Abstract
G protein-coupled receptor activation leads to the membrane recruitment and activation of G protein-coupled receptor kinases, which phosphorylate receptors and lead to their inactivation. We have identified a novel G protein-coupled receptor kinase-interacting protein, GIT1, that is a GTPase-activating protein (GAP) for the ADP ribosylation factor (ARF) family of small GTP-binding proteins. Overexpression of GIT1 leads to reduced β2-adrenergic receptor signaling and increased receptor phosphorylation, which result from reduced receptor internalization and resensitization. These cellular effects of GIT1 require its intact ARF GAP activity and do not reflect regulation of GRK kinase activity. These results suggest an essential role for ARF proteins in regulating β2-adrenergic receptor endocytosis. Moreover, they provide a mechanism for integration of receptor activation and endocytosis through regulation of ARF protein activation by GRK-mediated recruitment of the GIT1 ARF GAP to the plasma membrane.
G protein-coupled receptors control cellular responses through the activation of heterotrimeric G proteins, which alter the activity of intracellular messenger-generating enzymes such as adenylyl cyclase (1, 2). Agonist activation of G protein-coupled receptors, such as the prototypical β2-adrenergic receptor (β2AR), also leads to the activation and membrane recruitment of G protein-coupled receptor kinases (GRKs) (2, 3). The GRKs phosphorylate the activated receptor protein, promoting the binding of arrestin proteins to the phosphorylated receptor and the uncoupling of the receptor from further G protein activation (2, 3). This GRK/arrestin pathway thus promotes the desensitization of agonist-activated receptors. GRK phosphorylation of activated receptors followed by arrestin binding also promotes both the sequestration of the inactivated receptor proteins from the cell surface and their resensitization by dephosphorylation in an intracellular acidic vesicle compartment (2, 3).
Like the G proteins themselves, GRKs recognize and respond to the activation state of the receptors. First, GRKs “translocate” in response to receptor activation to the cellular membranes where the receptors are found (2, 3). Second, GRK kinase activity is directly activated by interaction with agonist-activated receptors (2, 3). Thus GRKs can be considered as effectors for G protein-coupled receptors, functioning in a negative feedback manner to regulate the receptors themselves. To investigate the potential for GRKs to serve a more direct signaling role by acting as adapters to bring associated proteins to the membrane in response to agonist activation of receptors, we searched for proteins that interact with GRK2 and identified a novel protein that appears to be involved in coordinating receptor activation and sequestration.
METHODS
Yeast Two-Hybrid Screening.
Bovine GRK2 cDNA was inserted into the pGBT9 vector (CLONTECH) to allow expression of a Gal4 DNA-binding domain–GRK2 fusion. Yeast strain HF7c was transformed with pGBT9-GRK2, fusion protein expression was confirmed by Western blotting by using a Gal4 antibody (Santa Cruz Biotechnology), and the yeast was transformed further with a rat brain cDNA library in pGAD10 (CLONTECH) following the CLONTECH MatchMaker protocol. Colonies were selected for growth on His−/Leu−/Trp− medium containing 10 mM 3-aminotriazole and for expression of β-galactosidase in filter assays. Plasmid DNA was isolated from yeast by phenol extraction and recovered in HB101 strain Escherichia coli by selection on minimal medium plates without Leu.
Cloning of Full-Length GIT1.
The full-length rat GIT1 cDNA sequence was obtained by two rounds of 5′ rapid amplification of cDNA ends (RACE) reactions by using specific primers and rat brain first-strand cDNA (CLONTECH, 5′RACE-ready cDNA). The full-length expression clone was reconstructed from a 5′ amplified cassette ligated with the pGAD10 insert into a modified pBK-CMV expression vector (Stratagene). The pBK-Flag-GIT1 carboxyl-terminal construct was prepared by inserting the EcoRI fragment of the original pGAD10 clone into a pBK-CMV vector containing a small sequence encoding an initiator Met codon followed by codons for a Flag epitope (DYKDDDDA). The pBK-Δ45-GIT1 construct was prepared by amplification using a 5′ primer that adds an initiator Met codon immediately before codon 46. All constructs were sequenced on both strands from specific primers by using automated dye terminator chemistry with AmpliTaq FS reagents (Applied Biosystems) and an ABI 377 instrument.
Northern blotting was performed on a rat multiple-tissue Northern blot (CLONTECH) by using random-primed labeled probe and ExpressHyb solution (CLONTECH) and following the manufacturer’s protocol. The human GIT1 gene was localized to a human chromosome region by amplification of the 3′ untranslated region (based on human expressed sequence tag sequences) from genomic DNA isolated from the Stanford G3 radiation hybrid cell line panel containing defined fragments of human chromosomes (Research Genetics) (4).
