Abstract
AIMS
To investigate the influence of the CYP2C19*17 allele on the pharmacokinetics of omeprazole, a commonly used CYP2C19 probe drug, in healthy volunteers.
METHODS
In a single-dose pharmacokinetic study, 17 healthy White volunteers genotyped as either CYP2C19*17/*17 or CYP2C19*1/*1 received an oral dose of 40 mg of omeprazole. Plasma was sampled for up to 10 h postdose, followed by quantification of omeprazole, 5-hydroxy omeprazole and omeprazole sulphone by high-performance liquid chromatography.
RESULTS
The mean omeprazole AUC∞ of 1973 h nmol l−1 in CYP2C19*17/*17 subjects was 2.1-fold lower [95% confidence interval (CI) 1.1, 3.3] than in CYP2C19*1/*1 subjects (4151 h nmol l−1, P = 0.04). A similar trend was observed for the sulphone metabolite with the CYP2C19*17/*17 group having a mean AUC∞ of 1083 h nmol l−1, 3.1-fold lower (95% CI 1.2, 5.5) than the CYP2C19*1/*1 group (3343 h nmol l−1, P = 0.03). A pronounced correlation (r2 = 0.95, P < 0.0001) was seen in the intraindividual omeprazole AUC∞ and omeprazole sulphone AUC∞ values.
CONCLUSIONS
The pharmacokinetics of omeprazole and omeprazole sulphone differ significantly between homozygous CYP2C19*17 and CYP2C19*1 subjects. For clinically important drugs that are metabolized predominantly by CYP2C19, the CYP2C19*17 allele might be associated with subtherapeutic drug exposure.
WHAT IS ALREADY KNOWN ABOUT THIS SUBJECT
The only existing study of CYP2C19*17-associated alterations in drug pharmacokinetics was retrospective and compared probe drug metabolic ratios.
The CYP2C19*17 allele had been associated with a two- and fourfold decrease in omeprazole and S/R-mephenytoin metabolic ratios.
WHAT THIS STUDY ADDS
This study characterized the single-dose pharmacokinetics of omeprazole, along with the 5-hydroxy and sulphone metabolites, in CYP2C19*17/*17 and CYP2C19*1/*1 subjects.
The observed differences in omeprazole AUC∞ suggest that the CYP2C19*17 allele is an important explanatory factor behind individual cases of therapeutic failure.
Keywords: 5-hydroxyomeprazole, depression, human, omeprazole sulphone, pantoprazole, pharmacogenetic, pharmacokinetics, SSRIs, ulcer
Introduction
Cytochrome P450 2C19 (CYP2C19) is an important enzyme in the metabolism of many clinically important drugs, including proton pump inhibitors (PPIs), antidepressants such as citalopram, amitriptyline and clomipramine, along with carisoprodol, diazepam, flunitrazepam, proguanil, phenytoin and mephenytoin [1]. Several genetic polymorphisms within the CYP2C19 gene have been identified (cf. http://www.cypalleles.ki.se/) and the clinical implications of these polymorphisms have been the focus of much investigation. The first and most extensively described CYP2C19 polymorphic variants cause the poor metabolizer phenotype (PM). Of the 19 variant CYP2C19 alleles, CYP219*2 and CYP2C19*3, which encode for nonfunctional proteins, are responsible for the vast majority of PM phenotypes. The frequency of the PM phenotype varies significantly between populations, ranging from 2 to 5% in White and Black populations to 13–23% in Asian populations [1]. The impact of these polymorphisms on the efficacy of several commonly used drug classes is well established. For example, studies have demonstrated dramatic CYP2C19-associated differences in diazepam disposition, with systemic drug exposure (AUC) varying more than sixfold between individuals [2, 3]. A similar trend has been observed with certain tricyclic antidepressants and serotonin reuptake inhibitors such as citalopram with a near 50% reduction of systemic clearance observed in PM subjects [4, 5]. For the PPIs omeprazole, lansoprazole and pantoprazole, CYP2C19 genotype is responsible for considerable interindividual differences in pharmacokinetics, with AUC values varying from five- to 12-fold [6, 7]. Furthermore, a prospective study of Japanese patients administered the standard 20-mg omeprazole dose in combination with amoxicillin for the treatment of Helicobacter pylori and peptic ulcer has demonstrated a clear association between genotype and treatment success. Cure rates for H. pylori infection were 29% in extensive metabolizer (EM) subjects compared with 100% in PM subjects, with a similar trend in the rates of ulcer healing [8, 9].
In contrast to the defective genes, we were able recently to identify a novel CYP2C19 gene variant associated with a more rapid metabolism phenotype [10]. Two single nucleotide polymorphisms (SNPs) in the 5′-flanking region (−3402C→T and −806C→T) of the CYP2C19 gene, which constitute the CYP2C19*17 allele (cf. http://www.cypalleles.ki.se/cyp2c19.htm), have been strongly linked with increased CYP2C19-dependent drug metabolism. The CYP2C19*17 allele has a frequency of 18% in both Swedes and Ethiopians, but only 4% in Chinese populations [10]. Phenotypic analysis of Ethiopian populations using the CYP2C19 probe drug mephenytoin has revealed a fourfold difference in median S/R-mephenytoin ratio between subjects homozygous for CYP2C19*1 and CYP2C19*17. A similar twofold difference in metabolic ratio (MR) has been observed in Swedish populations using omeprazole as a probe drug [10]. Functional genomic studies indicated that the −806C→T mutation caused increased nuclear factor binding and increased reporter gene transcription in mouse transfection studies. These observations suggest that the pharmacokinetics and clinical efficacy of CYP2C19 substrate drugs could be significantly affected in individuals possessing the CYP2C19*17 allele. The aim of the present study was to determine the pharmacokinetic differences of omeprazole and its two major metabolites 5-hydroxyomeprazole and omeprazole sulphone in subjects homozygous for the CYP2C19*17 and CYP2C19*1 alleles.
