Abstract
Mammalian cells defective in DNA end-joining are highly sensitive to ionizing radiation and are immunodeficient because of a failure to complete V(D)J recombination. By using cell-free extracts prepared from human lymphoblastoid cell lines, an in vitro system for end-joining has been developed. Intermolecular ligation was found to be accurate and to depend on DNA ligase IV/Xrcc4 and requires Ku70, Ku86, and DNA-PKcs, the three subunits of the DNA-activated protein kinase DNA-PK. Because these activities are involved in the cellular resistance to x-irradiation and V(D)J recombination, the development of this in vitro system provides an important advance in the study of the mechanism of DNA end-joining in human cells.
Keywords: DNA repair/recombination/Ku protein/DNA-PK/DNA ligase
Double-strand breaks are caused, directly or indirectly, by a variety of DNA damaging agents, including ionizing irradiation. To restore the integrity of the genome, cells repair these breaks by (i) homologous recombination, in which an intact chromosome serves as a template for repair, or (ii) nonhomologous end-joining (NHEJ). The latter pathway can be accurate, involving no sequence losses or additions at the joint, or may be inaccurate, a consequence of nucleolytic processing before religation. In simple eukaryotes, such as yeast, homologous recombination provides the dominant pathway for double-strand break repair. Mammalian cells, however, appear to repair the majority of breaks by homology-independent mechanisms.
Mutations in the XRCC4, XRCC5, XRCC6, and XRCC7 genes cause defects in NHEJ resulting in a sensitivity to ionizing radiation and immunodeficiency because of an inability to complete V(D)J recombination (1). XRCC4 encodes a 38-kDa protein that associates with DNA ligase IV and stimulates its ligase activity (2–4), XRCC5 and XRCC6 encode the 86- and 70-kDa subunits of Ku protein (5–7), and XRCC7 encodes the 460-kDa PI 3-like kinase DNA-PKcs (8, 9). DNA-PKcs is activated by association with the Ku heterodimer, which exhibits a high affinity for DNA ends (10–13). In Saccharomyces cerevisiae, homologues of Ku70 and Ku86 have been shown to play a role in NHEJ and telomere maintenance (14–20).
Mammalian cells contain three distinct joining activities, named DNA ligases I, III, and IV, which differ greatly in their ability to promote end-to-end ligation (21–23). Ku stimulates intermolecular end-ligation, but not nick-ligation, by all mammalian DNA ligases, presumably by providing a “bridge” between DNA ends without blocking them from the ligase (24, 25). Of the three ligases, DNA ligase IV in particular has been implicated in NHEJ because it interacts with the XRCC4 gene product (3, 4), and xrcc4 mutants are deficient in end-joining (2, 26, 27). However, the ability of ligase IV to promote end-joining in the presence of Ku and Xrcc4 is poor in vitro (24, 28). For it to promote NHEJ in vivo, ligase IV must, therefore, require other, as yet unknown factors. Unfortunately, efforts to study NHEJ in cell-free extracts so far have met with little success. Indeed, although several in vitro systems for DNA end-joining have been described (29–32), it has not been possible to show that these reactions depend on the factors implicated by genetic studies.
In this paper, we demonstrate that extracts prepared from human cells promote efficient intermolecular end-joining of complementary and blunt ends. The end-joining reaction is mostly accurate, occurring without nucleotide loss or addition, and is catalyzed by DNA ligase IV/Xrcc4. NHEJ by the human cell-free extract requires all activities known to be involved in cellular resistance to x-irradiation and V(D)J recombination, namely Ku70, Ku86, and DNA-PKcs. In addition, end-joining by fractions containing DNA-PK, DNA ligase IV, and Xrcc4 was found to be stimulated by other, as yet unidentified factors.
MATERIALS AND METHODS
Extract Preparation.
