Abstract
Cationic lipid-DNA (non-coding) complexes (CLDC) are activators of the innate immune response that increase survival of rodents with some acute viral infections and cancers. CLDC were evaluated for their ability to impact viral DNA levels in transgenic mice carrying an infectious clone of hepatitis B virus (HBV). Mice used in the studies were diet-restricted as nursing pups from solid food, because the expression of HBV DNA in the liver was increased above background levels in some mice with this restriction. Survival surgery was performed on these mice to obtain liver biopsies from which to select animals with suitable levels of liver HBV DNA for entry into the experimental protocols. Intravenous administration of 5 µg/mouse of CLDC on days 1, 7 and 13 reduced liver HBV DNA to similar low levels achieved with the positive control, adefovir dipivoxil. In a subsequent experiment, the same treatment schedule was used to determine that the minimal effective CLDC dose was between 0.5 to 0.05 µg/mouse. Selective cytokines were increased in the livers of CLDC- compared to placebo-treated mice in a dose-responsive manner. CLDC were effective in reducing liver HBV DNA and could be considered for further evaluation in other hepatitis models.
Introduction
Although the immune stimulatory properties of CpG oligonucleotides (ODN) and poly I:C are well-established, studies conducted over the past five years indicate that cationic lipid DNA complexes (CLDC) are more potent and effective inducers of type I and II interferons (Dow et al., 1999a, Dow et al., 1999b, Whitmore et al., 1999, Whitmore et al., 2001). The prevailing hypothesis to explain the innate immune enhancement observed with CLDC is that cationic liposomes facilitate endocytosis and direct delivery of the DNA to the endosomal compartment. The targeted delivery results in increased binding of nucleic acids to endosomally located TLRs, including TRL9, TLR7/8, and TLR3 molecules (Gursel et al., 2001), leading to enhanced innate immune activation and production of antiviral cytokines. However, the mechanism of action is not fully understood, and other pathways, such as a cytosolic DAI activator (Takaoka et al., 2007), may also be involved in up-regulation of innate immunity by CLDC.
The therapeutic potential of CLDC was first demonstrated in models of tumor immunotherapy (Dow et al., 1999b, Whitmore et al., 1999, Lanuti et al., 2000, Whitmore et al., 2001). The anti-tumor activity elicited by CLDC was found to be mediated primarily by IFN-γ released by activated NK cells, presumably in response to release of IL-12 and type I interferons.
The antiviral potential of CLDC immunotherapy has also been demonstrated in a lethal arbovirus infection model, using Punta Toro virus (PTV, Bunyaviridae, Phlebovirus) (Gowen et al., 2006). In these studies, treatment of mice with CLDC elicited high levels of IFN-α, IL-12, and IFN-γ production, and complete protective immunity when CLDC were administered by intraperitoneal (IP) injection 24h before or 24h after subcutaneous (SC) challenge with 10 × LD50 (2×104 PFU) PTV. Even low doses of CLDC (equivalent to 0.5 µg DNA per mouse) were protective by the IP route of administration. Intravenous (IV) administration of CLDC was even more potent in eliciting protective immunity, with DNA doses as low as 30 ng per mouse providing significant protection. Protection by both routes of CLDC administration was associated with significant suppression of viremia and virus replication in liver. Although the immunological mechanisms of protective immunity were not determined in these investigations, it was postulated that protection was mediated by type I IFNs. These studies, therefore, provided the rationale for investigating CLDC immunotherapy in other virus infection models.
Transgenic mouse lines (1.3.32 and 1.3.46) replicate HBV in the liver and produce measurable levels of HBV DNA in the serum comparable to levels found in chronically infected human patients (8). HBV DNA replicative intermediates (relaxed circular, double-stranded linear, and single-stranded) are identified in liver. Since these intermediates are normally produced in assembled virions as a part of the HBV life cycle, such data indicate that virus assembly is occurring in these mice. Indeed, Dane particles were identified in liver by electron microscopy (Guidotti et al., 1995). Infection of chimpanzees with the molecular clone that was used to generate the transgenic mice resulted in productive infection (Guidotti et al., 1999). HBV transgenic mice have proven valuable for evaluating therapeutic substances, such as adefovir dipivoxil (Julander et al., 2002), entecavir (Julander et al., 2003), lamivudine (Morrey et al., 1999), a dinucleoside analog (Iyer et al., 2004b), interleukin-12 (Cavanaugh et al., 1997), interleukin-18 (Kimura et al., 2002), Toll-like receptor ligands (Isogawa et al., 2005), and HBV-specific ribozyme (Morrissey et al., 2002). This study describes the therapeutic efficacy of CLDC measured by reduction in HBV DNA in the livers of transgenic mice. Interferon-α is an approved treatment for chronic HBV in humans; since CLDC induces interferon-α and other innate immune antiviral cytokines, this report may have relevance for the design of improved therapeutic regimens.
Materials and Methods
Animals
Homozygous female and male transgenic HBV mice were obtained from Dr. Frank Chisari (Scripps Research Institute, LaJolla, CA) (Guidotti et al., 1995) and were subsequently bred in the Biosafety Level 3 (BL-3) area of the AAALAC-accredited USU Laboratory Animal Research Center (LARC). The animals were derived from founder line 1.3.32. Animal use and care was in compliance with the Utah State University Institutional Animal Care and Use Committee.
CLDC
A sterile 10 mM solution of cationic liposomes composed of DOTIM [octadecenoyloxy (ethyl-2-heptadecenyl-3-hydroxyethyl) imidazolinium chloride] and cholesterol was prepared in a 1:1 molar ratio as previously described (Dow et al., 1999c, Gowen et al., 2006). Liposomes were extruded through a filter diameter of 200 nm rather than 100 nm. Prior to injection, cationic liposomes were gently mixed with plasmid DNA (pMB75.6 empty vector; 3 mg/mL) at a ratio of 16 nmol lipid per 1 µg DNA in 10% sucrose in water at room temperature. The plasmid DNA lacked a gene insert downstream of the HCMV promoter. The final plasmid DNA concentration was appropriate to deliver the dosage by injection in a 0.1 mL volume. Adefovir dipivoxil (ADV) was obtained from Gilead Sciences (Foster City, CA) as a positive control. The ADV was prepared in citric acid (0.05 M, pH 2.0), which was used to enhance the solubility of ADV in liquid suspension. It was administered by oral gavage in a 0.1 mL volume.
