irreversibly inactivates hAGT.
3.5.3. Developing More Potent Inhibitors
Since the discovery of O6-BG, many derivatives of O6-BG have been synthesized and tested for their ability to inhibit hAGT. Although O6-BG was unmatched in terms of simplicity and activity, some trends did emerge from this work.120 The ortho-substitution at the benzene ring renders the compound inactive, and para-substituents are better than meta-substituents. The recent crystal structure of hAGT bound to DNA containing a modified cytosine indicates that hydrogen bond donating groups substituted on the para-position of the benzene ring may aid binding of the modified base to the protein.109 Heterocyclic alkyl groups attached at the O6-position of guanine were developed and tested as well. One compound, O6-(4-bromoethenyl)guanine (PaTrin 2), had been tested in a phase I clinical trial in combination with the methylating agent temozolomide with positive results.120 In addition to tweaking the O6-alkyl group, the effects of substitutions elsewhere on the guanine base have been investigated. These substitutions may serve either as a method of increasing the affinity of the small molecule to the protein, or as a site to attach a cancer cell targeting moiety. The obvious N9-substitution, where the guanine connects to a sugar in DNA, proves to be the most tolerant site.127 So far substitution at this site has not produced an increase in activity, but recent work appending glucose to O6-alkylguanine derivatives may prove useful due to the compound’s ability to target tumor cells and its increased water solubility.128
3.6. Application of hAGT in Biotechnology
The unusual covalent linkage that can be established between the AGT protein and its substrates has been exploited for biological applications ranging from the specific quantification of AGT to using hAGT fusion proteins for in vivo fluorescent labeling. All of the uses are made possible due to the high tolerance of hAGT toward transfer of various large alkyl groups on the O6-position of guanine.
3.6.1. Quantification of AGT
AGT catalyzes a simple transfer of an alkyl group from O6-alkylguanine to its active site cysteine. The most straight-forward way to monitor this event is to radioactively label the alkyl group and track its addition to the protein. A DNA substrate (for example, calf thymus DNA) can be methylated with N-[3H]methyl-N′-nitrosourea.129 The methylated DNA is incubated with hAGT or extracts containing hAGT, precipitated, and hydrolyzed. The tritium-labeled O6-meG is then separated from other nucleosides by HPLC, and the radioactive counting is recorded. Because of the stoichiometric nature of AGT’s repair reaction, the amount of active protein can be assessed from the radioactivity remaining. This is the primary technique used to evaluate the potency of hAGT inhibitors. Other methods that exploit the unique alkyl transfer process of the AGT proteins have also been developed for the activity assays.130–133
3.6.2. Radioactive Imaging of Tumors
Since some types of tumors, especially those that have developed resistance to alkylating chemotherapeutic reagents, show increased production of hAGT, finding areas in tissues with high levels of hAGT may be useful as a way to monitor those tumors. A potential method for accomplishing this noninvasively may be through the recently developed technique of positron emission tomography (PET) where 11C or 18F are incorporated into tracer molecules. The distribution of this tracer is followed using a scanning instrument which detects the γ radiation emitted when the positron collides with an electron. The tracer can be a labeled alkyl group in an O6-BG derivative for transfer to hAGT. An in vivo study using O6-3-[131]iodobenzylguanine in xenographic mice bearing human rhabdomyosarcoma cells showed tumor uptake, but in amounts less than that of some normal tissues.134 Further work needs to be done before this will be a clinically useful technique for monitoring hAGT in vivo.
3.6.3. hAGT Fusion Proteins in Biotechnology
hAGT has been exploited for other applications completely removed from its DNA repairing function. Kai Johnsson’s group has pioneered the use of hAGT fusion proteins as in vitro and in vivo biotechnology tools.135 This group has evolved hAGT mutants that possess high reactivity for transferring large alkyl groups on the O6-position of guanine free base to the reactive Cys residue of the protein.135 Thus, bifunctional small molecules which contain O6-BG conjugated through the benzyl group to a probe molecule can be prepared as good substrates for the hAGT mutants.136 The mutant hAGT can be fused with other proteins of interest. Expressing the fusion protein inside cells followed by incubation with the bifunctional molecule leads to in vivo labeling of the fusion protein with the probe.137 Fluorescent probes have been used to determine the location of the target protein.138 The probe molecule can also be chosen to induce protein dimerization by capturing an endogenous protein with high affinity to the probe.139 The same concept has been used for in vitro immobilization of fusion proteins as well. In this case the alkyl group of O6-alkylguanine is a tether attached to a surface. Transfer of the alkyl group to hAGT anchors the fusion proteins to that surface.140 This offers a mild condition for fixing a wide range of proteins on a surface for evaluating their functions. Many of these hAGT-based techniques may become important tools in the biotechnology arsenal.
4. AlkB Proteins
4.1. Oxidative Dealkylation by AlkB
4.1.1. E. Coli AlkB
AlkB was first discovered in 1983 through genetic studies revealing that mutation of the alkB gene of E. coli specifically sensitizes the bacteria to the alkylating agent MMS.141 In experiments that followed, the alkylation sensitive mutant was found to be extremely responsive to the treatment of SN2 type alkylating agents,141,142 which uncovered the possibility that AlkB was involved in protecting bacteria from the lethal effects of alkylation damage.141,143,144 Further findings that MMS-treated λ phage survived better in wild type cells compared to alkB mutant cells suggested that AlkB influenced the way cells handle alkylated DNA. Specifically, AlkB did not seem to prevent MMS-induced DNA alkylation but appeared to be involved in repairing MMS-induced DNA damage.141 The AlkB protein, composed of 216 amino acids with a molecular weight of 23.9 kDa,143 was soon cloned and purified.144 This allowed E. coli AlkB to be expressed in alkylation sensitive human cell lines which conferred alkylation resistance and rescued cells from MMS-induced death, suggesting that increased alkylation resistance is intrinsic to AlkB.142 However, the explicit function of AlkB continued to remain unidentified for almost 20 years despite various efforts to discover its function.