GM00558, GM09820, and GM06315 were obtained from the NIGMS Human Genetic Mutant Cell Repository (Camden, NJ). They were grown in suspension to a density of 8 × 105 cells/ml and were harvested and washed once in DMEM containing 10% fetal calf serum, three times in ice-cold PBS, and once in hypotonic lysis buffer (10 mM Tris⋅HCl, pH 8.0/1 mM EDTA/5 mM DTT). Cells were resuspended in 2 vol of hypotonic buffer and, after 20 min at 0°C, were lysed by homogenization (20 strokes with an “A” pestle), and protease inhibitors were added (phenylmethylsulfonyl fluoride, 0.17 mg/ml; aprotinin, 0.01 trypsin inhibitor units/ml; pepstatin, 1 μg/ml; chymostatin, 1 μg/ml; leupeptin, 1 μg/ml). After 20 min on ice, 0.5 vol of high salt buffer (50 mM Tris⋅HCl, pH 7.5/1 M KCl/2 mM EDTA/2 mM DTT) was added, and the extract was centrifuged for 3 hours at 42,000 rpm in a Beckman SW50.1 rotor. The supernatant was dialyzed for 3 hours against E buffer [20 mM Tris⋅HCl, pH 8.0/0.1 M KOAc/20% (vol/vol) glycerol/0.5 mM EDTA/1 mM DTT] and was fast-frozen and stored at −70°C.
For fractionation studies, GM00558 extract (10 mg) was loaded onto a 1-ml phosphocellulose column equilibrated in E buffer, and proteins were eluted by using E buffer containing 0.15, 0.5, or 0.9 M KCl. Peak fractions (0.5 ml) designated PC-A (column flow through; 3.4 mg/ml), PC-B (0.15 M step; 1.8 mg/ml), PC-C (0.5 M step; 2.2 mg/ml), and PC-D (0.9 M step; 0.34 mg/ml) were dialyzed against E buffer.
Proteins and DNA.
Xrcc4/ligase IV (MonoQ fraction 11; ref. 22), DNA ligase III (33), and DNA-PK (8) generously were provided by the laboratories of T. Lindahl (Imperial Cancer Research Fund) and S. P. Jackson (Cambridge University). Form I pDEA-7Z duplex plasmid DNA (3.0 kilobases) (34), was linearized with BsaI (NEB), was dephosphorylated, and was 5′-32P-labeled by using polynucleotide kinase. Uniformly 32P-labeled form I pFB585 DNA (7.7 kilobases) was prepared by growth in the presence of [32P]orthophosphate.
End-Joining.
Reactions (10 μl) were carried out in 50 mM triethanolamine⋅HCl (pH 7.5), 0.5 mM Mg(OAc)2, 60 mM KOAc, 2 mM ATP, 1 mM DTT, and 100 μg/ml BSA. Cell-free extract was incubated for 5 min at 37°C before the addition of 32P-labeled DNA (10 ng). Incubation was for 1 hour at 37°C. 32P-labeled DNA products were deproteinized and analyzed by electrophoresis through 0.6% agarose gels followed by autoradiography. Quantification of joining efficiency was carried out by phosphorimaging. Experiments with the DNA-PK inhibitors wortmannin or LY294002 (Sigma) were carried out by incubation of the inhibitor with extract for 30 min on ice before a shift to 37°C (10 min) and addition of 32P-labeled DNA. Incubation then was continued for 1 hour.
Antibodies.
Rabbit polyclonal sera against Xrcc4 (3), ATM (35), and DNA ligase IV (S. Critchlow and S. P. Jackson, personal communication) were provided by S. P. Jackson. Antisera against DNA ligases I (36) and III (37) were provided by T. Lindahl, and those against Ku70, Ku86, and DNA-PKcs were purchased from Serotec. Purified antibodies against DNA-PKcs used for Western blotting were from Calbiochem.
For immunodepletion, GM00558 extract (50 μg) was incubated on a rotary wheel with polyclonal antiserum (3 μl) at 4°C for 90 min (50 μl total volume). The extract/antibody mixture was added to protein A-Sepharose beads and was incubated with rotation for 60 min. The beads then were removed by repeated centrifugation. Western blotting was carried out by standard procedures, and proteins were visualized by ECL (Amersham).
RESULTS
DNA End-Joining in Vitro.