Liver HBV DNA assay
Liver tissue was homogenized in a lysis buffer immediately upon necropsy. The tissue (approximately 0.1 g) was ground with a well-fitted pestle in a microcentrifuge tube containing lysis buffer (1 mM EDTA, 10 mM Tris, 10 mM NaCl, 0.5% SDS, proteinase K). After incubation for 5 – 10 min at room temperature, the tubes were snap-frozen in liquid nitrogen for storage. For extraction of the DNA, the samples were incubated at 55°C for 2 to 4 hours. Samples were poured into Phaselock™ Gel tubes containing 150 µL water, 350 to 500 µL phenol. After shaking and centrifuging at 12,000 rpm for 15 m, the contents were poured into a second tube containing 350 to 500 µL chloroform. After centrifuging, the DNA was precipitated with 50 µL 5 M NaCl and 650 µL isopropanol and washed with 70% ethanol. The dried pellets were suspended in 500 µL TE buffer containing 1 µL RNase and incubated overnight at 55°C with occasional shaking. A specified volume of the DNA solution typically containing 40 µg was digested with HindIII enzyme (New England Biolabs, Beverly, MA) for an incubation period of 3 h at 37°C. HindIII has been shown not to cut within the HBV transgene sequence. Digested DNA was separated by electrophoresis utilizing a 1% TAE-buffered agarose gel at 80 V for 3–4 h. The DNA was then transferred to a BioDyneTM B positive-charged nylon membrane by alkaline transfer method with the following modifications: 1) the gel was soaked in 0.4 M NaOH for 15–30 min, 2) the nylon membrane was soaked in water followed by 0.4 M NaOH for 5 min. The sponge used for transfer was also saturated in 0.4 M NaOH. The treated gel was placed (well-opening-side down) upon absorbent paper on the sponge. Transfer occurred over a 3 h period, after which the gel was discarded, and the nylon membrane was washed in a solution of 0.2 M Tris (pH 7.6), 2x SSC, and 0.1% SDS. The membrane was baked for >30 min at 80°C and UV- fixed using UV StratalinkerTM 1800 (Stratagene, La Jolla, CA). Prior to hybridization, the filter was rinsed twice for 30 min in a neutralizing solution of 0.1x SSC and 0.1% SDS. Hybridization using a [32P] CTP-labeled, HBV genomic probe (digested with Hae III) cloned into the pBluescript plasmid (gift of Dr. Luca Guidotti, The Scripps Institute, LaJolla, CA) occurred overnight at 60°C in a solution of 10% PEG-8000, 0.05 M NaPO4, 0.21 mg/ml salmon sperm DNA, and 7% SDS.
The radioactive signals were measured using a Phosphor Imaging method (Optiquant). An image of the radioactive filter was exposed overnight to a Cyclone™ Storage Phosphor Screen (Perkin Elmer Multisensitive Medium, Type MS PPN 7001723). The exposed screen was transferred to the Cyclone™ drum and read using the 600 dpi setting. The ratio of the viral DNA bands to the transgene band was used to determine the concentration of viral DNA per host DNA. This calculation was based upon the knowledge that there were 1.3 copies of the transgene present per host cell with this line of transgenic mice (personal communication, F. Chisari). The transgene was used as an internal indicator to calculate the pg of HBV DNA per µg of homozygous cellular host DNA.
Liver HBcAg assay
For detection of hepatitis B core antigen (HBcAg), liver biopsies were first paraffin-embedded. The paraffin was then removed from the sections by using two 5 min treatments with xylene. Tissues were fixed with two 3 min treatments with 95% ethanol. Sections were treated with deionized water for 3 min, exposed to 3% hydrogen peroxide for 5 min, and Biotin-block (#X0590, Dako Corporation) for 5 min. The primary antibody, rabbit anti-HBcAg (1:100 dilution) (#B0586, Dako Corporation), goat anti-rabbit secondary antibody (#k684 Dako, LSAB Peroxidase Kit), strepavidin peroxidase (#K684 Dako, LSAB Peroxidase Kit), and substrate-chromogen solution (3-amino-9-ethylcarbazole, AEC) were added for durations of 30, 30, 10 and 10 min, respectively. Sections were counterstained with Mayer's hematoxylin before being mounted.
Three different parameters were obtained from each tissue section. The first two measurements are based on the observation that cells surrounding the central veins of the liver are more strongly stained than are other areas of the liver (personal observation), and that a drug administered intraperitoneally should have ready access to the liver. The first two parameters were obtained from counting cells surrounding central veins as follows. The total number of cells, the number of cells with stained nuclei, and the number of cells with stained cytoplasms were counted around central veins. The stained nuclei counts or the stained cytoplasm counts were divided by the total cells. Three central vein areas were counted with each slide sample. For the third parameter, a field, not in a central vein area, was counted for the total number of stained nuclei. One quarter of the field was counted. Three such fields were counted per liver section. The identity of the samples were blinded to the person reading the slides.
Serrum HBeAg
Whole blood samples were obtained during necropsy by cardiac puncture. The whole blood was centrifuged for the collection of the serum component. Ten microliters of serum was then next diluted into 90 µL of negative control serum, resulting in a 1:10 dilution of each sample. Along with a serial dilution of a positive control and a calibrator, these samples were run on an HBeAg-specific ELISA (International Immuno Diagnostics, Foster City CA) per the manufacturer’s instructions. Using the known PEI units for the Calibrator, PEI units were formulated for the serial dilutions of the positive serum. A graph was generated and extrapolation was used to assign a PEI unit value for each sample with a high degree of confidence (R2 value of 0.9816). The ELISA manufactures cut-off was utilized.
Serum HBsAg
The same serum samples collected and prepared for the HBeAg ELISA were also used for the HBsAg ELISA. Fifteen microliters of serum were diluted in the same manner and ran on an HBsAg specific ELISA (International Immuno Diagnostics, Foster City CA) per the manufacturer’s instructions. Interpretations were then made from both the manufacture’s cut-off chart (with blank correction) and the manufacturer’s cut-off formulae (without blank correction).
Cytokine array – serum
The same serum samples collected and prepared for the HBeAg and HBsAg ELISAs were also used for the serum cytokine assay. Five microliters of serum were diluted into 120 µL of sample dilution buffer, resulting in a 1:25 dilution of each sample. Thirty microliters of this prepared sample was then run on a Q-Plex™ Mouse Cytokine Array ( Quansys Biosciences, Logan, UT) per the manufacturer’s instructions. The fully developed cytokine plate was then captured as a .tif image on a Fuji LAS-3000 Luminescent Image Analyzer (Fuji Life Sciences, Stamford, CT) and analyzed with Quansys Array Software™, version 1.3. Levels of serum IFN-α, IFN-γ, IL-6, and IL-10 were also determined with individual kits, not arrays (eBioSciences, Inc., San Diego, CA).