Methods
Design
The study was registered in the European Clinical Trials Database (EudraCT No2005-004717-15). The protocol was approved by the Regional Ethics Committee and the Swedish Medical Products Agency. The study was conducted in accordance with Good Clinical Practices and the Helsinki declaration. Previously genotyped volunteers were recruited from a database of former study participants at the Human Pharmacology Unit in Clinical Pharmacology, Karolinska University Hospital, Huddinge. All of the CYP2C19*17/*17 subjects in the database (ten) were asked to participate and CYP2C19*1/*1 subjects were recruited in a random fashion blinded to previous phenotypic data. Sample size calculations were based on the primary outcome represented by differences in AUC between CYP2C19*1/*1 and CYP2C19*17/*17 using our prior estimate of a 40% difference in omeprazole AUC between groups [10]. It was estimated that a true difference of 40% could be detected, given a one-sided level of significance (α) of 0.05 and a power (β) of 80%, using six CYP2C19*17/*17 and 12 CYP2C19*1/*1 subjects. Twelve volunteers previously genotyped as CYP2C19*1/*1 were included, but only five CYP2C19*17/*17 volunteers could participate. The study was conducted as an open label, one-phase trial where the volunteers were given a single oral dose of 40 mg omeprazole as two 20-mg tablets (Losec Mups®, AstraZeneca, Sweden). Peripheral blood samples were drawn from an i.v. cannula in a forearm vein into 10 ml sodium heparin collection tubes prior to drug administration and at the following times after omeprazole ingestion: 0.5, 1, 1.5, 2, 3, 4, 6, 8 and 10 h. After centrifugation for 10 min at 1500 g, plasma was separated and stored at −20°C until analysis.
Subjects
After oral and written explanation of the study, subjects gave written informed consent. Prior to starting the study, blood was obtained for clinical chemistry and genotype confirmation along with urine for drug screening and urinalysis. No medications other than paracetamol were allowed within 2 weeks before omeprazole administration and all drugs were prohibited within 24 h of omeprazole administration. Additional exclusion criteria included the use of Saint John's wort within 2 months, foods containing poppy seeds within 3 days or foods and drinks containing grapefruit within 2 days of omeprazole administration. Female volunteers were required to abstain from hormonal contraceptives for a minimum of 3 weeks. Subjects were excluded based on clinically significant abnormal ECG, blood chemistry or urine analysis, urine drug detection, or a positive pregnancy test. The subjects' sex, age, weight and smoking status are presented in Table 1. The volunteers were fasted from 8 h before to 2 h after omeprazole administration.
Table 1.
Demographic characteristics of the subjects
| CYP2C19*17/*17 | CYP2C19*1/*1 | |
|---|---|---|
| Number of subjects | 5 | 11 |
| Sex (male/female) | 2/3 | 2/9 |
| Age (years) | 37 (24–45) | 33 (21–60) |
| Weight (kg) | 74 (63–107) | 69 (60–109) |
| Number of smokers | 0 | 1 |
Age and weight are given as medians with ranges in parentheses.
Genotyping
Subjects were genotyped for CYP2C19*17 using a novel nested polymerase chain reaction (PCR) approach to introduce an allele-specific restriction site for restriction fragment length polymorphism analysis. Briefly, DNA was obtained from peripheral blood using a commercially available kit (DNeasy; Qiagen, West Sussex, UK). A 473 base pair (bp) fragment containing the CYP2C19*17 –806C→T mutation site was PCR-amplified using: 12.5 ng genomic DNA, 0.6 U Taq DNA polymerase (ABgene Epsom, UK), 2.5 μl 10× Buffer IV, 2 mM MgCl2, 0.2 mM dNTPs and 0.4 μM 2C19-1 forward primer (5′GCCCTTAGCACCAAATTCTC) and 2C19-1 reverse primer (5′ATTTAACCCCCTAAAAAAACACG). The thermoprofile consisted of an initial denaturation step at 95°C for 1 min followed by 35 cycles of 95°C for 30 s, 52°C for 30 s and 72°C for 30 s, followed by 72°C for 7 min. A 143-bp fragment with an allele-specific NsiI restriction site was created with the following: 0.05 μl of the initial PCR amplification, 0.6 U Taq DNA polymerase (ABgene Epsom, UK), 2.5 μl 10× Buffer IV, 2 mM MgCl2, 0.2 mM dNTPs and 0.25 μM 2C19-2 forward primer (5′AAATTTGTGTCTTCTGT TCTCAATG) and 2C19-2 reverse primer (5′AGACCCTGG GAGAACAGGAC). The thermoprofile consisted of 95°C for 1 min followed by 25 cycles of 95°C for 30 s, 51°C for 30 s and 72°C for 30 s, followed by 72°C for 7 min. Restriction digestion of the resultant PCR product was performed at 37°C for 8 h and contained the following: 15 μl PCR reaction, 2.5 μl 10× NEB reaction buffer 3 and 0.8 μl NsiI (New England BioLabs, Ipswich, MA, USA). Agarose gel electrophoresis revealed fragments of either 143 bp or 116 bp for the CYP219*17 and CYP2C19*1 alleles, respectively. Subjects were genotyped for the CYP2C19*2 allele using a TaqMan Allelic Discrimination Assay (Applied Biosystems, Foster City, CA, USA) on an ABI PRISM 7700 according to the manufacturer's recommendations.
