Abstract
Cellular expression of Mcl-1, an anti-apoptotic Bcl-2 family member, is tightly regulated. Recently, Bcl-2 expression was shown to be regulated by microRNAs, small endogenous RNA molecules that regulate protein expression through sequence-specific interaction with messenger RNA. By analogy, we reasoned that Mcl-1 expression may also be regulated by microRNAs. We chose human immortalized, but non-malignant, H69 cholangiocyte and malignant KMCH cholangiocarcinoma cell lines for these studies because Mcl-1 is dysregulated in cells with the malignant phenotype. In silico analysis identified a putative target site in the Mcl-1 mRNA for the mir-29 family, and we found that mir-29b was highly expressed in cholangiocytes. Interestingly, mir-29b was downregulated in malignant cells, consistent with Mcl-1 protein upregulation. Enforced mir-29b expression reduced Mcl-1 protein expression in KMCH cells. This effect was direct, as mir-29b negatively regulated expression of an Mcl-1 3’ untranslated region (UTR)-based reporter construct. Enforced mir-29b expression reduced Mcl-1 cellular protein levels and sensitized the cancer cells to TRAIL cytotoxicity. Transfection of non-malignant cells (that express high levels of mir-29) with a locked-nucleic acid antagonist of mir-29b increased Mcl-1 levels and reduced TRAIL-mediated apoptosis. Thus mir-29 is an endogenous regulator of Mcl-1 protein expression and, thereby, apoptosis.
Keywords: Apoptosis, post-transcriptional regulation, cholangiocarcinoma, microRNA, TRAIL
INTRODUCTION
Mcl-1 is a potent multidomain anti-apoptotic protein of the Bcl-2 family containing Bcl-2-homology domains BH1−3 that heterodimerizes with Bcl-2 family members. Specifically, Mcl-1 binds to the BH3-only proteins Bim, Bid, Bik, Noxa, and Puma (Chen et al., 2005), as well as Bak (Cuconati et al., 2003). Binding to Bid and Bim protects against TRAIL (tumor necrosis factor-related apoptosis-inducing ligand)-induced cell death. Either depletion of Mcl-1 (Nijhawan et al., 2003) or disruption of the interaction between Mcl-1 and Bak coincides with apoptosis following a cytotoxic stimulus (Leu et al., 2004). Still, the mechanism by which Bcl-2 family members modulate apoptosis remains incompletely understood (Willis & Adams, 2005).
Tight regulation of Mcl-1 protein levels is necessary; on one hand, insufficient Mcl-1 can result in inappropriate cell death and on the other hand, over-expression poses a risk of cellular transformation. For example, conditional deletion of Mcl-1 results in bone marrow failure due to inappropriate apoptosis of precursor cells (Opferman et al., 2003), whereas transgenic mice with constitutive Mcl-1 expression develop hematologic malignancies (Zhou et al., 2001). Mcl-1 overexpression is commonly observed in cancers (Konopleva et al., 2006), including cholangiocarcinoma (Kobayashi et al., 2005), and overexpression correlates with cancer prognosis and recurrence (Kaufmann et al., 1998; Wuilleme-Toumi et al., 2005). In cholangiocarcinoma, overexpression of Mcl-1 causes resistance to TRAIL-induced apoptosis while suppression sensitizes cells to apoptosis (Taniai et al., 2004).
The regulation of Mcl-1 protein levels is complex. Transcriptionally, Mcl-1 mRNA is upregulated by cytokines IL-3, IL-5, IL-6, IL-15, IL-22, and IFN-alpha, as well as by GM-CSF, and EGFR activation (reviewed by Michels et al., 2005). At the protein level, the half-life of Mcl-1 is approximately 2−4 hours. Ubiquitination by the E3-ligase Mule leads to rapid degradation by the proteasome, and prevention of ubiquitination increases Mcl-1 levels and enhances protection (Zhong et al., 2005). Alternatively, bile acids prevent Mcl-1 turnover through an EGFR-dependent pathway, again enhancing protection (Yoon et al., 2002). Thus Mcl-1 is regulated by multiple cellular processes.
Given the complexities of Mcl-1 regulation, it is highly likely that other mechanisms exist. Recent work has described a potential new mechanism for protein regulation, namely through microRNAs (Bartel, 2004). MicroRNAs act as endogenous sequence-specific suppressors of translation, and thus can decrease target protein expression (Lewis et al., 2005). The microRNA:target interaction occurs through base pairing of the “seed region” of the microRNA (nucleotides 2−7) to the cognate target (Lewis et al., 2005). While relatively few microRNA targets have been experimentally described, significantly the prototypical anti-apoptotic protein Bcl-2 is a target of microRNA regulation. mir-15 and mir-16 have base complementarity (including the seed region) to the Bcl-2 mRNA, and overexpression of mir-15/16 leads to decreased Bcl-2 protein and increased cell death (Cimmino et al., 2005). Although microRNAs potentially regulate expression of other Bcl-2 family proteins, this mechanism of Mcl-1 regulation has not been explored. The objective of the current study was to examine the role of microRNAs in regulation of Mcl-1 expression.