Cytokine array – liver
Liver punches (approximately 35 mg each) were collected during necropsy and quickly homogenized in sterile PBS containing 0.1% NP-40. These homogenized sample were then snap-frozen in liquid nitrogen until the assay was performed. Just prior to performing the assay, the sample were rapidly thawed and centrifuged at 3000 rpm’s for 20 minutes to remove any solid matter. The supernatant was then diluted 1:5 into sample dilution buffer and run on a Q-Plex™ Mouse Cytokine Array (Quansys Biosciences, Logan, UT) per the manufacturer’s instructions. The fully developed cytokine plate was then captured as a .tif image on a Fuji LAS-3000 Luminescent Image Analyzer (Fuji Life Sciences, Stamford, CT) and analyzed with Quansys Array Software™, version 1.3.
Serum chemistry panel
A VetScan® Chemistry, Electrolyte and T4 Analyzer, specifically designed for veterinary medicine, was used in our BL-2 laboratory to process samples for the “comprehensive diagnostic profile,” which consists of ALT, BUN, creatinine, total bilirubin, albumin, alkaline phosphatase, globulin, glucose, Na+, K+, phosphorous, total protein. Protocols for the instrument were used.
Liver HBV RNA
Real-time RT-PCR was used to assay HBV-specific RNA in liver biopsies. Total RNA from tissues was extracted using Trizol™ reagent. Primer-pairs (HBV3 forward ATAAAACGCCGCAGACACATC, HBV3 reverse AACCTCCAATCACTCACCAACC) and HBV3 Taq-man probe [6~FAM]-AGCGATAACCAGGACAAGTTGGAGGACA-[BHQ1a~6FAM] were used. A second primer-pair (HBV4 forward GGACAAACGGGCAACATACCT, HBV4 reverse TCTTCCTCTTCATCCTGCCTGCT) and HBV4 Taqman probe [6~FAM]TCCAGAAAGAACCAACAAGAAGATGAGGCA[BHQ1a~6FAM] was used, without a noticeable difference between the two sets, so the HBV3 probe/primer set was used. A duplex reaction was done with the internal control, mouse GAPDH primers/probe. The primers and probe were the forward GCATCTTGGGCTACACTGAGG, reverse GAAGGTGGAAGAGTGGGAGTTG, and probe [5~HEX]-ACCAGGTTGTCTCCTGCGACTTCAACAG-[BHQ1a~5HEX]. The one-step FullVelocity™ QRT-PCR Master Mix (Stratagene, La Jolla, CA) was used for RT and amplification of HBV RNA and mouse GAPDH with primers and probe at a final concentration of 0.1 µM. Two microliter of total cellular RNA, extracted from infected or control tissues was used. Samples were run on a DNA Engine Opticon 2 (MJ Research Inc, Waltham, MA). A 25 µL reaction consists of 12.5 µL FullVelocity™ QPCR Master Mix, 0.375 ul dilute reference dye (1:500), 0.25 ul Stratascript™ RT/RNase Block Enzyme Mixture, and 0.5 ul FullVelocity™ Enzyme. The reaction contained 0.25 µL of both HBV-primers, 0.25 µL of both GAPDH-primers, and 0.25 µL of both probes, all having a stock concentration of 10 µM. Reverse transcription of cellular RNA was performed for 30 min at 50°C followed by PCR, which consisted of 1 cycle of 2 minutes at 95°C, then 40 cycles of 10 sec at 95°C and 30 sec at 60°C. The assay was run with a series of 10-fold dilutions of pooled liver RNA from HBV transgenic mice to obtain a standard curve. The y axis was the log dilutions of the standard, and the x axis was the C(t) values. R2 values were used to measure the quality of the curve, which was always above 0.098. Mean C(t) values were obtained for duplicates of each sample. The mean C(t) values of each sample were used to obtain the log relative RNA value using a formula of the fit line of the standard curve.
Liver biopsies
The fur of ketamine and xylazine-anesthetized mice was clipped from the abdomen and scrubbed with Betadyne followed by alcohol. Within a sterile field and using sterile instruments, the ventral skin was cut longitudinally, followed by the peritoneum. While holding the liver with forceps, about half of the right lobe was cut with Teflon-coated scissors that had been heated in a bead-sterilizer at 250°C. The scissors immediately cauterized the liver. As quickly as possible, a second person slid ambient-temperature forceps along both sides of the scissors to easily separate the liver and biopsy from the hot scissors. The biopsy was processed as above for Southern blot hybridization. The peritoneum was closed with one suture (3.0 Vicryl) and the skin was joined with wound clips. The wound was saturated with Betadyne™.
Diet restriction experiments
Pups up to 3 weeks of age were restricted from solid food provided to the dam by placing the food in a solid tray at the top of the cage that could only be accessed by the dam. The small amount of food dropped by the dam into the bottom of the cage was not controlled. Pups that were raised without this diet restriction had access to solid food placed in the bottom of the cages.
CLDC experimental design
CLDC was administered IV on days 1, 7, and 13 wherein day 1 was the first day of the experiment. ADV was administered once daily for 14 days by oral gavage. The day after the last CLDC treatment and 4 hours after the last ADV treatment, necropsy was initiated. Mice were euthanized to collect tissues. Whole body weight was measured one day before initiation of treatments and on day 14 during necropsy. A dose of 5 µg DNA/mouse or serial doses of 5, 0.5, 0.05, or 0.005 µg DNA/mouse were used.
Statistical analysis
Either a one-way analysis of variance with a Newman-Keuls Multiple Comparison test or an unpaired t test was used with Prism 4 (GraphPad Software, Inc.).
Results
Liver biopsies to select mice for entry into experiments
To screen the liver HBV DNA for entry of mice into experiments, liver biopsies were obtained. The surgical procedure involving cauterization of the liver with hot scissors was successful in obtaining as much as half of the right lobe and in preventing death of mice. At 12 days after biopsy surgery, gross and microscopic examinations were done. The surgical site appeared to have healed normally. A thin pale band (1–2 mm wide) of tissue was noted on the biopsied liver lobe, which was immediately adjacent to the initial site of the biopsy. The remainder of the liver showed no gross abnormalities or abnormal histology from a hematoxylin/eosin stained section. The remainder of the abdomen was unremarkable upon gross examination. The serum ALT value was normal. Based on this, only mice recovered >3 weeks after surgery were used in experiments.