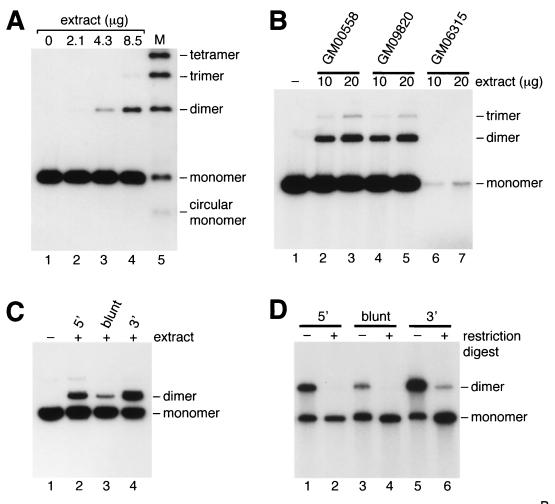
Nonhomologous end-joining catalyzed by extracts of human lymphoblastoid cell line GM00558 was observed by the ligation of 5′-32P-end-labeled linear duplex DNA (Fig. 1A, lanes 3 and 4). Approximately 10–20% of the input DNA molecules were ligated during a 1-hour incubation in a reaction that depended on the presence of ATP and Mg2+. End-joining activity was observed with extracts prepared from a range of lymphoblastoid cell lines (Fig. 1B), although some extracts (e.g., GM06315; see Fig. 1B, lanes 6 and 7) showed higher levels of nuclease activity, and end-joining could not be analyzed.
Figure 1.
DNA end-joining catalyzed by human cell-free extracts. (A) Protein extracts from GM00558 were incubated with 5′-32P-end-labeled BsaI-linearized pDEA-7Z DNA as described in Materials and Methods (lanes 1–4). Lane 5, DNA ligation ladder. (B) Extracts of three lymphoblastoid cell lines were analyzed for their ability to promote end-joining under standard assay conditions. (C) End-joining catalyzed by GM00558 extract (68 μg) was analyzed by using uniformly 32P-labeled pFB585 DNA (100 ng) linearized with BsaI (lanes 1 and 2), EcoRV (lane 3), or KpnI (lane 4). (D) Ligation products from reactions similar to those shown in C were purified by gel electrophoresis and treated with (+) or without (−) BsaI (lanes 1 and 2), EcoRV (lanes 3 and 4), or KpnI (lanes 5 and 6).
Double-strand breaks containing 3′-overhangs provided the best substrate for rejoining, as seen by comparing the ability of extract to rejoin linear DNA containing 5′-overhangs, blunt ends, or 3′-overhangs (Fig. 1C). Indeed, 27% of the 3′-overhang substrate was converted into dimers (Fig. 1C, lane 4), compared with 13% of the 5′-overhangs (Fig. 1C, lane 2) and 6% of the blunt-ended substrate (Fig. 1C, lane 3). The ligated products could be recut with the same restriction enzyme (Fig. 1D, lanes 2, 4 and 6), indicating that repair occurred without loss or addition of nucleotide sequences at the joint. Only in the case of the 3′-substrate was a fraction (≈5%) of the product resistant to cleavage (Fig. 1D, lane 6).
Requirement for DNA Ligase IV and Xrcc4.
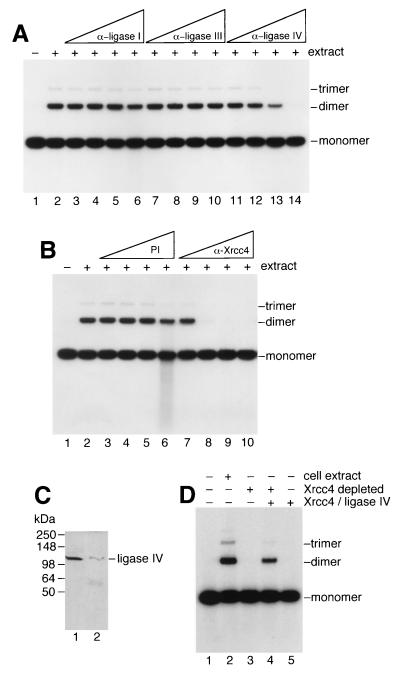
To determine whether the DNA end-joining activity could be attributed to a specific DNA ligase, extracts prepared from GM00558 were preincubated with polyclonal antisera raised against human DNA ligase I, ligase III, or ligase IV before addition of the 32P-end-labeled linear DNA and subsequent incubation. We found that antisera against DNA ligase IV inhibited end-joining (Fig. 2A, lanes 11–14) whereas antisera against either ligase I (Fig. 2A, lanes 3–6) or ligase III (Fig. 2A, lanes 7–10) did not.