Cell Culture and Immunoprecipitation.
A GIT1 antiserum was prepared by using GST-GIT1(375–770) fusion protein. The EcoRI fragment of the pGAD10 clone was inserted into the pGEX-4T-1 vector (Pharmacia), and fusion protein was expressed and purified on glutathione agarose as described previously (5) and used to immunize rabbits (Cocalico, Reamstown, PA). Immune serum was used for Western blots at 1:2,000.
COS-7 cells were transfected with 10 μg of plasmid DNAs together with 60 μl Lipofectamine (Life Technologies, Gaithersburg, MD). Flag β2AR and GRK expression clones and GRK antisera were described previously (5–7). Cells were harvested by scraping into 20 mM Tris, pH 7.4/1 mM EDTA/20 mM NaCl, lysed, and centrifuged at 14,000 × g. The soluble fraction was rotated for 1 h at 4°C after addition of 5 μg of M2 anti-Flag antibody (Kodak) and for an additional 1 h after addition of Protein G Plus/Protein A-agarose beads (Calbiochem). Beads were washed three times with lysis buffer, and bound proteins were eluted by boiling with SDS/PAGE sample buffer. Samples were separated in 10% polyacrylamide gels and transferred to nitrocellulose for immunoblotting as described previously (5).
Stable Overexpression of GIT1 and Cellular Signaling Assays.
HEK293 cells were transfected with pBK-GIT1 or pBK-Δ45-GIT1 vector and selected for 14 days in medium containing 400 μg/ml gentimycin (GIBCO). Surviving colonies were pooled and assayed for protein expression by Western blotting.
Cells labeled with [3H]adenine were incubated with 10 μM isoproterenol for the indicated times, and accumulated cAMP was assessed by column chromatography as described previously (7, 8). Data are normalized for total cellular [3H]adenine incorporation and for column recovery using [14C]cAMP.
Receptor phosphorylation was assessed after labeling of the intracellular ATP pool by incubating Flag-β2AR transfected cells with [32P]orthophosphate (100 μCi/ml) for 1 h in phosphate-free MEM/20 mM Hepes, pH 7.4, at 37°C as described previously (7). Cells were stimulated with 10 μM isoproterenol for 2 min and solubilized in 1 ml of Flag-RIPA (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) buffer. The receptor was immunoprecipitated from equivalent amounts of extract proteins (7–20 μg) with the M2 anti-Flag mAb (12 μg). Radioactive bands in dried gels were quantified by using a Molecular Dynamics PhosphorImager system and imagequant program, and then exposed to film.
Receptor sequestration was determined as described previously (6) by using Flag β2AR transiently transfected in HEK293 cells or in cells stably overexpressing GIT1. Receptor sequestration was defined as the fraction of cell surface receptor on naïve cells that was no longer accessible to antibodies outside the cells after 10-μM isoproterenol treatment. Baseline fluorescence from cells not transfected with the Flag β2AR was subtracted from each sample.
Purification of Recombinant GIT1.
GIT1 and Δ45-GIT1 sequences were modified to fuse 6xHis residues on the carboxyl terminal. Sf9 cells infected with recombinant baculoviruses were harvested in 20 mM Tris (pH 7.4) with a mixture of protease inhibitors. Cell lysates were prepared by using a Dounce homogenizer, and NaCl was added to 250 mM. The 20,000 × g supernatant was rotated for 2 h at 4°C with ProBond resin (Invitrogen). Beads were washed with 20 mM Tris, pH 7.4/250 mM NaCl and eluted with 20 mM Tris, pH 7.4/100 mM imidazole. The eluate was applied to a 5-ml HiTrap-Q column (Pharmacia) and eluted with a NaCl gradient in 20 mM Tris, pH 7.4/1 mM EDTA. Pooled eluate fractions were desalted, concentrated, and stored frozen at −120°C with 30% glycerol.
Kinase and Translocation Assays.
Bovine rod outer segment membranes, vesicle-reconstituted purified β2AR, bovine brain βγ-subunits, and recombinant GRK2 were prepared as described previously (5, 9). Rod outer segment membranes (20 μg) or vesicles containing β2AR (2 pmol) were mixed with 25 ng GRK2 and 250 ng GIT1 as indicated, in the presence of 100 μM [γ-32P]ATP in a 25-μl volume. β2AR assays also contained 150 nM βγ-subunits. Mixtures were incubated for 10 min at 30°C in the light or with 20 μM isoproterenol or 5 μM alprenolol. Proteins were separated by SDS/PAGE, and receptor phosphorylation was quantified by using the PhosphorImager.