Chemicals
Omeprazole, 5-hydroxyomeprazole, omeprazole sulphone, and internal standard H 259/36 were supplied by AstraZeneca (Mölndal, Sweden). All stock solutions were prepared in 5% methanol in sodium carbonate buffer 0.1 M, pH 9.3 and stored at −20°C. Unless otherwise stated, all other reagents were purchased from commercial vendors and were of reagent/analytical grade.
Sample analysis
Omeprazole and the two metabolites, 5-hydroxyomeprazole and omeprazole sulphone, were quantified using a reversed-phase high-performance liquid chromatography (R-HPLC) method based on previously published methodologies [11–13]. Duplicate 100-μl plasma samples were each combined with 800 μl methylene chloride:acetonitrile (9 : 1, v/v) and 21 μl 250 mM disodium phosphate buffer (pH 9.7) containing 10 nmol of internal standard. For samples near the limit of quantification, 400 μl of plasma was combined with 1.4 ml methylene chloride:acetonitrile and 21 μl of internal standard. All samples were extracted for 10 min using a vortex mixer followed by centrifugation (2000 g, 10 min) and aspiration of the plasma/aqueous phase. The organic phase was transferred to a new tube and evaporated to dryness at 60°C. Samples were reconstituted in 30 μl of methanol followed by 100 μl of 10 mM disodium phosphate buffer (pH 9.3). To minimize potential particulate carryover to R-HPLC analysis, the reconstituted samples were filtered using a 0.2-μm microcentrifuge spin column (Ultrafree-MC; Millipore, Nödinge, Sweden) and 77 μl of the resultant filtrate injected into the R-HPLC system.
Chromatography was performed using a Varian Prostar R-HPLC system, consisting of a Model 240 solvent delivery module, Model 310 UV-Vis detector and a Model 410 autosampler, combined with a LiChrospher 60 RP-select B 125 × 4 mm, 5 μm particle, column and an identical 4 × 4 mm precolumn (Merck, Whitehouse Station, NJ, USA). The mobile phases were 62.5 mM ammonium acetate pH 7.0 (A) and acetonitrile (B). The solvent gradient was as follows: initially 80% A:20% B with a linear ramp to 55% A:45% B from 2 min to 18 min, a linear return to initial condition from 23 min to 27 min followed by a 5-min equilibration before injection of the next sample. The flow rate was 1 ml min−1. Absorbance was monitored at 302 nm and peak areas determined using the Varian Star version 5.51 software package. Retention times were 7, 11, 12 and 13 min for 5-hydroxyomeprazole, internal standard, omeprazole and the sulphone metabolite, respectively. Standard curves were generated from spiking drug-free plasma with omeprazole and the two metabolites in the concentration range of 10–2500 nmol l−1 combined with internal standard followed by extraction and quantification as described above. The limit of quantification for all three analytes was 10 nmol l−1. The intraday and interday coefficients of variation for omeprazole and the two major metabolites were <10% and <15%, respectively.
Pharmacokinetic and statistical analysis
Pharmacokinetic parameters were determined using noncompartmental analysis and WinNonlin version 5.1 (Pharsight, Mountain View, CA, USA). The area under the concentration–time curve to infinity was calculated using the linear trapezoid rule and extrapolation to infinity using the log-linear terminal elimination phase (λΖ). Total body clearance was calculated as CL = molar dose/AUC∞ without normalization for the fraction of the administered dose reaching the central compartment (bioavailability). Data are reported as mean and 95% confidence interval (95% CI). Statistical calculations were performed using Graphpad Prism and Microsoft Excel. The pharmacokinetic parameters for CYP2C19*17/*17 and CYP2C19*1/*1 subjects were compared using an unpaired two-tailed heteroscedastic t-test.
Results
Omeprazole kinetics in healthy subjects
All subjects successfully completed the study in accordance with the protocol, and demographic information is shown in Table 1. No statistically significant differences in age, weight or smoking status were observed between genotype groups (P > 0.5). One subject displayed extremely aberrant and drastically reduced absorption kinetics (222 nM Cmaxvs. group mean 2109 nM Cmax) with first detectable drug observed at 6 h post administration (240 min Tlagvs. group mean 28 min Tlag) and was therefore excluded from the study.
The pharmacokinetic parameters for each study group are shown in Table 2. The high sensitivity and resolution of the analytical method combined with the 10-h duration of blood sampling resulted in a very limited extrapolated area contribution to the area under the plasma–concentration time curve (AUC∞). The mean extrapolated area for omeprazole, 5-hydroxyomeprazole and omeprazole sulphone were 2%, 3% and 19% of the total AUC∞, respectively. The mean omeprazole AUC∞ of 1973 h nmol l−1 in CYP2C19*17/*17 subjects was 2.1-fold lower (95% CI 1.1, 3.3) than in CYP2C19*1/*1 subjects (4151 h nmol l−1, P = 0.04), as shown in Figure 1. A similar trend was observed with the sulphone metabolite, with the CYP2C19*17/*17 group having a mean AUC∞ of 1083 h nmol l−1, 3.1-fold lower (95% CI 1.2, 5.5) than the CYP2C19*1/*1 group (3343 h nmol l−1, P = 0.03). There was no statistical difference in AUC∞ values for the 5-hydroxy metabolite between the CYP2C19*17/*17 and CYP2C19*1/*1 groups (2989 vs. 3359 h nmol l−1, P = 0.33). Comparison of omeprazole AUC∞/5-hydroxyomeprazole AUC∞ ratios also demonstrated a statistically significant difference between CYP2C19*17/*17 and CYP2C19*1/*1 groups (0.66 vs. 1.2, P = 0.04). Comparison of intraindividual omeprazole and metabolite AUC∞ values yielded an excellent correlation with omeprazole sulphone (r2 = 0.95), but a significantly weaker correlation with 5-hydroxyomeprazole (r2 = 0.51) (Figure 2).