Figure 2.
Involvement of DNA ligase IV and Xrcc4 in end-joining. (A) End-joining reactions were carried out as described for Fig. 1 by using GM00558 extract (34 μg) but contained polyclonal antisera (1 μl neat, lanes 6, 10, 14; diluted 1/5, lanes 5, 9, 13; diluted 1/25, lanes 4, 8, 12; or diluted 1/125, lanes 3, 7, 11) against DNA ligases I, III or IV as indicated. Lanes: 1, no extract; 2, complete reaction but without antiserum. Extract was incubated for 30 min on ice with antiserum, then was transferred to 37°C for 10 min followed by the addition of 32P-end-labeled DNA (10 ng). After 1-hour incubation, DNA products were analyzed by agarose gel electrophoresis. (B) Reactions were carried out as described in A by using preimmune serum (lanes 3–6) or anti-Xrcc4 antiserum (lanes 7–10). (C) Immunodepletion of Xrcc4/Ligase IV determined by Western analysis. Lanes 1 and 2, extract before and after immunodepletion of Xrcc4/DNA ligase IV by using anti-Xrcc4 antiserum. (D) Analysis of end-joining before and after Xrcc4/ligase IV immunodepletion. The presence of cell extract or immunodepleted extract is indicated. Purified Xrcc4/ligase IV was added where shown. End-joining assays were carried out as described for Fig. 1.
Because DNA ligase IV associates tightly with Xrcc4 (3, 4), we analyzed whether antiserum against Xrcc4 also could inhibit ligation. Fig. 2B shows that the anti-Xrcc4 serum abolished end-joining activity (Fig. 2B, lanes 7–10) whereas preimmune serum did not (Fig. 2B, lanes 3–6). None of the antisera used above affected the activity of T4 DNA ligase.
To confirm the requirement for Xrcc4, antiserum was used to immunodeplete Xrcc4 from the extract. As predicted, the Xrcc4-immunodepleted extract failed to promote end-joining (Fig. 2D, lane 3). Because Xrcc4/ligase IV complexes are highly stable (3, 4), immunodepletion also removed the majority of the ligase IV (Fig. 2C, compare lanes 1 and 2). Activity was reconstituted by the addition of purified Xrcc4/ligase IV complex (Fig. 2D, lane 4). Thus, DNA ligase IV and Xrcc4 are required for DNA end-joining catalyzed by human cell-free extracts. However, ligase IV/Xrcc4 were insufficient for the reaction because they failed to promote end-joining in the absence of extract (Fig. 2D, lane 5).
Inhibition by DNA-PK Antisera.
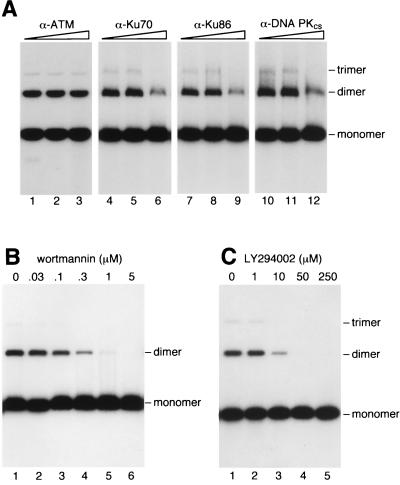
Antisera against the three subunits of DNA-PK (Ku70, Ku86, and DNA-PKcs) also reduced end-joining activity (Fig. 3A, lanes 4–12) whereas a control serum (anti-ATM) had no effect (Fig. 3A, lanes 1–3). Inhibition was, however, less than complete, because of either the large size of DNA-PK (>600 kDa) or the abundance of DNA-PK within the cell (38). Inhibitors of DNA-PKcs such as wortmannin (9) and LY294002 (G. M. Smith and S. P. Jackson, personal communication) also blocked DNA end-joining in vitro (Fig. 3 B and C).
