Abstract
The bovine papillomavirus type 1 (BPV-1) exonic splicing suppressor (ESS) is juxtaposed immediately downstream of BPV-1 splicing enhancer 1 and negatively modulates selection of a suboptimal 3′ splice site at nucleotide 3225. The present study demonstrates that this pyrimidine-rich ESS inhibits utilization of upstream 3′ splice sites by blocking early steps in spliceosome assembly. Analysis of the proteins that bind to the ESS showed that the U-rich 5′ region binds U2AF65 and polypyrimidine tract binding protein, the C-rich central part binds 35- and 54–55-kDa serine/arginine-rich (SR) proteins, and the AG-rich 3′ end binds alternative splicing factor/splicing factor 2. Mutational and functional studies indicated that the most critical region of the ESS maps to the central C-rich core (GGCUCCCCC). This core sequence, along with additional nonspecific downstream nucleotides, is sufficient for partial suppression of spliceosome assembly and splicing of BPV-1 pre-mRNAs. The inhibition of splicing by the ESS can be partially relieved by excess purified HeLa SR proteins, suggesting that the ESS suppresses pre-mRNA splicing by interfering with normal bridging and recruitment activities of SR proteins.
Alternative splicing is a major mechanism for controlling the expression of eukaryotic genes. Recent progress has been made in the identification of exonic sequences in viral and cellular genes that promote [exonic splicing enhancer (ESE)] (1–13) or repress [exonic splicing suppressor or silencer (ESS)] (13–21) utilization of alternative splice sites. These ESEs are usually purine-rich sequences located in an alternatively spliced exon downstream of a suboptimal 3′ splice site. Through their binding to serine/arginine-rich (SR) proteins (6–12, 22), the ESEs function by recruiting splicing factors, such as U2AF, to a suboptimal 3′ splice site at early stages of spliceosome assembly and thereby stimulating splicing of the upstream intron or inclusion of the alternative exon (23, 24). In contrast, the ESS elements recently described in several pre-mRNAs have considerable sequence variation although they all inhibit splicing (13–21). Although most ESSs have been found downstream of ESEs (13–17), this finding is not universal. The ESS in fibroblast growth factor receptor 2 K-SAM exon has no upstream ESE, but suppresses splicing of the exon (19, 20). On the other hand, the ESS in HIV-1 6D exon (21) and fibronectin EDA exon (18) normally functions upstream of an ESE. The mechanism(s) by which an ESS suppresses pre-mRNA splicing currently is unknown.
We previously have demonstrated that the BPV-1 nucleotide 3225 3′ splice site is a weak 3′ splice site with a nonconsensus branch point and a suboptimal polypyrimidine tract interrupted by three purines (13, 22). Utilization of this 3′ splice site in vitro and in vivo requires an ESE [splicing enhancer 1 (SE1)] in the downstream exon. In contrast, the BPV-1 ESS, which is juxtaposed immediately downstream of SE1, negatively modulates selection of the nucleotide 3225 3′ splice site. This 48-nt pyrimidine-rich ESS features a U-rich 5′ end, a C-rich central part, and an AG-rich 3′ end. Similar to other ESSs, the BPV-1 ESS also suppresses the splicing of other pre-mRNAs. This suppression requires an upstream suboptimal 3′ splice site, but not an upstream ESE (49). In this report we provide strong evidence that the BPV-1 ESS suppresses an early stage of spliceosome assembly through the binding of multiple splicing factors, most importantly SR proteins.
MATERIALS AND METHODS
BPV-1 and HIV-1 DNA Templates.
BPV-1 DNA templates used for in vitro runoff transcription were generated from plasmid pZMZ19–1 (13) by PCR using a common 5′ T7 primer (5′-ATTAATACGACTCACTATAG-3′) combined with antisense BPV-1 primers that have their 5′ ends at the positions indicated above each pre-mRNA 3′ end shown in Fig. 3A. Each pre-mRNA used for the mapping study contains a fixed size of exon 1 (187 nt), a truncated intron 1 (333 nt), and a variably sized exon 2 ranging from 81 to 129 nt depending on the deletion in the ESS region. The antisense ESS in Fig. 3B was cloned 20 nt downstream of the sense wild-type ESS at StuI and Asp 718 sites in pZMZ19–1. The resulting plasmid p3067 was linearized with Asp 718 and used as a template for in vitro transcription. The size of exon 2 in the resulting pre-mRNA is 202 nt. DNA templates used for in vitro runoff transcription of the pre-mRNAs shown in Figs. 3D and 4A also were generated from plasmid pZMZ19–1 as described above by using a 5′ T7 primer combined with an antisense primer containing either mutations in ESS (see Fig. 3D) or a functional ESS core with or without an ASF/SF2 binding site(s) downstream (see Fig. 4A).
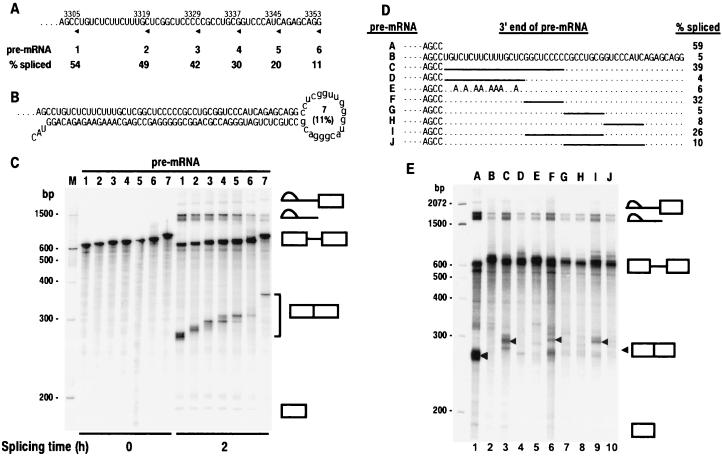
Figure 3.