RESULTS
Mcl-1 is a potential target of mir-29 microRNA regulation
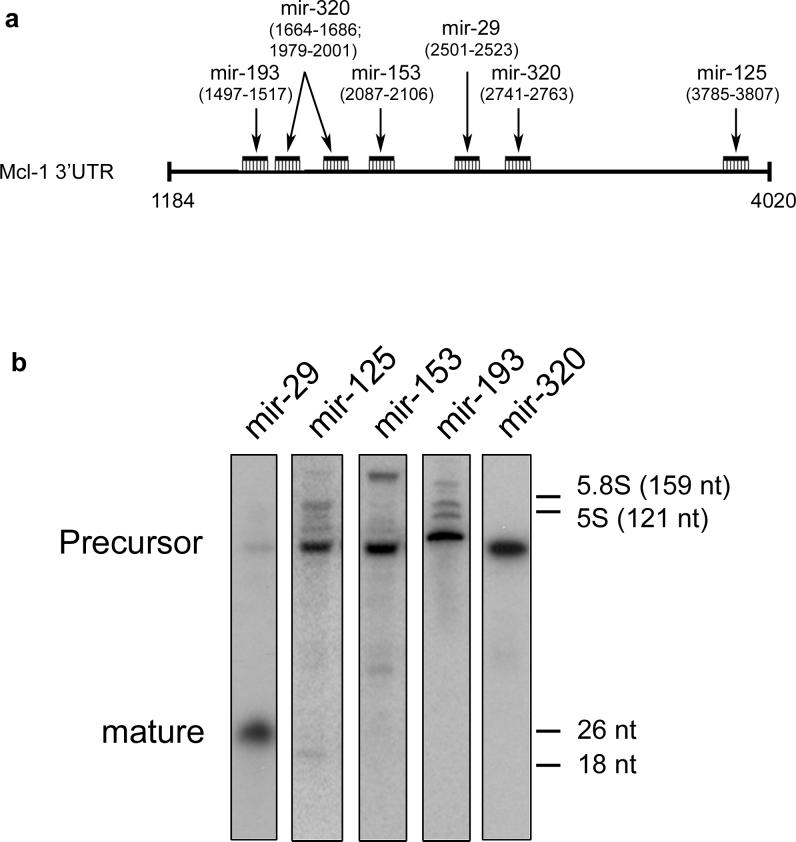
Predicted Mcl-1-binding microRNAs were located by computer search (Supplemental Material) and include mir-29b, mir-125b, mir-153, mir-193b, and mir320 (Figure 1a). Given the overexpression of Mcl-1 in malignant cholangiocarcinoma cells, we postulated the Mcl-1-inhibitory microRNA would be reduced in these cells, but more abundantly expressed in the H69 nonmalignant cholangiocyte cells which minimally express Mcl-1. Therefore, Northern blot analysis was performed for microRNAs in H69 cells. Only mir-29 was abundantly expressed in the mature form in H69 cholangiocytes (Figure 1b). Note that the precursor form was detected for all examined microRNAs. Several possible mechanisms could explain this regulation, including regulation of expression through the maturation or nuclear export process (Lee et al., 2002), or potentially through degradation of the mature microRNA. There is perfect complementarity between mir-29b and the 3’UTR of Mcl-1 over the first 9 nucleotides (including the 2−7 seed), consistent with a bona fide mir-29 binding site (Table 1, Supplemental Material). The three isoforms of mir-29 (a, b, c) share the first 9 nucleotides with minor divergence thereafter, suggesting all isoforms potentially target Mcl-1.
Figure 1. Putative microRNA binding sites on Mcl-1 messenger RNA.
Panel a: Schematic of the Mcl-1 3’UTR (#NM_021960) nucleotides 1184−4020. Five microRNAs were predicted to bind and the position of binding to the Mcl-1 transcript is shown in parentheses. Panel b: Northern blot of the five putative Mcl-1 binding microRNAs. RNA enriched for small RNA (<200nt's) from H69 cells was separated and probed for the indicated microRNAs. Note the robust expression of mir-29. Size markers are from SybrGreen-stained gels prior to transfer; 5.8S and 5S are endogenous rRNAs, 26 nt and 18 nt refer to synthetic oligodeoxynucleotides run in an adjacent lane.
mir-29 expression is reduced in cholangiocarcinoma cell line
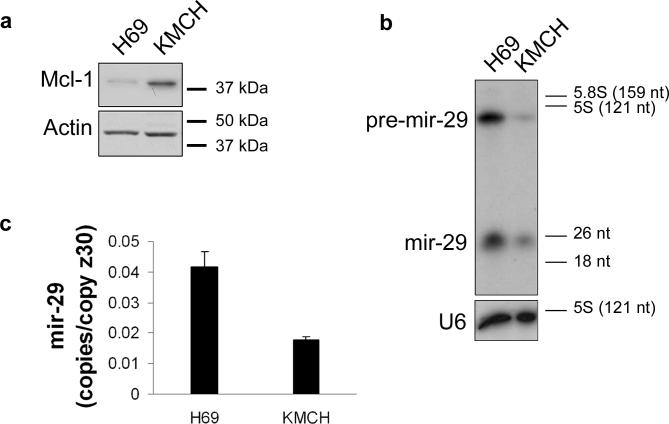
If mir-29b is a potent regulator of Mcl-1 protein expression, then mir-29 should be downregulated in cholangiocarcinoma cells which richly express Mcl-1 protein relative to the H69 cells which have minimal Mcl-1 protein expression (Figure 2a). Consistent with a role for mir-29b in Mcl-1 regulation, mir-29b is abundantly expressed in H69 cells, and is downregulated in the KMCH cholangiocarcinoma cells (Figure 2b). A microRNA microarray (Genosensor, Tempe AZ), demonstrated that the mir-29b isoform is the most abundant isoform in H69 and KMCH cells (data not shown). The probe employed for the Northern (mir-29a) should hybridize to all mir-29 isoforms under the present conditions; however, only a single band in the range of 18−26 nucleotides was observed (Figure 2b). Likely, the mir-29b product (23 nucleotides) predominates in this cell type. Further evidence that mir-29b is detectible with the mir-29a probe is the observation that transfection with pre-mir-29b results in an increased signal on Northern blot analysis using the mir-29a probe (Figure 3b). Consistent with the Northern blot analysis, the real-time RT-PCR assay demonstrates downregulation of mir-29b in KMCH cells (Figure 2c). In a subset (one-third) of cryopreserved human cholangiocarcinoma specimens (n=6), mir-29b was downregulated approximately 7-fold by real-time RT-PCR. Thus, mir-29b can be downregulated in human cholangiocarcinoma cells and tissue.