GIT1 translocation to vesicles was assessed as described previously for GRK2 (10). Reactions contained 3 μg of GIT1/4 μg of GRK2/1 μg of βγ-subunit as indicated, along with 80 μg of 99% phosphatidylcholine/1% PIP2 vesicles in a final volume of 90 μl. GIT1 in the pellet fraction was determined by immunoblotting.
ADP Ribosylation Factor GTPase-Activating Protein (ARF GAP) Assay.
Recombinant nonmyristoylated ARF1 was prepared from E. coli as described previously (11). ARF1 (0.5 μg) was incubated with [α-32P]GTP for 30 min at 30°C as described (12), except that the loading mixture contained 30 μg/ml of phosphatidylserine. Indicated concentrations of purified GIT1 (or buffer) were added, and incubation continued for 20 min. Unbound GTP was removed by vacuum filtration through nitrocellulose and washing five times. Filter-bound counts were eluted in 2 M formic acid, and GTP and GDP were separated by chromatography on polyethyleneimine-cellulose plates in 1 M formic acid/1 M LiCl. Dried plates were exposed to PhosphorImager screens to quantify the two nucleotides.
RESULTS
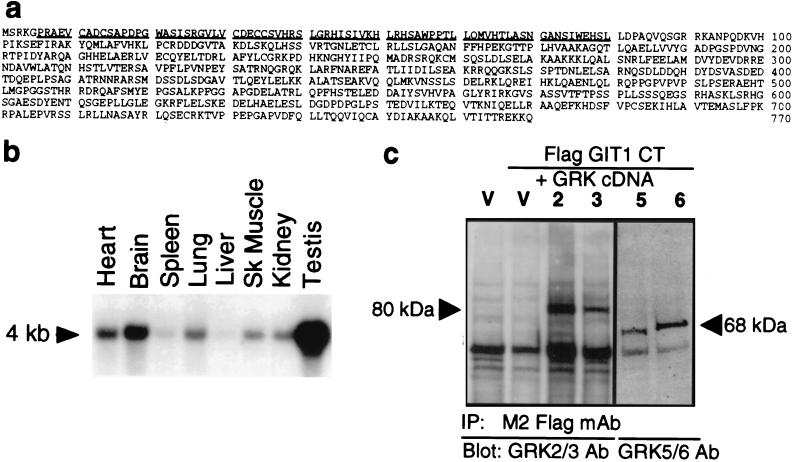
A rat brain cDNA library was screened for proteins interacting with GRK2 by using the yeast two-hybrid method. One clone positive for both His− cell growth and β-galactosidase expression was isolated and found to require the Gal4 DNA-binding domain–GRK2 fusion for activity. Expression of this clone with a GRK5 fusion also led to positive His− growth and β-galactosidase assays (data not shown). The isolated clone encodes the in-frame fusion with the carboxyl terminus of a novel protein, GRK-interactor 1 or GIT1. The full 3.2-kb rat GIT1 cDNA encodes an ORF of 770 aa (Fig. 1a). The carboxyl-terminal GRK-interaction domain of GIT1 has no similarity to other proteins of known function. The amino-terminal half of GIT1 has a zinc finger-like motif (CX2CX16CX2C) followed by ankyrin repeat units. Northern blotting of multiple rat tissue poly(A) RNAs with a GIT1 probe indicates that the 4-kb GIT1 mRNA is expressed widely at moderate levels, with quite high abundance in testis and low expression in liver and spleen (Fig. 1b).
Figure 1.
Sequence, expression, and GRK interactions of GIT1. (a) Deduced amino acid sequence of rat GIT1. GIT1 exhibits similarity with several putative proteins of unknown function (human D63482, D79989, D26069, and D30758, Caenorhabditis elegans F14F3.2), with yeast GCS1 and GLO3, and with rat ARF-GAP1, which share a putative zinc finger domain (underlined). D63482 is 75% identical to and colinear with GIT1, but terminates after 471 aa and lacks the GRK-interaction domain. D63482 is not a splice variant of GIT1, since the codon usage for human GIT1 determined from overlapping expressed sequence tag sequences differs substantially from that of D63482. Further, D63482 has been mapped to chromosome 12 (29), but we have localized the human GIT1 gene to chromosome band 17p11.2. (b) Tissue distribution of GIT1 mRNA in the indicated rat tissues. (c) Coimmunoprecipitation of GRKs with GIT1. COS7 cells were transiently transfected with Flag-tagged GIT1 CT and the indicated GRK cDNAs or empty pRK5 vector (V). Cytosol was incubated with M2 Flag antibody, and immune complexes were assayed for the presence of GRKs by immunoblotting (GRK2 and 3, 80 kDa; GRK5 and 6, 68 kDa).