4.1.2. Puzzle of AlkB
Through protein sequence comparisons, AlkB proteins were proposed to be a member of the oxidoreductase superfamily, converting alkylated bases to other forms.144 However, the activity of AlkB remained elusive because the alkB mutant did not appear to possess the functions of existing DNA modifying enzymes, such as DNA-methyltransferases, DNA-glycosylases, and DNA-nucleases,143–145 and its repair activity could not be observed in vitro. The pursuit to identify the function of AlkB was driven forward by the discovery that the alkB mutants were defective at repairing DNA methylation damage induced by SN2 type methylating agents146 and that AlkB preferentially binds ssDNA in vitro and specifically repairs ssDNA alkylation damage in vivo. In addition, AlkB was found to bind slightly more efficiently to the methylated form of ssDNA.146 These results indicated that AlkB may repair damage on ssDNA and that its target might be 1-meA and 3-meC, sites most vulnerable for attack by SN2 methylation in ssDNA (Figure 1).146
Another important lead was provided by results from a sequence alignment study of protein fold and sequence homology on AlkB.48 This study suggested that AlkB is a member of the α-ketoglutarate (αKG)- and iron(II)-dependent dioxygenases, a family of proteins that utilize iron(II) to activate dioxygen and perform oxidation of various substrates. The iron(II)/αKG-dependent dioxygenase superfamily is widespread in eukaryotes and bacteria48 and is the largest known family of oxidizing enzymes without a heme group.147–149 This sequence alignment study predicted that AlkB may use an unprecedented oxidative dealkylation mechanism to repair DNA base damage (Figure 3).48
4.1.3. The Unique Repair Function of AlkB
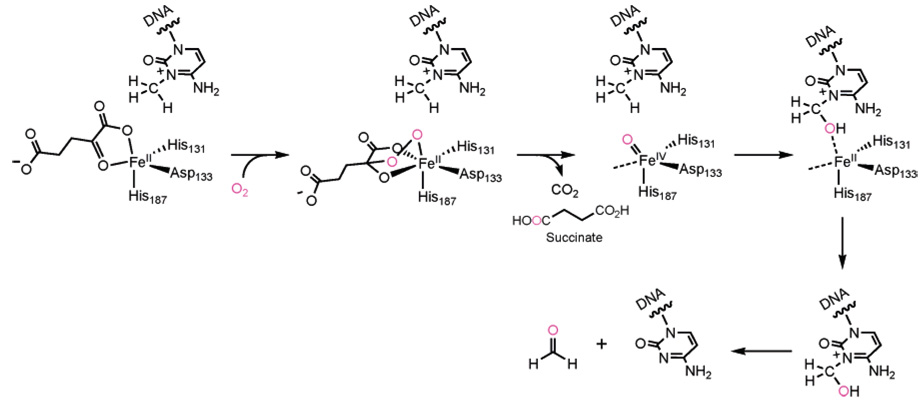
In 2002 it was discovered that AlkB catalyzes the direct reversion of 1-meA and 3-meC to adenine and cytosine, respectively, with the use of iron(II), αKG, and O2, through a proposed oxidative dealkylation mechanism (Figure 3 and Figure 12).39,40 Subsequent isolation and characterization of the native iron(II)-containing AlkB further confirmed that this protein is a member of the iron(II)/αKG-dependent dioxygenases.150 Almost all of the known members of this broadly studied superfamily have a conserved requirement for iron-(II) and catalyze two-electron oxidations.147,151 A possible general mechanism for these proteins is that they mediate a reaction between the active site iron(II) and O2 to first give a superoxo radical anion (O2−) bound to iron(III). Subsequently, the nucleophilic superoxide attacks at the α-keto carbon of an iron-bound αKG to give a bridged peroxo type intermediate. This bridged intermediate then undergoes a concerted decarboxylation of αKG and a heterolytic cleavage of the O–O bond to form the key active species, which is speculated to be a high-valent iron(IV)–oxo intermediate.147–149 A high-valent iron(IV)–oxo intermediate can be used by the dioxygenases to hydroxylate C–H bonds of various substrates depending on the function of each protein (Figure 12 and Figure 13). Thus, one of the oxygen atoms from O2 is incorporated into the succinate and the other is incorporated as a hydroxyl in the product. In the case of AlkB, the initial hydroxylation of the methyl group on the N1-position of adenine or the N3-position of cytosine by the iron(IV)–oxo intermediate leads to the resulting hydroxyl-methylated product decomposing to yield formaldehyde and an unmodified base (Figure 12). The overall effect leading to the direct repair of DNA damage is the same as that of AGT, but the mechanism of repair is quite different.
Figure 12.
Proposed mechanism for dioxygen activation and dealkylation repair of the AlkB proteins. The O2 reacts with the active site iron(II) and the α-keto carbon of an iron-bound αKG to give a bridged peroxo type intermediate. This intermediate undergoes a concerted decarboxylation and a heterolytic cleavage of the O–O bond to form the key active species, which is speculated to be a high-valent iron(IV)–oxo intermediate. This high-valent species oxidizes the alkyl group (3-meC is shown as an example) to afford an unstable alcohol which decomposes to an aldehyde and the repaired base.
Figure 13.
Change of the geometry of the active site iron(II) in E. coli AlkB from 5-coordinate to 6-coordinate with the addition of ssDNA.
4.1.4. Substrates
The lesions 1-meA and 3-meC are currently believed to be the physiological substrates for AlkB. The N1-adenine and N3-cytosine positions are involved in hydrogen bonding and are inaccessible in dsDNA. Once alkylation has occurred on either of these two sites, the damaged bases can no longer form Watson–Crick base pairs. Failure to repair these lesions in DNA leads to cell death in both E. coli and human cells.142
AlkB prefers 1-meA and 3-meC in a polynucleotide, but it is capable of demethylating smaller substrates as well.152 Although 1-methyldeoxyadenosine was reported to be very poorly or not detectably repaired,152 AlkB repairs 1-methyldeoxyadenosine 5′-triphosphate (1-medATP) as well as 1-meATP with low activity. However, 1-medAMP has been found to be the smallest substrate that can be efficiently repaired by AlkB.152 Studies with trimers have also shown efficient repair, comparable in effectiveness to those of polynucleotides. Thus, a polynucleotide is not essential for efficient repair; although a 5′-phosphate is required and essential for efficient repair,152 possibly to correctly position the methyl group for oxidation.
Since the initial discovery of substrates 1-meA and 3-meC, other modifications at these sites have been found to activate the repair function of AlkB. In vivo studies, which examined the survival of alkylated bacteriophage in an E. coli alkB mutant, have extended the AlkB substrate range to include methyl, ethyl, propyl, hydroxyethyl, and hydroxypropyl DNA adducts.50,145,152–154 In vitro studies have confirmed that 1-ethyladenine is repaired by AlkB to form adenine and acetaldehyde50 and have also successfully verified the substrate preferences of AlkB.
Methylations on the N3-position of thymine (3-meT) as well as the N1-position of guanine (1-meG) can also be repaired by AlkB.153–155 AlkB and its human homologues ABH2 and ABH3 are all able to demethylate 3-meT in DNA oligonucleotides, and 1-meG lesions introduced by chemical methylation on both DNA and tRNA were efficiently removed by AlkB. A study on the mutagenicity, cytotoxicity, and repair of 3-meT and 1-meG in wild type and alkB mutant strains of E. coli further indicates that these two lesions are indeed AlkB substrates in vivo.153 The physiological importance of these lesions and their repair remains to be unveiled. One study showed that, unlike 1-meA and 3-meC, these lesions did not stimulate the uncoupled AlkB-mediated decarboxylation of αKG, suggesting that these damages may be recognized differently by the enzyme.155
The N1-position of adenine and the N3-position of cytosine in most RNAs are exposed in a similar way as they are in ssDNA. In fact, 1-meA and 3-meC occur naturally in transfer RNA in both prokaryotes and eukaryotes.14,156 Repair of these lesions in RNA is required for correct RNA folding to allow efficient and accurate translation. AlkB and its human homologue ABH3 were found to effectively repair 1-meA and 3-meC lesions on RNA, a potentially important defense function for cells against alkylation damage.49,156
4.1.5. Side Reactions: Uncoupled Turnovers of αKG and Self-Hydroxylation
Enzymes in the iron(II)/αKG-dependent dioxygenase family can catalyze the reaction in which the substrate oxidation can be uncoupled from αKG decarboxylation, both with or without substrates.148,157 This uncoupled turnover of αKG results in the decomposition of αKG into succinate and CO2 and usually leads to enzyme deactivation. Furthermore, this decomposition of αKG may account for the oxidation of the iron(II) center to form the inactive iron-(III). This oxidation process can be reversed with addition of ascorbate39,158 as it is considered to act as an electron source in the uncoupled reaction.148 These uncoupled conversions possibly occur due to incorrect orientation of substrates in the active site, such that the substrates are never appropriately oxidized.157
The uncoupled reaction, leading to irreversible modifications of AlkB,159 was demonstrated as αKG decomposition was found to be partially uncoupled from DNA repair, accounting for the observed stimulation of AlkB activity in the presence of ascorbate.39,158 The uncoupled αKG turnover was also stimulated by binding of the small molecules 1-meA, 1-methyldeoxyadenosine (1-me(dA)), 3-meC, and 3-methyldeoxycytidine (3-me(dC)), but they were not repaired by AlkB.158 Unmethylated nucleosides did not stimulate αKG turnover, indicating that the presence of a methyl group in the substrate is important in initiating oxidation of αKG.158
Furthermore, in the absence of methylated DNA but in the presence of the other substrates (αKG, O2, and iron(II)) AlkB was found to catalyze self-oxidation of an amino acid side chain, Trp178. As O2 saturated water is added, the sample produces a blue chromophore at 590 nm, a peak attributed to the LMCT transition of OH-Trp coordinated to iron(III). The motivation for this aberrant reaction that hydroxylates Trp178159 is still uncertain, but the generation of a highly reactive intermediate in the absence of a substrate has been seen in other non-heme iron(II) dioxygenases as well.160–162
4.1.6. Characterization of Iron(II)–AlkB
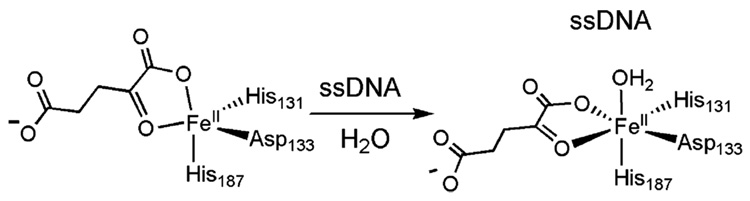
Recently, the overexpression and isolation of the native AlkB with bound cofactors directly from E. coli have facilitated the characterization of the iron(II) center.150 In all of the in vitro activity studies thus far, the cofactors, iron-(II) and αKG, have been added in excess to an apo-AlkB.39,40,49,152,154,155,158,159,163 The purified native AlkB protein exhibited an UV–vis band at 560 nm, which was assigned as the iron(II) to α-KG charge-transfer band.150 This peak shifted by 9 nm to a higher energy with the addition of excess ssDNA, suggesting a DNA-binding-induced geometry change of the active site. X-ray absorption spectra were collected on the native AlkB and its complex with ssDNA. The analysis of the data suggested that AlkB has a 5-coordinate iron-(II) center in the absence of DNA, which becomes 6-coordinate with the addition of ssDNA (Figure 14). The isolation of this protein form provides a means to further investigate the metal center and perform mechanistic investigations.
