Abstract
The plant pathogenic bacterium Erwinia chrysanthemi secretes pectate lyase proteins that are important virulence factors attacking the cell walls of plant hosts. Bacterial production of these enzymes is induced by the substrate polypectate-Na (NaPP) and further stimulated by the presence of plant extracts. The bacterial regulator responsible for induction by plant extracts was identified and purified by using a DNA-binding assay with the promoter region of pelE that encodes a major pectate lyase. A novel bacterial protein, called Pir, was isolated that produced a specific gel shift of the pelE promoter DNA, and the corresponding pir gene was cloned and sequenced. The Pir protein contains 272 amino acids with a molecular mass of 30 kDa and appears to function as a dimer. A homology search indicates that Pir belongs to the IclR family of transcriptional regulators. Pir bound to a 35-bp DNA sequence in the promoter region of pelE. This site overlaps that of a previously described negative regulator, KdgR. Gel shift experiments showed that the binding of either Pir or KdgR interfered with binding of the other protein.
Soft-rotting Erwinia species such as Erwinia chrysanthemi cause soft-rot diseases in a wide variety of host plants (1). The major virulence factors of these pathogens are pectate lyase (PL) enzymes that degrade the pectate fraction of the plant cell wall, a process classically called “maceration” (2). Several pel genes encoding these enzymes are induced by a metabolic product from the degradation of pectate [2-keto-3-deoxygluconate (KDG)], which inhibits the binding of a negative regulator protein, KdgR, at the KdgR-box in the promoter region (3, 4). This mechanism explains at least in part the induction of PL in the presence of pectic substances, a major component of the plant cell wall. Synthesis of PL also is affected by various environmental factors such as cell density (5), temperature, nitrogen starvation, oxygen concentration, osmolarity, the presence of rapidly metabolizable sugars (6), iron concentration (7), and the presence of plant extracts (8). Several mechanisms accounting for regulation by these factors have been elucidated, and because most of them also regulate genes other than those encoding pectic enzymes, they are called global regulatory mechanisms (9–12).
Among environmental factors affecting the synthesis of PL, plant signals other than pectate products are important. For example, in E. chrysanthemi 3937, it was reported that PL synthesis is induced 230-fold higher than the basal level by adding plant extract together with polypectate-Na (NaPP) into the bacterial growth medium (only a 9-fold induction occurred with NaPP alone) (8). In this paper, we describe the isolation of a plant inducible regulatory (Pir) protein and cloning of its structural gene (pir) from E. chrysanthemi EC16. Mutation of pir resulted in the loss of PL hyperinduction in response to plant signals and reduced bacterial virulence on plant tissues, but did not affect the regulation of other extracellular enzymes such as cellulases (Cel) or proteases (Prt).
MATERIALS AND METHODS
Bacterial Strains and Growth Media.
Strains of Erwinia chrysanthemi and of Escherichia coli were grown at 27°C in YP medium (1% polypeptone/0.5% yeast extract, pH 6.8) and at 37°C in Luria–Bertani medium (1% polypeptone/0.5% yeast extract/1% NaCl, pH 7.0), respectively. M63 medium (13) supplemented with a carbon source (0.2%) was used as minimal medium for both bacterial genera. Antibiotics were added at the following concentrations: ampicillin (100 μg/ml), kanamycin (150 μg/ml), streptomycin (25 μg/ml), and gentamycin (15 μg/ml). A crude potato extract was prepared by centrifugation at 10,000 × g for 10 min of homogenized extract of potato by grater followed by filtration through 0.45-μm nitrocellulose filters (Kurabo, Osaka, Japan). One hundredth volume of this extract was added to minimal medium.
DNA-Binding Assay.
The DNA-binding assay was performed as described by Ausubel et al. (13) with minor modifications. DNA fragments were labeled with 100 μCi of [α-32P]dCTP (4,000 Ci/mmol; 1 Ci = 37 GBq) by end-filling the perturbed ends with Klenow fragment of DNA polymerase I. The labeled DNA fragments were purified by using the Qiagen quick extraction kit. The reaction mixture consisted of 10% glycerol, 1 μg poly(dI-dC)-(dI-dC) (Pharmacia), 2 μg of BSA, 30 fmol of labeled DNA probe (≈5 × 104 cpm) and binding protein in 10 μl of 25 mM Hepes-potassium hydroxide (pH 7.9) buffer containing 50 mM KCl, 0.1 mM EDTA (pH 8.0), 0.5 mM DTT, and 0.5 mM phenylmethylsulfonyl fluoride. After incubation for 15 min at 27°C, the mixture was loaded onto a 4% polyacrylamide gel (15 cm × 15 cm) in high ionic strength (50 mM Tris⋅HCl/380 mM glycine/2.1 mM EDTA, pH 8.3) and electrophoresed in same buffer for 2.5 h at 20 mA. The gel was then vacuum-dried and exposed to HP film (Amersham).