The interaction of GIT1 with GRKs was verified by expressing a Flag epitope-tagged carboxyl-terminal domain of GIT1 in COS7 cells and testing for coimmunoprecipitation of GRKs. Immunoprecipitation of lysates of cells overexpressing individual GRKs using anti-FLAG antibody led to the appearance of immunoreactivity for GRKs 2, 3, 5, and 6 (Fig. 1c). GRK immunoreactivity was not seen in immunoprecipitates from cells not expressing this FLAG-GIT1 construct.
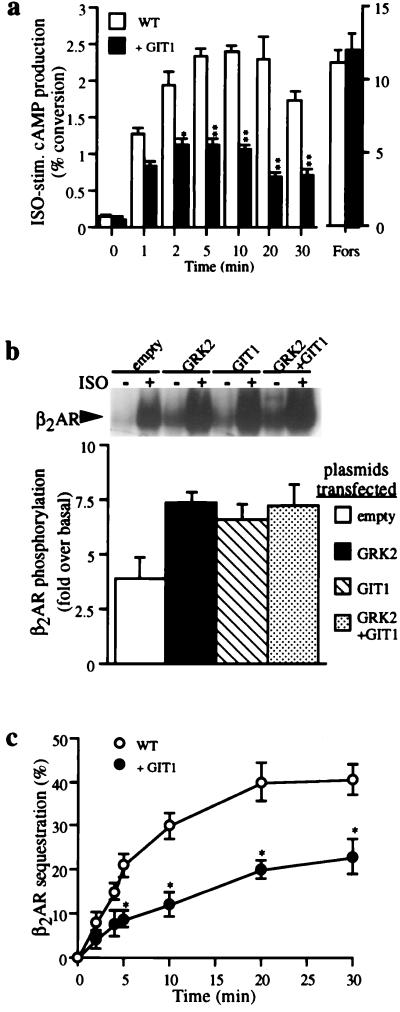
To examine the role of GIT1 in cellular signaling, we established HEK293 cells stably overexpressing GIT1. Cyclic AMP accumulation stimulated by isoproterenol through the endogenous β2AR was reduced markedly in cells overexpressing GIT1 compared with the parental HEK293 cells (Fig. 2a). However, overexpression of GIT1 did not alter the number of β2AR-binding sites on the cells (data not shown). Further, forskolin stimulation of cAMP accumulation was unaltered in cells overexpressing GIT1 (Fig. 2a), indicating that the effect of GIT1 on β2AR signaling occurs at the level of receptor or its coupling to G protein.
Figure 2.
Effects of GIT1 on β2AR function. (a) β2AR-stimulated cAMP accumulation is reduced by GIT1. Endogenous β2ARs were stimulated with 10 μM isoproterenol (“ISO”), and cAMP accumulation was evaluated at the indicated times in wild-type and GIT1-overexpressing HEK293 cells. Cells were also stimulated for 5 min with 100 μM forskolin (“Fors”). Values are means ± SEM for eight experiments done in triplicate. Results are expressed as percent conversion of [3H]ATP to [3H]cAMP. (b) Agonist-stimulated β2AR phosphorylation is increased in GIT1-overexpressing cells. Flag β2AR and GRK2 cDNAs were transfected into wild-type or GIT1-overexpressing HEK293 cells. Stimulation with isoproterenol (10 μM) was performed for 5 min (Inset). A representative gel demonstrating basal (−) and isoproterenol-stimulated (+) receptor protein phosphorylation (50–60 kDa) in cells transfected with the indicated DNAs. Values are expressed relative to basal in absence of isoproterenol. The graph shows the mean ± SEM of four experiments. (c) β2AR internalization is inhibited by GIT1. Wild-type or GIT1-overexpressing HEK293 cells transiently transfected with Flag-β2AR were incubated with Iso (10 μM) for the indicated times. Results are expressed as percentage of initial surface receptors. Values are mean ± SEM for eight experiments done in triplicate. Differences were assessed by using ANOVA followed by the Tukey–Kramer multiple comparison test. ∗, P < 0.01; ∗∗, P < 0.001.