Figure 14.
Sequence alignment of several homologues of AlkB, including the first three human homologues (ABH1–3), as well as mouse and viral homologues, generated using ClustalW. The accession numbers of the aligned sequences are NP_311128, NP_006011, NP_001001655, NP_631917, XP-132383, and AAA47787.
4.2. Homologues
4.2.1. Homologues of E. Coli AlkB
It is now known that the alkB gene is conserved from bacteria to human.48,164–166 Putative yeast genes have been shown to complement the MMS sensitivity of E. coli alkB mutant cells as well, but these yeast genes do not share any amino acid sequence homology with the AlkB protein.167 Thus, to date, no yeast homologue has been identified. The first human homologue, now called ABH1, is 52% similar and 23% identical to the E. coli AlkB. This 34 kDa protein was reported to convey MMS resistance to the E. coli mutant and partially complement the sensitivity of an E. coli alkB mutant toward an alkylating agent,164 although this activity could not be confirmed in a recent study.50 It was also shown through chemical cross-linking that ABH1 does not seem to interact with DNA.168 Two other human homologues, ABH2 and ABH3, discovered through sequence and fold similarity, were shown to function like AlkB and could complement the E. coli alkB mutant phenotype.49,50 Furthermore, five more human genes are predicted (ABH4–ABH8) to be phylogenetically and functionally related to the AlkB family48,166 The importance of AlkB homologues in RNA processing was further shown by its discovery in RNA viruses48 and the fact that AlkB-like domains are found in at least 22 different ssRNA positive-strand plant viruses.169 AlkB homologues were also classified into subfamilies based on phylogenetic properties, and a small number of bacteria have been found to have an additional AlkB homologue.51 The conservation of the AlkB proteins is indicative of their importance in cellular defense against alkylating agents.164
The essential residues for enzymatic activity are conserved for all the homologues. A HXD motif, a single H, and a RXXXXXR motif are conserved when the sequences of the homologues are aligned (Figure 14). The two histidines and the aspartic acid are ligands to the active site iron(II), whereas the first arginine is probably involved in binding to the αKG.48,50 Chemical cross-linking studies, in which the interactions of human and bacterial AlkB proteins with DNA were probed, also supported the three residues as the active site ligands.168,170 In these studies, each of these residues in AlkB was mutated to a cysteine, and the mutant proteins cross-link with DNA containing a modified thiol-tethered cytosine. The cross-linking was inhibited by the addition of metal, as the cross-linking site was also the metal binding site.170
4.2.2. Substrates for Homologues
Although the first human homologue ABH1 was initially thought to behave similarly to AlkB, the purified gene product showed no activity.49,50,168 However, ABH2 and ABH3 complement the alkB mutant phenotype, and they both show repair activities for both 1-meA and 3-meC with the same cofactors as AlkB. It has been shown that ABH2 prefers 1-meA while ABH3 prefers 3-meC.49,50 Perhaps due to their different subcellular localizations in human cells, these proteins may have unique cellular roles. ABH2 and ABH3 were also able to demethylate 3-meT in DNA oligonucleotides. Both homologues, however, seem to need longer polynucleotides, as they have a low activity or no activity with trimers.152 Another difference between these two human homologues is that ABH2 prefers dsDNA while ABH3 is more like AlkB; it prefers ssDNA and even RNA.143,149 It should be noted that ABH2 and ABH3, although optimized (for pH and cofactor requirements) previously, had only 0.7 and 2%, respectively, of AlkB’s activity in vitro, when assayed with 1-meA in poly(dA).152
4.2.3. Inhibitors
Since it has been shown that inhibition of the human AGT activity can help modulate the efficiency of alkylation chemotherapy, it is reasonable to speculate that inactivation of AlkB may increase the efficacies of therapeutic reagents. Developing in vivo inhibitors for AlkB and its homologues could also help reveal their physiological roles. AlkB was found to be inhibited by high concentrations of αKG, regardless of the concentration of iron(II), which suggests that this AlkB inhibition was unlikely a result of iron(II) chelation by αKG.158 Analogues of αKG, including 2-mercaptoglutarate, were found to specifically inhibit AlkB. The 2-keto group of αKG can be replaced with a thiol or an alcohol to eliminate the carbonyl group that reacts with dioxygen. Good inhibition was observed with the thiol-substituted form, but there was no inhibition with the C-2 alcohol form, up to a high concentration.
4.3. Questions Remaining
From the first genetic study identifying the alkB gene as being responsible for protecting cells against alkylation DNA damage,141 much has been discovered about this mystifying protein. Still, there are questions that need to be addressed with much anticipation.
4.3.1. Potential Oxidative Dealkylation Functions
So far the AlkB proteins have been shown to repair alkylation damage on the N1-positions of adenine and guanine and the N3-positions of cytosine and thymine (Figure 15). In principle, the oxidative dealkylation mechanism can be utilized to remove the alkyl adducts attached to most heteroatoms on DNA and RNA. As shown in Figure 15, methylations on N6-adenine, O6-guanine, N2-guanine, N4-cytosine, N4-thymine, or N2-thymine may all be subject to the oxidative dealkylation repair; the oxidized alcohol products are unstable and decompose to give back the unmodified bases. In principle, the exocyclic DNA adducts such as 1,N6-ethenoadenine and 3,N4-ethenocytosine could be oxidatively repaired (through a potential epoxide intermediate) in water as well. In fact, very recent studies indicate such a repair process of 1,N6-ethenoadenine can be catalyzed by the AlkB proteins.171,172 In addition, methylations ocurring on the N7-positions of adenine and guanine and the backbone phosphodiester may be oxidatively dealkylated in some organisms. These possibilities need to be considered when the biological oxidative dealkylation function is explored in the future.
Figure 15.
Sites that may also be subject to oxidative dealkylation repair. Exocyclic DNA adducts such as 1,N6-enthenoadenine could be repaired as well.
4.3.2. An Alternative Strategy
The oxidative dealkylation mechanism utilized by AlkB raises an interesting chemistry question. The heterocycles in 1-meA and 3-meC are good leaving groups. An alternative strategy involving a nucleophile to directly displace the alkyl adduct from the lesioned base can be envisioned (Figure 16). Metal ions are known to activate water to form a metal-bound hydroxide which may attack the methyl adduct to produce a methanol and the unmodified base. A thiol, bound or unbound to a metal, could also be activated to participate in such a process. Nature has already been utilizing this strategy in N-Ada and AGT proteins to directly reverse the damage; why doesn’t it use the same method to repair 1-meA and 3-meC? Perhaps this process exists somewhere. Conceivably, under anaerobic conditions cells may turn on such an alternative pathway or other potential pathways to remove alkylation damage. More mechanistic investigations on AlkB and the AlkB type proteins may provide further insight.