Figure 3.
Involvement of DNA-PK in end-joining. (A) End-joining reactions contained GM00558 cell-free extract (20 μg) and 32P-labeled linear DNA. Reactions were supplemented with dilutions of antisera and analyzed as described for Fig. 2. (B and C) Inhibition of end-joining by the DNA-PK inhibitors wortmannin and LY294002.
Fractionation and Reconstitution of End-Joining Activity.
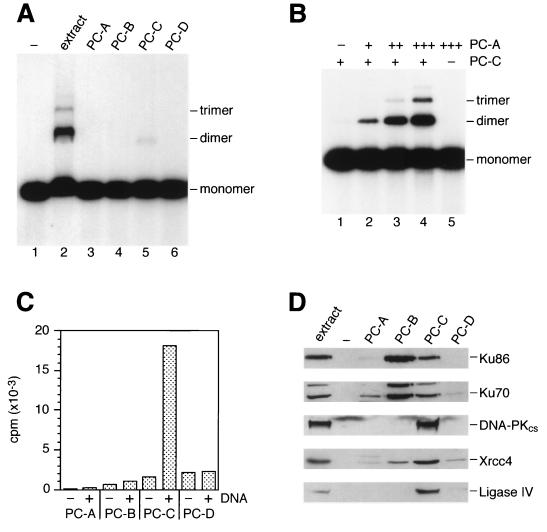
To determine whether factors other than those described above were required for DNA end-joining in vitro, the extract was fractionated by phosphocellulose chromatography. The unbound fraction (PC-A) and proteins that bound to the resin and eluted with three steps of increasing salt concentration (designated PC-B, PC-C, and PC-D) were tested for end-joining activity. Fraction PC-C promoted low levels of end-joining (Fig. 4A, lane 5) whereas the others did not (Fig. 4A, lanes 3, 4, and 6). When fractions PC-A and PC-C were mixed, however, efficient end-joining activity was reconstituted such that the specific activity was greater than that of the unfractionated extract (Fig. 4B, lanes 2–4). As expected, the reconstituted activity depended on DNA-PK and Xrcc4/ligase IV, as shown by using LY294002 and anti-Xrcc4 antiserum.
Figure 4.
Fractionation of end-joining activity. (A) GM00558 extract was fractionated by phosphocellulose chromatography as described in Materials and Methods. End-joining assays then were carried out by using crude extract (20 μg) or fractions PC-A, PC-B, PC-C, and PC-D (2 μl of each) as indicated. (B) Fraction PC-C (2.2 μg) was mixed on ice with 0, 0.3, 1.0, or 3.0 μg of fraction PC-A (lanes 1–4). After 5 min at 37°C, 32P-labeled linear DNA was added, and end-joining reactions were carried out for 1 hour. Lane 5, reaction contained PC-A (3 μg) only. (C) DNA-PK activity was measured by incubating 3.3 μg of the four fractions with biotinylated peptide substrate in the absence (−) and presence (+) of sonicated calf thymus DNA by using the Promega SignaTECT DNA-PK assay system. (D) Western blotting. Membranes probed with antisera against Ku70, Ku86, and DNA-PKcs contained 8 μg of total protein per gel lane, and those probed with anti-Xrcc4 and anti-ligase IV contained 30 μg of protein per lane.