Since GIT1 can interact with GRKs and can dramatically decrease β2AR function when overexpressed, the ability of GIT1 to alter GRK-mediated β2AR receptor phosphorylation was assessed by immunoprecipitation of the receptor protein. As described previously (7), phosphorylation of the β2AR increased after agonist treatment, and the extent of phosphorylation increased further in cells overexpressing GRK2 (Fig. 2b). Cells transfected with GIT1 exhibited greater agonist-dependent phosphorylation of the β2AR than did cells containing no exogenous GIT1. However, cotransfection of GRK2 and GIT1 did not lead to additive increases in receptor phosphorylation.
Since the β2AR undergoes rapid agonist-dependent sequestration from the cell surface through a clathrin-mediated process that is facilitated by GRK phosphorylation of the receptor (2, 3), we examined the ability of the β2AR to internalize in response to isoproterenol, with or without overexpressed GIT1. The extent of β2AR sequestration in GIT1-overexpressing cells was strikingly reduced compared with receptors in cells not overexpressing GIT1 (Fig. 2c). This effect of GIT1 to reduce receptor sequestration seemingly is at odds with the apparent augmentation of GRK action observed in cAMP accumulation or receptor phosphorylation assays, since enhancing GRK function would be expected to increase the rate and extent of receptor sequestration (2, 3).
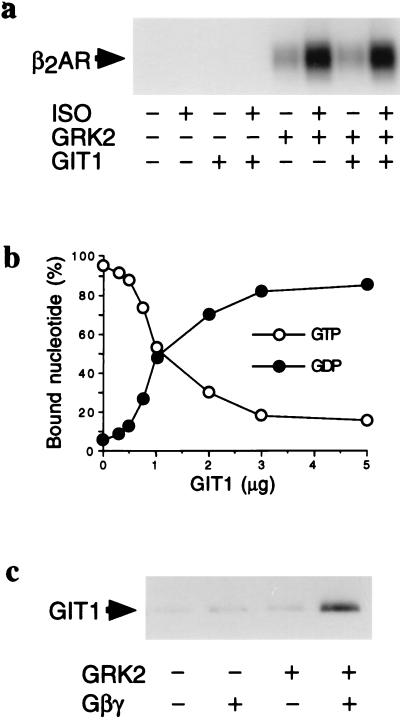
To address the mechanism underlying these alterations in β2AR function in whole cells, we assessed the effect of purified GIT1 protein on the kinase activity of GRK2. GIT1, at a 10:1 molar ratio over GRK2, did not significantly affect the ability of GRK2 to phosphorylate either rhodopsin (data not shown) or the β2AR (Fig. 3A). Further, GIT1 did not appear to be a significant phosphorylation substrate for GRK2 (data not shown).
Figure 3.
Biochemical effects of GIT1. (a) β2AR phosphorylation by GRK2 in the presence of GIT1. Vesicles containing purified β2AR were incubated with G protein βγ-subunits and with GRK2, GIT1, and 5 μM alprenolol (−) or 20 μM isoproterenol (+), as indicated. (b) GIT1 is an ARF GAP. The indicated amounts of purified GIT1 were added to recombinant ARF1 preloaded with [α-32P]GTP. The amount of GTP and GDP bound to ARF was quantified by PhosphorImager volume quantification after TLC separation of the two nucleotides. Results are expressed as percentage of bound nucleotide in the form of GTP or GDP. Data are shown as mean ± SEM of values from four experiments done in triplicate (error bars are within the symbols). (c) GIT1 binding to membranes is facilitated by GRK2. Purified GIT1 was incubated with vesicles composed of 99% PC/1% PIP2 in the presence of GRK2 and G protein βγ-subunits, as indicated. Vesicles were pelleted, and pellet-associated GIT1 was measured by immunoblotting. This assay was repeated five times with similar results, and a representative assay is shown.
GIT1 overexpression leads to increased GRK-mediated β2AR phosphorylation and decreased receptor signaling, yet it did not increase the kinase activity of GRK2 toward activated receptors. However, GIT1 also reduces internalization of the receptor, which has been shown previously to be required for β2AR dephosphorylation and resensitization within acidic endosomes (13). Since the regulatory effects of overexpressed GIT1 on β2AR function and phosphorylation appear to be mediated by inhibiting receptor sequestration and resensitization, we explored other possible mechanisms for these effects.