Mapping analysis of the BPV-1 ESS by deletion and point mutagenesis. (A) 3′ deletion of the ESS in a BPV-1 late pre-mRNA. The sequence of the ESS is shown. All of the pre-mRNAs also contain SE1. The 3′ end of each pre-mRNA is indicated by an arrowhead under the sequence and a number above the sequence that indicates the nucleotidet position of the 3′ end. The pre-mRNAs are labeled 1–6 and correspond to each pre-mRNA used for splicing in C. Splicing efficiency (% spliced) was calculated from the data in C as described in Materials and Methods and is shown under the pre-mRNA number. (B) The structure of pre-mRNA 7. It is a BPV-1 late pre-mRNA containing both a sense and an antisense ESS sequence downstream of SE1 in exon 2. The number in parentheses in the loop indicates the splicing efficiency of the pre-mRNA calculated from the data in C. (C) In vitro spliced products of pre-mRNAs shown in A and B. Positions of DNA size markers corresponding to a 100-bp ladder are shown at the left of the gel. The identity of the spliced products and intermediates is shown at the right. Unincubated pre-mRNA (0 h) was included as a control. (D) Sequence of wild-type or mutant ESS in BPV-1 late pre-mRNA. Small dashes on the left represent the sequences (including SE1) upstream of ESS. Periods and solid lines indicate unchanged nucleotides and deletions, respectively. Pre-mRNAs are identified at the left by the letters A–J. Splicing efficiency of each pre-mRNA was calculated based on the gel profile in E. (E) Splicing gel electrophoresis after in vitro splicing reactions of 32P-labeled pre-mRNAs in D. The identity of the spliced products and intermediates is shown at the right with an arrow for the fully spliced product. Positions of DNA size markers corresponding to a 100-bp ladder are shown at the left of the gel. The letters A–J at the top of the gel represent the pre-mRNAs in D that were used for the splicing assay.
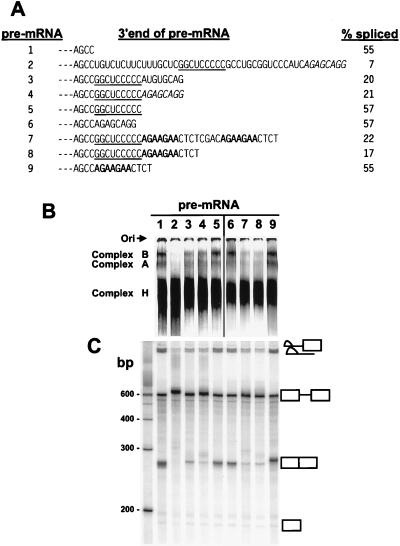
Figure 4.
Inhibition of BPV-1 late pre-mRNA splicing by the functional C-rich core of the ESS. (A) 3′ end sequences of the different pre-mRNAs that were used for in vitro spliceosome assembly (B) and splicing (C). The splicing efficiency of each pre-mRNA was calculated based on the gel profile in C. Small dashes on the left represent the sequences (including SE1) upstream. The functional C-rich core of the ESS is underlined. The ASF/SF2 binding site from the 3′ AG-rich part of the ESS and an optimal ASF/SF2 binding site octamer A3 (9) are indicated by italics and bold, respectively. Pre-mRNAs are identified at the left by the numbers 1–9. (B) Formation of spliceosome complexes on different pre-mRNA substrates with varied 3′ ends as shown in A. Spliceosome complex formation was performed under standard splicing condition (13, 22), stopped at 45-min incubation at 30°C by addition of heparin, and loaded and run immediately on a native 4% polyacrylamide gel. Spliceosome complexes A and B and the nonspecific complex H are indicated on the left of the gel. Lanes 1–5 were overexposed to compensate for loading variations of each pre-mRNA and to facilitate comparison of pre-mRNAs 3–5. Ori, origin. (C) Splicing gel electrophoresis of each correspondent pre-mRNA in B. The identity of the spliced products and intermediates is shown at the right. Positions of DNA size markers corresponding to a 100-bp ladder are shown at left of the gel.
Plasmid pHS2 (16), which contains a purine-rich ESE upstream of an ESS within tat-rev exon 3 of HIV-1, was used for introduction of the BPV-1 ESS by PCR. The plasmid pHS2 first was digested with HpaI and PvuII, and a restriction fragment of about 1.1 kb was gel-purified and used as a template for PCR using a 5′ sense T3 primer (oFD122: 5′-ATTAACCCTCACTAAAG-3′) combined with 3′ antisense primers HIV ESE (oZMZ94: 5′-TGTCTCTGTCTCTCTCTCC-3′) or chimeric HIV ESE/BPV-1 ESS (oZMZ93: 5′-CCTGCTCTGATGGGACCGCAGGCGGGGGAGCCGAGCAAAGAAGAGACA/TGTCTCTGTCTCTCTCTCCACCTTCTTCTTC-3′).
Pre-mRNA Preparation.
In vitro transcription was carried out with T7 (for BPV-1) or T3 (for HIV-1 tat/rev) RNA polymerase in the presence of the cap analog (m7GpppG) and [α-32P]rNTP as indicated in each experiment. Full-length transcripts were separated on 6% denaturing polyacrylamide gels and eluted from gel slices by overnight incubation in elution buffer as described (22). The eluted RNAs then were phenol-extracted, ethanol-precipitated, and resuspended in diethyl pyrocarbonate-treated water.