Figure 2. Reciprocal expression of mir-29b and Mcl-1.
Panel a: Total cellular protein from immortalized non-malignant H69 cells and malignant KMCH cells was probed using a Mcl-1 antibody. Actin was used as a loading control. Panel b: Northern blot for mir-29b using RNA enriched for small RNAs from H69 and KMCH cells. Note that both precursor and mature mir-29b are decreased in KMCH cells. As a loading control, an RNA probe against the small housekeeping RNA U6 (106 nt) was used. Panel c: Quantitative RT-PCR for mir-29b using total RNA from H69 and KMCH cells. Results expressed as copies of mir-29b per copy of Z30 RNA (mean +/− SEM; p < 0.05).
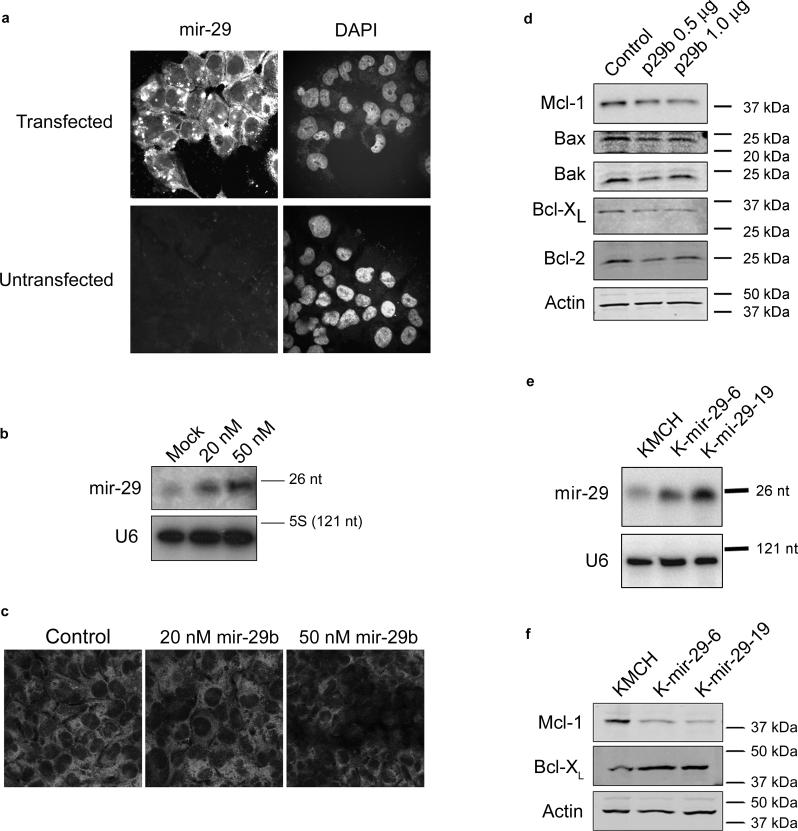
Figure 3. Mcl-1 protein level is negatively regulated by mir-29b.
Panel a: Fluorescently-labeled pre-mir-29b was generated by in vitro transcription (Supplemental Methods) and transfected into KMCH cells. After 24 hours, cells were counter-stained with the nuclear stain DAPI, and photographed. For comparison, untransfected cells are shown. Panel b: Northern blot using small RNA from KMCH cells transfected with 20 or 50 nM precursor mir-29b (not fluorescently-tagged) compared to mock-transfected cells. The mature, processed form (23 nt) is shown, demonstrating that the transfected RNA is processed. U6 was probed as a loading control. Panel c: Mcl-1 immunoreactivity in KMCH cells transfected with precursor mir-29b. A Texas red-conjugated secondary antibody was used, and cells were photographed under confocal microscopy. Panel d: Western blots of total cellular protein from KMCH cells transfected with a mir-29b expression vector (p29b). Control cells were transfected with 1 μg of pCDNA, experimentals were transfected with 0.5 or 1 μg p29b. Actin was used as a loading control. Panel e: Northern blot on total RNA isolated from KMCH cells stably transfected with p29b. Two lines were generated, K-mir-29−6 and K-mir-29−19. The housekeeping RNA U6 was probed as a loading control. Panel f: Western blot of total protein from KMCH, K-mir-29−6, and K-mir-29−19 (stably overexpress mir-29b). Actin was used as a loading control.