Diet restriction of pups
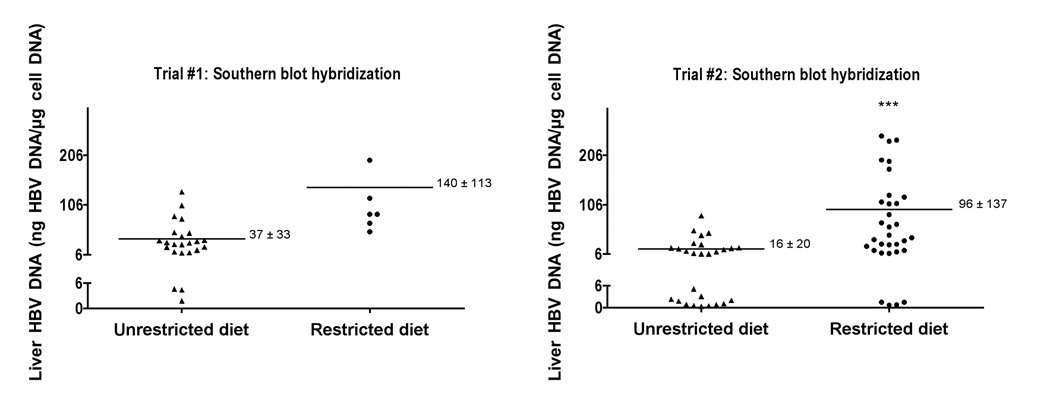
Since the liver HBV DNA is expressed after 3 weeks of age (Guidotti et al., 1995), we tested the hypothesis that early consumption of solid food by pups might affect liver HBV DNA expression. While solid food was provided to the dam, pups up to 3 weeks of age were restricted from solid food by placing the food in a solid tray at the top of the cage that could only be accessed by the dam. Pups that were raised without this diet restriction had access to solid food placed in the bottom of the cages. The livers of these mice were biopsied greater than 8 weeks of age and assayed for HBV DNA (Figure 1). In trial #1, the mean liver HBV DNA titer from mice that were restricted from solid food as pups was 140 ng /µg ± 113 as compared to 37 ng /µg ± 33 with an unrestricted diet. Since this difference was not statistically significant and since the numbers were low in the restricted diet group, trial #2 was conducted with larger numbers of animals. In this trial, the mean liver HBV DNA titer from mice that were restricted from solid food as pups was 96 ng /µg ± 137 as compared to 16 ng /µg ± 20 (p <0.001). Eleven of the twenty-seven samples (40.7%) from unrestricted-diet mice were at or below the limit of HBV DNA detection, as compared to 4/32 (12.5%) from restricted-diet mice. The restriction of diet did not eliminate the variability of DNA levels in positive mice, but it did reduce the proportion of mice at or below the limits of HBV DNA detection. Based on these results, transgenic mice used for CLDC experiments were restricted from solid food as pups and their liver biopsies were screened for adequate HBV DNA expression.
Figure 1.
Effect of restriction of solid food from <3 week-old pups on liver HBV DNA as adults (>8 weeks old). Two independent trials were conducted. Liver HBV DNA was assayed by Southern blot analysis. (***P ≤ 0.001 using unpaired t test).
CLDC treatment
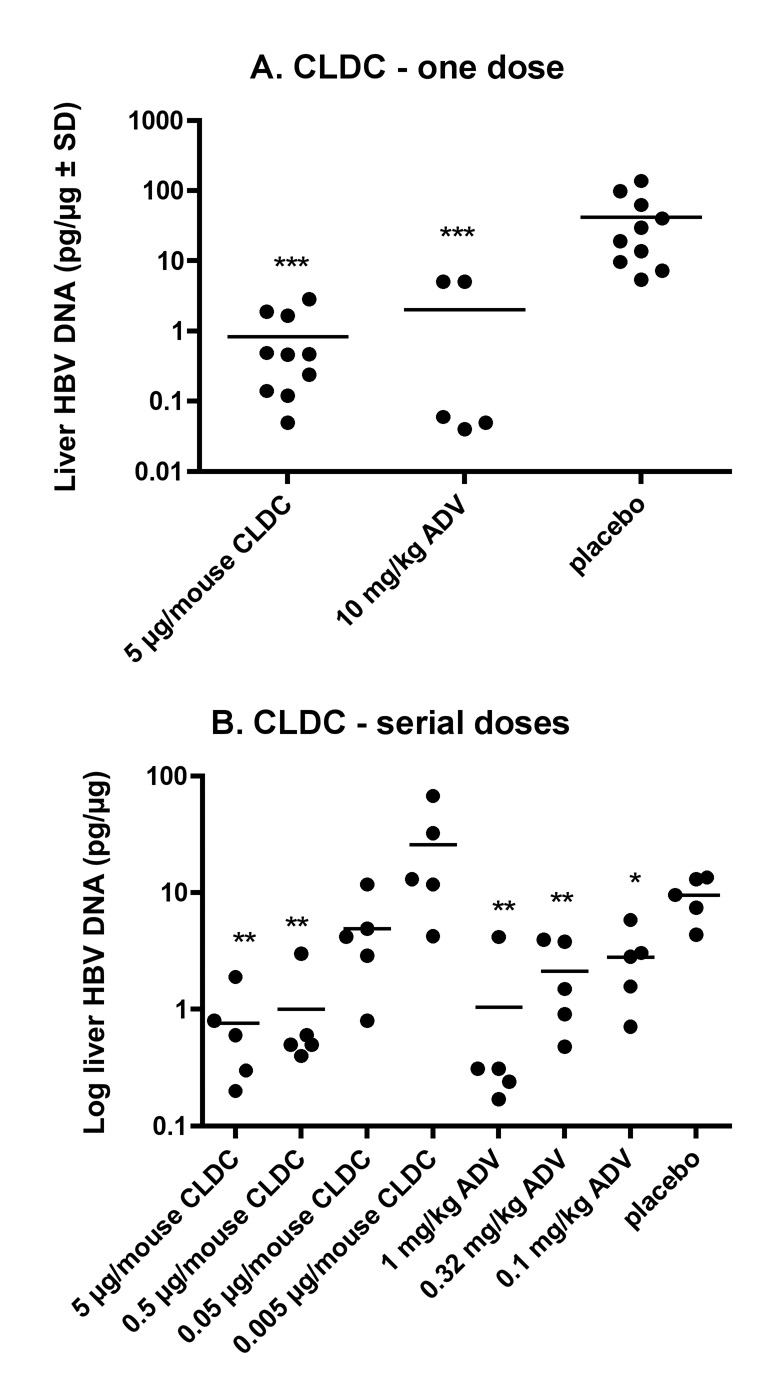
An IV treatment dose of 5 µg/mouse of CLDC on day 1, 7, and 13 IV was compared with ADV (10 mg/kg, once daily for 14 days) and the placebo; measurement of HBV DNA on day 14 was the primary outcome measure (Figure 2A). Both CLDC and ADV significantly reduced (P ≤ 0.001) liver HBV DNA. The values between the CLDC- and the ADV-treated animals were not statistically different. The liver HB core antigen or liver HBV RNA was not affected by CLDC or ADV treatment (data not shown).