In Vitro Splicing and Analysis of Splicing Products.
In vitro splicing reactions were performed at 30°C for 2 h using the Promega RNA splicing system with 4 ng of 32P-labeled pre-mRNA and 40% HeLa nuclear extracts (NE) as recommended by the manufacturer (13, 22). The reaction products were extracted with phenol, precipitated with ethanol, and analyzed by electrophoresis on denaturing 8% polyacrylamide-8 M urea gels followed by quantitation with a Molecular Dynamics PhosphorImager. The splicing efficiency for each pre-mRNA was calculated as the percentage of total splicing products (intermediates and fully spliced) divided by the sum of total splicing products plus the remaining pre-mRNA.
Spliceosome Assembly and Gel Electrophoresis.
Spliceosomal complex formation was performed under standard splicing conditions (13, 22) and stopped at scheduled time points by addition of heparin to a final concentration of 2 mg/ml. After incubation for an additional 10 min at 30°C, samples were loaded (no freezing and thawing) on a native 4% polyacrylamide gel (acrylamide/Bis = 80:1) and run at 200 V at room temperature for 4 h (25, 26).
UV Cross-Linking and Immunoprecipitation (IP).
ESS RNAs were transcribed from synthetic chimeric T7-ESS DNA templates by T7 RNA polymerase (27) and were used for UV cross-linking and IP analysis as described (22). The following proteins were used for UV cross-linking and/or IP. HeLa NE were purchased from Promega. Poly(U)-binding protein-depleted HeLa NE was prepared by using poly(U)-Sepharose 4 beads (Pharmacia) as described (28). HeLa SR proteins were prepared by using a two-step salt precipitation (ammonium sulfate and then magnesium chloride) (29). Human polypyrimidine tract binding protein (PTB) was expressed from PTB expression vector pET-PTB (30, 31). Polyclonal antibodies against U2AF65, U2AF35, and human PTB and mAbs against SR proteins, human PTB, and ASF/SF2 were used for IP. The techniques for UV cross-linking and IP have been detailed (22). All of the UV cross-linked RNA-protein mixtures and immunoprecipitated proteins were resolved on 12% SDS/PAGE gels.
RESULTS
Inhibition of Spliceosome Complex Formation by the BPV-1 ESS in Vitro.
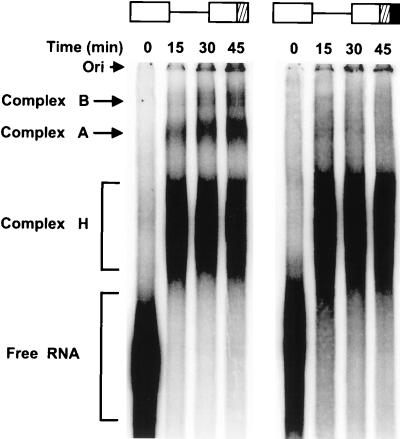
Processing of a pre-mRNA is a multistage, dynamic biochemical reaction carried out by the spliceosome. It has been well documented that assembly of the spliceosome proceeds through a highly ordered pathway of complex formation in the order E→A→B→C (25, 26, 32). To determine at what stage of spliceosomal assembly the ESS functions, splicing complex formation was studied by using BPV-1 late pre-mRNAs with or without the ESS in exon 2. Spliceosome complexes A and B readily formed on the pre-mRNA with no ESS, but were greatly reduced on the pre-mRNA with the ESS (Fig. 1). Thus the ESS functions at an early stage of spliceosome assembly before the formation of complex A.
Figure 1.
Inhibition of spliceosome complex formation by the BPV-1 ESS in vitro. The two pre-mRNAs used for the assay are shown diagrammatically at the top of the gel. The cross-hatched box in exon 2 indicates SE1; the black box in exon 2 indicates the ESS. Spliceosomal complex formation was carried out and analyzed as described in Materials and Methods. The 0 time indicates the pre-mRNA with no addition of HeLa NE. Spliceosomal complexes A and B, the nonspecific heterogeneous nuclear ribonucleoprotein complex H, and free pre-mRNA are indicated on the left of the gel. Ori, origin.
Identification of U2AF65, PTB, and SR Proteins Bound to the BPV-1 ESS.