Because PC-A and PC-C were sufficient for end-joining in vitro, each fraction was analyzed by immunoblotting for the presence of DNA-PK, ligase IV, and Xrcc4. Fraction PC-C contained Ku70, Ku86, DNA-PKcs, DNA ligase IV, and Xrcc4, although the majority of the Ku70/86 heterodimer was found in PC-B (Fig. 4D). DNA-dependent kinase activity was detected in fraction PC-C (Fig. 4C). Because substantial amounts of Ku70, Ku86, DNA-PKcs, DNA ligase IV, and Xrcc4 were in PC-C, the requirement for PC-A for end-joining was surprising and suggestive of a need for as yet unidentified factor(s). Whether these factor(s) are specific for DNA-PK-dependent end-joining is presently unknown, although specificity is indicated by observations showing that PC-A failed to stimulate end-joining by T4 DNA ligase or human DNA ligase III.
During these fractionation studies, we observed additional end-joining activities that were not inhibited by anti-Xrcc4 antibodies or DNA-PK inhibitors (data not shown). Similarly, when cell-free extracts were prepared by other protocols (39), joining activities were observed that did not show the same factor requirements as those reported here. Moreover, we found that rodent extracts promoted highly efficient end-joining reactions that were inhibited only partially by loss of DNA-PK function, either through mutation, antibody inhibition, or the use of chemical inhibitors (data not shown). These end-joining activities may be similar to those reported previously (29–32), and their relationship to DNA-PK-dependent reactions will require further investigation.
DISCUSSION
In this paper, we have described an in vitro system for NHEJ that has been shown to depend on factors implicated by genetic studies in DNA repair, namely the products of XRCC4 (Xrcc4 protein), XRCC5 (Ku86), XRCC6 (Ku70), and XRCC7 (DNA-PKcs). Genetic characterization of radiation-sensitive rodent cell lines has generated a large body of evidence suggesting crucial roles for DNA-PKcs, Ku70/86, and Xrcc4 in NHEJ. Ku binds duplex DNA ends and can transfer between two complementary DNA termini (7, 40–42). Indeed, direct imaging revealed that DNA termini bound by Ku associate with each other (25), indicating that Ku may provide a bridge between DNA molecules that (i) protects DNA ends from nucleolytic digestion and (ii) facilitates intermolecular DNA ligation. Consistent with a role as a DNA end alignment factor, Ku stimulates end-joining by purified DNA ligases (24), and cells lacking Ku exhibit a high frequency of imprecise end-joining (19, 43).
DNA-PKcs associates with the Ku heterodimer to form the DNA-activated protein kinase DNA-PK (11–13). The role of DNA-PKcs in end-joining is, however, presently unclear. In addition to binding termini via interaction with Ku (which again could serve an end-protection function), DNA-PKcs may regulate other components of the repair apparatus through its kinase activity. Because Xrcc4 forms a stable complex with DNA ligase IV and can be phosphorylated by DNA-PK in vitro (3, 44), it is tempting to speculate that interactions between Xrcc4 and DNA-PK result in the specific targeting of DNA ligase IV to the end-joining complex.
Although Ku stimulates all three DNA ligases in vitro (24), the x-ray sensitivity of xrcc4 mutant cell lines indicates that DNA ligase IV in particular promotes ligation during NHEJ. Here, we have shown that DNA ligase IV/Xrcc4 is indeed required for accurate end-joining by human cell-free extracts. Further evidence for an involvement of DNA ligase IV/Xrcc4 in end-joining recently was obtained in S. cerevisiae because disruption of LIG4 (ligase IV homologue) and LIF1 (Xrcc4 homologue) resulted in severe NHEJ defects similar to those observed in yku70 and yku86 mutants (45–48).
The in vitro system described here is a powerful tool that will help determine the role of DNA-PK in end-joining. Because our initial fractionation studies indicate that factors other than Ku70, Ku86, DNA-PKcs, DNA ligase IV, and Xrcc4 are required for efficient repair, this system should permit their isolation. Furthermore, we find that DNA end-joining activity is observed when a DNA-PK immunoprecipitate is incubated with ligase IV/Xrcc4 (unpublished data), raising the possibility that other components of the end-joining machinery might interact directly with DNA-PK and that DNA-PK itself could be used in their purification.