The amino terminus of GIT1 exhibits limited similarity to the rat ARF-GAP1 and yeast GCS1 proteins, which can act as GAPs for the ARF1 small GTP-binding protein (14, 15). We examined the potential of purified GIT1 to act as an ARF GAP. GIT1 protein added to [α-32P]GTP-bound ARF1 led to a concentration-dependent decrease in the ARF-bound GTP and a corresponding increase in the bound GDP, without altering the total amount of guanine nucleotide bound to ARF (Fig. 3B). This activity of GIT1 was constant for 30 min (data not shown). GIT1 had no effect on nucleotide binding to or release from ARF1, nor did GIT1 bind guanine nucleotides. Thus, GIT1 acts as a GTPase-activating protein for the small GTP-binding protein ARF1 in vitro. However, GRK2, with or without 100 μM ATP, had no effect on the ARF GAP activity of GIT1 (data not shown).
Since GIT1 had no effect on receptor phosphorylation activity of GRK2, and GRK2 had no effect on the ARF GAP activity of GIT1, we examined the potential for GRK2 to translocate GIT1 to a model membrane. GRK2 translocation to activated receptors is an important regulatory step and requires GRK2 interaction with both membrane PIP2 and heterotrimeric G protein βγ-subunits released upon receptor activation (9, 16). This translocation can be measured in vitro by assessing GRK2 interactions with phospholipid vesicles containing PIP2 and purified βγ-subunits (10). Purified GIT1 was found in the pellet fraction after incubation with GRK2, βγ-subunits, and phosphatidylcholine vesicles containing 1% PIP2 (Fig. 3C). Removal of either GRK2 or βγ-subunits led to decreased GIT1 in the pellet. Thus, GRK2 can anchor GIT1 to a model membrane in a βγ-subunit-dependent manner, which mimics receptor-dependent translocation of GRK2 (16).
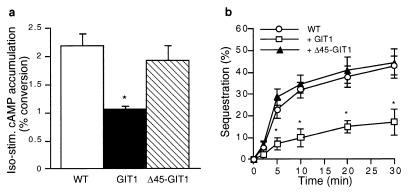
To test whether the ARF GAP activity of GIT1 is involved in reducing β2AR signaling in cells overexpressing GIT1, we deleted the putative zinc finger domain of GIT1 by removing the first 45 aa. Amino-terminal truncation of the putative zinc finger of the rat ARF-GAP1 protein ablated GAP activity (14). Purified Δ45-GIT1 failed to activate the GTPase activity of ARF1 (data not shown), indicating that the GIT1 GAP activity also requires the amino-terminal zinc finger-like domain.
Despite stable expression in HEK293 cells of the Δ45-GIT1 protein at a level equivalent to that of wild-type GIT1, Δ45-GIT1 was ineffective in diminishing β2AR-stimulated cAMP accumulation (Fig. 4A). Further, in HEK293 cells overexpressing Δ45-GIT1, agonist-stimulated internalization of the β2AR was indistinguishable from that in cells not overexpressing GIT1 (Fig. 4B). Thus, the reduction of β2AR activation of cAMP accumulation as well as the reduction of agonist-induced β2AR sequestration due to overexpression of GIT1 both appear to require the ARF GAP activity of GIT1.
Figure 4.
ARF GAP activity of GIT1 is required to regulate receptor function. (a) Δ45-GIT1 is ineffective in diminishing β2AR-stimulated cAMP accumulation. HEK 293 cells (wild-type or stably overexpressing GIT1 or Δ45-GIT1) were stimulated with isoproterenol (10 μM) for 10 min, and cAMP accumulation was measured. Data are mean ± SEM of values from four experiments done in triplicate. (b) Δ45-GIT1 does not affect β2AR internalization. HEK 293 cells were transiently transfected with Flag β2AR plus GIT1, Δ45-GIT1, or empty vector. Cell surface receptor number was determined by fluorescence-activated cell sorter analysis after the indicated time of isoproterenol (10 μM) stimulation. Results are expressed as percentage of initial surface receptors sequestered. Data are mean ± SEM of values from four experiments done in triplicate. Differences were assessed by using an ANOVA followed by the Tukey–Kramer multiple comparison test. ∗, P < 0.01.