Table 2.
Pharmacokinetic parameters of omeprazole, 5-hydroxyomeprazole and the sulphone metabolite after a single oral 40-mg dose of omeprazole to 16 healthy volunteers
| Units | CYP2C19*1/*1 Mean (95% CI) | CYP2C19*17/*17 Mean (95% CI) | P-value | |
|---|---|---|---|---|
| Omeprazole | ||||
| AUC∞ | nmol h l−1 | 4151 (2084, 6218) | 1973 (1476, 2469) | 0.04* |
| CL | l h−1 | 48 (22, 75) | 61 (44, 77) | 0.37† |
| Tmax | h | 2.1 (1.2, 3.1) | 2.1 (1.1, 3.1) | 0.95 |
| Cmax | nmol l−1 | 2109 (1169, 3049) | 1447 (1069, 1825) | 0.16 |
| λΖt1/2 | h | 1.2 (0.9, 1.5) | 0.9 (0.6, 1.2) | 0.18 |
| 5-Hydroxyomeprazole | ||||
| AUC∞ | nmol h l−1 | 3359 (2803, 3915) | 2989 (2273, 3706) | 0.33 |
| Tmax | h | 2.4 (1.2, 3.7) | 2.1 (1.1, 3.1) | 0.65 |
| Cmax | nmol l−1 | 1534 (1081, 1988) | 1527 (1234, 1819) | 0.97 |
| λΖt1/2 | h | 1.3 (0.9, 1.6) | 1.1 (1.0, 1.3) | 0.33 |
| Omeprazole sulphone | ||||
| AUC∞ | nmol h l−1 | 3343 (1301, 5384) | 1083 (690, 1476) | 0.03* |
| Tmax | h | 2.6 (1.4, 3.8) | 2.2 (1.3, 3.1) | 0.54 |
| Cmax | nmol l−1 | 563 (362, 765) | 336 (239, 433) | 0.04* |
| λΖt1/2 | h | 3.3 (1.7, 4.8) | 1.9 (1.4, 2.4) | 0.09 |
| OME AUC∞/5-OH AUC∞ | 1.2 (0.70, 1.6) | 0.66 (0.54, 0.79) | 0.04* | |
The lack of statistical significance, despite significant differences in AUC∞ values, is due to the transformation of AUC∞ values inherent in the calculation of clearance, CL = dose
(1/AUC∞). CYP2C19*17/*17 (n = 5) and CYP2C19*1/*1 (n = 11) groups were compared with an unpaired two-tailed heteroscedastic t-test. Asterisks denote P-values < 0.05. Values are means with 95% confidence interval in parentheses.
Figure 1.
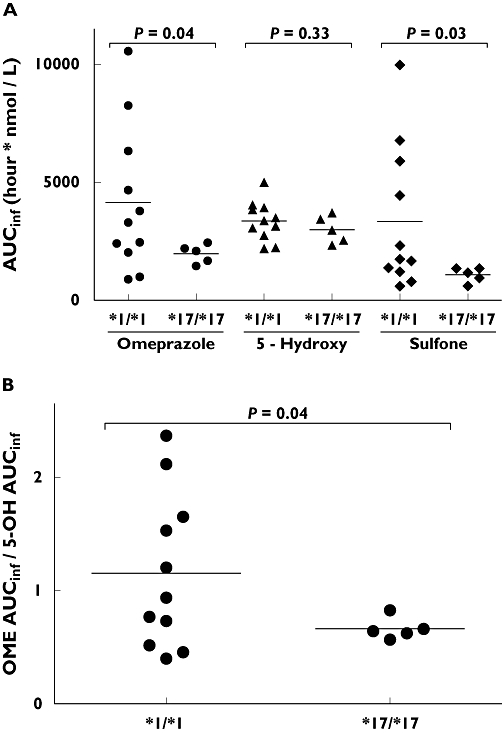
(A) Distribution of area under the concentration–time curve (AUC∞) values for omeprazole, 5-hydroxyomeprazole (5-hydroxy) and omeprazole sulphone (sulphone) in relation to CYP2C19*17/*17 (n = 5) and CYP2C19*1/*1 (n = 11) genotype. (B) Distribution of the ratio of area under the concentration–time curves of omeprazole (OME AUC∞) and 5-hydroxy omeprazole (5-OH AUC∞) in relation to CYP2C19*17/*17 (n = 5) and CYP2C19*1/*1 (n = 11) genotype. Bars represent mean values. Statistical analyses were performed with an unpaired two-tailed heteroscedastic t-test
Figure 2.