mir-29b downregulates Mcl-1 protein
MicroRNAs regulate gene expression through decreased translation, increased degradation of the target message, or both (Valencia-Sanchez et al., 2006). Despite substantial differences in protein expression, real-time PCR demonstrated Mcl-1 messenger RNA copy numbers were similar between H69 and KMCH cells. Specifically, H69 cells expressed 6.98 ± 2.6 copies of Mcl-1 messenger RNA for every 1000 copies of 18S RNA (internal control) and KMCH cells expressed 6.28 ± 0.9 copies of Mcl-1 mRNA per 1000 copies of 18S RNA. This disparity between message and protein expression is consistent with microRNA regulation of translation. To ascertain if mir-29b regulates Mcl-1, we transfected KMCH cells (which have reduced endogenous mir-29b levels) with single-stranded precursor mir-29b. Control experiments were performed in the same manner, except using mock-transfected cells, or cells transfected with an RNA of the same size, antisense to the precursor transfected in experimental cells. The transfection efficiency was examined by transfecting cells with a fluorescently-labeled precursor mir-29b RNA, and was >90% (Figure 3a). Cells were transfected with either 20 or 50 nM mir-29b precursor. Northern blot confirmed that the precursor is processed to mature mir-29b (Figure 3b). Increased expression of mir-29b following transfection was also observed by real-time RT-PCR (not shown). Immunofluorescence demonstrated decreased Mcl-1 reactivity in mir-29b transfected cells (Figure 3c). Similarly, KMCH cells transfected with the mir-29b expression plasmid p29b had reduced Mcl-1 protein by Western blot (Figure 3d). The reduction in Mcl-1 protein was specific as protein levels of Bax, Bak, Bcl-XL, and Bcl-2 were not altered.
To avoid the cellular stress of the transient transfection process, we also generated two KMCH cell lines stably transfected with mir-29b (K-mir-29−6 and K-mir-29−19). Northern blot analysis confirmed the overexpression of mir-29b compared to the parental KMCH cells (Figure 3e). Consistent with our other data, and with a role for mir-29b in regulating Mcl-1, these mir-29b overexpressing cells also manifest reduced steady-state levels of Mcl-1 protein (Figure 3f). Thus mir-29b acts as a regulator of Mcl-1 expression.
mir-29b does not affect Mcl-1 mRNA expression
To determine the effect of mir-29b on Mcl-1 mRNA levels, total RNA was isolated from KMCH cells stably transfected with the mir-29b expression construct p29b and parental cells. Mcl-1 message was quantitated by real-time RT-PCR. The two stable cell lines K-mir-29−6 and K-mir-29−19 had no decrease in Mcl-1 message, expressing 82% and 83% as much Mcl-1 mRNA as parental KMCH cells (p values not significant).
mir-29b acts directly at the Mcl-1 3’UTR
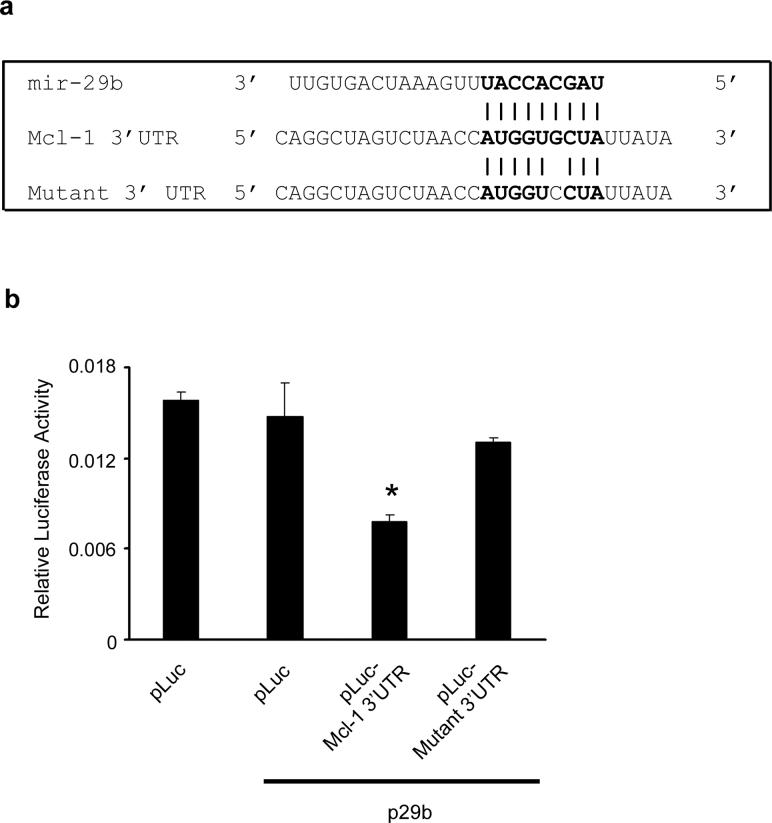
To demonstrate that the negative effect of mir-29b on Mcl-1 expression was direct, we used a luciferase reporter assay. The alignment of mir-29b with the 3’UTR insert is illustrated (Figure 4a). Cotransfection of HeLa cells with the parental luciferase construct (without the Mcl-1 3’UTR) plus the mir-29b expression vector does not significantly change expression of the reporter (Figure 4b). However, when the mir-29 target site from the Mcl-1 3’UTR is inserted into the luciferase construct, expression of luciferase is strongly decreased when cotransfected with mir-29b (Figure 4b). This suppression is relieved by a single base mutation in the binding site (Figure 4b). These data were replicated in KMCH cells (not shown), again the only effect was when both mir-29b and its target sequence were present. Thus, mir-29 directly inhibits expression of Mcl-1 by binding to its target sequence.
Figure 4. Effect of the putative mir-29 binding site derived from the Mcl-1 3’UTR on luciferase expression.