Figure 16.
Alternative strategies to repair a 1-meA or 1-meC lesion involving a nucleophile to directly displace the alkyl adduct from the damaged base.
4.3.3. Structure/Function Relationship
To date there has been no structure of an AlkB type protein. Despite the existence of many structures for the αRKG/iron(II)-dependent dioxygenases, our understanding of the structure/function relationship of the AlkB proteins is very limited. A high-resolution structure will help address many mechanistic issues. The AlkB proteins bind and repair damage on dsDNA, ssDNA, and ssRNA. How does the protein interact with different nucleic acid structures? How does the protein recognize the lesioned base? How does it search for the damaged base in both dsDNA and ssDNA? The crystal structures of this protein and of its DNA or RNA complex are highly anticipated; thus far, only the theoretical model of the enzyme structure has been reported.159
4.3.4. Physiological Roles
The function of ABH2 and ABH3 has been shown to be similar to that of AlkB. It was suggested that AlkB and ABH3 act predominantly on ssDNA or ssRNA, but AlkB was found to be 3-fold more reactive on dsDNA than ssDNA.39 What is the exact role of these proteins inside cells? After the oxidative repair, what happens to the formaldehyde that is released, since formaldehyde can also damage DNA? The exact functions of ABH1 and other human homologues ABH4–8 are still unknown. Some of them have been hypothesized to either be back-up enzymes for ABH2 and ABH3 or novel DNA or RNA repair/modification enzymes. What are their roles? Selective inhibition of these proteins may help reveal their roles inside human cells.
5. Concluding Remarks
Most of the DNA lesions processed by the direct repair proteins are either highly mutagenic or cytotoxic. Nature has evolved two different strategies to directly remove alkylation DNA damage: one is a stoichiometric transfer of the alkyl group, and the other is a catalytic oxidation of the adduct. These repair processes play essential roles to defend against alkylating agents in almost all organisms. In some bacteria the two direct removal strategies are used and regulated together in the same regulon. It is intriguing to think how the two pathways have evolved. The AlkB function was only recently elucidated. Studies on the structure, function, mechanism, and physiological roles of the AlkB proteins have attracted and will continue to attract extensive attention from both biologists and chemists. Even for O6-alkylguanine-DNA alkyltransferases, many important questions remain to be addressed. How do these proteins locate base lesions? What cellular factors might they interact with, and does the methylation of the protein serve a signaling role? Can we develop more potent inhibitors for the human protein which may improve the efficacy of alkylation chemotherapies in the clinic? Future efforts may lead to answers for these questions.
Among the four proteins activated during the adaptive methylation resistance response process in E. coli, three have been shown to repair alkylation damage on DNA or RNA.39 Two proteins use direct repair mechanisms to process the damage. The protein AidB is a homologue to a family of flavin-containing dehydrogenases that use a FAD or FMN cofactor to oxidize C–H bonds in various substrates. The function of AidB has not been revealed, but it has been shown to play a role in methylation resistance in bacteria.173 Potentially, AidB may also be involved in the oxidization of methyl or alkyl adducts on heteroatoms of proteins or nucleic acids to facilitate their removal, much like the function of AlkB. Considering the pervasiveness of dehydrogenases in biology, AidB may play a general role that has yet to be discovered.
6. Acknowledgments
We thank the W. M. Keck Foundation, The Searle Scholar Program, The Arnold and Marble Beckman Foundation, the G&P Foundation for Cancer Research, the Research Corporation, and the National Institutes of Health for supporting our research. E.M.D. is supported by the Burroughs Wellcome Fund Cross-Disciplinary Training Program (Grant No. 1001774C).
7. Abbreviations
- 1-meA
N1-methyladenine
- 1-me(dA)
1-methyldeoxyadenosine
- 1-medAMP
1-methyldeoxyadenosine 5′-monophosphate
- 1-medATP
1-methyldeoxyadenosine 5′-triphosphate
- 1-meG
N1-methylguanine
- 3-meC
N3-methylcytosine
- 3-me(dC)
3-methyldeoxycytidine
- 3-medCMP
3-methyldeoxycytidine 5′-monophosphate
- 3-meT
N3-methylthymine
- 8-oxoG
8-oxoguanine
- AGT
O6-alkylguanine-DNA alkyltransferase
- αKG
α-ketoglutarate
- BCNU
carmustine
- C-Ada
C terminal domain of Ada
- dsDNA
double-stranded DNA
- E. coli
Escherichia coli
- FTase
farnesyltransferase
- GTase
geranylgeranyltransferase
- hAGT
human AGT
- HPLC
high-performance liquid chromatography
- MeI
methyl iodide
- MNNG
N-methyl-N′-nitro-N′-nitrosoguanidine
- MNU
N-methyl-N′-nitrosourea
- N-Ada
N terminal domain of Ada
- NMR
nuclear magnetic resonance
- O6-BG
O6-benzylguanine
- O6-meG
O6-methylguanine
- PaTrin 2
O6-(4-bromothenyl)guanine
- PET
positron emission tomography
- ssDNA
single-stranded DNA
Biographies
Yukiko Mishina was born in Yokohama, Japan, in 1979. She received both her B.S. degree with honors in Chemistry (2001) and her M.S. degree in Chemistry (2002) from The University of Chicago. She is currently carrying out her graduate work in the lab of Professor Chuan He at The University of Chicago and hopes to obtain her Ph.D. in Inorganic Chemistry in 2006. Her research focuses on elucidating the structure and functions of DNA repair proteins with a particular emphasis on the AlkB protein.

Erica Duguid was born in Indiana in 1980. She received her B.S. in Chemistry and Biology in 2002 from Valparaiso University, where she was supported by the Dow Chemical Scholarship. She is currently at the University of Chicago, where she received her M.S. degree in 2003 and is now working towards her Ph.D. in Chemistry studying DNA repair proteins. She has been awarded the McCormick Fellowship, named a Burroughs Wellcome Interfaces Fellow, and participated in the Training Program at the Interface of Chemistry and Biology.

Chuan He was born in 1972 in P. R. China and graduated with a B.S. degree in Chemistry in 1994 from the University of Science and Technology of China, Hefei, China. Then he moved to the U.S. and obtained his Ph.D. in Chemistry at Massachusetts Institute of Technology in 2000, working with Professor Stephen J. Lippard. As a Damon-Runyon Cancer Research Foundation Postdoctoral Fellow, he worked in Professor Gregory L. Verdine’s group at Harvard University on DNA repair proteins. Dr. He started his independent research career as Assistant Professor in the Department of Chemistry at The University of Chicago in 2002. His research covers several different areas that include chemistry and catalysis with silver and gold; regulation of metals and virulence factors in bacteria; structure, function, and mechanism of direct DNA repair proteins; and proteomics on DNA repair and DNA modification proteins. He is a Searle Scholar, a G&P Foundation of Cancer Research Young Investigator, a W. M. Keck Foundation Distinguished Young Investigator, an Arnold and Mabel Beckman Foundation Young Investigator, a Cottrell Scholar from the Research Corporation, and a recipient of a Research Corporation Research Innovation Award and an Alfred P. Sloan Research Fellowship.