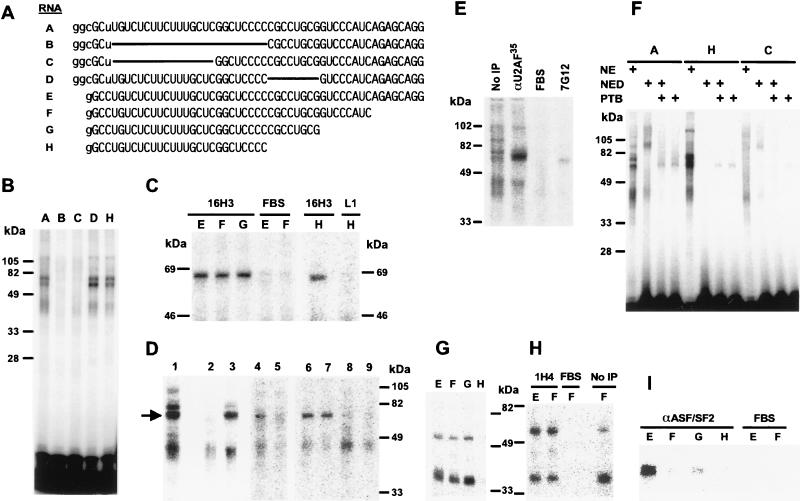
There are several mechanisms by which the ESS could inhibit spliceosome complex formation. One possibility is that the ESS could bind inhibitory cellular factors that are detrimental to spliceosomal assembly. An alternative possibility is that binding of splicing factors to the BPV-1 ESS may prevent spliceosome assembly by interfering with or reducing the availability of splicing factors required for utilization of the weak 3′ splice site. To look for cellular factors interacting with the BPV-1 ESS, we performed UV cross-linking and IP analyses. Small in vitro-transcribed RNAs containing just the ESS and short linker sequences were used in these assays (Fig. 2). ESS RNAs A and E are wild type, whereas ESS RNAs B–D have 5′ or internal deletions and RNAs F–H have successive 3′ deletions, but retain the 5′ U-rich region as well as the GGCUCCCC sequence (Fig. 2A). Two proteins in HeLa NE having apparent molecular masses of about 65 and 75 kDa appear to bind to the 5′ U-rich region of the ESS, because these proteins could be cross-linked to RNAs A, D, and H but not to RNAs B and C, which have this region deleted (Fig. 2B). The 65-kDa protein could be immunoprecipitated by using mAb 16H3, which recognizes U2AF65 as well as many SR proteins (Fig. 2 C and D) (33). The size of this protein, its binding specificity, and its absence in HeLa SR protein preparations (data not shown) suggested that it might be U2AF65, a common splicing factor that binds to the polypyrimidine tract at the 3′ splice site (34). This identity was further suggested by both IP and immunodepletion (ID) by using a polyclonal anti-U2AF35 antiserum that precipitates or depletes both subunits (U2AF65 and U2AF35) of the heterodimer protein U2AF (Fig. 2D, lanes 3 and 5, respectively) (24). However, we were unable to definitively identify this protein as U2AF65, because the anti-U2AF65 antiserum does not work for IP or ID (Fig. 2D, lanes 8 and 7, respectively) (24). Control antibodies were unable to immunoprecipitate the 65-kDa protein (Fig. 2D, lanes 2 and 9), demonstrating the specificity of the assays.
Figure 2.
Identification of U2AF65, PTB, and SR proteins bound to the BPV-1 ESS. (A) ESS RNAs used for protein binding assays. The sequences of the ESS RNAs are shown, with capital letters indicating BPV-1 sequences, lowercase letters indicating the flanking sequences from the T7 promoter region, and solid lines indicating internal deletions. (B) The 5′ U-rich region of the ESS binds two proteins in HeLa NE. 32P-rGTP-labeled RNAs A, B, and C, and 32P-rUTP-labeled RNAs D and H were used for UV cross-linking of HeLa NE (20 μg) and digested with RNase T2 before electrophoresis. (C) IP by mAb 16H3 of a 65-kDa protein cross-linked to the 5′ U-rich region of the ESS. ESS RNAs E, F, G, and H were uniformly labeled with all four 32P-rNTPs, UV cross-linked to the proteins in HeLa NE, digested with RNase A, and immunoprecipitated by using mAb 16H3 (33), anti-BPV-1 L1, or 20% fetal bovine serum (FBS) as indicated at the top. (D) Identification of the 65-kDa protein bound to the ESS as U2AF65 (arrow) by using polyclonal anti-U2AF35 antibody. ESS RNA G labeled with 32P-rUTP (lanes 1–3) or all four 32P-rNTPs (lanes 4–9) was UV cross-linked to proteins in either HeLa NE (lanes 1–4, 6, 8, and 9) or HeLa NE immunodepleted with anti-U2AF35 (lane 5) or anti-U2AF65 (lane 7). After digestion for 30 min with RNase A at 37°C, the RNA-protein mixtures were immunoprecipitated by using 20% FBS (lane 2), anti-U2AF35 (lane 3), mAb 16H3 (lanes 4–7), anti-U2AF65 (lane 8), or anti-BPV-1 L1 (lane 9) before loading for electrophoresis. Lane 1 is the UV cross-linked RNA-protein mixture without IP. (E) Identification of PTB bound to the 5′ U-rich region of the ESS by IP by using mAb 7G12. ESS RNA A labeled with 32P-rGTP was used for UV cross-linking of HeLa NE followed by digestion with RNase T2. The cross-linked RNA-protein mixtures were immunoprecipitated by using anti-U2AF35, anti-PTB mAb 7G12 (38), or 20% FBS. (F) Binding of recombinant PTB to the 5′ U-rich region of the ESS. HeLa NE (20 μg), poly (U)-depleted HeLa NE (NED) (20 μg), and/or recombinant PTB (1 μg) (30, 31) were used for UV cross-linking of ESS RNAs A, H, and C labeled with 32P-rGTP. After digestion with RNase T2, the cross-linked RNA-protein mixtures were resolved by electrophoresis. (G–I) Identification of SR proteins bound to the BPV-1 ESS by IP. HeLa SR proteins (20 μg) were cross-linked by UV irradiation to RNAs E, F, G, or H labeled with 32P-rGTP, digested with RNase A, and then resolved directly on a 12% SDS/PAGE gel (G, no IP) or subjected to IP by using mAb 1H4 (H) or mAb αASF2/SF2 (I) before electrophoresis. FBS (20%) was used as a mock antibody in both H and I.
Sequence analysis revealed that the 5′ U-rich region of the ESS has two overlapping core binding sites (UCUU) for PTB (31), a potent splicing repressor and alternative splicing modulator in mammalian systems (30, 35–37). Binding of PTB in HeLa NE to the ESS was confirmed by UV cross-linking and IP by using the PTB-specific antibody mAb 7G12 (Fig. 2E) (38). Proteins in the size range of PTB can be depleted from HeLa NE by using poly(U)-Sepharose beads (28)(see also NED lanes in Fig. 2F). Recombinant human PTB, either added to poly(U)-depleted NE (NED) or by itself, could be cross-linked to wild-type ESS or the 5′ portion of the ESS (RNAs A and H, Fig. 2F), but only very weakly to the ESS with a deleted 5′ U-rich region (RNA C, Fig. 2F). These studies confirm that the 5′ U-rich region of the ESS contains strong binding sites for PTB.