DNaseI Footprinting.
DNaseI footprinting was performed by using the method of Galas and Schmitz (14) with slight modification. The binding between protein and end-labeled DNA probe was carried out as done for DNA-binding assay. After incubation for 15 min at 27°C, DNaseI was added at the concentration of 1.5 10−1 mU/μl into the mixture and incubated for 2 min at 27°C. DNaseI digestion was stopped by adding the same volume (11 μl) of stop solution (100 mM EDTA, pH 8.0/200 μg/ml yeast tRNA). The volume of the reaction was brought to 100 μl with ice-cold TE buffer, pH 8.0. Then, DNA fragments were ethanol-precipitated after removal of proteins by extraction with phenol/chloroform. The pellet was redisolved in a dye mixture (0.05% bromophenol blue/0.05% xylen cyanol/0.01 mM EDTA, pH 8.0/95% formamide) and loaded onto a 6% polyacrylamide sequencing gel. Bands were detected by autoradiography onto HP film. Manual DNA sequencing was done with T7 polymerase sequencing kit (Pharmacia) and ran in the lanes next to those for footprinting.
Purification of Pir.
For purification of Pir, EC16 was grown in 5 liters minimal medium containing potato extract, NaPP, and glycerol, until the OD600 reached at 1.0. Cells were harvested by centrifugation at 4, 000 × g for 10 min, washed, and resuspended in 500 ml of buffer A (12 mM Hepes-KOH, pH 7.9/4 mM Tris⋅HCl, pH 7.9/0.1 mM EDTA, pH 8.0/0.5 mM phenylmethylsulfonyl fluoride/0.5 mM DTT/10% glycerol). Crude cell extracts were obtained by four times sonication of washed bacterial suspension by using Ultrasonic Disrupter UD-200 (Tommy, Tokyo, Japan) for 4 min in ice-cold condition. Then, sonicated crude extract was centrifuged at 12,000 × g for 30 min. The supernatant was fractionated by ammonium sulfate precipitation. The fraction of 25–40% saturation of ammonium sulfate was resuspended in 50 ml of buffer A and was dialyzed against 100 times vol of the same buffer for 16 h. After centrifugation and filtration of the dialysate through nitrocellulose filter (0.45 μm pore size), it was applied to a DEAE Sepharose Fast Flow column (16 mm × 100 mm, Pharmacia). The column was washed with buffer A and eluted with a linear gradient of KCl from 0 to 0.3 M at flow rate of 1.2 ml per min. The fractions were tested for binding activity by pelE-binding assay. Active fractions (eluted at ≈0.18 M of KCl) were pooled, diluted with equal volume of buffer A and applied to HEPARIN POROS column (4.6 mm × 100 mm, Boehringer). The column was washed with buffer A containing 0.1 M KCl and eluted with a linear gradient of KCl from 0.1 to 0.5 M at a flow rate of 0.6 ml per min. Active fractions identified by pelE-binding assay (eluted at ≈0.25 M) were pooled, diluted by adding equal volume of buffer A, and applied to a PI POROS column (4.6 mm × 100 mm, Boehringer). The column was equilibrated with buffer A containing 0.1 M KCl and eluted with a linear gradient of KCl from 0.1 to 1 M KCl at a flow rate of 0.6 ml/min. Active fractions (eluted at ≈0.45 M) were pooled and concentrated with Centricon 10 filtration device (Amicon). The treated solution was applied on Superdex 200HR 10/30 column (10 mm × 300 mm, Pharmacia) equilibrated with buffer A containing 0.15 M KCl. Elution with the same buffer was done at flow rate of 0.18 ml/min. Active fractions were pooled and concentrated with Centricon 10 filtration devices. The purity of the final preparation was checked by silver staining (Wako, Osaka, Japan) after SDS/PAGE. Purified Pir was stored at −20°C in buffer A containing 50% glycerol.
Protein Sequencing.