8. References
- 1.Friedberg EC, Walker GC, Siede W. DNA Repair and Mutagenesis. Washington, DC: ASM Press; 1995. [Google Scholar]
- 2.Lindahl T. Nature. Vol. 362. 1993. p. 709. [DOI] [PubMed] [Google Scholar]
- 3.Drabløs F, Feyzi E, Aas PA, Vaagbø CB, Kavli B, Bratlie MS, Peña-Diaz J, Otterlei M, Slupphaug G, Krokan HE. DNA Repair. 2004;3:1389. doi: 10.1016/j.dnarep.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Vaughan P, Lindahl T, Sedgwick B. Mutat. Res. 1993;293:249. doi: 10.1016/0921-8777(93)90076-s. [DOI] [PubMed] [Google Scholar]
- 5.Taverna P, Sedgwick B. J. Bacteriol. 1996;178:5105. doi: 10.1128/jb.178.17.5105-5111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldman R, Shields PG. J. Nutr. 2003;133:965S. doi: 10.1093/jn/133.3.965S. [DOI] [PubMed] [Google Scholar]
- 7.Hecht SS. Mutat. Res. 1999;424:127. doi: 10.1016/s0027-5107(99)00014-7. [DOI] [PubMed] [Google Scholar]
- 8.Hurley LH. Nat. Rev. Cancer. 2002;2:188. doi: 10.1038/nrc749. [DOI] [PubMed] [Google Scholar]
- 9.Sedgwick B. Nat. Rev. Mol. Cell Biol. 2004;5:148. doi: 10.1038/nrm1312. [DOI] [PubMed] [Google Scholar]
- 10.Rajski SR, Williams RM. Chem. Rev. 1998;98:2723. doi: 10.1021/cr9800199. [DOI] [PubMed] [Google Scholar]
- 11.Rydberg B, Lindahl T. EMBO J. 1982;1:211. doi: 10.1002/j.1460-2075.1982.tb01149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrows LR, Magee PN. Carcinogenesis. 1982;3:349. doi: 10.1093/carcin/3.3.349. [DOI] [PubMed] [Google Scholar]
- 13.Shooter KV, Howse R, Shah SA, Lawley PD. Biochem. J. 1974;137:303. doi: 10.1042/bj1370303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singer B, Grünberger D. Molecular Biology of Mutagens and Carcinogens. New York: Plenum Press; 1983. [Google Scholar]
- 15.Beranek DT. Mutat. Res. 1990;231:11. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 16.Sedgwick B, Lindahl T. Oncogene. 2002;21:8886. doi: 10.1038/sj.onc.1205998. [DOI] [PubMed] [Google Scholar]
- 17.Lindahl T, Sedgewick B, Sekiguchi M, Nakabeppu Y. Annu. Rev. Biochem. 1988;57:133. doi: 10.1146/annurev.bi.57.070188.001025. [DOI] [PubMed] [Google Scholar]
- 18.Carell T, Burgdorf LT, Kundu LM, Cichon M. Curr. Opin. Chem. Biol. 2001;5:491. doi: 10.1016/s1367-5931(00)00239-8. [DOI] [PubMed] [Google Scholar]
- 19.Sancar A. Chem. Rev. 2003;103:2203. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 20.Jamieson ER, Lippard SJ. Chem. Rev. 1999;99:2467. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 21.Philip PA, Souliotis VL, Harris AL, Salisbury A, Tates AD, Mitchell K, van Delft JH, Ganesan TS, Kyrtopoulos SA. Clin. Cancer Res. 1996;2:303. [PubMed] [Google Scholar]
- 22.Wilson DM, Barsky D. Mutat. Res. 2001;485:283. doi: 10.1016/s0921-8777(01)00063-5. [DOI] [PubMed] [Google Scholar]
- 23.Larson K, Sahm J, Shenkar P, Strauss B. Mutat. Res. 1985;150:77. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- 24.Labahn J, Scharer OD, Long A, Ezaz-Nikpay K, Verdine GL, Ellenberger TE. Cell. 1996;86:321. doi: 10.1016/s0092-8674(00)80103-8. [DOI] [PubMed] [Google Scholar]
- 25.Wyatt MD, Allan JM, Lau AY, Ellenberger TE, Samson LD. BioEssays. 1999;21:668. doi: 10.1002/(SICI)1521-1878(199908)21:8<668::AID-BIES6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 26.Kyrtopoulos SA, Anderson LM, Chhabra SK, Souliotis VL, Pletsa V, Valavanis C, Georgiadis P. Cancer Detect. Prev. 1997;21:391. [PubMed] [Google Scholar]
- 27.Kyrtopoulos SA. Mutat. Res. 1998;405:135. doi: 10.1016/s0027-5107(98)00130-4. [DOI] [PubMed] [Google Scholar]
- 28.Margison GP, Santibanez-Koref MF. BioEssays. 2002;24:255. doi: 10.1002/bies.10063. [DOI] [PubMed] [Google Scholar]
- 29.Pegg AE. Mutat. Res. 2000;462:83. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
- 30.Lindahl T, Demple B, Robins P. EMBO J. 1982;1:1359. doi: 10.1002/j.1460-2075.1982.tb01323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Demple B, Sedgwick B, Robin P, Totty N, Waterfield MD, Lindahl T. Proc. Natl. Acad. Sci. U.S.A. 1985;82:2688. doi: 10.1073/pnas.82.9.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleibl K. Mutat. Res. 2002;512:67. doi: 10.1016/s1383-5742(02)00025-x. [DOI] [PubMed] [Google Scholar]
- 33.Eisen JA, Hanawalt PC. Mutat. Res. 1999;435:171. doi: 10.1016/s0921-8777(99)00050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaworski S, Schoellhorn H, Eisenmann P, Thewalt U, Lippert B. Inorg. Chim. Acta. 1988;153:31. [Google Scholar]
- 35.Jang YH, Goddard WA, III, Noyes KT, Sowers LC, Hwang S, Chung DS. Chem. Res. Toxicol. 2002;15:1023. doi: 10.1021/tx010146r. [DOI] [PubMed] [Google Scholar]
- 36.Kampf G, Kapinos LE, Griesser R, Lippert B, Sigel HJ. Chem. Soc., Perkin Trans. 2002;2:1320. [Google Scholar]
- 37.Fujii T, Itaya T. Heterocycles. 1998;48:1673. [Google Scholar]
- 38.Stivers JT, L JY. Chem. Rev. 2003;103:2729. doi: 10.1021/cr010219b. [DOI] [PubMed] [Google Scholar]
- 39.Trewick S, Henshaw TF, Hausinger RP, Lindahl T, Sedgwick B. Nature. 2002;419:174. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- 40.Falnes PO, Johansen RF, Seeberg E. Nature. 2002;419:178. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 41.Sedgwick B, Robins P, Totty N, Lindahl T. J. Biol. Chem. 1988;263:4430. [PubMed] [Google Scholar]
- 42.Yarosh DB. Mutat. Res. 1985;145:1. doi: 10.1016/0167-8817(85)90034-3. [DOI] [PubMed] [Google Scholar]
- 43.Pegg AE. Cancer Res. 1990;50:6119. [PubMed] [Google Scholar]
- 44.Mitra S, Kaina B. Prog. Nucleic Acid Res. Mol. Biol. 1993;44:109. doi: 10.1016/s0079-6603(08)60218-4. [DOI] [PubMed] [Google Scholar]
- 45.Zak P, Kleibl K, Laval F. J. Biol. Chem. 1994;269:730. [PubMed] [Google Scholar]
- 46.Myers LC, Terranova MP, Ferentz AE, Wagner G, Verdine GL. Science. 1993;261:1164. doi: 10.1126/science.8395079. [DOI] [PubMed] [Google Scholar]
- 47.Matthews RG, Goulding CW. Curr. Opin. Chem. Biol. 1997;1:332. doi: 10.1016/s1367-5931(97)80070-1. [DOI] [PubMed] [Google Scholar]
- 48.Aravind L, Koonin EV. Genome Biol. 2001;2:007.1. doi: 10.1186/gb-2001-2-3-research0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aas PA, Otterlei M, Falnes Pø, Vågbo CB, Skorpen F, Akari M, Sundheim O, Bjørås M, Slupphaug G, Seeberg E, Krokan H. Nature. 2003;421:859. doi: 10.1038/nature01363. [DOI] [PubMed] [Google Scholar]