Figure 2.
Effect of cationic liposome-DNA complexes (CLDC) on hepatitis B virus in transgenic mice. For both experiments (A, B), CLDC were administered IV on days 1, 7, and 13. ADV was administered once daily for 14 days by oral gavage. The day after the CLDC and 4 hours after the ADV treatment, necropsy was initiated. A.) A dose of 5 µg DNA/mouse or B.) serial doses of 5, 0.5, 0.05, or 0.005 µg DNA/mouse were used (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 compared to placebo using one-way analysis of variance with a Newman-Keuls Multiple Comparison test).
Since the dynamic range for measuring the liver HBV DNA in these transgenic mice was not large enough to compare the activities of CLDC and ADV, graded doses of each drug were evaluated to determine end-points of activity (Figure 2B). There was a dose-dependent decrease in liver HBV DNA. With only 5 animals per group, a statistically significant reduction in HBV DNA was observed in the 5 µg and 0.5 µg/mouse CLDC-dose groups (P ≤ 0.01). Even with the 0.05 µg/mouse-treated group, there was a decrease, but not with the 0.005 µg/mouse-treated group. The minimal effective dose, therefore, was between 0.5 and 0.05 µg CLDC/mouse. The minimal effective concentration for the positive control, ADV, was determined to be <0.1 mg/kg dosage, but we did not reach an ineffective dosage. None of the treatments statistically affected the liver HBV RNA levels, serum HBe or HBs antigens (data not shown).
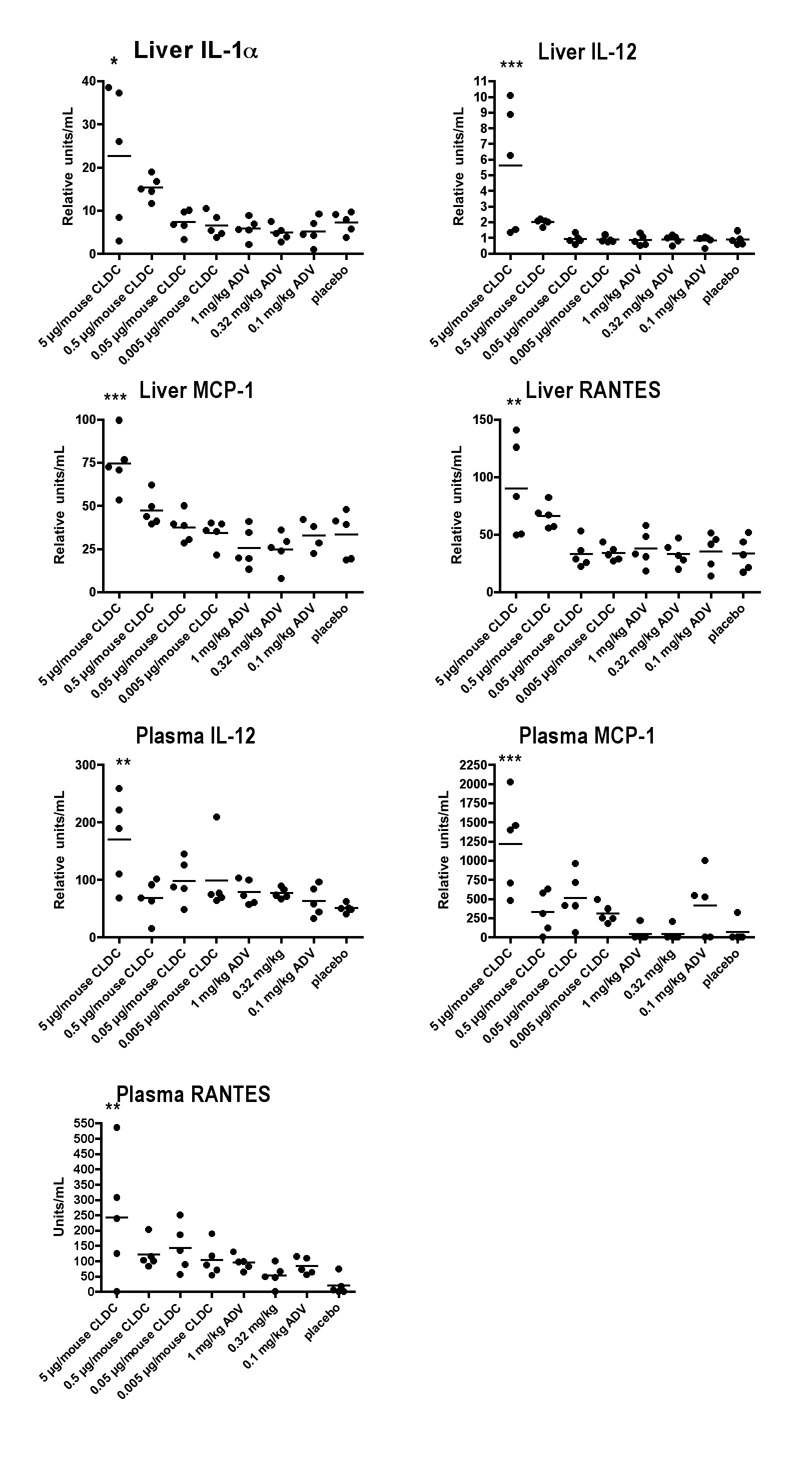
CLDC treatment increased the expression of some cytokines in liver homogenates and serum in a dose-dependent manner (Figure 3). From all of the cytokines in the array (IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12, MCP-1, TNF-α, MIP-1, GM-CSF, and RANTES), the inflammatory cytokines IL-1α, MCP-1, RANTES, and the TH1 cytokine IL-12 were statistically increased in the liver the day after the last treatment. IL-12, MCP-1 and RANTES were also statistically increased dose responsively in the serum.