Purified Pir protein was visualized by staining with Coomassie Brilliant Blue R250 after SDS/PAGE. The protein in the gel was blotted onto a poly(vinylidene difluoride) membrane (PVDF, Amersham). The stained band on the membrane was cut and used for determination of the amino acid sequence from the N-terminal end by using an automatic protein sequencer (Applied Biosystems).
Recombinant DNA Techniques.
Preparation of total and plasmid DNA, restriction digestion, ligation, DNA electrophoresis, Southern and colony blot hybridization and electrophoresis were done as described by Sambrook et al. (15). Nucleotide sequence analysis was performed by DNA-Autosequencer (model 4000, Li-Cor, Lincoln, NE). Restriction and modifying enzymes were obtained from Nippon Gene (Toyama, Japan) and Pharmacia.
Introduction of Mutation into pir in EC16 by Marker-Exchange.
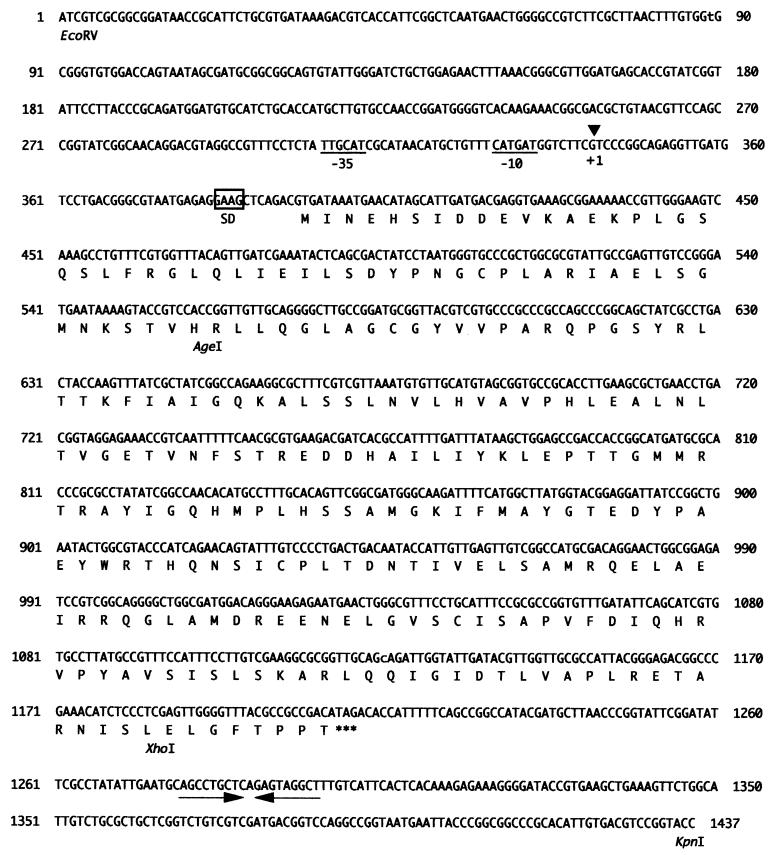
pir in EC16 was inactivated by insertion with both 2.1-kbp SmaI–SalI fragment containing promoter-less β-glucuronidase gene from pBI101 (16) and 1.3-kbp SalI fragment containing kanamycin resistance gene from pUC4K (Pharmacia) into between the AgeI site, which was blunted with Klenow fragment, and the XhoI site of pIEC-1 (Fig. 6) to construct pIEC-GK. This plasmid was introduced into E. chrysanthemi EC16 by electroporation using Cell-Porator (set at 9.4 kV/cm, 160 μF, and 4 ohms, Bethesda Research Laboratories). Transformants were selected on YP plate containing kanamycin and ampicillin. After transferring the culture of the tranformants 10 times in YP medium without antibiotics, marker-exchanged strains were selected as kanamycin resistant, ampicillin sensitive, and β-glucuronidase positive colonies. The marker-exchanged pir minus mutants (K2367) were confirmed by Southern analysis by using PIRO-2 (5′-dCAGGCTTTGACTTCCCAACGG-3′, complementary to the sequence from 458 to 438 nt in pir-coding region, Fig. 6) as the probe.
Figure 6.
Nucleotide sequence of the pir gene. Translational start site (GTG) and stop site (TAG) are shown at positions 393 nt and 1,209 nt, respectively. The possible −35 and −10 boxes of σ70 type promoter are underlined. The boxed sequence indicates a possible ribosome binding site. A possible ρ-independent transcriptional termination site is shown by inverted arrows.
