Abstract
Several proteins secreted by enteric bacteria are thought to contribute to virulence by disturbing the signal transduction of infected cells. Here, we report that SopB, a protein secreted by Salmonella dublin, has sequence homology to mammalian inositol polyphosphate 4-phosphatases and that recombinant SopB has inositol phosphate phosphatase activity in vitro. SopB hydrolyzes phosphatidylinositol 3,4,5-trisphosphate, an inhibitor of Ca2+-dependent chloride secretion. In addition, SopB hydrolyzes inositol 1,3,4,5,6 pentakisphosphate to yield inositol 1,4,5,6-tetrakisphosphate, a signaling molecule that increases chloride secretion indirectly by antagonizing the inhibition of chloride secretion by phosphatidylinositol 3,4,5-trisphosphate [Eckmann, L., Rudolf, M. T., Ptasznik, A., Schultz, C., Jiang, T., Wolfson, N., Tsien, R., Fierer, J., Shears, S. B., Kagnoff, M. F., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 14456–14460]. Mutation of a conserved cysteine that abolishes phosphatase activity of SopB results in a mutant strain, S. dublin SB c/s, with decreased ability to induce fluid secretion in infected calf intestine loops. Moreover, HeLa cells infected with S. dublin SB c/s do not accumulate high levels of inositol 1,4,5,6-tetrakisphosphate that are characteristic of wild-type S. dublin-infected cells. Therefore, SopB mediates virulence by interdicting inositol phosphate signaling pathways.
Keywords: phosphatidylinositol/cell signaling/chloride channels
Enteric Salmonella infections are accompanied by marked intestinal inflammation and secretory diarrhea. Bacterial proteins translocated into host cells by the Salmonella Inv/Spa type III secretion system are essential for pathogenesis (1). Mutants of Salmonella typhimurium defective in the Inv/Spa type III system do not cause an inflammatory response or fluid secretion, suggesting that proteins of this system are essential for virulence (2). A number of proteins secreted by the type III systems contribute to virulence by altering host cell signal transduction pathways. For example, YopH of Yersinia and SptP of Salmonella are protein tyrosine phosphatases (3, 4); YpkA of Yersinia is a Ser/Thr protein kinase (5).
A secreted protein of Salmonella dublin, SopB, recently was shown to be essential for enteropathogenicity. Deletion of sopB does not affect the invasiveness of the organism but does abrogate fluid secretion and neutrophil accumulation into infected calf ileal loops (6). Virulent strains of S. dublin perturb inositol phosphate metabolism within 30 min of infection of colonic epithelial cells with a 14-fold rise in the levels of inositol 1,4,5,6-tetrakisphosphate (Ins 1,4,5,6-P4) (7). This inositol tetrakisphosphate has been shown to block inhibition of chloride transport that is mediated by phosphatidylinositol 3,4,5-trisphosphate (PtdIns 3,4,5-P3) after epidermal growth factor stimulation of these cells. Thus, it was proposed that the production of Ins 1,4,5,6-P4 during infection might allow increased chloride secretion and diarrhea (7).
MATERIALS AND METHODS
Enteropathogenesis Assay.
Bovine ligated ileal loop assays were as described else-where (6). Experiments were performed in the mid-ileum of 28-day-old Friesial calves. Ileal loops were prepared as follows. Calves were anesthetized for the duration of the experiment by using pentobarbitone (0.44 ml/kg). The abdominal wall was opened, and the distal ileum was exteriorized. The lumen of the distal ileum proximal to the continuous Peyer’s patch was flushed with saline, and 6- to 8-cm loops with 1-cm spacers were ligated by using surgical silk. Loops were inoculated with 1 ml of test strains or 1 ml of Luria–Bertani medium broth as a negative control. Loops were replaced within the abdominal cavity, and the wound was repaired. Glucose (500 ml, 5% wt/vol) in saline was administered i.v. by drip.
Twelve hours after challenge, the loops were sampled, and enteropathogenesis was assessed with respect to fluid accumulation and neutrophil infiltration. Animals were killed by barbiturate overdose. Fluid secretion was measured as a ratio of volume of fluid accumulated per loop length (V/L ratio). Neutrophils were labeled with 111indium (vide infra), and infiltration was measured as a ratio of 111indium activity in test loops vs. control loops inoculated with sterile Luria–Bertani medium.