Figure 3.
Effect of cationic liposome-DNA complexes (CLDC) on cytokine expression in the liver and serum of HBV transgenic mice. Treatment schedule and mice were the same as in Figure 2. The cytokine array included IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12, MCP-1, TNF-α, MIP-1, GM-CSF, and RANTES. Only those cytokines statistically affected by treatment are shown. (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 compared to placebo using one-way analysis of variance with a Newman-Keuls Multiple Comparison test).
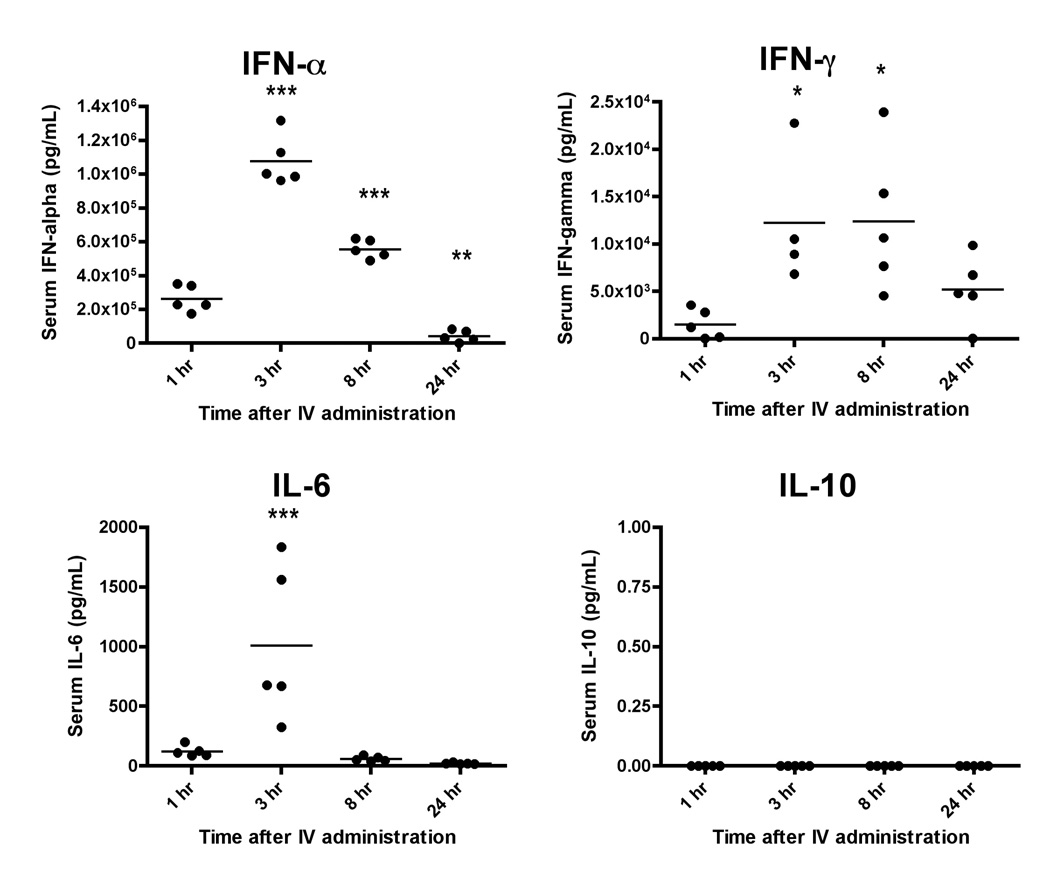
To better understand the temporal expression of the cytokines in response to CLDC, the serum of C57BL/6 mice at 1, 3, 8 or 24 hr after a single IV injection of CLDC (5 µg/mouse) was assayed for IFN-α, IFN-γ, IL-6, and IL-10 (Figure 4). All cytokines were increased with a peak at 3 hr after CLDC administration, except for IL-10, which was not affected. The levels of IFN-α and IL-6 reached baseline levels by 24 hr. IFN-γ levels were declining by 24 hr but had not yet reached baseline levels.
Figure 4.
Temporal serum cytokine expression in response to CLDC injected IV once into C57BL/6 mice at 1, 3, 8 and 24 hr with five mice per time period.
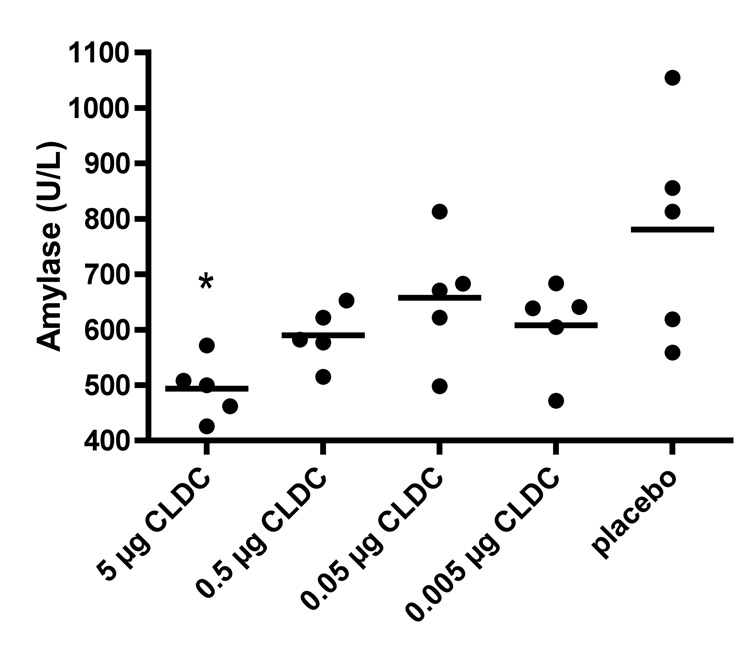
A chemistry panel was performed on serum samples. All serum chemistry values were normal, except for amylase being slightly reduced by CLDC treatment (P ≤ 0.05) (Figure 5).
Figure 5.