Insertion of pelE Promoter-lux Fusion into Chromosome of E. chrysanthemi.
A 260-bp NdeI–MluI fragment containing promoter region of pelE (−150 nt to +110 nt, Fig. 1) was converted to blunt end by Klenow fragment and ligated into the SmaI site of pHSK728 (17), which contains the promoter-less lux cassette from Vibrio fischeri and IR of Tn7 so that the promoter-lux cassettes are mobile in the presence of the genes for Tn7 transposases (18). The direction of lux insert was confirmed by sequencing using PNL-2 (5′-dATCCACCTTGCCGAGGGACA-3′, from −9 to +11 of pelE promoter region, Fig. 1) as the primer. This construct was electroporated into EC16 or K2367 cells containing the helper plasmid pMON7181, which supplies Tn7 transposases. The transformants were selected by plating on YP plate containing gentamycin and streptomycin. After four times transferring the transformant in YP broth without antibiotics, the cell suspension was plated onto YP plate containing streptomycin. Each streptomycin colonies were tested for the loss of resistance to ampicillin and gentamycinr to isolate the strain inserted with pelE-lux construct.
Figure 1.
Sites of mutations in promoter region of pelE gene of E. chrysanthemi EC16. Numbering is based on the transcriptional start site determined by primer extension (data not shown). The translational start site is at +96 nt. The NdeI–MluI fragment (260 bp) was used for the binding assay and for the construction of lux fusion. The −35 and −10 regions of σ70 type promoter are underlined. Two KdgR boxes are double underlined and designated as operator-1 (op-1) and operator-2 (op-2). ED-OP-1 and ED-OP-2 are deletion mutants obtained by site-directed mutagenesis at these operators (23). Boxed sequence indicates the protect region by Pir from digestion with DNaseI (−1 to +34 nt). Protect region by KdgR from DNaseI was shown with overhead line (+6 to +55 nt).
Bioluminescence Assay.
Light production by a pelE-lux construct on the chromosome was measured 10 times for 10 sec every 1 h by Chemiluminescence Detector CLD-100 (Tohoku Electric, Sendai, Japan). The mean of these readings (cpm) in 1 ml was divided by OD600 were used as the specific luciferase activity (cpm/ml/OD600).
RESULTS
DNA-Binding Assay Using the Promoter Region of pelE.
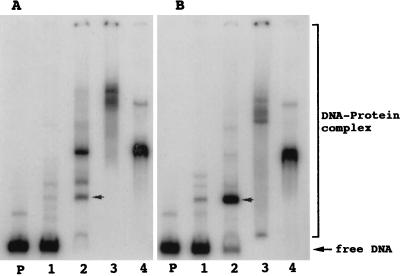
A 260-bp NdeI–MluI fragment containing the promoter region of pelE (−150 to +110 nt, Fig. 1) was used in binding assays with crude extracts from EC16 cells grown in the presence of NaPP and glycerol with or without potato extract. When the extract prepared after growth in the presence of potato extract in addition to NaPP and glycerol was used, the intensity of one of the shifted bands increased (arrow in Fig. 2 A and B) compared with when the extract from cells grown in the presence of NaPP and glycerol was used. The protein accounting for this shifted band was called Pir.
Figure 2.
DNA-binding assay using promoter region of pelE. NdeI–MluI fragment of pelE (Fig. 1) as the target DNA. Protein source was obtained by ammonium sulfate (AmS) fractionation of the sonicated extract of EC16 cells grown in minimal salts + glycerol + NaPP (A) and minimal salts + glycerol + NaPP + potato extract (B). Lanes: P, target DNA only; 1, DNA + 0–25% AmS fraction; 2, DNA + 25–40% AmS fraction; 3, DNA + 40–55% AmS fraction; 4, DNA + 55–80% AmS fraction.
Purification of Pir Protein.
Pir was purified by ammonium sulfate precipitation followed by chromatography through four different columns (DEAE Sepharose Fast Flow, PI POROS, HEPARIN POROS, and Superdex 200HR 10/30 columns). After these purification steps, a single 30-kDa band was observed on SDS/PAGE (Fig. 3). Because the molecular mass was estimated to be 60 kDa by gel filtration (data not shown), Pir probably exists as a dimer in the bacterial extracts.
Figure 3.