Panel a: Alignment of mir-29b with the insert derived from the Mcl-1 3’UTR. Note the complementarity at the 5’ end of mir-29b, where the crucial seed region is located. A single-base mutant insert was also synthesized, as shown. Inserts were cloned into the 3’ UTR of the p-Mir-Report vector. Panel b: Luciferase activity in HeLa cells transiently transfected with the luciferase construct alone, or cotransfected with an expression plasmid for mir-29b (p29b). Luciferase vectors were parental (pLuc), luciferase with the Mcl-1-derived 3’UTR insert (pLuc-Mcl-1 3’UTR), or luciferase with the mutated insert (pLuc-Mutant 3’UTR). Mean ± SEM, *p<0.01.
mir-29b sensitizes cells to apoptosis
KMCH cholangiocarcinoma cells are resistant to TRAIL-induced cell death compared to their non-malignant counterpart H69 cells due to Mcl-1 expression (Taniai et al., 2004). Therefore, we postulated that transfection of KMCH with mir-29b, which downregulates cellular Mcl-1 levels, would sensitize the cells to TRAIL cytotoxicity. Indeed, transfection with 50 nM precursor mir-29b RNA sensitized KMCH cells to TRAIL-induced apoptosis (Figure 5). Enhanced apoptosis, as assessed by morphology (Figure 5a) or caspase 3/7 activity (Figure 5b) was observed in TRAIL-treated cells following mir-29b transfection. These data suggest that mir-29b downregulation of Mcl-1 is functional as it sensitizes cells to apoptosis. If this concept is correct, then inhibition of mir-29b should increase Mcl-1 expression and increase cellular resistance to apoptosis.
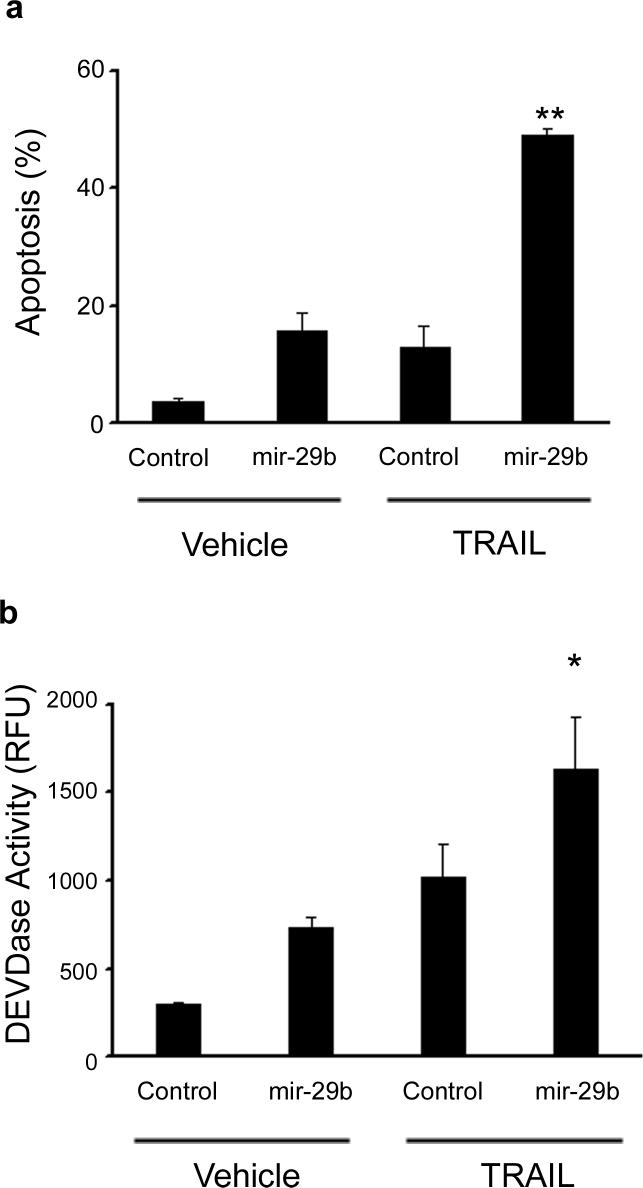
Figure 5. Sensitivity to apoptosis is increased after transfection of mir-29b.
Panel a: KMCH cells were transfected with pre-mir-29b RNA or control RNA of the same length (antisense) at 50 nM. After 22 hours, TRAIL was added where indicated at 2 ng/mL in fresh media and the cells were incubated for 4 hours. Cells were then stained with DAPI and cells with apoptotic morphology were counted. Mean ± SEM, **p<0.001. Panel b: In parallel, cells were transfected and treated with TRAIL as in panel a, but after 4 hours caspase 3/7-like (DEVDase) activity was measured. Mean ± SEM, *p<0.01.
To test this postulate, H69 cells were employed as they abundantly express mir-29 with a corresponding relative reduction of cellular Mcl-1 protein levels. mir-29b function was inhibited by transfecting cells with a locked nucleic acid (LNA) oligonucleotide that binds tightly to mir-29b. Transfection with a mir-29b-targeting LNA increases Mcl-1 cellular protein levels (Figure 6a). The increased cellular Mcl-1 levels due to antagonism of mir-29b by LNA protected the H69 cells from TRAIL cytotoxicity (Figure 6b). Because KMCH cells only weakly express mir-29b, antagonism of mir-29b by LNA transfection in these cells was similar to control, and did not further alter Mcl-1 overexpression (Figure 6a), nor suppress TRAIL cytotoxicity (Figure 6b). Consistent with the morphologic assessment of cell death, LNA transfection protected H69 cells from caspase 3/7 activation upon TRAIL treatment (Figure 6c). KMCH cells were already resistant to TRAIL-induced caspase activation and did not enjoy further protection upon LNA transfection (Figure 6c). Taken together, the data support a role for mir-29b-mediated Mcl-1 regulation that modulates cellular sensitivity to TRAIL.