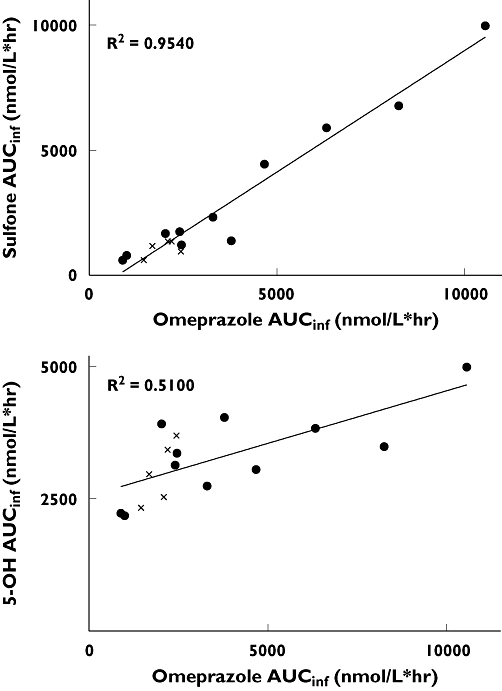
Correlation of intraindividual omeprazole AUC∞ and metabolite AUC∞ values in CYP2C19*17/*17 (,n = 5) and CYP2C19*1/*1 (•, n = 11) subjects. (A) Relationship between omeprazole AUC∞ and omeprazole sulphone (Sulphone) AUC∞. Linear regression yields r2 = 0.9540 (P < 0.0001). (B) Relationship between omeprazole AUC∞ and 5-hydroxy omeprazole (5-OH) AUC∞. Linear regression yields r2 = 0.5100
Omeprazole and omeprazole sulphone Tmax, Cmax and λΖt1/2-values demonstrate a consistent trend towards an increased rate of elimination in the CYP2C19*17 group; however, only the omeprazole sulphone Cmax reached the level of statistical significance (Table 2). Conversely, there were few if any genotype-associated differences in the pharmacokinetics of the 5-hydroxy metabolite.
Discussion
Our results indicate that individuals homozygous for the CYP2C19*17 allele experience significantly reduced systemic exposure to the CYP2C19 probe drug omeprazole. On average, subjects with the CYP2C19*17/*17 genotype have a 2.1-fold reduction in mean omeprazole AUC∞vs. CYP2C19*1/*1 individuals. To our knowledge, this is the first prospective investigation of the impact of the CYP2C19*17 allele on drug pharmacokinetics.
This study is in excellent accord with previous CYP2C19*17 phenotyping studies utilizing either single time point evaluations of circulating drug levels or urinary metabolite ratios [10, 14]. In a very recent study, Rudberg et al.[14] have investigated both the frequency and clinical impact of the CYP2C19*17 allele on escitalopram serum levels in 166 psychiatric patients during a routine patient drug monitoring programme. Significantly, they observed a 42% reduction in mean escitalopram serum concentrations in CYP2C19*17/*17 subjects compared with CYP2C19*1/*1 subjects. They therefore postulated that homozygous CYP2C19*17 carriers would require a 50% increase in escitalopram dose to attain drug exposures similar to homozygous CYP2C19*1 carriers. In our initial investigation of the CYP2C19*17 allele, omeprazole and mephenytoin metabolic ratio determinations led us also to postulate that homozygous carriers for this allele could potentially experience ≈ 40% decline in systemic drug exposure [10]. These prior reports, combined with the 52% (2.1-fold) reduction in omeprazole AUC∞ observed in the present study, demonstrate a consistent relationship between the CYP2C19*17/*17 genotype and a significant reduction in circulating drugs levels for these three CYP2C19 substrate drugs.
The impact of the CYP2C19*17 allele was less obvious in a report published by Kurzakski et al., as the authors were unable to observe any association between the presence of the CYP2C19*17 allele and a decline in the effectiveness of H. pylori eradication in patients treated for peptic ulcer using a triple therapy comprised of pantoprazole, amoxicillin and metronidazole [15]. It is noteworthy that the majority of studies demonstrating a clear association between increased H. pylori eradication and the PM phenotype have used clarithromycin instead of metronidazole [16]. Clarithromycin is an established substrate and mechanism-based inactivator of CYP3A4 and co-administration would therefore decrease CYP3A4-mediated PPI metabolism and potentiate any existing PPI dependencies on CYP2C19-mediated metabolism [17]. This is supported by studies demonstrating clarithromycin to increase significantly the differences in systemic exposure (AUC) to either omeprazole or lansoprazole observed in EM, intermediate metabolizer and PM subjects [18, 19]. In contrast, metronidazole is not a substrate for CYP3A4 and is instead believed to be a substrate for CYP2C9, making a similar additive effective with the PM phenotype unlikely [20, 21]. Taken together, these factors could provide an explanation as to why the effectiveness of triple therapy using pantoprazole, amoxicillin and metronidazole for the eradication H. pylori is not significantly altered in individuals possessing the CYP219*17 allele.