SDS/PAGE of active fractions during purification of Pir protein. Samples were load onto a SDS/PAGE and detected by silver staining. Lanes: M, dalton marker; 1, EC16 crude extract; 2, active fraction from DEAE Sepharose Fast Flow column; 3, active fraction from HEPARIN POROS column; 4, active fraction from Superdex 200HR 10/30 column.
Cloning of the Gene Encoding Pir.
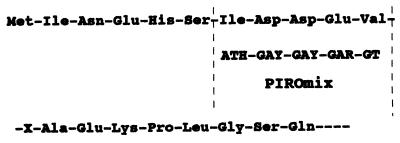
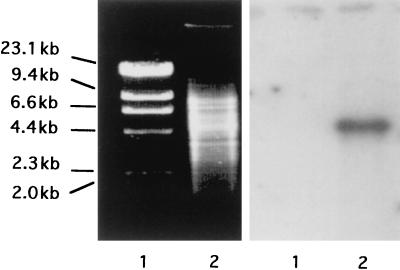
N-terminal amino acid sequence of purified Pir was determined, and mixed DNA oligomers (PIROmix) were designed (Fig. 4) and then used as the probe in colony hybridization. Genomic DNA of EC16 was shown to contain a single 4.5-kbp EcoRV fragment hybridizing with this probe (Fig. 5). By colony hybridization using PIROmix as the probe, five cosmid clones were obtained. The 4.5-kbp EcoRV fragment from one of the cosmid clones was cloned in pUC118 to generate pIEC-EV.
Figure 4.
Amino acid sequence of N-terminal end of Pir protein and designed mixture of oligomers (PIROmix) used as the probe for searching the clone of the structural gene for Pir. H, Y, and R indicated A + C + T mixture, C + T mixture, and A + G mixture, respectively.
Figure 5.
Southern blot analysis using PIROmix as the probe. Total DNA of EC16 after digestion with EcoRV was electrophoresed (Left), and PIROmix labeled with 32P-ATP was used as the probe (Right). Lanes: 1, HindIII-digested λ DNA (size markers); 2, EC16 total DNA.
DNA Sequencing of Region Containing pir.
The nucleotide sequence of a hybridizing subclone, pIEC-1, containing a 1.45-kbp EcoRV–KpnI fragment from pIEC-EV was determined (Fig. 6). An ORF was found from 393 nt to 1208 nt of this fragment. This ORF was predicted to encode a protein of 272 amino acid residues with a molecular mass of 30.1 kDa and an isoelectric point of 6.18. Because the molecular mass was close to that of the purified Pir protein and the N-terminal amino acid sequence of this ORF was identical to Pir (Fig. 4), it was concluded to be the pir gene. A potential ribosome binding site (SD sequence, AGGA) was found 8 nt upstream of the putative translational start site (GTG). The transcriptional start site was determined by primer extension analysis to be the guanine residue at 52 nt upstream from the translational start site (data not shown). A possible σ70 promoter, TTGCAT in the −35 region and CATGAT in the −10 region, was found at position 306–311 nt and 329–334 nt of this fragment, respectively. A possible ρ-independent transcriptional termination site was observed 76 nt downstream from the translational stop site (TAG). Homology search for the deduced amino acid sequence (Fig. 6) by using GenBank database indicated that the Pir protein showed high homology to the YiaJ protein (71.4% identity), a hypothetical transcriptional regulator in E. coli (ref. 19 and GenBank nucleotide sequence database accession no. AJ223475), which belongs to the IclR family (20). Pir also showed 29.4% identity in 254 aa residues to KdgR of E. chrysanthemi, the major negative regulator of PL synthesis and thought to belong to the IclR family (21).
Molecular Mass of Pir Determined by pelE-Binding Assay.
When purified Pir protein after overproduction was used for binding assays by using the promoter region of pelE, a single shifted band was observed at Pir concentrations between 30 and 70 nM (Fig. 7A). The molecular mass of the band in this native PAGE was calculated to be ≈60 kDa by using the method of Bading (22). Because the molecular mass of the translational product was estimated to be ≈30 kDa (Fig. 3), Pir seems to bind to the promoter region of pelE as a dimer.
Figure 7.
DNA-binding assay using purified Pir or KdgR onto promoter region of pelE. Labeled NdeI–MluI fragment of promoter region of pelE (30 fmol) (free, lane P) was mixed with 30 nM (lane 1), 50 nM (lane 2), 70 nM (lane 3) of purified Pir (A), and with 50 nM (lane 1), 70 nM (lane 2), 90 nM (lane 3) of purified KdgR (B).