111Indium-Labeling of Neutrophils.
Approximately 4 hr after loop inoculation, 60 ml of blood was sampled, from which neutrophils were isolated (8). Neutrophils finally were resuspended in 2 ml of Ca2+-free Tyrodes buffer and were labeled by using 111indium oxinate (Mallinckrodt) as described (9).
Preparation of Ins(1[32PO4], 3, 4, 5, 6)P5.
Ins 3,4,5,6-P4 1-kinase was prepared from 1-day-old chicken erythrocytes by precipitation of a crude homogenate in 50% ammonium sulfate. The dialyzed enzyme (0.34 mg) was incubated in 50 mM Hepes (pH 7.0), 5 mM ATP, 6 mM MgCl2, 2 mM EGTA, 100 mM KCl, and 25 μCi [γ32P]-ATP (30 Ci/mmol) in 1.5 ml. After 45 min at 37°C, the reaction was stopped with 10 ml of water. The Ins 1[32PO4], 3,4,5,6-P5 product was separated from ATP on a 2-ml Dowex formate (Bio-Rad) column. ATP was eluted by 1 M ammonium formate (pH 3.5), and the product was eluted with 1.6 M of the same salt. The product was desalted by lyophilization and was verified by HPLC chromatography on absorbosphere SAX with an internal standard of 3H Ins 1,3,4,5,6-P5 as described (10).
Construction of S. dublin SB c/s.
The C460S SopB mutant was constructed by site-directed mutagenesis that converted TGT(C460) to TCG(S). This also created an EcoRI site in the mutant allele. Two DNA fragments were amplified by PCR with S. dublin 2229 chromosomal DNA as template by using bmut1/bmut2 and bmut3/bmut4, respectively. The two DNA fragments were isolated, mixed, and used as a template in a PCR reaction with bmut1 and bmut4. The resulting DNA was cloned into the suicide plasmid vector pDM4 to yield pCS. pCS was conjugated from Escherichia coli s17.1 λpir into S. dublin 2229, and chloramphenicol-resistant transconjugates were obtained. The suicide plasmid then was excised by a second recombination event (11). The chloramphenicol-sensitive recombinants were obtained and screened for a mutated allele by PCR of sopB followed by digestion of products with EcoRI. Several EcoRI-sensitive clones were identified, and one of these was sequenced to confirm the mutation; it was used for further studies and was designated S. dublin SB c/s. The glutathione S-transferase–SopB and glutathione S-transferase–SopB c/s mutant constructs were created by using pGEX-2T plasmid vector resulting in fusion proteins lacking the amino-terminal 29 amino acids of SopB. Recombinant proteins were expressed in E. coli and were purified on glutathione-agarose.
HeLa Cell Infections.
HeLa cells were grown to 50% confluence in DMEM containing 1/10th of the normal concentration of inositol supplemented with 10% heat-inactivated fetal calf serum containing 10 μCi 3H-inositol/ml for 3 days. S. dublin or S. dublin SB c/s (5 × 107/ml medium) were added for 1 hr at 37°C. Growth medium was removed, and cells were washed twice with cold PBS and then were scraped from the dish in 2 ml of PBS. The cells were extracted with 3 ml of chloroform/methanol (1:1). The aqueous layer was removed, was dried under nitrogen, and was resuspended in 0.5 ml of water containing an internal standard of 32PO4 Ins 1,3,4,5-P4 (10). Samples were analyzed by HPLC on adsorbosphere SAX (7).
Miscellaneous Procedures.
Preparation of 3-phosphate containing inositol lipids was prepared as described (12), and their hydrolysis was assayed as described (13). Inositol triphosphate and inositol tetrakisphosphate hydrolysis assays were performed as described (14).