The implications of these findings on the therapeutic outcome for a specific therapeutic must, however, be evaluated within the context of both the therapeutic window and the magnitude and exclusivity for CYP2C19-mediated metabolism. These specific issues are especially relevant to the existing challenges of patient dose titration in the area of antidepressant drug therapy. To begin to provide a potential framework for genotype-based dosing adjustments, Kirchheiner et al. have analysed the available pharmacogenetic data pertinent to antidepressant and antipsychotic therapy, making the tentative recommendation to dose adjust PMs to 60% and EMs to 110% of the standard dose for certain selective serotonin reuptake inhibitors or tricyclic antidepressants [22]. Perhaps even more poignant examples of critical dose titration involve the chemotherapeutics cyclophosphamide and thalidomide, which are activated by CYP2C19. Not surprisingly, the CYP2C19*2 allele has been linked to a decrease in side-effects but also a decrease in response to cyclophosphamide treatment [23]. Similarly, response rates following thalidomide treatment for multiple myeloma were nearly 50% lower in carriers of two defective CYP2C19 alleles compared with individuals with at least one functional CYP2C19 allele [24]. By extension, it could be expected that carriers of the CYP2C19*17 allele would experience the converse, generating increased levels of the reactive metabolites and having an increased likelihood of treatment-related side-effects. Future clinical studies correlating CYP2C19*17 with the efficacy of CYP2C19 substrates such as escitalopram, cyclophosphamide, thalidomide and omeprazole would extend the existing relationships between genotype and pharmacokinetics and provide the critical rationale and justification for drug dose adjustments in the future.
Although not a primary study objective, characterization of omeprazole sulphone kinetics revealed a slightly larger statistical difference in sulphone AUC∞ values between CYP2C19*17 and CYP2C19*1 homozygous individuals than similar comparisons using omeprazole AUC∞ values (Figure 1). This is not entirely unexpected and can be explained as the result of two additive processes. Omeprazole is metabolized by two competing enzymes, CYP2C19, producing the 5-hydroxy metabolite, and CYP3A4, generating the sulphone and 3-hydroxymetabolites [25]. Therefore, subjects with elevated CYP2C19 activity will generate more of the 5-hydroxy and less of the sulphone metabolites. Additionally, prior studies of secondary omeprazole metabolism have shown omeprazole sulphone metabolism to be primarily mediated via CYP2C19 to generate the hydroxysulphone, thereby compounding the observed reduction in sulphone AUC∞ in CYP2C19*17/*17 individuals [26, 27]. Correlation of intraindividual omeprazole and omeprazole sulphone AUC∞ values revealed a very strong linear relationship between subjects (r2 = 0.95), with the CYP2C19*17/*17 subjects clustering closest to the origin (Figure 2). Given that omeprazole sulphone has been previously employed as a measure of CYP3A4 activity, we would have therefore anticipated individual differences in CYP3A4 activity to have manifested more prominently in Figure 2[28]. These observations serve as a second independent measure of CYP2C19 activity and lend further support to the relationship between the CYP2C19*17 allele and increased CYP2C19-associated metabolism.
CYP2C19 metabolic activity can be assessed using different probe drugs and a variety of different end-points. CYP2C19-mediated metabolism of omeprazole is commonly quantified using either the AUC ratio of omeprazole/5-hydroxyomeprazole or, alternatively, the plasma concentration ratio between omeprazole and the 5-hydroxy metabolite 3 h after drug administration [29]. We have demonstrated a nearly equivalent statistically significant difference between genotypes using omeprazole AUC∞, omeprazole sulphone AUC∞ and the ratio of omeprazole AUC∞/5-hydroxyomeprazole AUC∞. The interindividual variability in absorption kinetics made a meaningful comparison of omeprazole plasma concentration ratios impossible.
As a group, CYP2C19*17 homozygous individuals have a relatively homogeneous phenotype, and conversely, CYP2C19*1/*1 individuals display significant heterogeneity in rates of CYP2C19-associated metabolism (Figure 1, f = 0.001). The scatter plots from both Rudberg et al.[14] and Sim et al.[10], along with Figure 1 from the present study, demonstrate a very similar distribution pattern for each genotype group irrespective of end-point. The explanation for both these phenomena could lie within the genetics and structure of the CYP2C gene locus. Previously, we have shown via electrophoretic mobility shift analysis that the −806C→T mutation directly affects the binding of nuclear proteins, indicating a direct effect on CYP2C19 transcription [10]. However, it stands to reason that the −3402C→T and −806C→T SNPs could also be genetic markers for a haplotype block extending significantly beyond the region from −3.5 kb to the 3′UTR of the CYP2C19 gene influencing the expression of the CYP2C19 gene. Consistent with this, genotyping studies have shown CYP2C19*17 and CYP2C9*1 to be in 100% linkage, and additional information indicates that CYP2C18, CYP2C19 and the exonic regions of CYP2C9 reside within the same haplotype block [14, 30]. The existence of unidentified polymorphisms could thus account for the heterogeneity of CYP2C19-associated metabolic activity observed in the group currently defined as ‘CYP2C19*1’. It could also be postulated that drugs which are substrates for both CYP2C19 and CYP2C9 could be influenced by the suggested genetic linkage between these two genes.
In summary, we have further substantiated and also directly quantified the impact of the CYP2C19*17 allele on the pharmacokinetics of omeprazole relative to the CYP2C19*1 allele. Homozygous CYP2C19*17 carriers experience a mean 2.1-fold reduction in omeprazole systemic drug exposure. Taken within the broader context of clinically important drugs that are metabolized predominantly by CYP2C19, the CYP2C19*17 allele has the potential to be a critical factor behind individual cases of therapeutic failure. Future studies will help to address the value of genotyping either prospectively or in instances of treatment resistance or failure.
Competing interests
None to declare.