Binding Sites for Pir and KdgR.
DNaseI footprinting experiments were performed on the 260-bp NdeI–MluI fragment of pelE by using purified Pir and KdgR, an established negative regulator for pectic enzymes (4). The region from −1 to +34 nt from its transcriptional start site was protected by addition of Pir from digestion with DNaseI, whereas the region from +6 nt to +55 nt was protected by addition of KdgR (Fig. 1).
Previously, analyses of induction pattern of site directed mutants had predicted op-1 (from +9 to +25 nt) and op-2 (from +31 to +47 nt) (Fig. 1) as KdgR-boxes (23). Binding assays by using these mutant promoters as the target DNA and purified KdgR confirmed this conclusion (Fig. 7B). In this case, mutation in either of the two putative KdgR binding sites prevented binding of KdgR at the remaining binding site. In the case of Pir, mutation at op-1 (ED-OP-1) but not op-2 (ED-OP-2) resulted in loss of the shift (Fig. 7A). Thus, although the binding sites for Pir and KdgR are close to each other in the promoter region of pelE, they are distinguishable.
Competition Between KdgR and Pir at the Promoter Region of pelE.
Competition analysis between Pir and KdgR on the NdeI–MluI pelE promoter fragment was examined by the binding assay. When increasing concentrations of KdgR were added to a constant concentration of Pir, the intensity of the Pir-DNA band decreased and that of the KdgR-DNA complex increased (Fig. 8A). On the other hand, when increasing concentrations of Pir were added to a constant concentration of KdgR, the intensity of the KdgR-DNA band decreased and that of the Pir-DNA complex increased (Fig. 8B). This result suggests that the binding sites for Pir and KdgR overlap and that they compete for binding. Thus, in the hyperinducing condition (in the presence of NaPP and potato extract), release of KdgR from the KdgR-box caused by accumulation of KDG, a catabolic product of pectin, may enhance the binding of Pir. This may therefore partially account for hyperinduced transcription of pelE.
Figure 8.
Competition of binding between Pir and KdgR at promoter region of pelE. Purified Pir and KdgR are mixed with 32P-labeled NdeI–MluI fragment of pelE (30 fmol) before DNA-binding assay. (A) Increasing concentrations of KdgR (0–100 nM) were added to a constant concentration of Pir (50 nM). (B) Increasing concentrations of Pir (0–75 nM) were added to a constant concentration of KdgR (120 nM).
Induction Pattern of PLe in a pir Mutant.
The promoter region of pelE (from −150 to +110 nt) was cloned in front of the luxA-E cassette in plasmid pHSK728. The cassette was inserted into the chromosome of the EC16 wild-type bacteria and pir-deficient mutant, K2367. It was confirmed that in the wild type, expression of pelE-lux was inducible in the presence of NaPP and glycerol and it was hyperinducible by further addition of potato extract (Fig. 9). In K2367, however, hyperinduction in the presence of potato extract with NaPP and glycerol was not observed. When the total activity of PL was compared in EC16 and in K2367, hyperinduction of PL also was not observed in the pir-deficient mutant (data not shown). Thus, pir may be responsible for hyperinduction of not only PLe but also of other PL isozymes.
Figure 9.
pelE expression in the pir mutant. Light production by EC16 (A) and K2367 (B) carrying the pelE-lux on their chromosome were measured by using Chemiluminescence detector CLD 100 (Tohoku Electric, Sendai, Japan). The specific activity of pelE-lux was expressed as cpm/ml/OD600. Background values of this detector was about 103 cpm/ml/OD600. □, minimal salts + glycerol; ⋄, minimal salts + glycerol + NaPP; ○, minimal salts + glycerol + potato extract; ▵, minimal salts + glycerol + NaPP + potato extract. NaPP (2 g/l), potato extract (1%) were added into the minimal salts containing glycerol (2 g/l). All cultures were grown on shaker at 27°C.
When Cel, Prt and NaPP-degrading activities were compared between EC16 and K2367 grown under several conditions, Cel and Prt activity was identical between these strains under all tested growth conditions whereas hyperinduction of NaPP-degrading activity was observed only in the wild type. DNA-binding assays using the promoter region of PL genes (pelA-E) and pectin-catabolizing genes (ogl, kdgK) as the target DNA for purified Pir, shifted bands were observed. However when the celY and prtC genes were used as target DNA, no shifted bands were observed (unpublished results). Thus, Pir seems to be unique among the described regulators because it acts positively and appears to affect only the synthesis of pectate degrading and pectin catabolizing enzymes.