When purified HeLa SR protein preparations (29) were used for binding studies, two proteins with apparent molecular masses of approximately 35 and 55 kDa were cross-linked strongly to ESS RNAs E, F, and G, but only very weakly to ESS RNA H (Fig. 2G) even though the latter RNA was able to bind to U2AF65 from HeLa NE (Fig. 2C, RNA H). The 35- and 55-kDa proteins could be immunoprecipitated by using mAb 1H4, an IgG-type, pan-SR protein antibody (12), indicating that they are SR proteins (Fig. 2H). Data suggest that binding of these proteins requires the central C-rich part of the ESS, including the adjacent 3′ sequences (compare RNAs G and H in Fig. 2 G and A). The identities of these SR proteins were not further pursued because of unavailability of specific antibody against individual SRp30b (SC35), SRp30c, SRp54, and SRp55 during this study. A 35-kDa protein immunoprecipitated with mAb αASF/SF2 bound strongly to RNA E and only weakly to shorter RNAs (Fig. 2I), indicating that the sequence AGAGCAGG at the 3′ end of the ESS probably is required for binding of the SR protein ASF/SF2 (SRp30a) (39–42).
Mutational Analysis of the BPV-1 ESS.
RNA structure analyses predicted a secondary structure in which the ESS is base-paired with SE1. To determine whether the ESS interferes with SE1 function through an antisense mechanism, an antisense ESS was cloned 20 nt downstream of the wild-type ESS in pZMZ19–1, a BPV-1 plasmid containing a mini-gene for the late mRNA (13, 22). An RNA structure in which the antisense ESS is base-paired with the ESS will be energetically favored over the structure in which the ESS is base-paired with SE1. Thus the antisense ESS should prevent the ESS from base-pairing with SE1. As expected, splicing of the BPV-1 late pre-mRNA containing the wild-type ESS (pre-mRNA 6) was significantly inhibited (about a 5-fold reduction) in a 2-h splicing reaction when compared with splicing of the pre-mRNA containing no ESS (pre-mRNA 1) (Fig. 3 A and C). The presence of an antisense ESS had no significant effect on splicing efficiency (compare pre-mRNAs 6 and 7 in Fig. 3 A–C), suggesting that inhibition of splicing by the ESS is not caused by an antisense mechanism. This finding is consistent with the observation that an upstream ESE is not required for ESS function (49).
To determine the functional relevance of the binding of each protein identified above, we determined which binding sites are essential. BPV-1 pre-mRNAs containing SE1 and various mutated ESSs were tested for in vitro splicing. Successive 3′ deletions in the ESS gradually eliminated the inhibition of splicing (Fig. 3 A and C, pre-mRNAs l-6). Deletion of just the 3′ AG-rich ASF/SF2-binding region increased splicing 2-fold (compare pre-mRNAs 5 and 6), suggesting that the binding of this protein is functionally important. An additional 3′ deletion into the C5 sequence also increased splicing 2-fold (pre-mRNA 3). This deletion also eliminated binding of the 30- and 55-kDa SR proteins (Fig. 2 G and H), suggesting that these proteins also are required for maximal suppression of splicing. Although the 5′ half of the ESS retained some inhibitory activity (Fig. 3 A and C, pre-mRNA 3), deletion or mutation of just the 5′ U-rich region of the ESS didn’t significantly affect the suppression of splicing (Fig. 3 D and E, pre-mRNAs D and E). Because U2AF65 and PTB bind to this region, this result suggests that their binding is not essential for ESS activity. In contrast, any deletion that involved the sequence GGCUCCCC in the central part of the ESS gave a dramatic increase in splicing, indicating that this region is essential for ESS function (Fig. 3 D and E, pre-mRNAs C, F, and I). Thus, the most critical regions of the ESS were mapped to the SR protein binding central C-rich and 3′ AG-rich portions of the ESS.
Examination of the sequence of the mutant suppressor in pre-mRNA J (Fig. 3) suggests that a functional suppressor consists of the core sequence GGCUCCCC and an SR protein binding site. To test this hypothesis we built a synthetic splicing suppressor by using the core sequence GGCUCCCCC and known SR protein binding sites. For SR protein binding sites we used either the 3′ AG-rich ASF/SF2 binding site from the ESS or one or two copies of the optimal ASF/SF2 binding site determined by SELEX (9). These synthetic suppressors then were tested for their ability to inhibit BPV-1 late pre-mRNA spliceosome assembly and splicing in vitro. Although neither the core nor any of the ASF/SF2 binding sites had any significant effect by themselves (pre-mRNAs 5, 6, and 9 in Fig. 4), the core inhibited spliceosome assembly and splicing approximately 3-fold when placed upstream of any of the ASF/SF2 binding sites (pre-mRNAs 4, 7, and 8). Surprisingly, the sequences 3′ to the core may not need to be an ASF/SF2 binding site because a mutant 3′ AG-rich region gave similar results (pre-mRNA 3). Although the mutant AG-rich region has not been tested for ASF/SF2 binding, these results suggest that ASF/SF2 binding per se may not be essential. The observation that additional 3′ nucleotides are required for the function of the core is consistent with the observation that deletion of the 3′ half of the ESS up to the core eliminated the binding of 35- and 55-kDa SR proteins (RNA H in Fig. 2G). Although the C-rich core plus additional nonspecific downstream sequences strongly inhibited splicing, these sequences were not sufficient for full suppressor activity (compare pre-mRNAs 3, 4, 7, and 8 with pre-mRNA 2 in Fig. 4). One explanation is that full suppressor activity of the ESS requires the binding of additional SR proteins to sequences downstream of the core. Alternatively, the U-rich region of the ESS may play a redundant role with these downstream sequences.