RESULTS AND DISCUSSION
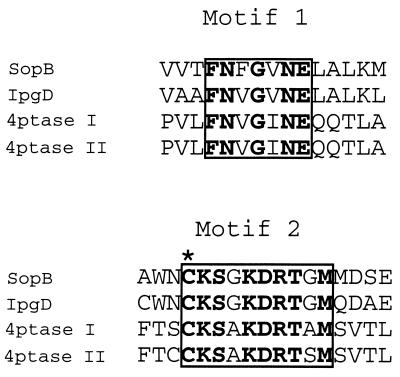
Our analysis of the amino acid sequence of SopB and its Shigella orthologue IpgD (6) revealed that these two proteins contain two motifs that are related to sequences in inositol polyphosphate 4-phosphatases, enzymes that hydrolyze the lipid second messenger PtdIns 3,4-P2 (12, 15, 16) (Fig. 1). Motif 2 represents the putative active site of 4-phosphatases in which the cysteine is thought to be the catalytic residue (16). Site-directed mutation of this cysteine to serine inactivates 4-phosphatase (F.A.N. and P.W.M., unpublished observation). The presence of these motifs suggested that SopB might be an inositol phosphate phosphatase.
Figure 1.
Conserved amino acid sequence motifs present in SopB, IpgD, and human inositol polyphosphatase types I and II. The regions of sequence similarity are boxed, with identical amino acids indicated by bold type. Motif 2 contains a consensus sequence Cys-Xaa5-Arg characteristic of Mg2+-independent phosphatases. An asterisk indicates the Cys residue that is essential for catalysis.
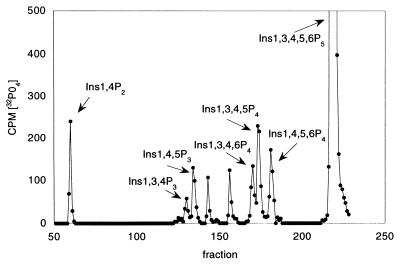
We expressed recombinant glutathione S-transferase–SopB and found that it hydrolyzed several inositol phosphates (Table 1). An attractive hypothesis is that SopB hydrolyses Ins 1,3,4,5,6-P4 to produce Ins 1,4,5,6-P4. This isomer accumulates rapidly in cells infected with S. dublin and has been implicated in enhancing Cl− secretion by antagonizing PtdIns 3,4,5-P3-mediated ion channel closing. We synthesized Ins 1[32PO4],3,4,5,6-P5 to test the hypothesis that SopB hydrolyzes this substrate to form Ins 1,4,5,6P4. We analyzed the products of this reaction and found that several InsP4 isomers were formed, including Ins 1,4,5,6-P4 (Fig. 2). Because the label is in the 1-position and no 32PO4 was released, we can exclude the enantiomer Ins 3,4,5,6-P4 as the product. The other major products formed include Ins 1,3,4,5-P4, Ins 1,4,5-P3, and Ins 1,4-P2, indicating that SopB can hydrolyze phosphates at the 3-, 5-, and 6-positions. SopB also hydrolyzes the 4-position phosphate of Ins 1,3,4-P3 and PtdIns 3,4-P2. SopB does not have 1-phosphatase activity, and no hydrolysis of Ins1,4-P2, PtdIns 4-P, PtdIns 4,5-P2, InsP6, p-nitrophenylphosphate, or raytide, a phosphopeptide substrate of protein tyrosine phosphatases, was detected. In addition, glutathione S-transferase–SopB c/s, a protein with the conserved cysteine mutated to serine, had no detectable phosphatase activity. Although recombinant SopB has broad substrate specificity, the activities observed are very low compared with those of the specific cellular enzymes that normally metabolize these substrates. The cellular enzymes all catalyze micromoles of substrate/minute/milligram protein. The affinity of SopB for substrates is relatively low compared with cellular enzymes. For example, the apparent km values for SopB hydrolysis of Ins1,3,4P3 and 1,4,5P3 were 23 and 178 μM, values greater than those of cellular phosphatases.
Table 1.