DISCUSSION
We have isolated a new regulatory protein that accounts for the hyperinduction of pectin catabolizing enzymes production by E. chrysanthemi cells in the presence of potato extract, NaPP, and glycerol. This protein, called Pir, appears to be a positive regulator of pelE gene expression for the following reasons: (i) mutation of pir no longer allows hyperinduction of pelE expression in the presence of potato extract, NaPP and glycerol; and (ii) enhanced binding of Pir at the promoter region of pelE was observed with extracts from cells grown in the hyperinduced state. As such, Pir appears to have considerable significance to the production of PLe by the bacteria. It is especially noteworthy that pir mutant bacteria were markedly reduced in virulence against potato, celery, and Chinese cabbage tissues when low concentrations of cells (≈106 cells/ml) were inoculated (data not shown). This result is consistent with the importance of pel genes, especially pelE, in E. chrysanthemi EC16 virulence (24) and further indicates that hyperinduction of these enzymes by Pir is required for maximal virulence.
In E. carotovora ssp. carotovora, aep genes activate the expression of several genes for extracellular enzymes (pel, peh, cel, and prt) in response to plant signals (10). Unlike the negative regulators PecS, PecT, and KdgR (9) in E. chrysanthemi, Pir behaves as a positive factor similar to Aep. Also unlike PecS, PecT, Aep, and RsmA (12), Pir is not a global regulator of several unrelated genes but appears to specifically regulate genes encoding pectinolysis genes.
KdgR is a well characterized negative regulator of pel and other genes in E. chrysanthemi and interacts with a defined sequence, called the KdgR-box (ref. 23 and Fig. 7B), called op-1 and op-2 in Fig. 1. Footprinting experiments (not shown) also have indicated that Pir binds a sequence that overlaps the KdgR-binding region in the pelE promoter (Fig. 1). However, Pir did not bind to the promoter region with a mutation in op-1, but normal binding occurred with a mutation in op-2. These results clearly distinguish the effects of Pir and KdgR. The overlapping binding sites for Pir and KdgR suggest that a significant part of PL activity in hyperinduction may be mediated through competition of its binding to the promoter by the KdgR regulator.
Pir exhibited 71.4% identity to the YiaJ protein of E. coli. YiaJ was recently identified from genome sequencing and is suspected to be a regulatory protein for expression of the yiaK-S operon for carbohydrate utilization (ref. 19 and GenBank nucleotide sequence database accession no. AJ223475). Pir also showed 29.4% identity to KdgR. YiaJ and KdgR belong to the IclR family, which has a conserved region in the C-terminal domain ([GA]-X(3)-[DS]-X(2)-E-X(6)-[CSA]-[LIVM]-[GSA]-X(2)-[LIVM]-[FYH]-[DN]) (20). Pir exactly contained this consensus region at amino acid position 204–225.
Mutation of pir resulted in a decrease of ≈100-fold in pelE-lux activity compared with that of wild-type EC16 when cells were grown in the presence of potato extract, NaPP, and glycerol (Fig. 9). This confirmed its role in hyperinduction. However, when cells were grown in minimal salts with glycerol or in minimal salts with NaPP and glycerol, the expression of pelE-lux in K2367 was ≈10-fold higher than in EC16. Therefore, Pir also may repress PL synthesis under these conditions.
There was an unique gel shift band that completely disappeared only when the extract from the culture grown in the presence of potato extract, NaPP, and glycerol was used in the DNA-binding assay (Fig. 2). Because this unique protein-DNA complex was detected even when mutants in the KdgR box (op-1 and op-2) of pelE were used as the target DNA (data not shown), this regulator should be distinguishable from KdgR. Further study on this apparently negative regulator protein should enhance understanding of the molecular basis of hyperinduction of pectate degradative and catabolic enzymes by the Pir protein in responding to plant extracts.
Acknowledgments
We are very grateful to Noel T. Keen at University of California, Riverside, for carefully reading the manuscript and for providing the lux fusion system, pET overexpression system, and cosmid vector. We also appreciate Y. Ichiwaki for providing the genomic library of EC16. This research was supported in part by a Scientific promotion grant from the Ministry of Education, Japan (09460026), by a grant in Plant Biotechnology from the Ministry of Agriculture, Forestry and Fisheries of Japan, and by a grant for Japan-France Exchange Program from the Japanese Society for the Promotion of Science (JSPS).