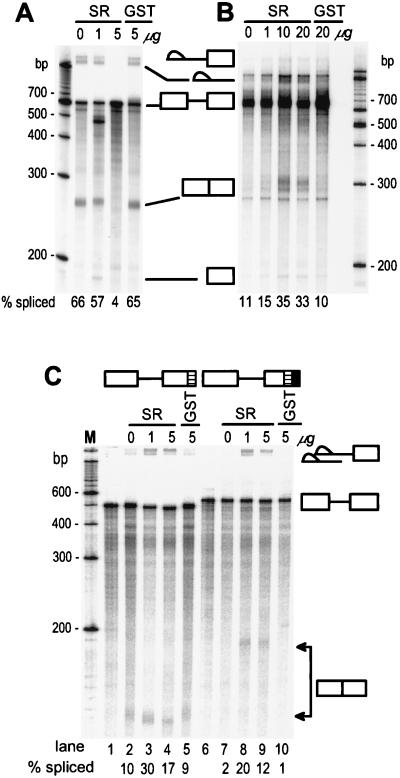
Restoration of ESS-Suppressed Splicing of BPV-1 Late and HIV-1 tat/rev pre-mRNAs by Exogenous SR Proteins.
The observation that the C-rich and 3′ AG-rich regions of the ESS are required for both strong suppression of splicing (Figs. 3 and 4) and SR protein binding (Fig. 2) suggests that these regions might limit the amount of SR proteins available for 3′ splice site and 5′ splice site interaction during pre-mRNA splicing. If this hypothesis is correct, addition of excess exogenous HeLa SR proteins to the splicing reaction should overcome the deficiency and relieve this inhibition of splicing. The results in Fig. 5B show that increasing SR protein concentrations in HeLa NE significantly eliminated the suppression by the ESS on splicing of BPV-1 pre-mRNAs B (Fig. 3D), supporting this hypothesis. In contrast, excess SR proteins in the HeLa NE inhibited the splicing of BPV-1 pre-mRNA A (Fig. 3D), which contains no ESS (Fig. 5A). Similar experiments were conducted on an HIV tat/rev pre-mRNA with or without the BPV-1 ESS. In this situation, addition of SR proteins to HeLa NE increased the splicing efficiency of both pre-mRNAs at all SR protein concentrations tested (Fig. 5C). This finding confirms that excess SR proteins overcome the effect of the ESS and also demonstrates that the suppression seen at high SR protein levels with BPV-1 late pre-mRNA is substrate specific.
Figure 5.
Restoration of ESS-suppressed splicing of BPV-1 late and HIV-1 tat/rev pre-mRNA by addition of excess exogenous SR proteins. BPV-1 late pre-mRNAs A (no ESS) and B (ESS +) in Fig. 3D and HIV-1 tat/rev pre-mRNA were used in this study. All of the pre-mRNAs were prepared by in vitro transcription and labeled by incorporation of 32P-rGTP (13). Different concentrations of purified HeLa SR proteins or recombinant glutathione S-transferase (GST) protein were added to each standard splicing reaction. Addition of 10 μg of purified SR proteins approximately doubles the amount of SR proteins in the splicing reaction. The spliced products of each pre-mRNA were resolved by electrophoresis on an 8% polyacrylamide-8 M urea gel and shown in A (BPV-1 late pre-mRNA A in Fig. 3D), B (BPV-1 late pre-mRNA B in Fig. 3D), and C (HIV-1 tat/rev pre-mRNA). The structures of HIV-1 tat/rev pre-mRNAs are diagramed at the top of C with the HIV-1 tat/rev ESE indicated as a hatched box and the BPV-1 ESS as a filled box. Lanes 1–5 are the ESS (−) HIV-1 tat/rev pre-mRNA and lanes 6–10 are the ESS (+) HIV-1 tat/rev pre-mRNA. Lanes 1 and 6 are the unspliced pre-mRNA controls. The splicing efficiency of each pre-mRNA was calculated based on the gel profile in each panel and shown at the bottom of each splicing gel.
DISCUSSION
The 5′ U-rich region of the BPV-1 ESS is 67% uridine within the first 12 nt at the 5′ end. This region contains two overlapping core PTB binding sites UCUU (31). Our study demonstrated that this region binds both PTB and U2AF65, which is consistent with the observation that PTB binds with a similar affinity to a multimerized PTB core as to the optimal U2AF binding site (31). PTB is a negative regulator of splicing that mediates 3′ splice site repression by preventing the binding of U2AF65 to the polypyrimidine tract (36). U2AF65, an essential splicing factor, binds the polypyrimidine tract in the first step of splicing and recruits the U2 small nuclear ribonucleoprotein to the branch point (43). Although both PTB and U2AF65 bind the 5′ U-rich region of the ESS, it is unclear what, if any, role these interactions have in ESS function. Clearly they are not essential for ESS function. However, they may play a role that is redundant with that of factors that bind to the 3′ half of the ESS. Comparing the splicing efficiency of pre-mRNA J in Fig. 3D to that of pre-mRNA 4 in Fig. 4A supports this assumption.