Effect of cationic liposome-DNA complexes (CLDC) on serum amylase. Treatment schedule and mice were the same as in Figure 2. The chemistry panel included albumin, alkaline phosphatase, alanine transferase, amylase, total bilirumin, blood-urea-nitrogen, calcium, phosphorous, creatinine, sodium, potassium, total protein, globulin, and glucose. Only amylase was possibly affected by CLDC treatment. (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 compared to placebo using one-way analysis of variance with a Newman-Keuls Multiple Comparison test).
Discussion
CLDC at 5 µg DNA/mouse administered intravenously on days 1, 7 and 13 reduced liver HBV DNA to similar low levels achieved with daily treatments of the positive control, adefovir dipivoxil. In a subsequent experiment, the same treatment schedule was use to determine that the minimal effective dose was between 0.5 to 0.05 µg/mouse. Liver HBV RNA, serum HBe and HBs, and liver HB core were not significantly affected by the CLDC treatment. The HBV RNA in the transgenic mice is produced primarily from an HBV transgene, unlike a natural infection wherein it is expressed from HBV covalently closed circular DNA (cccDNA) (Raney et al., 2001). Unlike the natural infection, HBV produced from the transgene in mice cannot infect mouse cells for successive rounds of viral replication. In the natural infection, HBV RNA and HBe and HBs proteins are derived from successive rounds of replication to produce increasing levels of cccDNA, and consequently, increasing levels of HBV RNA and proteins. In this regard the use of the HBV transgenic mice is analogous to HBV stably transfected, cells such as Hep G 2.2.15 cells (Iyer et al., 2004a). It was not unexpected, therefore, that a selective reduction of HBV DNA would not necessarily affect HBV RNA or protein levels, since these levels can be independently derived from the HBV RNA transcribed from the transgene.
The mechanism of action for CLDC in the HBV model is likely a result of cytokine induction and innate immunity, as it is for other viral diseases (Gowen et al., 2006). Moreover, HBV DNA is reduced in transgenic mice upon treatment with interleukin-12 (Cavanaugh et al., 1997), interleukin-18 (Kimura et al., 2002), interferon (Guidotti et al., 1994), IL-2 (Guidotti et al., 1994), and Toll-like receptor ligands (Isogawa et al., 2005). Selective cytokines were increased in the livers and serum of CLDC- compared to placebo-treated mice in a dose-responsive manner when assayed on day 14, one day after the last CLDC administration. Inflammatory cytokines (IL-1α, MCP-1, RANTES) were increased in the liver, including a TH1 IL-12 response. Serum cytokines at 14 days did not appear to be as sensitive to the effects of CLDC since not all four cytokines elevated in the liver were elevated in serum; nevertheless, the increased serum IL-12, MCP-1 and RANTES reflected the increases in the liver.
Since transgenic mice are immuno-tolerant to HBV antigens, the CLDC mechanism for reduction of HBV DNA does not involve acquired immunity, such as the killing of infected hepatocytes by virus-specific cytotoxic T cells (Robek et al., 2007). Another known mechanism of viral clearance is the noncytolytic blockage of HBV DNA replication mediated by both IFN-α/β and IFN-γ and IFN-related protein IL28/IFN (Guidotti and Chisari, 1999). The cellular IFN response inhibits HBV DNA replication in hepatocytes without affecting expression of viral mRNA by inhibiting the assembly of viral pregenomic RNA-containing capsids, as demonstrated in cell culture and in transgenic mice (Wieland et al., 2000, Wieland et al., 2005). The antiviral effect of IFN in HBV transgenic mice requires proteasome activity, although this inhibition occurs independently of the IFN-inducible proteasome catalytic subunits, (LMP2, LMP7) (Robek et al., 2007). Induction of Toll-like receptors (TLR), such as TLR3, TLR4, TLR5, TLR7, and TLR9, but not TLR2, non-cytopathically inhibit HBV DNA in transgenic mice in an alpha/beta interferon-dependent manner (Isogawa et al., 2005). In this study, serum IFN-α and IFN-γ, in addition to IL-6, were significantly increased at 3 to 8 hr after IV administration of CLDC; therefore, CLDC may reduce HBV DNA by induction of TLRs or IFN responses.
Since the expression of HBV DNA is very low at birth in the liver of these transgenic mice but increases to varying levels by 3–4 weeks of age (Guidotti et al., 1995), we hypothesized that the time of weaning and consumption of solid food could elicit cellular development in the liver to express varying levels of HBV DNA. Indeed, liver HBV DNA levels in adult mice raised as pups with a restricted diet were statistically increased as compared to mice that had no restriction on their diets. The physiological basis for this is not known.
All serum chemistry values were normal, except for amylase being slightly reduced by CLDC treatment. Increased amylase is typically considered diagnostic, but lower levels of amylase with no other abnormalities in other clinical chemistry parameters, as observed here, is typically not considered diagnostic of pathology, although the reason for the decreased levels of amylase are not known.
Currently therapeutic strategies for chronic hepatitis B are limited in efficacy, the development of resistance, toxicity, the requirement for prolonged treatment, high cost and limited availability. Here we describe a new immunotherapeutic approach that deserves further study as a potential addition to the range of products available for treatment of this important disease.