ABBREVIATIONS
- PL
pectate lyase
- NaPP
polypectate-Na
- KDG
2-keto-3-deoxygluconate
- Pir
plant inducible regulatory protein
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The nucleotide sequence data reported in this paper has been deposited in the DNA Data Bank of Japan (DDBJ)/European Molecular Biology Laboratory (EMBL)/GenBank nucleotide sequence databases (accession no. AB017637).
References
- 1.Perombelon M C M, Kelman A. Annu Rev Phytopathol. 1980;18:361–387. [Google Scholar]
- 2.Collmer A, Keen N T. Annu Rev Phytopathol. 1986;24:383–409. [Google Scholar]
- 3.Nasser W, Reverchon S, Robert-Baudouy J. Mol Microbiol. 1992;6:257–265. doi: 10.1111/j.1365-2958.1992.tb02007.x. [DOI] [PubMed] [Google Scholar]
- 4.Nasser W, Reverchon S, Condemine G, Robert-Baudouy J. J Mol Biol. 1994;236:427–440. doi: 10.1006/jmbi.1994.1155. [DOI] [PubMed] [Google Scholar]
- 5.Hogouvieux-Cotte-Pattat N, Reverchon S, Condemine G, Robert-Baudouy J. J Gen Microbiol. 1986;132:2099–2106. [Google Scholar]
- 6.Hogouvieux-Cotte-Pattat N, Dominguez H, Robert-Baudouy J. J Bacteriol. 1992;174:7807–7818. doi: 10.1128/jb.174.23.7807-7818.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sauvage C, Expert D. Mol Plant–Microbe Interact. 1994;7:71–77. doi: 10.1094/MPMI.1999.12.2.119. [DOI] [PubMed] [Google Scholar]
- 8.Bourson C, Favey S, Reverchon S, Robert-Baudouy J. J Gen Microbiol. 1993;139:1–9. doi: 10.1099/00221287-139-1-1. [DOI] [PubMed] [Google Scholar]
- 9.Hogouvieux-Cotte-Pattat N, Condemine G, Nasser W, Reverchon S. Annu Rev Microbiol. 1996;50:213–257. doi: 10.1146/annurev.micro.50.1.213. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Murata H, Chatterjee A, Chatterjee A K. Mol Plant–Microbe Interact. 1993;6:299–308. doi: 10.1094/mpmi-6-299. [DOI] [PubMed] [Google Scholar]
- 11.Murata H, Chatterjee A, Liu Y, Chatterjee A K. Appl Environ Microbiol. 1994;60:3150–3159. doi: 10.1128/aem.60.9.3150-3159.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatterjee A, Cui Y, Liu Y, Dumenyo C K, Chatterjee A K. Appl Environ Microbiol. 1995;61:1959–1967. doi: 10.1128/aem.61.5.1959-1967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ausbel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhi K. Current Protocols in Molecular Biology. New York: Wiley-Interscience; 1987. [Google Scholar]
- 14.Galas D J, Schmitz A. Nucleic Acids Res. 1981;5:3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 16.Jefferson R A, Kavanagh T A, Bevan M W. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen H, Gold S E, Tamaki S J, Keen N T. Gene. 1992;122:27–34. doi: 10.1016/0378-1119(92)90028-n. [DOI] [PubMed] [Google Scholar]
- 18.Barry G F. Gene. 1988;71:75–84. doi: 10.1016/0378-1119(88)90079-0. [DOI] [PubMed] [Google Scholar]
- 19.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 20.Sofia H J, Burland V, Daniels D L, Plunkett G, Blattner F R. Nucleic Acids Res. 1994;22:2576–2586. doi: 10.1093/nar/22.13.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reverchon S, Nasser W, Robert-Baudouy J. Mol Microbiol. 1991;5:2203–2216. doi: 10.1111/j.1365-2958.1991.tb02150.x. [DOI] [PubMed] [Google Scholar]
- 22.Bading H. Nucleic Acids Res. 1988;16:5241–5248. doi: 10.1093/nar/16.12.5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gold S, Nishio S, Tsuyumu S, Keen N T. Mol Plant–Microbe Interact. 1992;2:170–178. [PubMed] [Google Scholar]
- 24.Payne J H, Schoedel C, Keen N T, Collmer A. Appl Environ Microbiol. 1987;53:2315–2320. doi: 10.1128/aem.53.10.2315-2320.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]