The C-rich region of the ESS is essential for both splicing suppression and binding of SR proteins. The most important part of this region is the core sequence GGCUCCCCC. This core is insufficient for the inhibition of spliceosome complex formation and splicing when it is at the 3′ end of the pre-mRNA and requires additional nonspecific sequences downstream for its function in vitro. This observation may represent an in vitro artifact because the ESS normally resides in the middle of a pre-mRNA. RNA sequences 3′ to the core may stabilize the binding of splicing factors or protect the core from 3′ exoribonucleolytic digestion. The first possibility is supported by the observation that binding of SR proteins to the C-rich region also requires some additional sequences downstream of the C-rich core (compare RNAs G and H in Fig. 2G). It is not known what proteins other than SR proteins bind the C-rich region. One possible candidate might be poly(C)-binding proteins such as PCBP1, PCBP2, and heterogeneous nuclear ribonucleoprotein K (44). Other possibilities include cellular splicing inhibitory factors such as ROX21-like proteins (45). Rox21, also called repressor splicing factor 1 (RSF1), binds to ESEs and inhibits in vitro splicing at the level of spliceosome assembly (H. M. Bourbon and J. Tazi, personal communication). However, the mechanism of splicing inhibition mediated by ROX21 differs from that mediated by the BPV-1 ESS. ROX21 suppresses splicing by preventing efficient 5′ splice site selection by U1 small nuclear ribonucleoprotein and ASF/SF2 (H. M. Bourbon and J. Tazi, personal communication), whereas the BPV-1 ESS inhibits splicing by blocking selection of suboptimal 3′ splice sites (49). The C-rich and 3′ AG-rich regions of the ESS contain, respectively, an ACE-like motif similar to the in vivo-selected sequences 4.11.7, 4.11.10, and 5.25.1 (46) and an ASF/SF2 binding site. ACE motifs are A/C-rich ESEs that may interact with at least some of the same proteins that bind purine-rich ESEs (46). Although our results showed that the C-rich region binds 35-and 55-kDa SR proteins and the 3′ AG-rich region binds ASF/SF2, the BPV-1 ESS doesn’t function as an ESE (49). Why the binding of SR proteins to the ESS suppresses splicing rather that stimulates splicing remains to be elucidated. One possibility is that additional SR protein(s) may be required for stimulation of splicing. We also have hypothesized that the ESS might compete with the upstream 3′ splice site or 5′ splice site for SR protein binding or limit the amount of SR proteins available for 3′ splice site and 5′ splice site interaction during pre-mRNA splicing. The observation that the inhibition of splicing by the ESS can be relieved with excess purified SR proteins supports this model. Alternatively, excess SR proteins may overcome the effect of an unidentified cellular inhibitory factor(s) recruited by the ESS and reinforce the SR protein-mediated selection of suboptimal 3′ splice site.
Surprisingly, the addition of excess SR proteins to HeLa NE inhibited splicing of ESS(−) BPV-1 late pre-mRNA, but increased splicing of both ESS(−) and ESS(+) HIV-1 tat/rev pre-mRNA. This opposite effect on two different pre-mRNAs was clearly not caused by contaminants in our SR protein preparations, but rather by a SR protein-dependent, substrate-specific phenomenon. One possible interpretation is that the BPV-1 pre-mRNA may have additional low affinity SR protein binding sites that block recognition of splice sites when occupied at high SR protein levels. Excess SR proteins can either activate or inhibit pre-mRNA splicing, depending on different RNA substrates. Interestingly, two recent reports indicate that in vivo overexpression of SR proteins ASF/SF2 or SC35 suppresses both simian virus 40 large T and small t pre-mRNA splicing (47) and even at physiological levels, SR protein ASF/SF2 inhibits HIV-1 tat pre-mRNA splicing in vivo (48).
In summary, we have demonstrated that the BPV-1 ESS blocks splicing at an early stage of spliceosome assembly (before A complex). Furthermore, the C-rich and 3′ AG-rich regions of the ESS are critical for ESS function and for binding SR proteins. Binding of SR proteins to the ESS may reduce the amount of SR proteins available for spliceosome complex formation or interfere with the normal bridging and recruitment activities of SR proteins. Although U2AF65 and PTB bind the 5′ U-rich region of the ESS, it is unclear how these interactions contribute to splicing suppression. Nevertheless, the ESS, as a whole, may interfere with exon definition at an early stage of spliceosome assembly through binding these splicing factors and SR proteins as well as an unidentified inhibitory factor(s).
Acknowledgments
We gratefully acknowledge K. M. Neugebauer and M. Roth for mAb 16H3 and mAb 1H4, A. R. Krainer for mAb αASF/SF2, J. G. Patton for PTB expression vector pET-PTB and polyclonal rabbit anti-human PTB antibody, G. Dreyfuss for mAb 7G12, P. Zuo and T. Maniatis for polyclonal rabbit anti-U2AF65 and U2AF35 antibodies, C. M. Stoltzfus for HIV-1 tat/rev plasmid pHS2, and R. Gontarek, D. Derse, M. McNally, and H. Ge for providing reagents and helpful discussions. We also thank Steve Mount, Susan Haynes, and Hui Ge for valuable comments on the manuscript.