- 50.Duncan T, Trewick SC, Koivisto P, Bates PA, Lindahl T, Sedgwick B. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16660. doi: 10.1073/pnas.262589799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Falnes PO, Rognes T. Res. Microbiol. 2003;154:531. doi: 10.1016/S0923-2508(03)00150-5. [DOI] [PubMed] [Google Scholar]
- 52.Margison G. DNA Repair. 2002;1:1057. doi: 10.1016/s1568-7864(02)00169-6. [DOI] [PubMed] [Google Scholar]
- 53.Jiricny J. Curr. Biol. 2002;12:R846. doi: 10.1016/s0960-9822(02)01350-7. [DOI] [PubMed] [Google Scholar]
- 54.Samson L, Cairns J. Nature. 1977;267:281. doi: 10.1038/267281a0. [DOI] [PubMed] [Google Scholar]
- 55.Seeberg E, Berdal KG. In: Base Excision Repair of DNA Damage. Hickson ID, editor. Austin, TX: Landes Bioscience; 1999. [Google Scholar]
- 56.Landini P, Volkert MR. J. Bacteriol. 2000;182:6543. doi: 10.1128/jb.182.23.6543-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teo I, Sedgwick B, Kilpatrick MW, McCarthy TV, Lindahl T. Cell. 1986;45:315. doi: 10.1016/0092-8674(86)90396-x. [DOI] [PubMed] [Google Scholar]
- 58.He C, Hus J-C, Sun LJ, Zhou P, Norman DPG, Dötsch V, Gross JD, Lane WS, Wagner G, Verdine GL. Mol. Cell. 2005;20:117. doi: 10.1016/j.molcel.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 59.Sakumi K, Sekiguchi M. J. Mol. Biol. 1989;205:373. doi: 10.1016/0022-2836(89)90348-3. [DOI] [PubMed] [Google Scholar]
- 60.Myers LC, Jackow F, Verdine GL. J. Biol. Chem. 1995;270:6664. doi: 10.1074/jbc.270.12.6664. [DOI] [PubMed] [Google Scholar]
- 61.Akimaru H, Sakumi K, Yoshikai T, Anai M, Sekiguchi M. J. Mol. Biol. 1990;216:261. doi: 10.1016/S0022-2836(05)80318-3. [DOI] [PubMed] [Google Scholar]
- 62.Landini P, Busby SJW. J. Biol. Chem. 1999;181:1524. doi: 10.1128/jb.181.5.1524-1529.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Landini P, Volkert MR. J. Biol. Chem. 1995;270:8285. doi: 10.1074/jbc.270.14.8285. [DOI] [PubMed] [Google Scholar]
- 64.Myers LC, Terranova MP, Nash HM, Markus MA, Verdine GL. Biochemistry. 1992;31:4541. doi: 10.1021/bi00134a002. [DOI] [PubMed] [Google Scholar]
- 65.Sakashita H, Sakuma T, Ohkubo T, Kainosho M, Sakumi K, Sekiguchi M, Morikawa K. FEBS Lett. 1993;323:252. doi: 10.1016/0014-5793(93)81351-y. [DOI] [PubMed] [Google Scholar]
- 66.Myers LC, Wagner G, Verdine GL. J. Am. Chem. Soc. 1995;117:10749. [Google Scholar]
- 67.Myers LC, Verdine GL, Wagner G. Biochemistry. 1993;32:14089. doi: 10.1021/bi00214a003. [DOI] [PubMed] [Google Scholar]
- 68.Wilker JJ, Lippard SJ. J. Am. Chem. Soc. 1995;117:8682. [Google Scholar]
- 69.Wilker JJ, Lippard SJ. Inorg. Chem. 1997;36:969. doi: 10.1021/ic961082o. [DOI] [PubMed] [Google Scholar]
- 70.Blake PR, Park JB, W AMW, Summers MF. J. Am. Chem. Soc. 1992;114:4931. [Google Scholar]
- 71.Maynard AT, Covell DG. J. Am. Chem. Soc. 2001;123:1047. doi: 10.1021/ja0011616. [DOI] [PubMed] [Google Scholar]
- 72.Allen JR, Clark DD, Krum JG, Ensign SA. Proc. Natl. Acad. Sci. U.S.A. 1999;96:8432. doi: 10.1073/pnas.96.15.8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hightower KE, Fierke CA. Curr. Opin. Chem. Biol. 1999;3:176. doi: 10.1016/s1367-5931(99)80030-1. [DOI] [PubMed] [Google Scholar]
- 74.LeClerc GM, Grahame DA. J. Biol. Chem. 1996;271:18725. doi: 10.1074/jbc.271.31.18725. [DOI] [PubMed] [Google Scholar]
- 75.He C, Verdine GL. Chem. Biol. 2002;9:1297. doi: 10.1016/s1074-5521(02)00283-1. [DOI] [PubMed] [Google Scholar]
- 76.Lin Y, Dötsch V, Wintner T, Peariso K, Myers LC, Penner-Hahn JE, Verdine GL, Wagner G. Biochemistry. 2001;40:4261. doi: 10.1021/bi002109p. [DOI] [PubMed] [Google Scholar]
- 77.He C, Wei H, Verdine GL. J. Am. Chem. Soc. 2003;125:1450. doi: 10.1021/ja028046a. [DOI] [PubMed] [Google Scholar]
- 78.Graves RJ, Li BF, Swann PF. Carcinogenesis. 1989;10:661. doi: 10.1093/carcin/10.4.661. [DOI] [PubMed] [Google Scholar]
- 79.Lips J, Kaina B. Mutat. Res. 2001;487:59. doi: 10.1016/s0921-8777(01)00105-7. [DOI] [PubMed] [Google Scholar]
- 80.Sibghat U, Day RS., III Biochemistry. 1992;31:7998. doi: 10.1021/bi00149a034. [DOI] [PubMed] [Google Scholar]
- 81.Goodtzova K, Kanugula S, Edara S, Pauly GT, Moschel RC, Pegg AE. J. Biol. Chem. 1997;272:8332. doi: 10.1074/jbc.272.13.8332. [DOI] [PubMed] [Google Scholar]
- 82.Sassanfar M, Dosanjh MK, Essigmann JM, Samson L. J. Biol. Chem. 1991;266:2767. [PubMed] [Google Scholar]
- 83.Paalman SR, Sung C, Clarke ND. Biochemistry. 1997;36:11118. doi: 10.1021/bi970740t. [DOI] [PubMed] [Google Scholar]
- 84.Moore MH, Gulbis JM, Dodson E, Demple B, Moody PC. EMBO J. 1994;13:1495. doi: 10.1002/j.1460-2075.1994.tb06410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chan CL, Wu Z, Ciardelli T, Eastman A, Bresnick E. Arch. Biochem. Biophys. 1993;300:193. doi: 10.1006/abbi.1993.1027. [DOI] [PubMed] [Google Scholar]
- 86.Takahashi M, Sakumi K, Sekiguchi M. Biochemistry. 1990;29:3431. doi: 10.1021/bi00466a002. [DOI] [PubMed] [Google Scholar]
- 87.Wilkinson MC, Potter PM, Cawkwell L, Georgiadis P, Patel D, Swann PF, Margison GP. Nucleic Acids Res. 1989;17:8475. doi: 10.1093/nar/17.21.8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hashimoto H, Inoue T, Nishioka M, Fujiwara S, Takagi M, Imanaka T, Kai Y. J. Mol. Biol. 1999;292:707. doi: 10.1006/jmbi.1999.3100. [DOI] [PubMed] [Google Scholar]
- 89.Xiao W, Samson L. Nucleic Acids Res. 1992;20:3599. doi: 10.1093/nar/20.14.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kooistra R, Zonneveld JB, Watson AJ, Margison GP, Lohman PH, Pastink A. Nucleic Acids Res. 1999;27:1795. doi: 10.1093/nar/27.8.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zaboikin M, Srinivasakumar N, Zaboikina T, Schuening F. Hum. Gene. Ther. 2004;15:383. doi: 10.1089/104303404322959533. [DOI] [PubMed] [Google Scholar]
- 92.Shiraishi A, Sakumi K, Nakatsu Y, Hayakawa H, Sekiguchi M. Carcinogenesis. 1992;13:289. doi: 10.1093/carcin/13.2.289. [DOI] [PubMed] [Google Scholar]