We thank Tommy Andersson, Kjell Andersson and Lars Weidolf from AstraZeneca for their assistance, expertise and for graciously providing analytical standards. We also thank Jolanta Widen for her assistance and technical expertise. We also wish to acknowledge Lilleba Bohman and Susanne Virding for their roles in patient genotyping, and Peter Johansson and Ilona Skilving for their invaluable assistance in the clinic. The clinical component of this work was supported by a grant from The Stockholm County Council (ALF 20050551/20060420). Additional funding was provided by The Swedish Research Council, The Danish Agency for Science, Technology and Innovation, The Lundbeck Foundation and from Torsten och Ragnar Söderbergs stiftelser. E.E. is an associate professor at Karolinska Institutet, but currently employed by AstraZeneca R & D Södertälje. However, at the time of the study E.E. was employed by Karolinska University Hospital. E.E. was the recipient of a clinical pharmacology research stipend from Pfizer Sweden AB.
REFERENCES
- 1.Desta Z, Zhao X, Shin JG, Flockhart DA. Clinical significance of the cytochrome P450 2C19 genetic polymorphism. Clin Pharmacokinet. 2002;41:913–58. doi: 10.2165/00003088-200241120-00002. [DOI] [PubMed] [Google Scholar]
- 2.Bertilsson L, Henthorn TK, Sanz E, Tybring G, Sawe J, Villen T. Importance of genetic factors in the regulation of diazepam metabolism: relationship to S-mephenytoin, but not debrisoquin, hydroxylation phenotype. Clin Pharmacol Ther. 1989;45:348–55. doi: 10.1038/clpt.1989.40. [DOI] [PubMed] [Google Scholar]
- 3.Qin XP, Xie HG, Wang W, He N, Huang SL, Xu ZH, Ou-Yang DS, Wang YJ, Zhou HH. Effect of the gene dosage of CYP2C19 on diazepam metabolism in Chinese subjects. Clin Pharmacol Ther. 1999;66:642–6. doi: 10.1016/S0009-9236(99)90075-9. [DOI] [PubMed] [Google Scholar]
- 4.Herrlin K, Yasui-Furukori N, Tybring G, Widen J, Gustafsson LL, Bertilsson L. Metabolism of citalopram enantiomers in CYP2C19/CYP2D6 phenotyped panels of healthy Swedes. Br J Clin Pharmacol. 2003;56:415–21. doi: 10.1046/j.1365-2125.2003.01874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sindrup SH, Brosen K, Hansen MG, Aaes-Jorgensen T, Overo KF, Gram LF. Pharmacokinetics of citalopram in relation to the sparteine and the mephenytoin oxidation polymorphisms. Ther Drug Monit. 1993;15:11–7. doi: 10.1097/00007691-199302000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Klotz U, Schwab M, Treiber G. CYP2C19 polymorphism and proton pump inhibitors. Basic Clin Pharmacol Toxicol. 2004;95:2–8. doi: 10.1111/j.1600-0773.2004.pto950102.x. [DOI] [PubMed] [Google Scholar]
- 7.Andersson T, Holmberg J, Rohss K, Walan A. Pharmacokinetics and effect on caffeine metabolism of the proton pump inhibitors, omeprazole, lansoprazole, and pantoprazole. Br J Clin Pharmacol. 1998;45:369–75. doi: 10.1046/j.1365-2125.1998.t01-1-00702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furuta T, Ohashi K, Kamata T, Takashima M, Kosuge K, Kawasaki T, Hanai H, Kubota T, Ishizaki T, Kaneko E. Effect of genetic differences in omeprazole metabolism on cure rates for Helicobacter pylori infection and peptic ulcer. Ann Intern Med. 1998;129:1027–30. doi: 10.7326/0003-4819-129-12-199812150-00006. [DOI] [PubMed] [Google Scholar]
- 9.Klotz U. Clinical impact of CYP2C19 polymorphism on the action of proton pump inhibitors: a review of a special problem. Int J Clin Pharmacol Ther. 2006;44:297–302. doi: 10.5414/cpp44297. [DOI] [PubMed] [Google Scholar]
- 10.Sim SC, Risinger C, Dahl ML, Aklillu E, Christensen M, Bertilsson L, Ingelman-Sundberg M. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther. 2006;79:103–13. doi: 10.1016/j.clpt.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi K, Chiba K, Sohn DR, Kato Y, Ishizaki T. Simultaneous determination of omeprazole and its metabolites in plasma and urine by reversed-phase high-performance liquid chromatography with an alkaline-resistant polymer-coated C18 column. J Chromatogr. 1992;579:299–305. doi: 10.1016/0378-4347(92)80395-7. [DOI] [PubMed] [Google Scholar]
- 12.Tybring G, Bottiger Y, Widen J, Bertilsson L. Enantioselective hydroxylation of omeprazole catalyzed by CYP2C19 in Swedish white subjects. Clin Pharmacol Ther. 1997;62:129–37. doi: 10.1016/S0009-9236(97)90060-6. [DOI] [PubMed] [Google Scholar]
- 13.Uno T, Niioka T, Hayakari M, Yasui-Furukori N, Sugawara K, Tateishi T. Absolute bioavailability and metabolism of omeprazole in relation to CYP2C19 genotypes following single intravenous and oral administrations. Eur J Clin Pharmacol. 2007;63:143–9. doi: 10.1007/s00228-006-0251-7. [DOI] [PubMed] [Google Scholar]