Acknowledgments
We are indebted to the Jackson and Lindahl laboratories for the generous gifts of antibodies and proteins, and to Deborah Barnes, Richard Bowater, Steve Jackson, Tomas Lindahl, Peter Robins, and Graeme Smith for helpful discussions. This work was supported by the Imperial Cancer Research Fund and the Human Frontiers Science Program.
ABBREVIATION
- NHEJ
nonhomologous end-joining
References
- 1.Thompson L H, Jeggo P A. Mutat Res. 1995;337:131–137. doi: 10.1016/0921-8777(95)00018-f. [DOI] [PubMed] [Google Scholar]
- 2.Li Z Y, Otevrel T, Gao Y J, Cheng H L, Seed B, Stamato T D, Taccioli G E, Alt F W. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 3.Critchlow S E, Bowater R P, Jackson S P. Curr Biol. 1997;7:588–598. doi: 10.1016/s0960-9822(06)00258-2. [DOI] [PubMed] [Google Scholar]
- 4.Grawunder U, Wilm M, Wu X T, Kulesza P, Wilson T E, Mann M, Lieber M R. Nature (London) 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 5.Errami A, Smider V, Rathmell W K, He D M, Hendrickson E A, Zdzienicka M Z, Chu G. Mol Cell Biol. 1996;16:1519–1526. doi: 10.1128/mcb.16.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singleton B K, Priestley A, Steingrimsdottir H, Gell D, Blunt T, Jackson S P, Lehmann A R, Jeggo P A. Mol Cell Biol. 1997;17:1264–1273. doi: 10.1128/mcb.17.3.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Getts R C, Stamato T D. J Biol Chem. 1994;269:15981–15984. [PubMed] [Google Scholar]
- 8.Blunt T, Finnie N J, Taccioli G E, Smith G C M, Demengeot J, Gottlieb T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A, et al. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 9.Hartley K O, Gell D, Smith G C M, Zhang H, Divecha N, Connelly M A, Admon A, Leesmiller S P, Anderson C W, Jackson S P. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 10.Mimori T, Hardin J A. J Biol Chem. 1986;261:10375–10379. [PubMed] [Google Scholar]
- 11.Gottlieb T M, Jackson S P. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 12.Yaneva M, Kowalewski T, Lieber M R. EMBO J. 1997;16:5098–5112. doi: 10.1093/emboj/16.16.5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammarsten O, Chu G. Proc Natl Acad Sci USA. 1998;95:525–530. doi: 10.1073/pnas.95.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feldmann H, Driller L, Meier B, Mages G, Kellermann J, Winnacker E L. J Biol Chem. 1996;271:27765–27769. doi: 10.1074/jbc.271.44.27765. [DOI] [PubMed] [Google Scholar]
- 15.Mages G J, Feldmann H M, Winnacker E L. J Biol Chem. 1996;271:7910–7915. doi: 10.1074/jbc.271.14.7910. [DOI] [PubMed] [Google Scholar]
- 16.Milne G T, Jin S F, Shannon K B, Weaver D T. Mol Cell Biol. 1996;16:4189–4198. doi: 10.1128/mcb.16.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siede W, Friedl A A, Dianova I, Eckardt-Schupp F, Friedberg E C. Genetics. 1996;142:91–102. doi: 10.1093/genetics/142.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boulton S J, Jackson S P. Nucleic Acids Res. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boulton S J, Jackson S P. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 20.Boulton S J, Jackson S P. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindahl T, Barnes D E. Annu Rev Biochem. 1992;61:251–281. doi: 10.1146/annurev.bi.61.070192.001343. [DOI] [PubMed] [Google Scholar]
- 22.Robins P, Lindahl T. J Biol Chem. 1996;271:24257–24261. doi: 10.1074/jbc.271.39.24257. [DOI] [PubMed] [Google Scholar]
- 23.Tomkinson A E, Levin D S. BioEssays. 1997;19:893–901. doi: 10.1002/bies.950191009. [DOI] [PubMed] [Google Scholar]