- 93.Shiota S, von Wronski MA, Tano K, Bigner DD, Brent TP, Mitra S. Biochemistry. 1992;31:1897. doi: 10.1021/bi00122a001. [DOI] [PubMed] [Google Scholar]
- 94.Sakumi K, Shiraishi A, Hayakawa H, Sekiguchi M. Nucleic Acids Res. 1991;19:5597. doi: 10.1093/nar/19.20.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rydberg B, Spurr N, Karran P. J. Biol. Chem. 1990;265:9563. [PubMed] [Google Scholar]
- 96.Kanugula S, Pegg AE. Environ. Mol. Mutagen. 2001;38:235. doi: 10.1002/em.1077. [DOI] [PubMed] [Google Scholar]
- 97.Kanugula S, Pauly GT, Moschel RC, Pegg AE. Proc. Natl. Acad. Sci. U.S.A. 2005;102:3617. doi: 10.1073/pnas.0408719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pegg AE, Dolan ME, Moschel RC. Prog. Nucleic Acid Res. Mol. Biol. 1995;51:167. doi: 10.1016/s0079-6603(08)60879-x. [DOI] [PubMed] [Google Scholar]
- 99.Bender K, Federwisch M, Loggen U, Nehls P, Rajewsky MF. Nucleic Acids Res. 1996;24:2087. doi: 10.1093/nar/24.11.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Scicchitano D, Jones RA, Kuzmich S, Gaffney B, Lasko DD, Essigmann JM, Pegg AE. Carcinogenesis. 1986;7:1383. doi: 10.1093/carcin/7.8.1383. [DOI] [PubMed] [Google Scholar]
- 101.Toorchen D, Lindamood C, III, Swenberg JA, Topal MD. Carcinogenesis. 1984;5:1733. doi: 10.1093/carcin/5.12.1733. [DOI] [PubMed] [Google Scholar]
- 102.Wibley JEA, Pegg AE, Moody PCE. Nucleic Acids Res. 2000;28:393. doi: 10.1093/nar/28.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Daniels DS, Mol CD, Arvai AS, Kanugula S, Pegg AE, Tainer JA. EMBO J. 2000;19:1719. doi: 10.1093/emboj/19.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Daniels DS, Tainer JA. Mutat. Res. 2000;460:151. doi: 10.1016/s0921-8777(00)00024-0. [DOI] [PubMed] [Google Scholar]
- 105.Guengerich FP, Fang Q, Liu L, Hachey DL, Pegg AE. Biochemistry. 2003;42:10965. doi: 10.1021/bi034937z. [DOI] [PubMed] [Google Scholar]
- 106.Rasimas JJ, Kanugula S, Dalessio PM, Ropson IJ, Fried MG, Pegg AE. Biochemistry. 2003;42:980. doi: 10.1021/bi026970b. [DOI] [PubMed] [Google Scholar]
- 107.Verdemato PE, Brannigan JA, Damblon C, Zuccotto F, Moody PCE, Lian LY. Nucleic Acids Res. 2000;28:3710. doi: 10.1093/nar/28.19.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Daniels DS, Woo TT, Luu KX, Noll DM, Clarke ND, Pegg AE, Tainer JA. Nat. Struct. Biol. Mol. Biol. 2004;11:714. doi: 10.1038/nsmb791. [DOI] [PubMed] [Google Scholar]
- 109.Duguid EM, Rice PS, He C. J. Mol. Biol. 2005;350:657. doi: 10.1016/j.jmb.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 110.Xu-Welliver M, Pegg AE. Carcinogenesis. 2002;23:823. doi: 10.1093/carcin/23.5.823. [DOI] [PubMed] [Google Scholar]
- 111.Rasimas JJ, Pegg AE, Fried MG. J. Biol. Chem. 2003;278:7973. doi: 10.1074/jbc.M211854200. [DOI] [PubMed] [Google Scholar]
- 112.Teo AK, Oh HK, Ali RB, Li BF. Mol. Cell Biol. 2001;21:7105. doi: 10.1128/MCB.21.20.7105-7114.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Verdine GL, Bruner SD. Chem. Biol. 1997;4:329. doi: 10.1016/s1074-5521(97)90123-x. [DOI] [PubMed] [Google Scholar]
- 114.Banerjee A, Yang W, Karplus M, Verdine GL. Nature. 2005;434:612. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 115.Chen L, Haushalter KA, Lieber CM, Verdine GL. Chem. Biol. 2002;9:345. doi: 10.1016/s1074-5521(02)00120-5. [DOI] [PubMed] [Google Scholar]
- 116.Cao C, Jiang YL, Stivers JT, Song F. Nat. Struct. Mol. Biol. 2004;11:1230. doi: 10.1038/nsmb864. [DOI] [PubMed] [Google Scholar]
- 117.Krosky DJ, Schwarz FP, Stivers JT. Biochemistry. 2004;43:4188. doi: 10.1021/bi036303y. [DOI] [PubMed] [Google Scholar]
- 118.Duguid EM, Mishina Y, He C. Chem. Biol. 2003;10:827. doi: 10.1016/j.chembiol.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 119.Zhang H, Fang Q, Pegg AE, Guengerich FP. J. Biol. Chem. 2005;280:30873. doi: 10.1074/jbc.M505283200. [DOI] [PubMed] [Google Scholar]
- 120.McElhinney RS, McMurry TB, Margison GP. Mini-Rev. Med. Chem. 2003;3:471. doi: 10.2174/1389557033487980. [DOI] [PubMed] [Google Scholar]
- 121.Gerson SL, Willson JK. Hematol. Oncol. Clin. North Am. 1995;9:431. [PubMed] [Google Scholar]
- 122.Gorbacheva LB, Kukushkina GV, Elksne SJ, Miniker TD, Melnik S, Preobrazhenskaya MN. Anticancer Drugs. 1991;2:185. doi: 10.1097/00001813-199104000-00009. [DOI] [PubMed] [Google Scholar]
- 123.Yarosh DB, Hurst-Calderone S, Babich MA, Day RS., III Cancer Res. 1986;46:1663. [PubMed] [Google Scholar]
- 124.Mineura K, Izumi I, Watanabe K, Kowada M, Kohda K, Koyama K, Terashima I, Ikenaga M. Int. J. Cancer. 1994;58:706. doi: 10.1002/ijc.2910580515. [DOI] [PubMed] [Google Scholar]
- 125.Friedman HS, Pluda J, Quinn JA, Ewesuedo RB, Long L, Friedman AH, Cokgor I, Colvin OM, Haglund MM, Ashley DM, Rich JN, Sampson J, Pegg AE, Moschel RC, McLendon RE, Provenzale JM, Stewart ES, Tourt-Uhlig S, Garcia-Turner AM, Herndon JE, Bigner DD, Dolan ME. J. Clin. Oncol. 2000;18:3522. doi: 10.1200/JCO.2000.18.20.3522. [DOI] [PubMed] [Google Scholar]
- 126.Quinn JA, Pluda J, Dolan ME, Delaney S, Kaplan R, Rich JN, Friedman AH, Reardon DA, Sampson JH, Colvin OM, Haglund MM, Pegg AE, Moschel RC, McLendon RE, Provenzale JM, Gururangan S, Tourt-Uhlig S, Herndon JE, Bigner DD, Friedman HS. J. Clin. Oncol. 2002;20:2277. doi: 10.1200/JCO.2002.09.084. [DOI] [PubMed] [Google Scholar]
- 127.Pegg AE, Chung L, Moschel RC. Biochem. Pharmacol. 1997;53:1559. doi: 10.1016/s0006-2952(97)00060-9. [DOI] [PubMed] [Google Scholar]
- 128.Kaina B, Muhlhausen U, Piee-Staffa A, Christmann M, Garcia Boy R, Rosch F, Schirrmacher R. J. Pharmacol. Exp. Ther. 2004;311:585. doi: 10.1124/jpet.104.071316. [DOI] [PubMed] [Google Scholar]
- 129.Dolan ME, Moschel RC, Pegg AE. Proc. Natl. Acad. Sci. U.S.A. 1990;87:5368. doi: 10.1073/pnas.87.14.5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wilson BD, Strauss M, Stickells BJ, Hoal-van Helden EG, van Helden P. Carcinogenesis. 1994;15:2143. doi: 10.1093/carcin/15.10.2143. [DOI] [PubMed] [Google Scholar]
- 131.Klein S, Oesch F. Anal. Biochem. 1992;205:294. doi: 10.1016/0003-2697(92)90438-d. [DOI] [PubMed] [Google Scholar]
- 132.Moser AM, Patel M, Yoo H, Balis FM, Hawkins ME. Anal. Biochem. 2000;281:216. doi: 10.1006/abio.2000.4554. [DOI] [PubMed] [Google Scholar]