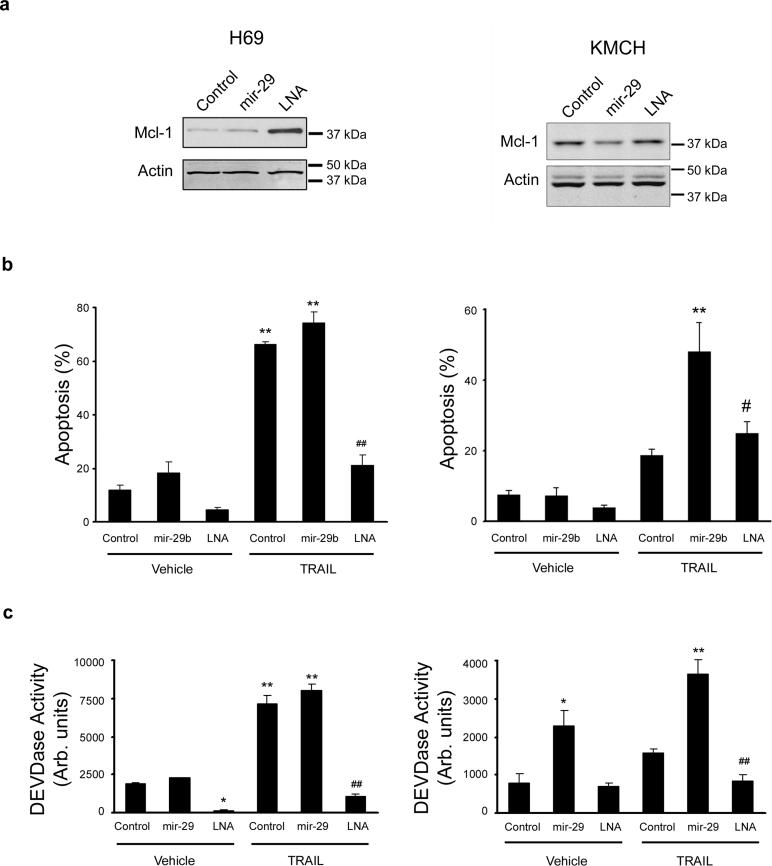
Figure 6. Antagonism of mir-29b protects cells from apoptosis.
Panel a: Western blot of total protein isolated from non-malignant H69 cells and malignant KMCH cells transfected with control RNA, pre-mir-29b, or the mir-29 antagonist oligonucleotide, LNA (50nM each). Panel b. Apoptotic morphology measured as in Figure 5 on H69 and KMCH cells transfected as above. Note that H69 cells are sensitive to TRAIL killing, as noted in previous studies, while KMCH cells are relatively resistant unless mir-29b is transfected. Mean ± SEM, **p<0.0001 compared to vehicle treated control; # p<0.01 compared to TRAIL-treated mir-29; ## p<0.0001 compared to TRAIL-treated mir-29. Panel c: Caspase 3/7-like activity (DEVDase) on cells transfected as above. Caspase activity parallels the apoptosis counts using morphology. Mean ± SEM, *p<0.01 compared to vehicle treated control; **p<0.0001 compared to vehicle treated control; ## p<0.0001 compared to TRAIL-treated mir-29.
DISCUSSION
Several observations suggest that mir-29b regulates Mcl-1 expression. For example, enforced mir-29b expression reduces cellular Mcl-1 protein levels while LNA inhibition of mir-29b function increases Mcl-1 protein expression. The ability of mir-29b to regulate Mcl-1 protein expression is likely direct as it binds to the 3’UTR region of Mcl-1 messenger RNA, with complementarity to the mir-29b seed region. mir-29b appears to inhibit Mcl-1 protein translation as steady state Mcl-1 mRNA levels are similar between KMCH and H69 cells despite their disparate expression of mir-29b, and Mcl-1 mRNA levels are not different in KMCH cells stably overexpressing mir-29b versus parental cells.
Our studies have implications for cancer biology. Increasing evidence suggests that microRNAs play a role in carcinogenesis by altering transcription of oncogenes and tumor suppressor genes (McManus, 2003). Our data extend these concepts by suggesting that decreased mir-29b expression also helps cells evade cell death, a cardinal feature of cancer cells. The decreased expression of mir-29b with concomitant enhancement of Mcl-1 expression may explain in part the up-regulation of Mcl-1 in this cancer. Indeed, a convincing precedent for microRNA regulation of Bcl-2 proteins has been established (Cimmino et al., 2005). Our limited data demonstrating a down-regulation of mir-29b in a subset of human cholangiocarcinomas also supports this concept.
mir-29 may have a broader role in tumor biology in addition to a potential role in cholangiocarcinoma suggested in the current study. For example, mir-29 is also down-regulated in aggressive CLL, colon, and breast cancer (Calin et al., 2005; Cummins et al., 2006; Yanaihara et al., 2006). mir-29a and mir-29b1 are expressed on the same transcript from a locus at chromosome 7q32 which coincides with the common fragile site FRA7H, and may allow for frequent mir-29 down-regulation in these cancers (Calin et al., 2004). Loss of heterozygosity at this site is associated with more aggressive prostate cancer (Neville et al., 2002). Iorio et al. found that mir-29 was down-regulated in aggressive breast cancer specimens; those that lacked estrogen and progestin receptors (mir-29b), and those with vascular invasion (Iorio et al., 2005). The relationship between loss of mir-29b expression in human cancer and regulation of target proteins such as Mcl-1 merits further examination.