Phosphatase activity of recombinant SopB*
| Substrate | nmol/min/mg protein |
|---|---|
| PtdIns 3-P | 47 |
| PtdIns 3,4-P2 | 51 |
| PtdIns 3,4,5-P3 | 68 |
| Ins 1,3,4-P3 | 9.1 |
| Ins 1,4,5-P3 | 13 |
| Ins 1,3,4,5-P4 | 24 |
| Ins 1,3,4,5,6-P5 | 4.5 |
Assays were performed with 10 μM substrate in 50 mM Mops (pH 7.0), 10 mM EDTA, and 1 mM DTT.
Figure 2.
HPLC of products of hydrolysis of Ins1 [32PO4],3,4,5,6-P5 by SopB. Recombinant SopB (0.6 μg) was incubated with Ins 1[32PO4],3,4,5,6-P5 in 50 mM Mops (pH 7.0), 10 mM EDTA, and 1 mM DTT for 30 min at 37°C. HPLC on Adsorbosphere SAX was performed as described (10) by using 3H-labeled internal standards of Ins 1,4-P2, Ins 1,3-P2, Ins 3,4-P2, Ins 1,3,4-P3, and Ins 1,3,4,5-P4.
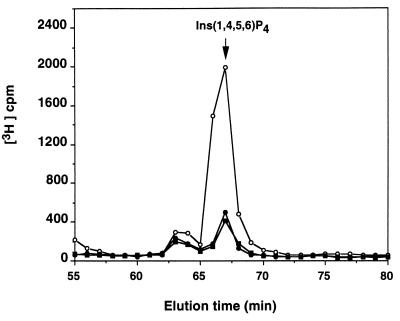
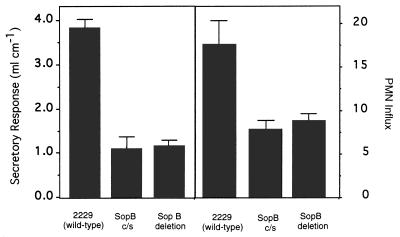
To evaluate the importance of SopB phosphatase activity in vivo, we constructed the mutant S. dublin SB c/s. This mutant strain expressed and secreted the SopB c/s protein at levels indistinguishable to those of the wild type (data not shown). We infected HeLa cells with wild-type S. dublin or S. dublin SB c/s and found that only wild-type organisms induced production of Ins 1,4,5,6-P4 (Fig. 3). No increases in other InsP4 isomers were observed. This experiment demonstrates that accumulation depends on SopB enzyme activity. We next tested the enteropathogenicity of these organisms in a calf intestinal loop infection assay. The point mutation abrogated the ability of the mutant strain to induce both fluid secretion and neutrophil accumulation to the same extent as a sopB deletion (Fig. 4). These results indicate that SopB contributes to Salmonella virulence by subverting cellular inositol phosphate signaling reactions.
Figure 3.
Effect of wild-type and mutant S. dublin SB c/s infection on the accumulation of Ins 1,4,5,6-P4 in HeLa cells. HeLa cells were infected for 30 min with wild-type S. dublin (open circles) or mutant S. dublin SB c/s (closed circles) or were not infected (closed squares). The inositol phosphates were extracted and separated by HPLC as described (7).
Figure 4.
Effect of SopB mutations on S. dublin infection-induced fluid secretion and polymorphonuclear leukocyte influx in bovine calf ligated ileal loops. Bars indicate the average of at least three intestinal sections. Error bars indicate the SEM of each sample.
The experiments with S. dublin SB c/s, wherein the SopB phosphatase activity is absent because of a single base change at the active site of the enzyme, indicate that inositol phosphate phosphatase activity per se is required for virulence as measured by fluid and neutrophil accumulation in infected intestinal loops. Based on the in vitro enzyme activity of SopB, two mechanisms for production of diarrhea are apparent. Thus, SopB hydrolyzes PtdIns 3,4,5-P3, which is a messenger molecule that inhibits chloride secretion. It also results in formation of Ins 1,4,5,6-P4 from Ins 1,3,4,5,6-P5, the isomer that antagonizes the action of PtdIns 3,4,5-P3 in closing chloride channels. These actions may combine to produce fluid secretion in infected epithelia. We cannot explain why SopB in vitro produces several inositol tetraphosphates whereas, in infected cells, only Ins 1,4,5,6-P4 appears to accumulate. Although several other examples in which bacteria subvert host signaling reactions to mediate virulence have been described, in this case, virulence results from derangement of the phosphatidylinositol signaling pathway. The broad specificity of SopB in hydrolyzing inositol phosphates and lipids suggests that other signaling reactions are likely to be disrupted in cells infected with virulent organisms. The homology of SopB to the Shigella virulence factor IpgD suggests that similar disruption of inositol phosphate metabolism are likely to contribute to pathogenic responses in infections with that organism.