Acknowledgements
Funding: HHSN266200500036C, Enteric and Hepatic Diseases, NIAID, NIH (JDM)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cavanaugh VJ, Guidotti LG, Chisari FV. Interleukin-12 inhibits hepatitis B virus replication in transgenic mice. Journal of Virology. 1997;71:3236–3243. doi: 10.1128/jvi.71.4.3236-3243.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow SW, Elmslie RE, Fradkin LG, Liggitt DH, Heath TD, Willson AP, Potter TA. Intravenous cytokine gene delivery by lipid-DNA complexes controls the growth of established lung metastases. Hum Gene Ther. 1999a;10:2961–2972. doi: 10.1089/10430349950016375. [DOI] [PubMed] [Google Scholar]
- Dow SW, Fradkin LG, Liggitt DH, Willson AP, Heath TD, Potter TA. Lipid-DNA complexes induce potent activation of innate immune responses and antitumor activity when administered intravenously. J Immunol. 1999b;163:1552–1561. [PubMed] [Google Scholar]
- Dow SW, Schwarze J, Heath TD, Potter TA, Gelfand EW. Systemic and local interferon gamma gene delivery to the lungs for treatment of allergen-induced airway hyperresponsiveness in mice. Hum Gene Ther. 1999c;10:1905–1914. doi: 10.1089/10430349950017266. [DOI] [PubMed] [Google Scholar]
- Gowen BB, Fairman J, Smee DF, Wong MH, Jung KH, Pace AM, Heiner ML, Bailey KW, Dow SW, Sidwell RW. Protective immunity against acute phleboviral infection elicited through immunostimulatory cationic liposome-DNA complexes. Antiviral Res. 2006;69:165–172. doi: 10.1016/j.antiviral.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Guidotti LG, Chisari FV. Cytokine-induced viral purging--role in viral pathogenesis. Current opinion in microbiology. 1999;2:388–391. doi: 10.1016/s1369-5274(99)80068-x. [DOI] [PubMed] [Google Scholar]
- Guidotti LG, Guilhot S, Chisari FV. Interleukin-2 and alpha/beta interferon down-regulate hepatitis B virus gene expression in vivo by tumor necrosis factor-dependent and -independent pathways. Journal of Virology. 1994;68:1265–1270. doi: 10.1128/jvi.68.3.1265-1270.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti LG, Matzke B, Schaller H, Chisari FV. High-level hepatitis B virus replication in transgenic mice. Journal of Virology. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti LG, Rochford R, Chung J, Shapiro M, Purcell R, Chisari FV. Viral clearance without destruction of infected cells during acute HBV infection. Science. 1999;5415:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- Gursel I, Gursel M, Ishii KJ, Klinman DM. Sterically stabilized cationic liposomes improve the uptake and immunostimulatory activity of CpG oligonucleotides. J Immunol. 2001;167:3324–3328. doi: 10.4049/jimmunol.167.6.3324. [DOI] [PubMed] [Google Scholar]
- Isogawa M, Robek MD, Furuichi Y, Chisari FV. Toll-like receptor signaling inhibits hepatitis B virus replication in vivo. J Virol. 2005;79:7269–7272. doi: 10.1128/JVI.79.11.7269-7272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer RP, Jin Y, Roland A, Morrey JD, Mounir S, Korba B. Phosphorothioate di-and tri-nucleotides as a novel class of anti-HBV agents. Antimicrobial Agents and Chemotherapy. 2004a;48:2199–2205. doi: 10.1128/AAC.48.6.2199-2205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer RP, Roland A, Jin Y, Mounir S, Korba B, Julander JG, Morrey JD. Anti-hepatitis B virus activity of ORI-9020, a novel phosphorothioate dinucleotide, in a transgenic mouse model. Antimicrobial Agents and Chemotherapy. 2004b;48:2318–2320. doi: 10.1128/AAC.48.6.2318-2320.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julander JG, Colonno RJ, Sidwell RW, Morrey JD. Characterization of antiviral activity of entecavir in transgenic mice expressing hepatitis B virus. Antiviral Research. 2003;59:155–161. doi: 10.1016/s0166-3542(03)00109-8. [DOI] [PubMed] [Google Scholar]
- Julander JG, Sidwell RW, Morrey JD. Characterizing antiviral activity of adefovir dipivoxil in transgenic mice expressing hepatitis B virus. Antiviral Research. 2002;55:27–40. doi: 10.1016/s0166-3542(01)00223-6. [DOI] [PubMed] [Google Scholar]
- Kimura K, Kakimi K, Wieland S, Guidotti LG, Chisari FV. Interleukin-18 inhibits hepatitis B virus replication in the livers of transgenic mice. J Virol. 2002;76:10702–10707. doi: 10.1128/JVI.76.21.10702-10707.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanuti M, Rudginsky S, Force SD, Lambright ES, Siders WM, Chang MY, Amin KM, Kaiser LR, Scheule RK, Albelda SM. Cationic lipid:bacterial DNA complexes elicit adaptive cellular immunity in murine intraperitoneal tumor models. Cancer research. 2000;60:2955–2963. [PubMed] [Google Scholar]
- Morrey JD, Bailey KW, Korba BE, Sidwell RW. Utilization of transgenic mice replicating high levels of hepatitis B virus for antiviral evaluation of lamivudine. Antiviral Research. 1999;42:97–108. doi: 10.1016/s0166-3542(99)00009-1. [DOI] [PubMed] [Google Scholar]
- Morrissey DV, Lee PA, Johnson DA, Overly SL, McSwiggen JA, Beigelman L, Mokler VR, Maloney L, Vargeese C, Bowman K, O'Brien JT, Shaffer CS, Conrad A, Schmid P, Morrey JD, Macejak DG, Pavco PA, Blatt LM. Characterization of nuclease-resistant ribozymes directed against hepatitis B virus RNA. J Viral Hepat. 2002;9:411–418. doi: 10.1046/j.1365-2893.2002.00383.x. [DOI] [PubMed] [Google Scholar]
- Raney AK, Eggers CM, Kline EF, Guidotti LG, Pontoglio M, Yaniv M, McLachlan A. Nuclear covalently closed circular viral genomic DNA in the liver of hepatocyte nuclear factor 1 alpha-null hepatitis B virus transgenic mice. Journal of Virology. 2001;75:2900–2911. doi: 10.1128/JVI.75.6.2900-2911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robek MD, Garcia ML, Boyd BS, Chisari FV. Role of immunoproteasome catalytic subunits in the immune response to hepatitis B virus. J Virol. 2007;81:483–491. doi: 10.1128/JVI.01779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Whitmore M, Li S, Huang L. LPD lipopolyplex initiates a potent cytokine response and inhibits tumor growth. Gene Ther. 1999;6:1867–1875. doi: 10.1038/sj.gt.3301026. [DOI] [PubMed] [Google Scholar]
- Whitmore MM, Li S, Falo L, Jr, Huang L. Systemic administration of LPD prepared with CpG oligonucleotides inhibits the growth of established pulmonary metastases by stimulating innate and acquired antitumor immune responses. Cancer Immunol Immunother. 2001;50:503–514. doi: 10.1007/s002620100227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland SF, Eustaquio A, Whitten-Bauer C, Boyd B, Chisari FV. Interferon prevents formation of replication-competent hepatitis B virus RNA-containing nucleocapsids. Proc Natl Acad Sci U S A. 2005;102:9913–9917. doi: 10.1073/pnas.0504273102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland SF, Guidotti LG, Chisari FV. Intrahepatic induction of alpha/beta interferon eliminates viral RNA-containing capsids in hepatitis B virus transgenic mice. J Virol. 2000;74:4165–4173. doi: 10.1128/jvi.74.9.4165-4173.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]