- 24.Ramsden D A, Gellert M. EMBO J. 1998;17:609–614. doi: 10.1093/emboj/17.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pang D, Yoo S, Dynan W S, Jung M, Dritschilo A. Cancer Res. 1997;57:1412–1415. [PubMed] [Google Scholar]
- 26.Jeggo P A. Mutat Res. 1990;239:1–16. doi: 10.1016/0165-1110(90)90028-a. [DOI] [PubMed] [Google Scholar]
- 27.Taccioli G E, Rathbun G, Oltz E, Stamato T, Jeggo P A, Alt F W. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 28.Ramsden D A, Paull T T, Gellert M. Nature (London) 1997;388:488–491. doi: 10.1038/41351. [DOI] [PubMed] [Google Scholar]
- 29.Bøe S-O, Sodroski J, Helland D E, Farnet C M. Biochem Biophys Res Commun. 1995;215:987–993. doi: 10.1006/bbrc.1995.2561. [DOI] [PubMed] [Google Scholar]
- 30.Fairman M P, Johnson A P, Thacker J. Nucleic Acids Res. 1992;20:4145–4152. doi: 10.1093/nar/20.16.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson A P, Fairman M P. Mutat Res. 1996;364:103–116. doi: 10.1016/0921-8777(96)00028-6. [DOI] [PubMed] [Google Scholar]
- 32.Mason R M, Thacker J, Fairman M P. Nucleic Acids Res. 1996;24:4946–4953. doi: 10.1093/nar/24.24.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nash R A, Caldecott K W, Barnes D E, Lindahl T. Biochemistry. 1997;36:5207–5211. doi: 10.1021/bi962281m. [DOI] [PubMed] [Google Scholar]
- 34.Shah R, Bennett R J, West S C. Nucleic Acids Res. 1994;22:2490–2497. doi: 10.1093/nar/22.13.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lakin N D, Weber P, Stankovic T, Rottinghaus S T, Taylor A M, Jackson S P. Oncogene. 1996;13:2707–2716. [PubMed] [Google Scholar]
- 36.Lasko D D, Tomkinson A E, Lindahl T. J Biol Chem. 1990;265:12618–12622. [PubMed] [Google Scholar]
- 37.Caldecott K W, Aoufouchi S, Johnson P, Shall S. Nucleic Acids Res. 1996;24:4387–4394. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson C W, Carter T H. In: Molecular Analysis of DNA Rearrangements in the Immune System. Jessberger R, Lieber M R, editors. Heidelberg: Springer; 1996. pp. 91–112. [Google Scholar]
- 39.Manley J L, Fire A, Cano A, Sharp P A, Gefter M L. Proc Natl Acad Sci USA. 1980;77:3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taccioli G E, Gottlieb T M, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann A R, Alt F W, Jackson S P, Jeggo P A. Science. 1994;265:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 41.Bliss T M, Lane D P. J Biol Chem. 1997;272:5765–5773. doi: 10.1074/jbc.272.9.5765. [DOI] [PubMed] [Google Scholar]
- 42.Cary R B, Peterson S R, Wang J T, Bear D G, Bradbury E M, Chen D J. Proc Natl Acad Sci USA. 1997;94:4267–4272. doi: 10.1073/pnas.94.9.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang F, Jasin M. J Biol Chem. 1996;271:14405–14411. doi: 10.1074/jbc.271.24.14405. [DOI] [PubMed] [Google Scholar]
- 44.Leber R, Wise T W, Mizuta R, Meek K. J Biol Chem. 1998;273:1794–1801. doi: 10.1074/jbc.273.3.1794. [DOI] [PubMed] [Google Scholar]
- 45.Schär P, Herrmann G, Daly G, Lindahl T. Genes Dev. 1997;11:1912–1924. doi: 10.1101/gad.11.15.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teo S H, Jackson S P. EMBO J. 1997;16:4788–4795. doi: 10.1093/emboj/16.15.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herrmann G, Lindahl T, Schär P. EMBO J. 1998;17:4188–4198. doi: 10.1093/emboj/17.14.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson T E, Grawunder U, Lieber M R. Nature (London) 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]