Acknowledgments
We thank Mike Wood, Samantha Hedges, Pat Watson, and Sue Paulin for technical assistance. This research was supported by grants from the National Institutes of Health, the American Heart Association, the Biotechnology and Biological Sciences Research Council, and the Ministry of Agriculture, Fisheries, and Food.
ABBREVIATIONS
- Ins 1
4,5,6-P4, inositol 1,4,5,6-tetrakisphosphate
- PtdIns 3
4,5-P3, phosphatidylinositol 3,4,5-triphosphate
Footnotes
A Commentary on this article begins on page 14006.
References
- 1.Galán J E. Mol Microbiol. 1996;20:263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 2.Watson P R, Galyov E E, Paulin S M, Jones P W, Wallis T S. Infect Immun. 1998;66:1432–1438. doi: 10.1128/iai.66.4.1432-1438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan K, Dixon J E. Science. 1990;249:553–555. doi: 10.1126/science.2166336. [DOI] [PubMed] [Google Scholar]
- 4.Fu Y, Galán J E. Mol Microbiol. 1998;27:359–368. doi: 10.1046/j.1365-2958.1998.00684.x. [DOI] [PubMed] [Google Scholar]
- 5.Galyov E E, Håkansson S, Forsberg Å, Wolf-Watz H. Nature (London) 1993;361:730–732. doi: 10.1038/361730a0. [DOI] [PubMed] [Google Scholar]
- 6.Galyov E E, Wood M W, Rosqvist R, Mullan P B, Watson P R, Hedges S, Wallis T S. Mol Microbiol. 1997;25:903–912. doi: 10.1111/j.1365-2958.1997.mmi525.x. [DOI] [PubMed] [Google Scholar]
- 7.Eckmann L, Rudolf M T, Ptasznik A, Schultz C, Jiang T, Wolfson N, Tsien R, Fierer J, Shears S B, Kagnoff M F, et al. Proc Natl Acad Sci USA. 1997;94:14456–14460. doi: 10.1073/pnas.94.26.14456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson G P, Kaneko J J. Proc Soc Exper Biol Med. 1973;142:853–855. doi: 10.3181/00379727-142-37131. [DOI] [PubMed] [Google Scholar]
- 9.Wallis T S, Hawker R J H, Candy D C A, Qi G M, Clarke G J, Worton K J, Osborne M P, Stephen J. J Med Microbiol. 1989;30:149–156. doi: 10.1099/00222615-30-2-149. [DOI] [PubMed] [Google Scholar]
- 10.Wilson M P, Majerus P W. J Biol Chem. 1996;271:11904–11910. doi: 10.1074/jbc.271.20.11904. [DOI] [PubMed] [Google Scholar]
- 11.Milton D L, O’Toole R, Horstedt P, Wolf-Watz H. J Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norris F A, Majerus P W. J Biol Chem. 1994;269:8716–8720. [PubMed] [Google Scholar]
- 13.Caldwell K K, Lips D L, Bansal V S, Majerus P W. J Biol Chem. 1991;266:18378–18386. [PubMed] [Google Scholar]
- 14.Zhang X, Jefferson A B, Auethavekiat V, Majerus P W. Proc Natl Acad Sci USA. 1995;92:4853–4856. doi: 10.1073/pnas.92.11.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norris F A, Auethavekiat V, Majerus P W. J Biol Chem. 1995;270:16128–16133. doi: 10.1074/jbc.270.27.16128. [DOI] [PubMed] [Google Scholar]
- 16.Norris F A, Atkins R C, Majerus P W. J Biol Chem. 1997;272:23859–23864. doi: 10.1074/jbc.272.38.23859. [DOI] [PubMed] [Google Scholar]