In summary, Mcl-1 protein expression can be regulated by mir-29b. These studies extend our knowledge of Mcl-1 expression, a key anti-apoptotic protein which has been referred to as a “linchpin in the network of anti-apoptotic regulators” (van Delft & Huang, 2006). Thus, in addition to transcriptional and post-transcriptional regulation, Mcl-1 is also regulated at the translational level by microRNA, an emerging paradigm for many proteins (Bartel, 2004). Based on these concepts, we propose that the regulation of mir-29b may be a key process in Mcl-1 biology and pathobiology. In particular, mechanisms to enhance mir-29b expression which would reduce cellular Mcl-1 levels could potentially be of value in cancer therapy.
MATERIALS AND METHODS
The Methods for cell lines and human tissue, mir-29 binding site sequencing, transient transfection, microRNA array, fluorescent microRNA transfection, Northern blot conditions, reporter gene assay, and Western blot antibodies are available as Supplemental Materials.
MicroRNA target predictions
Computer-based programs were used to predict microRNAs that potentially bind Mcl-1. Using “MCL1” as a search term, we queried Miranda (John et al., 2004) (http://www.microrna.org/), Pictar (Krek et al., 2005) (http://pictar.bio.nyu.edu/), and miRBase Targets (Griffiths-Jones et al., 2006) (http://microrna.sanger.ac.uk/). Prediction algorithms and known microRNAs change over time, and the analysis included here is from June 2006.
Northern blotting
Total RNA isolation was performed per the manufacturers instructions, and for some experiments, small RNA species were enriched (mirVana kit, Ambion, Austin TX). For Northern blotting, the fraction enriched for small RNAs (1 microgram per well) or total RNA (5 micrograms) was run on a 15% polyacrylamide-urea gel, transferred to a Zeta-probe membrane (BioRad, Hercules CA), and UV-crosslinked.
Real-time Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Real-time RTPCR for human Mcl-1 was performed as described (Yoon et al.) using a capillary thermocycler (Lightcycler, Roche, Mannheim, Germany). For microRNA RT-PCR the dominant family member was determined by the microRNA array (below) and mir-29b, -125b, -153, -193b, and -320 were detected (Applied Biosystems, Foster City CA). As a control, the small housekeeping RNA Z30 was amplified and quantitated. RT-PCR was performed on 50 ng total RNA. The mir-29b reaction was tested on 0.01 ng to 100 ng of target RNA (H69 cells) and the plot of cycle number versus log RNA concentration was linear over this range. The PCR products for mir-29b, -125b, -153, -193b, and -320 were cloned (pCR2.1 TOPO, Invitrogen, Carlsbad CA) and sequenced to confirm the product identity. Products for mir-29b and Z30 were gel purified (single visible band) and quantitated by absorbance at 260 nm to generate a standard curve (linear at least between 103 and 109 copies).
RNA transfection
In vitro transcribed RNA (MegaScript; Ambion, Austin TX) was generated corresponding to approximately the precursor stem-loop of mir-29b-1. This was synthesized using a gel-purified PCR-generated amplicon containing the precursor for mir-29b-1 using genomic DNA as template (forward primer 5’GGTACCGGTTGTCTTGGGTTTATTG; reverse 5’GAATTCAAATACTTCAGAGCTG). The T7 promoter was added at the 5’ end of the forward primer by a second PCR. Control RNA (antisense to the pre-mir-29 sequence) was generated from the same amplicon except that the T7 promoter was separately added to the 5’ end of the reverse primer by PCR.
Antagonism of mir-29b was through transfection of a chimeric locked nucleic acid (LNA)/DNA oligonucleotide designed to hybridize to mir-29. The sequence used was 5’ AACACT *G*A*T*T TCAATGGT *G*C*T*A (Proligo, Boulder CO) where * before a base denotes an LNA base (2’O-4’C-methylene modified).
Luciferase assay
The p-MIR-Report plasmid (Ambion, Austin TX) was modified by insertion of the Mcl-1-derived mir-29b binding site or a single-base mutant thereof into the 3’UTR. These reporters were transfected into HeLa or KMCH cells, and transfection efficiency corrected by a renilla luciferase vector (pRL-CMV, Promega, Madison WI). The p29b expression plasmid was co-transfected where indicated. HeLa cells were chosen because they are readily transfected, and they express low levels of endogenous mir-29 (Lagos-Quintana et al., 2001). Comparable results were observed in KMCH cells.
Western blotting
Proteins were resolved on 12% polyacrylamide gels, transferred to nitrocellulose membrane (Biorad, Hercules CA), and blocked with 5% nonfat dairy milk in Tris-buffered saline (20 mM Tris, 150 mM NaCl, pH 7.4) with 0.1% Tween-20.
Immunofluorescence
Phosphate buffered saline (PBS; 0.9% sodium chloride, 20 mM sodium phosphate, pH 7.4) was used for all wash steps (three times five minutes each). Cells grown on glass coverslips were washed, and then fixed with paraformaldehyde (3% paraformaldehyde in 100 mM PIPES pH 6.95, 1 mM EGTA, 3 mM magnesium sulfate). Cells were washed of fixative and permeabilized with 0.0125% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid (CHAPS; in PBS). Non-specific protein binding was blocked with 5% normal goat serum (Sigma, St. Louis MO), 5% glycerol in PBS. Mcl-1 was detected using a monoclonal antibody (1:250 dilution in blocking buffer, Epitomics, Burlingame CA), washed and detected with Texas-red conjugated secondary antibody (Molecular Probes, Eugene OR). Finally cells were washed with PBS, then rinsed with water and mounted onto slides using ProLong Antifade Kit (Molecular Probes, Eugene OR). Images were captured by confocal microscopy using a Zeiss LSM 510 inverted microscope (Carl Zeiss Inc., Thornwood, New Jersey).