- 14.Rudberg I, Mohebi B, Hermann M, Refsum H, Molden E. Impact of the ultrarapid CYP2C19*17 allele on serum concentration of escitalopram in psychiatric patients. Mol Ther. 2008;83:322–7. doi: 10.1038/sj.clpt.6100291. [DOI] [PubMed] [Google Scholar]
- 15.Kurzawski M, Gawronska-Szklarz B, Wrzesniewska J, Siuda A, Starzynska T, Drozdzik M. Effect of CYP2C19*17 gene variant on Helicobacter pylori eradication in peptic ulcer patients. Eur J Clin Pharmacol. 2006;62:877–80. doi: 10.1007/s00228-006-0183-2. [DOI] [PubMed] [Google Scholar]
- 16.Padol S, Yuan Y, Thabane M, Padol IT, Hunt RH. The effect of CYP2C19 polymorphisms on H. pylori eradication rate in dual and triple first-line PPI therapies: a meta-analysis. Am J Gastroenterol. 2006;101:1467–75. doi: 10.1111/j.1572-0241.2006.00717.x. [DOI] [PubMed] [Google Scholar]
- 17.Rodrigues AD, Roberts EM, Mulford DJ, Yao Y, Ouellet D. Oxidative metabolism of clarithromycin in the presence of human liver microsomes. Major role for the cytochrome P4503A (CYP3A) subfamily. Drug Metab Dispos. 1997;25:623–30. [PubMed] [Google Scholar]
- 18.Furuta T, Ohashi K, Kobayashi K, Iida I, Yoshida H, Shirai N, Takashima M, Kosuge K, Hanai H, Chiba K, Ishizaki T, Kaneko E. Effects of clarithromycin on the metabolism of omeprazole in relation to CYP2C19 genotype status in humans. Clin Pharmacol Ther. 1999;66:265–74. doi: 10.1016/S0009-9236(99)70034-2. [DOI] [PubMed] [Google Scholar]
- 19.Saito M, Yasui-Furukori N, Uno T, Takahata T, Sugawara K, Munakata A, Tateishi T. Effects of clarithromycin on lansoprazole pharmacokinetics between CYP2C19 genotypes. Br J Clin Pharmacol. 2005;59:302–9. doi: 10.1111/j.1365-2125.2004.02329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roedler R, Neuhauser MM, Penzak SR. Does metronidazole interact with CYP3A substrates by inhibiting their metabolism through this metabolic pathway? Or should other mechanisms be considered? Ann Pharmacother. 2007;41:653–8. doi: 10.1345/aph.1H401. [DOI] [PubMed] [Google Scholar]
- 21.Blyden GT, Scavone JM, Greenblatt DJ. Metronidazole impairs clearance of phenytoin but not of alprazolam or lorazepam. J Clin Pharmacol. 1988;28:240–5. doi: 10.1002/j.1552-4604.1988.tb03139.x. [DOI] [PubMed] [Google Scholar]
- 22.Kirchheiner J, Nickchen K, Bauer M, Wong ML, Licinio J, Roots I, Brockmoller J. Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol Psychiatry. 2004;9:442–73. doi: 10.1038/sj.mp.4001494. [DOI] [PubMed] [Google Scholar]
- 23.Takada K, Arefayene M, Desta Z, Yarboro CH, Boumpas DT, Balow JE, Flockhart DA, Illei GG. Cytochrome P450 pharmacogenetics as a predictor of toxicity and clinical response to pulse cyclophosphamide in lupus nephritis. Arthritis Rheum. 2004;50:2202–10. doi: 10.1002/art.20338. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Hou J, Jiang H, Wang D, Fu W, Yuan Z, Chen Y, Zhou L. Polymorphisms of CYP2C19 gene are associated with the efficacy of thalidomide based regimens in multiple myeloma. Haematologica. 2007;92:1246–9. doi: 10.3324/haematol.11319. [DOI] [PubMed] [Google Scholar]
- 25.Andersson T, Miners JO, Veronese ME, Tassaneeyakul W, Tassaneeyakul W, Meyer UA, Birkett DJ. Identification of human liver cytochrome P450 isoforms mediating omeprazole metabolism. Br J Clin Pharmacol. 1993;36:521–30. doi: 10.1111/j.1365-2125.1993.tb00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Regardh CG, Andersson T, Lagerstrom PO, Lundborg P, Skanberg I. The pharmacokinetics of omeprazole in humans – a study of single intravenous and oral doses. Ther Drug Monit. 1990;12:163–72. doi: 10.1097/00007691-199003000-00010. [DOI] [PubMed] [Google Scholar]
- 27.Andersson T, Miners JO, Veronese ME, Birkett DJ. Identification of human liver cytochrome P450 isoforms mediating secondary omeprazole metabolism. Br J Clin Pharmacol. 1994;37:597–604. doi: 10.1111/j.1365-2125.1994.tb04310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bottiger Y. Use of omeprazole sulfone in a single plasma sample as a probe for CYP3A4. Eur J Clin Pharmacol. 2006;62:621–5. doi: 10.1007/s00228-006-0156-5. [DOI] [PubMed] [Google Scholar]
- 29.Chang M, Dahl ML, Tybring G, Gotharson E, Bertilsson L. Use of omeprazole as a probe drug for CYP2C19 phenotype in Swedish Caucasians: comparison with S-mephenytoin hydroxylation phenotype and CYP2C19 genotype. Pharmacogenetics. 1995;5:358–63. doi: 10.1097/00008571-199512000-00004. [DOI] [PubMed] [Google Scholar]
- 30.Walton R, Kimber M, Rockett K, Trafford C, Kwiatkowski D, Sirugo G. Haplotype block structure of the cytochrome P450 CYP2C gene cluster on chromosome 10. Nat Genet. 2005;37:915–6. doi: 10.1038/ng0905-915. author reply 916. [DOI] [PubMed] [Google Scholar]