DISCUSSION
The functional effects of GIT1 on β2AR function appear to be a result of its ability to stimulate the GTPase activity of ARF, thereby attenuating ARF activity. Although originally identified as cofactors required for cholera toxin-catalyzed ADP ribosylation of Gαs (17), the ARF proteins now are thought to function primarily as GTP-dependent regulators of vesicular trafficking (18, 19). ARF proteins also have been demonstrated to mediate GTP-dependent activation of phospholipase D (18, 19), and it has been proposed that ARF proteins are involved in regulating plasma membrane endocytosis and transferrin receptor internalization and trafficking (20–22). Thus, a plasma membrane-targeted ARF GAP would be expected to prevent or slow the accumulation of plasma membrane-associated GTP-bound ARF, leading to decreased internalization of vesicles. Because efficient dephosphorylation of GRK-phosphorylated β2-adrenergic receptors requires their internalization followed by acidification of the intracellular vesicles (13), slowing the rate of receptor internalization would result in the accumulation of phosphorylated, inactive receptors on the cell surface. Since GIT1 appears to alter receptor internalization by a general mechanism and is widely expressed in rat tissues, it may be a common component of G protein-coupled receptor signaling cascades and play a role in coordinating internalization of a variety of cell surface proteins. That the GIT1 ARF GAP activity can alter G protein-coupled receptor internalization is consistent with a role for ARF proteins in coordinating G protein-coupled receptor internalization. GIT1 also might have functional effects on other cellular processes that require GTP-bound ARF proteins, such as activation of phospholipase D (23, 24).
GIT1 is a member of a subfamily of ARF GAPs that interact with GRKs, since a highly similar GIT2 protein is widely expressed in tissues and cell lines (R.T.P., unpublished observation). Additional ARF GAP family members identified in database searches contain ARF GAP-like domains connected to a variety of distinct carboxyl-terminal sequences (25–27). We speculate that the modular nature of these ARF GAP-like proteins allows distinct signaling networks to access ARF pathways by controlling the activity or localization of these presumed ARF GAP proteins. The importance of localization for ARF GAP activity has been highlighted by a recent study in which rat ARF-GAP1 was shown to require interaction with the KDEL receptor protein ERD2 for endoplasmic reticulum and Golgi membrane localization and function (28). Agonist-dependent translocation of GRKs to receptors may bring GIT1 to the plasma membrane and allow ARF protein functions to be modulated by G protein-coupled receptors. One result of GIT1 recruitment is to slow β2AR internalization and resensitization, thus enhancing receptor phosphorylation and desensitization. This indicates a role for GRKs in coordinating two distinct receptor regulatory pathways: β-arrestin-dependent receptor uncoupling and receptor sequestration. This mechanism also provides a means for GRKs to serve directly as signaling intermediates independent of their role as inactivators of G protein-coupled receptor signaling.
Acknowledgments
We thank Millie McAdams and Judy Phelps for DNA sequencing, Grace Irons for cell culture, Humphrey Kendall for technical assistance, Helena Abushamaa and the Genome Center of the Duke University Medical Center Comprehensive Cancer Center for gene mapping, Drs. Rick Kahn and Dan Cassel for helpful discussions, Drs. Randy Hall and Louis Luttrell for a critical reading of the manuscript, and Donna Addison and Mary Holben for secretarial assistance. A.C. is a recipient of a postdoctoral fellowship from the Heart and Stroke Foundation of Canada. This work was supported by National Institutes of Health Grant HL16037 (R.J.L.).
ABBREVIATIONS
- ARF
ADP ribosylation factor
- β2AR
β2-adrenergic receptor
- GAP
GTPase-activating protein
- GIT
GRK interactor
- GRK
G protein-coupled receptor kinase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank/EMBL databases [accession no. AF085693 (rat GIT1)].
References