Cell death assays
Cells plated at least one day prior were transfected with pre-mir-29b and incubated 22 hours. TRAIL (R&D Systems, Minneapolis MN) was then added (2 ng/mL final) in fresh media and the cells were treated for 4 hours. Cells were stained with 4’,6-diamidino-2-phenylindole dihydrochloride (DAPI, 1000X stock = 5 mg/mL in water; Sigma, St. Louis MO) and observed under fluorescence microscopy. Apoptosis was quantitated by counting nuclei with characteristic apoptotic changes. Media was changed for untreated cells at the same time TRAIL was added to treated cells to keep conditions the same. Caspase 3/7-like activity (DEVD-ase activity) was measured using the ApoOne Kit (Promega, Madison WI).
Statistical analysis
Data are expressed as mean ± S.E.M., all blots represent at least three separate experiments. Cell death data shown were done in triplicate for each experiment, and are representative of three or more experiments. Comparison of the expression of mir-29b using RT-PCR was made using the Students T-test, for all other analyses multiple comparisons were possible so ANOVA was used with Bonferroni post hoc correction.
ACKNOWLEDGEMENTS
The authors thank Erin Nystuen-Bungum for secretarial assistance. Grant support was from the National Institutes of Health DK59427 (G.J.G).
Supplementary Material
REFERENCES
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuconati A, Mukherjee C, Perez D, White E. DNA damage response and MCL-1 destruction initiate apoptosis in adenovirus-infected cells. Genes Dev. 2003;17:2922–32. doi: 10.1101/gad.1156903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr., Sjoblom T, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–92. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann SH, Karp JE, Svingen PA, Krajewski S, Burke PJ, Gore SD, et al. Elevated expression of the apoptotic regulator Mcl-1 at the time of leukemic relapse. Blood. 1998;91:991–1000. [PubMed] [Google Scholar]
- Kobayashi S, Werneburg NW, Bronk SF, Kaufmann SH, Gores GJ. Interleukin-6 contributes to Mcl-1 up-regulation and TRAIL resistance via an Akt-signaling pathway in cholangiocarcinoma cells. Gastroenterology. 2005;128:2054–65. doi: 10.1053/j.gastro.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Konopleva M, Contractor R, Tsao T, Samudio I, Ruvolo PP, Kitada S, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10:375–88. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. Embo J. 2002;21:4663–70. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu JI, Dumont P, Hafey M, Murphy ME, George DL. Mitochondrial p53 activates Bak and causes disruption of a Bak-Mcl1 complex. Nat Cell Biol. 2004;6:443–50. doi: 10.1038/ncb1123. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- McManus MT. MicroRNAs and cancer. Semin Cancer Biol. 2003;13:253–8. doi: 10.1016/s1044-579x(03)00038-5. [DOI] [PubMed] [Google Scholar]
- Michels J, Johnson PW, Packham G. Mcl-1. Int J Biochem Cell Biol. 2005;37:267–71. doi: 10.1016/j.biocel.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Neville PJ, Conti DV, Paris PL, Levin H, Catalona WJ, Suarez BK, et al. Prostate cancer aggressiveness locus on chromosome 7q32-q33 identified by linkage and allelic imbalance studies. Neoplasia. 2002;4:424–31. doi: 10.1038/sj.neo.7900254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhawan D, Fang M, Traer E, Zhong Q, Gao W, Du F, et al. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev. 2003;17:1475–86. doi: 10.1101/gad.1093903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC, Korsmeyer SJ. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 2003;426:671–6. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- Taniai M, Grambihler A, Higuchi H, Werneburg N, Bronk SF, Farrugia DJ, et al. Mcl-1 mediates tumor necrosis factor-related apoptosis-inducing ligand resistance in human cholangiocarcinoma cells. Cancer Res. 2004;64:3517–24. doi: 10.1158/0008-5472.CAN-03-2770. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- van Delft MF, Huang DC. How the Bcl-2 family of proteins interact to regulate apoptosis. Cell Res. 2006;16:203–13. doi: 10.1038/sj.cr.7310028. [DOI] [PubMed] [Google Scholar]
- Willis SN, Adams JM. Life in the balance: how BH3-only proteins induce apoptosis. Curr Opin Cell Biol. 2005;17:617–25. doi: 10.1016/j.ceb.2005.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuilleme-Toumi S, Robillard N, Gomez P, Moreau P, Le Gouill S, Avet-Loiseau H, et al. Mcl-1 is overexpressed in multiple myeloma and associated with relapse and shorter survival. Leukemia. 2005;19:1248–52. doi: 10.1038/sj.leu.2403784. [DOI] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Yoon J-H, Werneburg NW, Higuchi H, Canbay AE, Kaufmann SH, Akgul C, et al. Bile Acids Inhibit Mcl-1 Protein Turnover via an Epidermal Growth Factor Receptor/Raf-1-dependent Mechanism. Cancer Res. 2002;62:6500–6505. [PubMed] [Google Scholar]
- Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–95. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Zhou P, Levy NB, Xie H, Qian L, Lee CY, Gascoyne RD, et al. MCL1 transgenic mice exhibit a high incidence of B-cell lymphoma manifested as a spectrum of histologic subtypes. Blood. 2001;97:3902–9. doi: 10.1182/blood.v97.12.3902. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.