- 133.Nagel G, Brenner W, Johnsson K, Kaina B. Anal. Biochem. 2003;321:38. doi: 10.1016/s0003-2697(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 134.Vaidyanathan G, Affleck DJ, Norman J, Welsh P, Liu W, Johnson SP, Friedman HS, Zalutsky MR. Bioconjugate Chem. 2004;15:402. doi: 10.1021/bc0341977. [DOI] [PubMed] [Google Scholar]
- 135.Juillerat A, Gronemeyer T, Keppler A, Gendreizig S, Pick H, Vogel H, Johnsson K. Chem. Biol. 2003;10:313. doi: 10.1016/s1074-5521(03)00068-1. [DOI] [PubMed] [Google Scholar]
- 136.Damoiseaux R, Keppler A, Johnsson K. ChemBioChem. 2001;2:285. doi: 10.1002/1439-7633(20010401)2:4<285::AID-CBIC285>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 137.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. Nat. Biotechnol. 2003;21:86. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 138.Keppler A, Pick H, Arrivoli C, Vogel H, Johnsson K. Proc. Natl. Acad. Sci. U.S.A. 2004;101:9955. doi: 10.1073/pnas.0401923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gendreizig S, Kindermann M, Johnsson K. J. Am. Chem. Soc. 2003;125:14970. doi: 10.1021/ja037883p. [DOI] [PubMed] [Google Scholar]
- 140.Kindermann M, George N, Johnsson N, Johnsson K. J. Am. Chem. Soc. 2003;125:7810. doi: 10.1021/ja034145s. [DOI] [PubMed] [Google Scholar]
- 141.Kataoka H, Yamamoto Y, Sekiguchi M. J. Bacteriol. 1983;153:1301. doi: 10.1128/jb.153.3.1301-1307.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chen BJ, Carroll P, Samson L. J. Bacteriol. 1994;176:6255. doi: 10.1128/jb.176.20.6255-6261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kataoka H, Sekiguchi M. Mol. Gen. Genet. 1985;198:263. doi: 10.1007/BF00383004. [DOI] [PubMed] [Google Scholar]
- 144.Kondo H, Nakabeppu Y, Kataoka H, Kuhara S, Kawabata S, Sekiguchi M. J. Biol. Chem. 1986;261:15772. [PubMed] [Google Scholar]
- 145.Dinglay S, Gold B, Sedgwick B. Mutat. Res. 1998;407:109. doi: 10.1016/s0921-8777(97)00065-7. [DOI] [PubMed] [Google Scholar]
- 146.Dinglay S, Trewick SC, Lindahl T, Sedgwick B. Genes Dev. 2000;14:2097. [PMC free article] [PubMed] [Google Scholar]
- 147.Schofield CJ, Zhang Z. Curr. Opin. Struct. Biol. 1999;9:722. doi: 10.1016/s0959-440x(99)00036-6. [DOI] [PubMed] [Google Scholar]
- 148.Solomon EI, Brunold TC, Davis MI, Kemsley JN, Lee S-K, Lehnert N, Neese F, Skulan AJ, Yang Y-S, Zhou J. Chem. Rev. 2000;100:235. doi: 10.1021/cr9900275. [DOI] [PubMed] [Google Scholar]
- 149.Que LJ. Nat. Struct. Biol. 2000;7:182. doi: 10.1038/73270. [DOI] [PubMed] [Google Scholar]
- 150.Mishina Y, Chen LX, He C. J. Am. Chem. Soc. in press. [Google Scholar]
- 151.Hausinger RP. Crit. Rev. Biochem. Mol. Biol. 2004;39:21. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 152.Koivisto P, Duncan T, Lindahl T, Sedgwick B. J. Biol. Chem. 2003;278:44348. doi: 10.1074/jbc.M307361200. [DOI] [PubMed] [Google Scholar]
- 153.Delaney JC, Essigmann JM. Proc. Natl. Acad. Sci. U.S.A. 2004;101:14051. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Koivisto P, Robins P, Lindahl T, Sedgwick B. J. Biol. Chem. 2004;279:40470. doi: 10.1074/jbc.M407960200. [DOI] [PubMed] [Google Scholar]
- 155.Falnes PO. Nucleic Acids Res. 2004;32:6260. doi: 10.1093/nar/gkh964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ougland R, Zhang CM, Liiv A, Johansen RF, Seeberg E, Hou YM, Remme J, Falnes PO. Mol. Cell. 2004;16:107. doi: 10.1016/j.molcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 157.Lee H-J, Lloyd MD, Harlos K, Clifton IJ, Baldwin JE, Schofield CJ. J. Mol. Biol. 2001;308:937. doi: 10.1006/jmbi.2001.4649. [DOI] [PubMed] [Google Scholar]
- 158.Welford RWD, Schlemminger I, McNeill LA, Hewitson KS, Schofield CJ. J. Biol. Chem. 2003;278:10157. doi: 10.1074/jbc.M211058200. [DOI] [PubMed] [Google Scholar]
- 159.Henshaw TF, Feig M, Hausinger RP. J. Inorg. Biochem. 2004;98:856. doi: 10.1016/j.jinorgbio.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 160.Ryle MJ, Koehntop KD, Liu A, Que L, Jr, Hausinger RP. Proc. Natl. Acad. Sci. U.S.A. 2003;100:3790. doi: 10.1073/pnas.0636740100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Liu A, Ho RYN, Que L, Jr, Ryle MJ, Phinney BS, Hausinger RP. J. Am. Chem. Soc. 2001;123:5126. doi: 10.1021/ja005879x. [DOI] [PubMed] [Google Scholar]
- 162.Ryle MJ, Liu A, Muthukumaran RB, Ho RYN, Koehntop KD, McCracken J, Que L, Jr, Hausinger RP. Biochemistry. 2003;42:1854. doi: 10.1021/bi026832m. [DOI] [PubMed] [Google Scholar]
- 163.Falnes PO, Bjoras M, Aas PA, Sundheim O, Seeberg E. Nucleic Acids Res. 2004;32:3456. doi: 10.1093/nar/gkh655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wei Y-F, Carter KC, Wang R-P, Shell BK. Nucleic Acids Res. 1996;24:931. doi: 10.1093/nar/24.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Colombi D, Gomes SL. J. Bacteriol. 1997;179:3139. doi: 10.1128/jb.179.10.3139-3145.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kurowski MA, Bhagwat AS, Papaj G, Bujnicki JM. BMC Genomics. 2003;4:48. doi: 10.1186/1471-2164-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wei YF, Chen BJ, Samson L. J. Bacteriol. 1995;177:5009. doi: 10.1128/jb.177.17.5009-5015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mishina Y, Lee C-HJ, He C. Nucleic Acids Res. 2004;32:1548. doi: 10.1093/nar/gkh319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bratlie MS, Drablos F. BMC Genomics. 2005;6:1. doi: 10.1186/1471-2164-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mishina Y, He C. J. Am. Chem. Soc. 2003;125:8730. doi: 10.1021/ja034636c. [DOI] [PubMed] [Google Scholar]
- 171.Mishina Y, Yang C-G, He C. J. Am. Chem. Soc. 2005;127:14594. doi: 10.1021/ja055957m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Delaney JC, Smeester L, Wong C, Frick LE, Taghizadeh K, Wishnok JS, Drennan CL, Samson LD, Essigmann JM. Nat. Struct. Mol. Biol. 2005;12:855. doi: 10.1038/nsmb996. [DOI] [PubMed] [Google Scholar]
- 173.Landini P, Hajec LI, Volkert MR. J. Bacteriol. 1994;176:6583. doi: 10.1128/jb.176.21.6583-6589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]