ABBREVIATIONS
- BPV-1
bovine papillomavirus type 1
- ESS
exonic splicing suppressor
- ESE
exonic splicing enhancer
- SE1
splicing enhancer 1
- SR
serine/arginine-rich
- NE
nuclear extracts
- IP
immunoprecipitation
- PTB
polypyrimidine tract binding protein
- ASF/SF2
alternative splicing factor/splicing factor 2
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Watakabe A, Tanaka K, Shimura Y. Genes Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- 2.Xu R, Teng J, Cooper T A. Mol Cell Biol. 1993;13:3660–3674. doi: 10.1128/mcb.13.6.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeakley J M, Hedjran F, Morfin J-P, Merillat N, Rosenfeld M G, Emeson R B. Mol Cell Biol. 1993;13:5999–6011. doi: 10.1128/mcb.13.10.5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dirksen W P, Hampson R K, Sun Q, Rottman F M. J Biol Chem. 1994;269:6431–6436. [PubMed] [Google Scholar]
- 5.Tanaka K, Watakabe A, Shimura Y. Mol Cell Biol. 1994;14:1347–1354. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lavigueur A, La Branche H, Kornblihtt A R, Chabot B. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 7.Sun Q, Mayeda A, Hampson R K, Krainer A R, Rottman F M. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 8.Lynch K W, Maniatis T. Genes Dev. 1995;9:284–293. doi: 10.1101/gad.9.3.284. [DOI] [PubMed] [Google Scholar]
- 9.Tacke R, Manley J L. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gontarek R R, Derse D. Mol Cell Biol. 1996;16:2325–2331. doi: 10.1128/mcb.16.5.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeakley J M, Morfin J-P, Rosenfeld M G, Fu X-D. Proc Natl Acad Sci USA. 1996;93:7582–7587. doi: 10.1073/pnas.93.15.7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramchatesingh J, Zahler A M, Neugebauer K M, Roth M B, Cooper T A. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng Z M, He P, Baker C C. J Virol. 1996;70:4691–4699. doi: 10.1128/jvi.70.7.4691-4699.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caputi M, Casari G, Guenzi S, Tagliabue R, Sidoli A, Melo C A, Baralle F E. Nucleic Acids Res. 1994;22:1018–1022. doi: 10.1093/nar/22.6.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amendt B A, Hesslein D, Chang L-J, Stoltzfus C M. Mol Cell Biol. 1994;14:3960–3970. doi: 10.1128/mcb.14.6.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amendt B A, Si Z-H, Stoltzfus C M. Mol Cell Biol. 1995;15:4606–4615. doi: 10.1128/mcb.15.8.4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staffa A, Cochrane A. Mol Cell Biol. 1995;15:4597–4605. doi: 10.1128/mcb.15.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Staffa A, Acheson N H, Cochrane A. J Biol Chem. 1997;272:33394–33401. doi: 10.1074/jbc.272.52.33394. [DOI] [PubMed] [Google Scholar]
- 19.Del Gatto F, Breathnach R. Mol Cell Biol. 1995;15:4825–4834. doi: 10.1128/mcb.15.9.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Del Gatto F, Gesnel M C, Breathnach R. Nucleic Acids Res. 1996;24:2017–2021. doi: 10.1093/nar/24.11.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wentz M P, Moore B E, Cloyd M W, Berget S M, Donehower L A. J Virol. 1997;71:8542–8551. doi: 10.1128/jvi.71.11.8542-8551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Z M, He P J, Baker C C. J Virol. 1997;71:9096–9107. doi: 10.1128/jvi.71.12.9096-9107.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staknis D, Reed R. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuo P, Maniatis T. Genes Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]
- 25.Jamison S F, Crow A, Garcia-Blanco M A. Mol Cell Biol. 1992;12:4279–4287. doi: 10.1128/mcb.12.10.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konarska M M, Sharp P A. Cell. 1986;46:845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- 27.Milligan J F, Groebe D R, Witherell G W, Uhlenbeck O C. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dietrich-Goetz W, Kennedy I M, Levins B, Stanley M A, Clements J B. Proc Natl Acad Sci USA. 1997;94:163–168. doi: 10.1073/pnas.94.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zahler A M, Lane W S, Stolk J A, Roth M B. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 30.Lin C-H, Patton J G. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- 31.Perez I, Lin C-H, McAfee J G, Patton J G. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- 32.Sharp P A. Cell. 1994;77:805–816. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 33.Neugebauer K M, Stolk J A, Roth M B. J Cell Biol. 1995;129:899–908. doi: 10.1083/jcb.129.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zamore P D, Green M R. Proc Natl Acad Sci USA. 1989;86:9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patton J G, Mayer S A, Tempst P, Nadal-Ginard B. Genes Dev. 1991;5:1237–1251. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- 36.Singh R, Valcarcel J, Green M R. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 37.Ashiya M, Grabowski P J. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- 38.Ghetti A, Pinol-Roma S, Michael W M, Morandi C, Dreyfuss G. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ge H, Manley J L. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- 40.Ge H, Zuo P, Manley J L. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- 41.Krainer A R, Conway G C, Kozak D. Cell. 1990;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- 42.Krainer A R, Mayeda A, Kozak D, Binns G. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- 43.Valcarcel J, Gaur R K, Singh R, Green M R. Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 44.Leffers H, Dejgaard K, Celis J E. Eur J Biochem. 1995;230:447–453. [PubMed] [Google Scholar]
- 45.Brand S F, Pichoff S, Noselli S, Bourbon H-M. Gene. 1995;154:187–192. doi: 10.1016/0378-1119(94)00840-o. [DOI] [PubMed] [Google Scholar]
- 46.Coulter L R, Landree M A, Cooper T A. Mol Cell Biol. 1997;17:2143–2150. doi: 10.1128/mcb.17.4.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J, Manley J L. RNA. 1995;1:335–346. [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J, Xiao S H, Manley J L. Genes Dev. 1998;12:2222–2233. doi: 10.1101/gad.12.14.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng, Z. M., He, P. J. & Baker, C. C. (1999) J. Virol., in press.