- 1.Dohlman H G, Thorner J, Caron M G, Lefkowitz R J. Annu Rev Biochem. 1991;60:653–688. doi: 10.1146/annurev.bi.60.070191.003253. [DOI] [PubMed] [Google Scholar]
- 2.Lefkowitz R J. J Biol Chem. 1998;273:18677–18680. doi: 10.1074/jbc.273.30.18677. [DOI] [PubMed] [Google Scholar]
- 3.Pitcher J A, Freedman N J, Lefkowitz R J. Annu Rev Biochem. 1998;67:653–692. doi: 10.1146/annurev.biochem.67.1.653. [DOI] [PubMed] [Google Scholar]
- 4.Stewart E A, McKusick K B, Aggarwal A, Bajorek E, Brady S, Chu A, Fang N, Hadley D, Harris M, Hussain S, et al. Genome Res. 1997;7:422–433. doi: 10.1101/gr.7.5.422. [DOI] [PubMed] [Google Scholar]
- 5.Premont R T, Koch W J, Inglese J, Lefkowitz R J. J Biol Chem. 1994;269:6832–6841. [PubMed] [Google Scholar]
- 6.Barak L S, Tiberi M, Freedman N J, Kwatra M M, Lefkowitz R J, Caron M G. J Biol Chem. 1994;269:2790–2795. [PubMed] [Google Scholar]
- 7.Freedman N J, Liggett S B, Drachman D E, Pei G, Caron M G, Lefkowitz R J. J Biol Chem. 1995;270:17953–17961. doi: 10.1074/jbc.270.30.17953. [DOI] [PubMed] [Google Scholar]
- 8.Salomon Y. Methods Enzymol. 1991;195:22–28. doi: 10.1016/0076-6879(91)95151-9. [DOI] [PubMed] [Google Scholar]
- 9.Pitcher J A, Touhara K, Payne E S, Lefkowitz R J. J Biol Chem. 1995;270:11707–11710. doi: 10.1074/jbc.270.20.11707. [DOI] [PubMed] [Google Scholar]
- 10.Touhara K, Koch W J, Hawes B E, Lefkowitz R J. J Biol Chem. 1995;270:17000–17005. doi: 10.1074/jbc.270.28.17000. [DOI] [PubMed] [Google Scholar]
- 11.Hong J X, Haun R S, Tsai S C, Moss J, Vaughan M. J Biol Chem. 1994;269:9743–9745. [PubMed] [Google Scholar]
- 12.Vitale N, Moss J, Vaughan M. J Biol Chem. 1996;272:3897–3904. doi: 10.1074/jbc.272.7.3897. [DOI] [PubMed] [Google Scholar]
- 13.Krueger K M, Daaka Y, Pitcher J A, Lefkowitz R J. J Biol Chem. 1997;272:5–8. doi: 10.1074/jbc.272.1.5. [DOI] [PubMed] [Google Scholar]
- 14.Cukierman E, Huber I, Rotman M, Cassel D. Science. 1995;270:1999–2002. doi: 10.1126/science.270.5244.1999. [DOI] [PubMed] [Google Scholar]
- 15.Poon P P, Wang X, Rotman M, Huber I, Cukierman E, Cassel D, Singer R A, Johnston G C. Proc Natl Acad Sci USA. 1996;93:10074–10077. doi: 10.1073/pnas.93.19.10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pitcher J A, Inglese J, Higgins J B, Arriza J L, Casey P J, Kim C, Benovic J L, Kwatra M M, Caron M G, Lefkowitz R J. Science. 1992;257:1264–1267. doi: 10.1126/science.1325672. [DOI] [PubMed] [Google Scholar]
- 17.Kahn R A, Gilman A G. J Biol Chem. 1984;259:6235–6240. [PubMed] [Google Scholar]
- 18.Boman A L, Kahn R A. Trends Biochem Sci. 1995;20:147–150. doi: 10.1016/s0968-0004(00)88991-4. [DOI] [PubMed] [Google Scholar]
- 19.Moss J, Vaughan M. J Biol Chem. 1995;270:12327–12330. doi: 10.1074/jbc.270.21.12327. [DOI] [PubMed] [Google Scholar]
- 20.Zhang C-J, Rosenwald A G, Willingham M C, Skuntz S, Clark J, Kahn R A. J Cell Biol. 1994;124:289–300. doi: 10.1083/jcb.124.3.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D’Souza-Schorey C, Li G, Colombo M I, Stahl P D. Science. 1995;267:1175–1178. doi: 10.1126/science.7855600. [DOI] [PubMed] [Google Scholar]
- 22.Radhakrishna H, Donaldson J G. J Cell Biol. 1997;139:49–61. doi: 10.1083/jcb.139.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell R, McCulloch D, Lutz E, Johnson M, MacKenzie C, Fennell M, Fink G, Zhou W, Sealfon S C. Nature (London) 1998;392:411–414. doi: 10.1038/32937. [DOI] [PubMed] [Google Scholar]
- 24.Moss J, Vaughan M. J Biol Chem. 1998;273:21431–21434. doi: 10.1074/jbc.273.34.21431. [DOI] [PubMed] [Google Scholar]
- 25.Bogerd H P, Fridell R A, Madore S, Cullen B R. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- 26.Hammonds-Odie L P, Jackson T R, Profit A A, Blader I J, Turck C W, Prestwich G D, Thiebert A B. J Biol Chem. 1996;271:18859–18868. doi: 10.1074/jbc.271.31.18859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang C-J, Cavenagh M M, Kahn R A. J Biol Chem. 1998;273:19792–19796. doi: 10.1074/jbc.273.31.19792. [DOI] [PubMed] [Google Scholar]
- 28.Aoe T, Cukierman E, Lee A, Cassel D, Peters P J, Hsu V W. EMBO J. 1997;16:7305–7316. doi: 10.1093/emboj/16.24.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagase T, Seki N, Tanaka A, Ishikawa K-I, Nomura N. DNA Res. 1995;2:167–174. doi: 10.1093/dnares/2.4.167. [DOI] [PubMed] [Google Scholar]